Abstract
Rad23 and cell division cycle protein 48 (Cdc48), two key regulators of postubiquitylation events, act on distinct and overlapping sets of substrates. The principle underlying their division of labor and cooperation in proteolysis remains elusive. Both Rad23 and Cdc48 bind a ubiquitin protein ligase ubiquitin fusion degradation-2 (Ufd2), and regulate the degradation of Ufd2 substrates. With its ability to bind ubiquitin chains directly and the proteasome via different domains, Rad23 serves as a bridge linking ubiquitylated substrates to the proteasome. The significance and specific role of the Ufd2–Cdc48 interaction are unclear. Here, we demonstrate that mutations in Ufd2 alter its interaction with Cdc48 and impair its function in substrate proteolysis but not in ubiquitylation. Furthermore, Cdc48 promotes the disassembly of the Ufd2–Rad23 complex in an manner that is dependent on ATP and Ufd2 binding, revealing a biochemical role for Cdc48. Rad23 was shown to bind separately to Ufd2 and to the proteasome subunit Rpn1, which define two distinct steps in proteolysis. The action of Cdc48 could free Rad23 from Ufd2 to allow its subsequent association with Rpn1, which in turn may facilitate the orderly transfer of the substrate from the ubiquitylation apparatus to the proteasome.
Keywords: ATPase, protein degradation, complex disassembly
The mechanism by which a ubiquitylated substrate is transferred to the proteasome is largely undefined (1, 2). Rad23-like ubiquitin (Ub)-binding proteins are emerging as key regulators of this process (1, 3). Rad23 belongs to a family of proteins that contains both the Ub-associated (UBA) domain and a Ub-like (UBL) motif that directly bind to Ub conjugates and the proteasome, respectively (1, 4–9). These biochemical properties allow UBA/UBL proteins to act as a bridge between ubiquitylated proteins and the proteasome (1, 3). Indeed, yeast Rad23 and its human homologs are required for the degradation of a subset of substrates, including an artificial model substrate UbV76–V– β-galactosidase (β-gal) (10), the cyclin-dependent kinase (CDK) inhibitor Far1 (11), malfolded proteins (12, 13), and the tumor suppressor p53 (14). However, our understanding of the mechanism underlying the action of Rad23 in vivo remains rudimentary. For example, how does Rad23 recognize specific substrates? How are substrates transferred from the ubiquitylation machinery to Rad23? How are substrates unloaded from Rad23 to the proteasome?
A key to unraveling the mechanism underlying the substrate transfer process is to understand the protein–protein interaction network that postubiquitylation regulators (e.g., Rad23) engage. We found previously that Rad23 directly binds ubiquitin fusion degradation-2 (Ufd2) (15), an E4 enzyme essential for Ub chain assembly onto a subset of proteasomal substrates (16), suggesting that the Ufd2–Rad23 complex links the substrate ubiquitylation step directly to the substrate delivery process. The region of Rad23 responsible for Ufd2 binding was mapped to the UBL element (15), which also is known to bind the proteasome subunit Rpn1 (7). Ufd2 and Rpn1 compete for the binding of Rad23, suggesting that the UBL domain of Rad23 binds to Ufd2 and the proteasome separately (15). This result then raises an interesting question: How is the Ufd2–Rad23 complex disassembled to allow Rad23 to bind Rpn1?
One potential regulator of Ