Abstract
Basic ideas about the constancy and randomness of mutagenesis that drives evolution were challenged by the discovery of mutation pathways activated by stress responses. These pathways could promote evolution specifically when cells are maladapted to their environment (i.e., are stressed). However, the clearest example—a general stress-response–controlled switch to error-prone DNA break (double-strand break, DSB) repair—was suggested to be peculiar to an Escherichia coli F′ conjugative plasmid, not generally significant, and to occur by an alternative stress-independent mechanism. Moreover, mechanisms of spontaneous mutation in E. coli remain obscure. First, we demonstrate that this same mechanism occurs in chromosomes of starving F− E. coli. I-SceI endonuclease-induced chromosomal DSBs increase mutation 50-fold, dependent upon general/starvation- and DNA-damage-stress responses, DinB error-prone DNA polymerase, and DSB-repair proteins. Second, DSB repair is also mutagenic if the RpoS general-stress-response activator is expressed in unstressed cells, illustrating a stress-response–controlled switch to mutagenic repair. Third, DSB survival is not improved by RpoS or DinB, indicating that mutagenesis is not an inescapable byproduct of repair. Importantly, fourth, fully half of spontaneous frame-shift and base-substitution mutation during starvation also requires the same stress-response, DSB-repair, and DinB proteins. These data indicate that DSB-repair-dependent stress-induced mutation, driven by spontaneous DNA breaks, is a pathway that cells usually use and a major source of spontaneous mutation. These data also rule out major alternative models for the mechanism. Mechanisms that couple mutagenesis to stress responses can allow cells to evolve rapidly and responsively to their environment.
Keywords: antibiotic resistance, cancer, genome evolution, rapid evolution, stress-induced mutagenesis
How, when, and where mutations form underpins understanding pathogen–host interactions, antibiotic resistance, aging, cancer progression and therapy resistance, and evolution generally. Initial models of mutagenesis that drives evolution imagined random stochastic processes, roughly constant with time, and blind to selective environments (1). In contrast, bacterial, yeast, and human cells appear to possess mechanisms that induce mutation pathways specifically during stress, under the control of stress responses (2) (stress-induced mutagenesis or SIM). These pathways suggest mechanisms by which genetic diversity could be generated preferentially when cells are maladapted to their environment (i.e., are stressed), potentially accelerating evolution responsively to environments, a major departure from classic views (1). However, the significance of such mechanisms has been debated, as have the mechanisms themselves.
First, mutagenesis associated with the DNA-damage response has been argued to be an unavoidable consequence of induced DNA repair (3, 4), not an evolutionary engine (5, 6). Second, the strongest support for the idea of increased mutation rate during a general stress, and the most detailed understanding of a molecular mechanism, comes from starving Escherichia coli in an assay, the mechanism behind which has been debated. This mechanism appears to entail a switch from high-fidelity to error-prone DNA double-strand break (DSB) repair under stress, controlled by the SOS DNA-damage and the RpoS-general/starvation stress responses (7). Each stress response is necessary but not sufficient for the switch to mutagenic repair. Both confer up-regulation of the DinB error-prone DNA polymerase, which then makes errors that become mutations in acts of DSB repair via homologous recombination (HR) (7). Whereas up-regulation of DinB is the sole role of SOS in DSB-dependent SIM (8), RpoS additionally licenses the use of DinB (7, 9, 10) and other low-fidelity DNA polymerases (10) in DSB repair by a mechanism not yet elucidated. This process causes a switch from high-fidelity to mutagenic DSB repair under RpoS-inducing stress (7). RpoS is activated by many general stressors, including starvation, osmotic shock, cold shock, and oxidative stress (11), making DSB-dependent SIM potentially important for producing genetic diversity when cells are maladapted to many stressing environments. SOS is induced in most or all acts of DSB repair (12), making RpoS the switch to error-prone repair during stress. However, it has been argued that the mutations could form via an alternative stress-independent mechanism. Although similar phenomena are seen in other bacteria, yeast, and human cells (2), the mechanisms are less well understood.
Both synchrony with stress and the DSB repair-dependence of DSB-dependent SIM are nonrandom aspects of mutation (7), and both were argued to be peculiar to the assay system, caused by a stress-independent mechanism, and not generally relevant to spontaneous mutagenesis underlying evolution. First, DSB-dependent SIM controlled by RpoS was studied mostly in assays for reversion by frame-shift mutation of defective lac (13) or tet (e.g., ref. 7) genes in an F′ conjugative plasmid, which has engendered longstanding concerns about its significance. The mutagenesis appeared to be peculiar to F-located alleles (14–17) and to the specific F′ used (18, 19), which carries an extra copy of dinB, calling into question the general relevance of this mechanism (16). Although chromosomal mutations were observed (20, 21), these were postulated to be an indirect consequence of the peculiar dinB-expressing F′ and independent of stress (19). Second, because dinB is near lac in this specific F′, when Lac+ mutants are selected, spontaneous amplifications of the weakly functional lac gene can be selected for increased lac function and might include and overexpress the nearby dinB. The mutagenesis was argued to be independent of stress and merely a byproduct of growth of a cell carrying the selected gene amplification (16, 19), and although other studies failed to repeat (22) and contradicted (e.g., refs. 22 and 23) key predictions of the amplification growth-and-mutation model in the Lac assay, the model persists as an explanation for DSB-dependent SIM (16, 19). Third, DSB-dependent mutation was discovered in E. coli (24, 25) and then demonstrated and detailed in yeast (e.g., refs. 26–28), the latter not known to require stress or stress responses. DSB-coupling can make mutation nonrandom in genomic space (7) in addition to temporal nonrandomness. However, in both E. coli and yeast, DSB-dependent mutagenesis requires induction of DSBs by endonucleases, either encoded by F (7) or engineered into cells (7, 26–28). Because spontaneous DSBs are far rarer (12), DSB-dependent mutation pathways might contribute insignificantly to most spontaneous mutation. Mechanisms of spontaneous mutation in bacteria are virtually unknown (29, 30), as was the possible contribution of DSB-dependent mutation or SIM. Here we show that DSB-dependent SIM occurs in the bacterial chromosome of plasmid-free cells, does not result from growth of selected gene amplifications promoting mutation independently of stress and, moreover, represents about half of spontaneous frame-shift and base-substitution mutagenesis in starving cells. These data indicate the generality of DSB-dependent stress-induced mutation, rule out major alternative models for the mechanism, and define a mechanism underlying a major component of spontaneous mutation in E. coli.
Results and Discussion
Chromosomal DSB-Dependent Stress-Induced Mutation in F− Cells.
We show that DSB repair-dependent SIM is not peculiar to lac, selection for Lac+ phenotype, amplification, genes near lac, conjugative plasmids, or cells carrying them. We developed a mutation assay for reversion of a chromosomal tet +1-bp frame-shift allele in starving F− cells. The tet gene was placed 8.5 kb from an I-SceI double-strand endonuclease cutsite (I-site A, tet2, Fig. 1A) engineered into cells with or without a chromosomal, regulatable I-SceI gene, which we created and used previously to show that DSBs promote SOS-, RpoS-, DinB-, and DSB-repair protein-dependent mutation in the F′ (7). The cells are grown in liquid minimal medium with glucose both as a carbon source and to repress the PBAD promoter controlling I-SceI. When glucose is exhausted, the cells enter stationary phase and derepress PBADI-SceI slightly: enough to generate some DSBs, but not enough to cut all sites, so that repair by HR using an uncleaved sister replicon is possible (7). Most stationary cells contain only one chromosome, but about 40% carry two (31). Presumably, successful repair events occur in that subfraction. E. coli lacks efficient nonhomologous end-joining, and does little DSB repair without homologous sequence from a sister chromosome (12). Although possible, repair by HR using a duplicated chromosome segment is less likely because duplications are less frequent [≤10−3 (16)] than sister chromosomes [∼40% (31)].
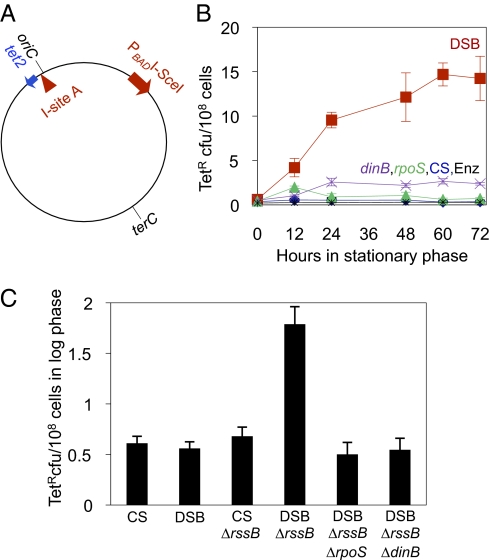
Fig. 1.
Stationary-phase and DSB activation of stress-induced mutation in the E. coli chromosome. (A) Mutation assay. A cleavage site for I-SceI endonuclease (I-site A, red triangle), and tetA allele with a +1-bp frame-shift mutation (tet2, blue arrow) are engineered into the chromosome of cells carrying the chromosomal PBAD-promoter-regulated I-SceI endonuclease gene (red arrow), which expresses I-SceI slightly (SI Appendix, Fig. S1A) in the absence of glucose (7), the condition used here. TetR mutants, caused by compensatory frame-shift mutations (SI Appendix, Fig. S5), occur during starvation in liquid medium without tetracycline (B) and are scored as cfu on rich glucose tetracycline plates after rescue from starvation. oriC and terC, origin and terminus of chromosomal DNA replication. Arrows, 5′ to 3′ orientation of genes. (B) I-SceI–mediated DSBs promote TetR reversion in prolonged stationary phase. Strains: CS (cutsite only) (◆); Enz, (enzyme only) (□); DSB (enzyme and cutsite) (■); DSB ΔdinB (×); DSB ΔrpoS (▲). (C) I-SceI–induced DSBs are not mutagenic in unstressed growing cells unless RpoS is up-regulated artificially by deletion of its negative regulator, rssB. The mutagenicity requires RpoS and DinB. In all figures, mutant frequencies are mean ± SEM for at least three independent experiments each, with three cultures per strain per experiment. SI Appendix, Table S5 shows the data in each figure.
Fig. 1B shows that frequencies of tetracycline-resistant (TetR) mutants increase with time in stationary phase when both I-SceI enzyme and cutsite are present. After 72 h in stationary phase, TetR mutant frequency was 55 ± 10-fold higher (mean ± SEM, three experiments) in the DSB strain than the “cutsite-only” control strain lacking I-SceI endonuclease (CS) (Figs. 1B, 2, and 3A) and “enzyme-only” control strain (Enz) (Fig. 1B), demonstrating that DSBs in the E. coli chromosome promoted mutation (Figs. 1B, 2, and 3A).
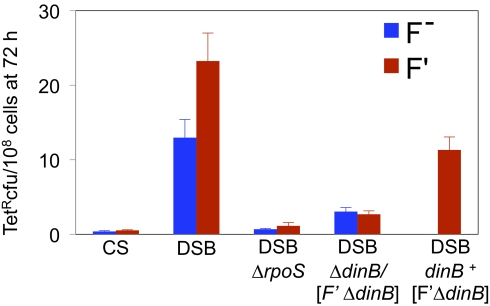
Fig. 2.
Proportional contribution of the F′ 128 episome to DSB-induced chromosomal mutation because of the F′ dinB copy. Cultures were assayed for TetR mutants after 72 h in stationary phase. Isogenic strains with (red) or without (blue) the F′ 128 plasmid. CS, cutsite only; DSB, enzyme plus cutsite.
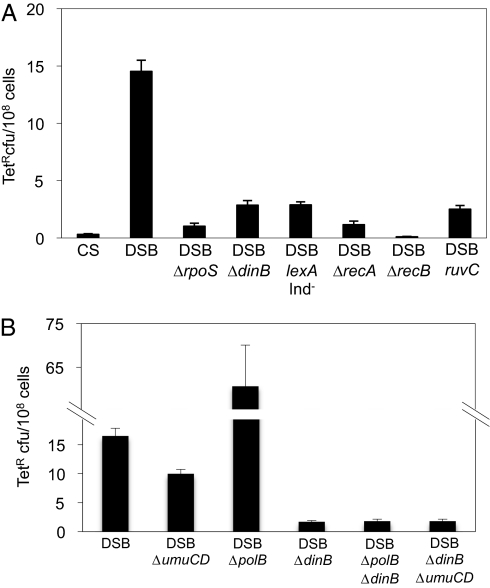
Fig. 3.
Genetic requirements of chromosomal DSB-dependent stress-induced mutagenesis in F− cells. (A) DSB-repair/HR proteins, the SOS DNA-damage response, the RpoS general stress response, and DinB/Pol IV are required. Apart from the cutsite-only control (CS), all strains have enzyme and cutsite present (can make DSBs) and are isogenic to the DSB strain except for the mutations indicated. Fold-decreases in mutant frequency, compared with the DSB strain: 58 ± 10 (CS), 25 ± 12 (ΔrpoS), 5.5 ± 0.75 (ΔdinB), 5.2 ± 0.54 (lexAInd−, an SOS-induction-defective mutant), 16 ± 3.6 (ΔrecA), 220 ± 130 (ΔrecB), 5.8 ± 0.5 (ruvC), and these genes act in a single pathway (SI Appendix, Fig. S3). (B) DNA Pol II inhibits Pol IV/DinB-dependent mutation, and small Pol V (UmuD′C) requirement for chromosomal TetR mutation. The DSB ΔumuCD strain is significantly different from the DSB strain (P = 0.025).
We quantified the number of cells with DSBs (SI Appendix, SI Materials and Methods) at the 72-h starvation time point used in most of the experiments here. Between 7% and 16% of the cells have a DSB, 47 times more than in no-DSB controls (SI Appendix, Fig. S1A).
Isogenic strains with deletions of rpoS or dinB did not exhibit increased mutant frequencies (Figs. 1B, 2, and 3). Cells carrying the rpoS or dinB mutations and a functional tet gene form colonies normally on glucose tetracycline plates under exact reconstructions of experimental conditions (SI Appendix, Table S1). Thus, rpoS and dinB mutations did not depress TetR colony (cfu) counts by slowing colony formation of TetR mutants, once formed. We conclude that the DSB-provoked mutation process itself requires RpoS and DinB functions. The requirement for an RpoS stress response indicates that these are stress-induced mutants.
The stress that induces mutagenesis appears to be starvation, not exposure to the antibiotic tetracycline, because mutagenesis relates to the time starving in the absence of tetracycline (Fig. 1B), not to exposure to selective tetracycline plates, which did not occur until after cells were returned to growth medium. This finding rules out the possibility that selection of amplification of the tet gene was required for mutation. Subsequent experiments were performed at 72 h in stationary phase.
RpoS Activates DSB-Dependent Mutation in Unstressed Growing Cells.
We show that DSBs are not always mutagenic but rather become mutagenic only when cells are stressed and induce their RpoS-controlled general stress response. We examined unstressed growing cells in which DSBs were induced by a brief pulse of arabinose [to induce I-SceI transcription (7)] and find that DSB repair is not mutagenic because RpoS is not expressed at that time (Fig. 1C). This finding is true also in cells carrying the F′ with dinB+ (7). However, if RpoS is expressed inappropriately in unstressed log-phase cells by deletion of the rssB-encoded negative regulator of RpoS, then the DSBs promote mutations, and these are both RpoS- and DinB-dependent (Fig. 1C). Note that ΔrssB abrogates only one (posttranslational) of the at least three levels of repression of RpoS in log phase; the transcriptional and translational repression (32, 33) remain intact such that we have increased RpoS levels less than is expected in stationary phase, yet still restore some mutagenesis. We conclude that DSB-promoted RpoS-dependent mutation in F− E. coli cells is regulated temporally to times of stress via its coupling to the RpoS response. Furthermore, stress is not required; activation of the stress response is sufficient.
Proportional Contribution of F′128 via dinB.
When added back, the F′ plasmid increased chromosomal mutagenesis in 72-h starved cells only approximately twofold compared with isogenic F− cells, and mutagenesis still required RpoS and DinB (Fig. 2). The contribution of F′128 to mutagenesis results from an extra dinB gene carried by this F′, in that it is abolished in a strain deleted for the F′ copy of dinB (Fig. 2, Rightmost bar). Therefore, the chromosomal dinB copy is sufficient for DSB-promoted SIM. Cells with the F′ dinB+ and chromosomal ΔdinB mutate similarly to F− dinB+ cells (SI Appendix, Fig. S2), showing that location of dinB in F or chromosome is equivalent. A major alternative model in which mutation is not stress-induced, but rather requires selection of cells carrying amplification of the lac gene with the F′ dinB (16, 19) when dinB is next to lac in the F′, is ruled out because the cells in these experiments carry both a lac deletion and no F′ dinB, and also are not starved in lactose, which would be needed for selection of lac amplification.
DSB Repair Proteins, SOS Response, and Sequences of Chromosomal DSB-Dependent Stress-Induced Mutations.
Chromosomal DSB-provoked tet frame-shift reversion in F− cells requires functional DSB-repair and HR proteins RecA, RecB, and RuvC, an inducible SOS DNA-damage response, RpoS, and DinB (Fig. 3A) functioning in a single pathway (SI Appendix, Fig. S3), but not RecQ or RecJ (SI Appendix, Fig. S4), as shown for mutation in F-borne genes (2, 7, 24, 34). These data imply that chromosomal DSB-provoked mutation occurs by a similar or the same mechanism as described for lac in the F′. In addition, as with F′ Lac+ reversion (e.g., ref. 35), Pol II appears to compete with DinB/Pol IV such that Pol II− cells show increased DSB-dependent SIM that is DinB-dependent (Fig. 3B). Furthermore, a weak but significant Pol V (umuCD) requirement (Fig. 3B) (P = 0.025) resembles chromosomal DSB-dependent SIM in F′-carrying cells (21). We sequenced the tet2 gene from 12 DSB-provoked TetR mutants from the 72-h time point and 12 Tet-sensitive cells. All of the TetR and none of the TetS isolates carry a true reversion: a −1-G deletion in the 5-G repeat at position 331 of this tet gene (SI Appendix, Fig. S5A). Deletions of −1 bp in mononucleotide repeats are common errors of DinB/Pol IV (36) and dominate DSB-associated SIM of a lac frame-shift allele (7, 25, 37), although base substitutions are also abundant in a nonframe-shift assay (21). We conclude that chromosomal DSB-dependent SIM occurs by a similar or the same mechanism, using the same proteins and causing similar mutation sequences, as DSB-dependent SIM at lac in the F′.
RpoS and DinB Are Not Required for DSB Survival.
Of the genes required for DSB-dependent mutation (Fig. 3A), only the DSB repair genes are required for survival of the DSB (SI Appendix, Fig. S1B); the rest promote mutagenesis (Fig. 3A) but are not required for DSB repair during starvation (SI Appendix, Fig. S1B). This finding indicates that use of the error-prone DNA polymerase is not an unavoidable requisite for repair (as suggested in refs. 3 and 4), but rather a regulated decision not needed for DSB survival.
DSB-Dependent Stress-Induced Mutagenesis in Wild-Type Strain MG1655.
Chromosomal DSB-provoked SIM is not peculiar to the FC36 strain background (13) used here. Sequenced “wild-type” K12 strain MG1655 also shows RpoS- and DinB-dependent DSB-provoked tet mutation during starvation, with a 13 ± threefold increase in the DSB- versus the cutsite-only control strain (SI Appendix, Fig. S6). These results imply that DSB-provoked chromosomal SIM is general to RpoS-competent E. coli rather than specific to a particular strain. The difference in mutant frequency in FC36 versus MG1655 might reflect the variability of the general stress response in different E. coli K12 strains (38).
Spontaneous Frame-Shift and Base-Substitution Mutation via Similar SIM Mechanism.
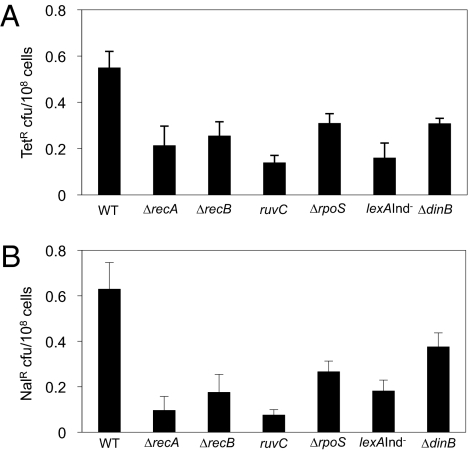
We show that much of spontaneous frame-shift and base-substitution mutation, in the absence of induced DSBs, during starvation is DSB-repair protein-, DinB-, and stress-response–dependent. We find that about half of spontaneous frame-shift (TetR) and base-substitution (NalR) mutagenesis (no I-SceI) in 72-h stationary cultures of F− cells requires the RpoS and SOS stress responses, DinB, and DSB-repair proteins RecA, RecB, and RuvC (Fig. 4). From the NalR mutants, we sequenced the gyrA gene, which is frequently mutated in cells that have acquired NalR (39), and confirmed that the NalR mutants carry base-substitution mutations of similar sequence and position in the gyrA gene, as observed for NalR mutations previously (mostly C to T and A to G) (SI Appendix, Fig. S5B) (39). We conclude that about half of spontaneous frame-shift and base-substitution mutagenesis in starving cells proceeds via a pathway that requires the DSB-repair and DinB proteins, SOS, and RpoS general stress responses. These data imply that these spontaneous mutations result from a mechanism similar to or the same as DSB-dependent SIM.
Fig. 4.
Much of spontaneous frame-shift and base-substitution mutation during starvation, in the absence of induced DSBs, is DSB repair-protein DinB- and stress-response–dependent. (A) Frame-shift reversion. The DSB-repair–dependent SIM pathway, requiring RecA, RecB, RuvC, SOS/DinB, and RpoS, constitutes about half of spontaneous TetR frame-shift–reversion mutation. Mutant frequencies after 72 h in stationary phase in strains with no I-SceI, and so with only spontaneous DSBs. The mutant strains differ from wild-type significantly: ΔrecA P = 0.001, ΔrecB P = 0.004, ruvC P < 0.001, lexAInd− (SOS−) P = 0.001, ΔrpoS P = 0.006, and ΔdinB P = 0.004, Wilcoxon-Mann–Whitney U Test. (B) Half of spontaneous base-substitution mutation (SI Appendix, Fig. S5B) to nalidixic-acid resistance during starvation is DSB-repair protein, DinB- and stress-response–dependent. The mutants differ from wild-type significantly: ΔrecA P < 0.001, ΔrecB P = 0.001, ruvC P < 0.001, lexAInd− (SOS−) P = 0.001, ΔrpoS P = 0.01, and ΔdinB P = 0.074.
Conclusions.
The data presented show that DSB-dependent stress-induced mutagenesis is a general feature of E. coli biology. It is not, as suggested (14–19), peculiar to the F′ replicon, to cells carrying an F plasmid, or to cells with more than one dinB copy. These data rule out stress-independent mechanisms caused by selecting for revertants of a leaky lac allele that can be amplified and generate the mutations near lac, or be peculiar to cells with coamplification of lac and a nearby dinB gene (16, 19). Rather, I-SceI–induced DSBs provoke nearby mutations in chromosomes of F− E. coli during generic starvation conditions, independently of selection for amplification of the mutated gene or dinB (Figs. 1–3), in two different E. coli K12 strain backgrounds (Figs. 3 and SI Appendix, Fig. S6). Although it might seem possible that a stress-independent amplification, selection, growth and mutation model might dominate DSB-dependent Lac but not Tet reversion, first, previous data failed to support amplification-mutagenesis at Lac (22 and 23, and reviewed in 2 and 40), and second, the congruence of proteins used for Lac and Tet mutagenesis (Figs. 1 and 3, and SI Appendix, Figs. S3 and S4) (2, 7, 24) and similar sequences of the reversions (SI Appendix, Fig. S5) (7, 21, 25, 37) are most simply interpreted as a similar or the same mutation mechanism dominating in both circumstances. The mutagenesis occurs only in stressed cells (Fig. 1B) but can occur in unstressed growing cells if the RpoS response is activated artificially (Fig. 1C), illustrating the RpoS-controlled switch from high-fidelity to mutagenic DSB repair (7). Moreover, stress is not required for mutation; activation of the stress response is sufficient (Fig. 1C). This finding excludes models in which RpoS was proposed to promote recovery of mutants by allowing survival of stress (16). Importantly, the DSB-dependent stress-induced mutation pathway, or one like it in its requirements for the RpoS and SOS responses, DinB and DSB-repair proteins, appears to account for fully half of spontaneous frame-shift and base-substitution mutation in starving E. coli (Fig. 4). These results demonstrate that DSB-dependent SIM is a major mutation route in E. coli.
Generality of RpoS-Dependent and Stress-Inducible Genomic Instability.
Whereas our results imply the generality of DSB-, RpoS-, and DinB-dependent SIM, recent work suggested that RpoS- and DinB-dependent SIM did not occur in a relative of E. coli, Salmonella enterica strain LT2, even when the LT2 rpoS gene was overexpressed (41). This finding probably reflects the choice of Salmonella strain. LT2 is a nonpathogenic natural variant that is nonpathogenic because it is RpoS-deficient (42, 43), having a UUG rather than AUG start codon (43). Thus, RpoS-dependent mutagenesis would not be expected to occur in LT2. RpoS is present or functional heterogeneously both among bacterial species and between isolates of a species (44), and variants that lack RpoS evolve other control of genes/pathways normally regulated by RpoS (45). These facts could explain why RpoS does not affect mutagenesis in LT2. In contrast, a pathogenic S. enterica strain, expected to express RpoS normally (42, 43), displayed DSB repair-protein DinB-, SOS-, and RpoS-dependent SIM when exposed to bile (a stressor) and induced bile-resistant mutants (46, 47). Thus, use of DSB repair-protein- RpoS-, SOS-, and DinB-dependent mutation appears to occur in Salmonella but, as expected, to be confined to strains with functional RpoS.
Other pathways of stress-inducible mutation that are less defined molecularly also require RpoS, including transposition/excision of phage-μ (48, 49), stress-inducible point mutation (50), and transposition (51) in Pseudomonas putida, DSB-independent SIM in aging colonies of an E. coli natural isolate (52), and DSB-dependent (7, 53) stress-induced gene amplifications in E. coli (54). The importance of coupling inducible mutagenesis pathways to a broad general stress response like RpoS might be that genetic diversity may be generated responsively to many different stressors and environments.
Other bacterial starvation and general stress responses also promote mutagenesis during stress (8). These processes include the stringent and the competence starvation-stress responses in Bacillus subtilis (55), the stringent (56, 57) cAMP (49, 58) responses to starvation, and RpoE membrane-protein stress response (40) in E. coli. These processes promote base-substitutions (56, 58), frame-shift mutations (40), amplification (40), mobile-intron movement (57), and transposon excision (49, 57). These examples illustrate the apparently multiple evolutions of mechanisms that couple genomic instability pathways with stress responses and stress. The importance of all of these is that genetic diversity is generated preferentially when cells are maladapted to their environment: when stressed. This finding contrasts with early ideas about the constancy of mutation fueling evolution (1) and recent arguments that growth of selected amplifications can explain apparent SIM independently of stress-inducibility (16).
When Amplification-Mutagenesis Applies.
Although models with selection for amplification, growth, replication, mutagenesis, then loss of the nonmutant copies (amplification-mutagenesis) cannot explain DSB-dependent SIM (above), they may pertain to Salmonella selected for resistance to protamine, a membrane-destabilizing peptide antibiotic (59). Mutations in eight genes conferred resistance but caused slow growth without antibiotic. Of the eight loci, hemC lies between rDNA repeats. Only this locus reverted rapidly to faster growth without antibiotic, and some of the revertants were heterogenotes with duplications of hemC. If the duplication strains were grown for 30 to 70 (59) or 210 generations (60) without antibiotic, faster-growing revertants with one mutated hemC gene were obtained. Thus, amplification-mutagenesis may predominate when a partial-functional allele can be selected for amplification, and lies between repeats that promote duplication, and when the many generations required for postmutational loss of amplification are allowed. In DSB-dependent stress-induced Lac and Tet reversion, the many generations required are unlikely or even impossible, and neither gene lies between repeats (and tet amplification is not selected).
Mutagenesis Is Not Needed for DSB Repair.
It has been argued that mutagenesis associated with SOS induction is an unavoidable consequence of the DNA repair/survival functions of SOS, not an enhancer of evolution (3, 4), as was suggested initially (5, 6). Importantly, we find that neither RpoS nor DinB confers any detectable boost in DSB survival, even under RpoS-inducing stress conditions (SI Appendix, Fig. S1B) (7). Previously, RpoS and DinB slightly decreased DSB-survival (7). Thus, RpoS and DinB do not improve—and may slightly reduce—DSB-repair efficiency. We conclude that RpoS/DinB-induced mutagenesis is not an unavoidable consequence of the DSB repair to which the mutagenesis is coupled. Both RpoS and DinB allow survival of problems other than DSBs (11, 61). However, the roles of RpoS and DinB in DSB repair are not needed for the repair and rather might confer the survival advantage of generating rare better-adapted mutants.
DSB-Dependent Spontaneous Mutation.
For evolution, cancer, infectious disease, and biology, the most important mutation mechanisms are those that produce “spontaneous” mutations: the mutations that cells accrue without experimental manipulations designed to induce mutation pathways, which might not predominate otherwise. However, spontaneous mutation mechanisms are poorly understood (29, 30). DSB-dependent mutagenesis was discovered in E. coli (24, 25), then demonstrated and elaborated in yeast (e.g., refs. 26–28), but whether DSB-dependent mutagenesis occurred only in cells with artificially induced DSBs was unclear. In E. coli and yeast, DSB-dependent mutagenesis required induction of DSBs by endonucleases, either encoded by F (7) or engineered into cells (7, 26–28). Remarkably, we found that the proteins of DSB-dependent SIM are required for half of spontaneous frame-shift and base-substitution mutation in starving F− cells (Fig. 4), implying a major contribution of this pathway to spontaneous mutation during starvation. Given the large number of mechanisms of spontaneous mutation that operate simultaneously (29), it is impressive that fully half can be accounted for by this particular SIM pathway. Previous screens for proteins required for spontaneous mutation uncovered error-prone DNA polymerases in yeast (62, 63), and in E. coli, some of the SOS-response and DSB-repair proteins found here (RecA, RecB, SOS) (64–66). Although, with the exception of ref. 66, these authors did not study starvation or other stress conditions knowingly, it is possible that most of the spontaneous mutations in those assays also arose from occasional stressed cells, possibly by DSB-dependent SIM or a similar mechanism.
Stress-Inducible Mutation, Evolution, and Disease.
Starvation, stationary-phase, and stress are normal experiences of microbes in the wild and are encountered frequently by cells of multicellular organisms. Mechanisms that promote mutation during stress by coupling mutagenesis to stress responses can enhance the ability of cells to evolve rapidly, responsively to their environment, specifically when they are maladapted. This responsive gene-diversification strategy is similar to how protein diversity is maximized during stress when chaperones are less available, potentially promoting evolvability (67). Generation of both phenotypic and genetic variation may be means by which organisms accelerate evolution responsively to their environments. Both stress-induced mutagenesis and phenotypic diversification (67) could contribute to observations of climate/heat stress-induced enhanced expression of genetic variation, leading to rapid evolution (68).
Several antibiotics induce resistance mutations (69–71) and may well do so via DSB repair-coupled SIM, as suggested for ciprofloxacin, which induces resistance DSB-repair-protein-, SOS- and DinB-dependently (69). The involvement of oxygen radicals in some antibiotic-induced mutagenesis (71) might be via DSB-production, or perhaps activation of the RpoS response, which is activated by oxidative stress. Similarly in cancer cells, stressors including hypoxia do (72) and chemotherapies could provoke the genomic instability that drives all stages of progression of cancer and resistance. With better understanding of SIM mechanisms, these problems might be addressable therapeutically in infections and cancer.
Materials and Methods
A brief summary of methods used follows. For full details, see SI Appendix, SI Materials and Methods. E. coli strains used in this study are given in SI Appendix, Table S2. The tet reporter gene, chromosomal I-SceI expression system, and sites are described in SI Appendix, SI Materials and Methods, and Tables S2 to S4. Bacteria were grown on LBH or M9 minimal medium supplemented with 10 μg/mL thiamine (vitamin B1) and 0.1% glucose as carbon source. Other additives were used at the following concentrations (μg/mL): ampicillin, 100; chloramphenicol, 25; kanamycin, 50; tetracycline, 10; sodium citrate 20 mM. For mutation assays, single colonies from M9 glucose vitamin B1 (B1) plates that had been incubated for ∼22 h at 37 °C were inoculated each into 5 mL of M9 glucose B1 broth and grown for 12 h shaking, three independent cultures per genotype per experiment. These cultures were diluted 1:100 into the same medium and grown 8 to 10 h, diluted 1:100, and grown 12 h to saturation (time “0”) (Fig. 1B), then incubated further for 72 h. Mutant frequencies were determined by plating on LBH glucose tetracycline (TetR mutant cfu) and LBH glucose plates (total cfu), and are expressed as TetR per total cfu. Data from which plots were generated are given in SI Appendix, Table S5. For log-phase experiments (Fig. 1C), cells were grown in M9 glycerol with 0.000005% arabinose (to induce I-SceI expression slightly per ref. 7) to midlog phase, and then plated on LBH glucose with and without Tet to score TetR-mutant and total cfu.
Supplementary Material
Acknowledgments
We thank D. Bates, H. Dierick, R. Frisch, R. Galhardo, J. Halliday, P. J. Hastings, C. Herman, G. Ira, J .D. Wang, J. W. Drake and reviewer 2 for helpful comments on the manuscript. This work was supported by Public Health Service Grant R01-GM53158.
Footnotes
The authors declare no conflict of interest.
*This Direct Submission article had a prearranged editor.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1104681108/-/DCSupplemental.
References
- 1.Mayr E. The Growth of Biological Thought: Diversity, Evolution, and Inheritance. The Growth of Biological Thought: Diversity, Evolution, and Inheritance. Cambridge, MA: Harvard University Press; 1985. [Google Scholar]
- 2.Galhardo RS, Hastings PJ, Rosenberg SM. Mutation as a stress response and the regulation of evolvability. Crit Rev Biochem Mol Biol. 2007;42:399–435. doi: 10.1080/10409230701648502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Andersson DI, Koskiniemi S, Hughes D. Biological roles of translesion synthesis DNA polymerases in eubacteria. Mol Microbiol. 2010;77:540–548. doi: 10.1111/j.1365-2958.2010.07260.x. [DOI] [PubMed] [Google Scholar]
- 4.Lynch M. Evolution of the mutation rate. Trends Genet. 2010;26:345–352. doi: 10.1016/j.tig.2010.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Radman M. SOS repair hypothesis: Phenomenology of an inducible DNA repair which is accompanied by mutagenesis. Basic Life Sci. 1975;5A:355–367. doi: 10.1007/978-1-4684-2895-7_48. [DOI] [PubMed] [Google Scholar]
- 6.McPartland A, Green L, Echols H. Control of recA gene RNA in E. coli: Regulatory and signal genes. Cell. 1980;20:731–737. doi: 10.1016/0092-8674(80)90319-0. [DOI] [PubMed] [Google Scholar]
- 7.Ponder RG, Fonville NC, Rosenberg SM. A switch from high-fidelity to error-prone DNA double-strand break repair underlies stress-induced mutation. Mol Cell. 2005;19:791–804. doi: 10.1016/j.molcel.2005.07.025. [DOI] [PubMed] [Google Scholar]
- 8.Galhardo RS, et al. DinB upregulation is the sole role of the SOS response in stress-induced mutagenesis in Escherichia coli. Genetics. 2009;182:55–68. doi: 10.1534/genetics.109.100735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Storvik KA, Foster PL. RpoS, the stress response sigma factor, plays a dual role in the regulation of Escherichia coli's error-prone DNA polymerase IV. J Bacteriol. 2010;192:3639–3644. doi: 10.1128/JB.00358-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Frisch RL, et al. Separate DNA Pol II- and Pol IV-dependent pathways of stress-induced mutation during double-strand-break repair in Escherichia coli are controlled by RpoS. J Bacteriol. 2010;192:4694–4700. doi: 10.1128/JB.00570-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hengge-Aronis R. Signal transduction and regulatory mechanisms involved in control of the sigma(S) (RpoS) subunit of RNA polymerase. Microbiol Mol Biol Rev. 2002;66:373–395. doi: 10.1128/MMBR.66.3.373-395.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Pennington JM, Rosenberg SM. Spontaneous DNA breakage in single living Escherichia coli cells. Nat Genet. 2007;39:797–802. doi: 10.1038/ng2051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cairns J, Foster PL. Adaptive reversion of a frameshift mutation in Escherichia coli. Genetics. 1991;128:695–701. doi: 10.1093/genetics/128.4.695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Radicella JP, Park PU, Fox MS. Adaptive mutation in Escherichia coli: A role for conjugation. Science. 1995;268:418–420. doi: 10.1126/science.7716545. [DOI] [PubMed] [Google Scholar]
- 15.Foster PL, Trimarchi JM. Adaptive reversion of an episomal frameshift mutation in Escherichia coli requires conjugal functions but not actual conjugation. Proc Natl Acad Sci USA. 1995;92:5487–5490. doi: 10.1073/pnas.92.12.5487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Roth JR, Kugelberg E, Reams AB, Kofoid E, Andersson DI. Origin of mutations under selection: The adaptive mutation controversy. Annu Rev Microbiol. 2006;60:477–501. doi: 10.1146/annurev.micro.60.080805.142045. [DOI] [PubMed] [Google Scholar]
- 17.Slechta ES, Harold J, Andersson DI, Roth JR. The effect of genomic position on reversion of a lac frameshift mutation (lacIZ33) during non-lethal selection (adaptive mutation) Mol Microbiol. 2002;44:1017–1032. doi: 10.1046/j.1365-2958.2002.02934.x. [DOI] [PubMed] [Google Scholar]
- 18.Godoy VG, Gizatullin FS, Fox MS. Some features of the mutability of bacteria during nonlethal selection. Genetics. 2000;154:49–59. doi: 10.1093/genetics/154.1.49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Slechta ES, et al. Adaptive mutation: General mutagenesis is not a programmed response to stress but results from rare coamplification of dinB with lac. Proc Natl Acad Sci USA. 2003;100:12847–12852. doi: 10.1073/pnas.1735464100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bull HJ, Lombardo MJ, Rosenberg SM. Stationary-phase mutation in the bacterial chromosome: Recombination protein and DNA polymerase IV dependence. Proc Natl Acad Sci USA. 2001;98:8334–8341. doi: 10.1073/pnas.151009798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Petrosino JF, Galhardo RS, Morales LD, Rosenberg SM. Stress-induced beta-lactam antibiotic resistance mutation and sequences of stationary-phase mutations in the Escherichia coli chromosome. J Bacteriol. 2009;191:5881–5889. doi: 10.1128/JB.00732-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Stumpf JD, Poteete AR, Foster PL. Amplification of lac cannot account for adaptive mutation to Lac+ in Escherichia coli. J Bacteriol. 2007;189:2291–2299. doi: 10.1128/JB.01706-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hastings PJ, Slack A, Petrosino JF, Rosenberg SM. Adaptive amplification and point mutation are independent mechanisms: Evidence for various stress-inducible mutation mechanisms. PLoS Biol. 2004;2:e399. doi: 10.1371/journal.pbio.0020399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Harris RS, Longerich S, Rosenberg SM. Recombination in adaptive mutation. Science. 1994;264:258–260. doi: 10.1126/science.8146657. [DOI] [PubMed] [Google Scholar]
- 25.Rosenberg SM, Longerich S, Gee P, Harris RS. Adaptive mutation by deletions in small mononucleotide repeats. Science. 1994;265:405–407. doi: 10.1126/science.8023163. [DOI] [PubMed] [Google Scholar]
- 26.Strathern JN, Shafer BK, McGill CB. DNA synthesis errors associated with double-strand-break repair. Genetics. 1995;140:965–972. doi: 10.1093/genetics/140.3.965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Yang Y, Sterling J, Storici F, Resnick MA, Gordenin DA. Hypermutability of damaged single-strand DNA formed at double-strand breaks and uncapped telomeres in yeast Saccharomyces cerevisiae. PLoS Genet. 2008;4:e1000264. doi: 10.1371/journal.pgen.1000264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hicks WM, Kim M, Haber JE. Increased mutagenesis and unique mutation signature associated with mitotic gene conversion. Science. 2010;329:82–85. doi: 10.1126/science.1191125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Drake JW. General antimutators are improbable. J Mol Biol. 1993;229:8–13. doi: 10.1006/jmbi.1993.1002. [DOI] [PubMed] [Google Scholar]
- 30.Smith KC. Spontaneous mutagenesis: Experimental, genetic and other factors. Mutat Res. 1992;277:139–162. doi: 10.1016/0165-1110(92)90002-q. [DOI] [PubMed] [Google Scholar]
- 31.Akerlund T, Nordström K, Bernander R. Analysis of cell size and DNA content in exponentially growing and stationary-phase batch cultures of Escherichia coli. J Bacteriol. 1995;177:6791–6797. doi: 10.1128/jb.177.23.6791-6797.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hengge R. The two-component network and the general stress sigma factor RpoS (sigma S) in Escherichia coli. Adv Exp Med Biol. 2008;631:40–53. doi: 10.1007/978-0-387-78885-2_4. [DOI] [PubMed] [Google Scholar]
- 33.McCullen CA, Benhammou JN, Majdalani N, Gottesman S. Mechanism of positive regulation by DsrA and RprA small noncoding RNAs: pairing increases translation and protects rpoS mRNA from degradation. J Bacteriol. 2010;192:5559–5571. doi: 10.1128/JB.00464-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.He AS, Rohatgi PR, Hersh MN, Rosenberg SM. Roles of E. coli double-strand-break-repair proteins in stress-induced mutation. DNA Repair (Amst) 2006;5:258–273. doi: 10.1016/j.dnarep.2005.10.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hastings PJ, et al. Competition of Escherichia coli DNA polymerases I, II and III with DNA Pol IV in stressed cells. PLoS ONE. 2010;5:e10862. doi: 10.1371/journal.pone.0010862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Tang M, et al. Roles of E. coli DNA polymerases IV and V in lesion-targeted and untargeted SOS mutagenesis. Nature. 2000;404:1014–1018. doi: 10.1038/35010020. [DOI] [PubMed] [Google Scholar]
- 37.Foster PL, Trimarchi JM. Adaptive reversion of a frameshift mutation in Escherichia coli by simple base deletions in homopolymeric runs. Science. 1994;265:407–409. doi: 10.1126/science.8023164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Spira B, Hu X, Ferenci T. Strain variation in ppGpp concentration and RpoS levels in laboratory strains of Escherichia coli K-12. Microbiology. 2008;154:2887–2895. doi: 10.1099/mic.0.2008/018457-0. [DOI] [PubMed] [Google Scholar]
- 39.Sáenz Y, Zarazaga M, Briñas L, Ruiz-Larrea F, Torres C. Mutations in gyrA and parC genes in nalidixic acid-resistant Escherichia coli strains from food products, humans and animals. J Antimicrob Chemother. 2003;51:1001–1005. doi: 10.1093/jac/dkg168. [DOI] [PubMed] [Google Scholar]
- 40.Gibson JL, et al. The sigma(E) stress response is required for stress-induced mutation and amplification in Escherichia coli. Mol Microbiol. 2010;77:415–430. doi: 10.1111/j.1365-2958.2010.07213.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Koskiniemi S, Hughes D, Andersson DI. Effect of translesion DNA polymerases, endonucleases and RpoS on mutation rates in Salmonella typhimurium. Genetics. 2010;185:783–795. doi: 10.1534/genetics.110.116376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Swords WE, Cannon BM, Benjamin WH., Jr Avirulence of LT2 strains of Salmonella typhimurium results from a defective rpoS gene. Infect Immun. 1997;65:2451–2453. doi: 10.1128/iai.65.6.2451-2453.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Lee IS, Lin J, Hall HK, Bearson B, Foster JW. The stationary-phase sigma factor sigma S (RpoS) is required for a sustained acid tolerance response in virulent Salmonella typhimurium. Mol Microbiol. 1995;17:155–167. doi: 10.1111/j.1365-2958.1995.mmi_17010155.x. [DOI] [PubMed] [Google Scholar]
- 44.Ferenci T. What is driving the acquisition of mutS and rpoS polymorphisms in Escherichia coli? Trends Microbiol. 2003;11:457–461. doi: 10.1016/j.tim.2003.08.003. [DOI] [PubMed] [Google Scholar]
- 45.Stoebel DM, Hokamp K, Last MS, Dorman CJ. Compensatory evolution of gene regulation in response to stress by Escherichia coli lacking RpoS. PLoS Genet. 2009;5:e1000671. doi: 10.1371/journal.pgen.1000671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Prieto AI, Ramos-Morales F, Casadesús J. Bile-induced DNA damage in Salmonella enterica. Genetics. 2004;168:1787–1794. doi: 10.1534/genetics.104.031062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Prieto AI, Ramos-Morales F, Casadesús J. Repair of DNA damage induced by bile salts in Salmonella enterica. Genetics. 2006;174:575–584. doi: 10.1534/genetics.106.060889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Gómez-Gómez JM, Blázquez J, Baquero F, Martínez JL. H-NS and RpoS regulate emergence of Lac Ara+ mutants of Escherichia coli MCS2. J Bacteriol. 1997;179:4620–4622. doi: 10.1128/jb.179.14.4620-4622.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Lamrani S, et al. Starvation-induced Mucts62-mediated coding sequence fusion: A role for ClpXP, Lon, RpoS and Crp. Mol Microbiol. 1999;32:327–343. doi: 10.1046/j.1365-2958.1999.01352.x. [DOI] [PubMed] [Google Scholar]
- 50.Saumaa S, Tover A, Kasak L, Kivisaar M. Different spectra of stationary-phase mutations in early-arising versus late-arising mutants of Pseudomonas putida: Involvement of the DNA repair enzyme MutY and the stationary-phase sigma factor RpoS. J Bacteriol. 2002;184:6957–6965. doi: 10.1128/JB.184.24.6957-6965.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Ilves H, Hõrak R, Kivisaar M. Involvement of sigma(S) in starvation-induced transposition of Pseudomonas putida transposon Tn4652. J Bacteriol. 2001;183:5445–5448. doi: 10.1128/JB.183.18.5445-5448.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Bjedov I, et al. Stress-induced mutagenesis in bacteria. Science. 2003;300:1404–1409. doi: 10.1126/science.1082240. [DOI] [PubMed] [Google Scholar]
- 53.Slack A, Thornton PC, Magner DB, Rosenberg SM, Hastings PJ. On the mechanism of gene amplification induced under stress in Escherichia coli. PLoS Genet. 2006;2:e48. doi: 10.1371/journal.pgen.0020048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Lombardo MJ, Aponyi I, Rosenberg SM. General stress response regulator RpoS in adaptive mutation and amplification in Escherichia coli. Genetics. 2004;166:669–680. doi: 10.1534/genetics.166.2.669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Robleto EA, Yasbin R, Ross C, Pedraza-Reyes M. Stationary phase mutagenesis in B. subtilis: A paradigm to study genetic diversity programs in cells under stress. Crit Rev Biochem Mol Biol. 2007;42:327–339. doi: 10.1080/10409230701597717. [DOI] [PubMed] [Google Scholar]
- 56.Wright BE, Longacre A, Reimers JM. Hypermutation in derepressed operons of Escherichia coli K12. Proc Natl Acad Sci USA. 1999;96:5089–5094. doi: 10.1073/pnas.96.9.5089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Coros CJ, Piazza CL, Chalamcharla VR, Smith D, Belfort M. Global regulators orchestrate group II intron retromobility. Mol Cell. 2009;34:250–256. doi: 10.1016/j.molcel.2009.03.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Taddei F, Matic I, Radman M. cAMP-dependent SOS induction and mutagenesis in resting bacterial populations. Proc Natl Acad Sci USA. 1995;92:11736–11740. doi: 10.1073/pnas.92.25.11736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Pränting M, Lööv C, Burman R, Göransson U, Andersson DI. The cyclotide cycloviolacin O2 from Viola odorata has potent bactericidal activity against Gram-negative bacteria. J Antimicrob Chemother. 2010;65:1964–1971. doi: 10.1093/jac/dkq220. [DOI] [PubMed] [Google Scholar]
- 60.Pränting M, Andersson DI. Escape from growth restriction in small colony variants of Salmonella typhimurium by gene amplification and mutation. Mol Microbiol. 2011;79:305–315. doi: 10.1111/j.1365-2958.2010.07458.x. [DOI] [PubMed] [Google Scholar]
- 61.Nohmi T. Environmental stress and lesion-bypass DNA polymerases. Annu Rev Microbiol. 2006;60:231–253. doi: 10.1146/annurev.micro.60.080805.142238. [DOI] [PubMed] [Google Scholar]
- 62.Quah SK, von Borstel RC, Hastings PJ. The origin of spontaneous mutation in Saccharomyces cerevisiae. Genetics. 1980;96:819–839. doi: 10.1093/genetics/96.4.819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Lawrence CW. Cellular roles of DNA polymerase zeta and Rev1 protein. DNA Repair (Amst) 2002;1:425–435. doi: 10.1016/s1568-7864(02)00038-1. [DOI] [PubMed] [Google Scholar]
- 64.Sargentini NJ, Smith KC. Much of spontaneous mutagenesis in Escherichia coli is due to error-prone DNA repair: Implications for spontaneous carcinogenesis. Carcinogenesis. 1981;2:863–872. doi: 10.1093/carcin/2.9.863. [DOI] [PubMed] [Google Scholar]
- 65.Albertini AM, Hofer M, Calos MP, Miller JH. On the formation of spontaneous deletions: The importance of short sequence homologies in the generation of large deletions. Cell. 1982;29:319–328. doi: 10.1016/0092-8674(82)90148-9. [DOI] [PubMed] [Google Scholar]
- 66.Bhamre S, Gadea BB, Koyama CA, White SJ, Fowler RG. An aerobic recA-, umuC-dependent pathway of spontaneous base-pair substitution mutagenesis in Escherichia coli. Mutat Res. 2001;473:229–247. doi: 10.1016/s0027-5107(00)00155-x. [DOI] [PubMed] [Google Scholar]
- 67.Rutherford SL, Lindquist S. Hsp90 as a capacitor for morphological evolution. Nature. 1998;396:336–342. doi: 10.1038/24550. [DOI] [PubMed] [Google Scholar]
- 68.Gross L. Climate change could change rates of evolution. PLoS Biol. 2011;9:e1001015. doi: 10.1371/journal.pbio.1001015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Cirz RT, et al. Inhibition of mutation and combating the evolution of antibiotic resistance. PLoS Biol. 2005;3:e176. doi: 10.1371/journal.pbio.0030176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Cohen SE, Walker GC. The transcription elongation factor NusA is required for stress-induced mutagenesis in Escherichia coli. Curr Biol. 2010;20:80–85. doi: 10.1016/j.cub.2009.11.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Kohanski MA, DePristo MA, Collins JJ. Sublethal antibiotic treatment leads to multidrug resistance via radical-induced mutagenesis. Mol Cell. 2010;37:311–320. doi: 10.1016/j.molcel.2010.01.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Huang LE, Bindra RS, Glazer PM, Harris AL. Hypoxia-induced genetic instability—A calculated mechanism underlying tumor progression. J Mol Med (Berl) 2007;85:139–148. doi: 10.1007/s00109-006-0133-6. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.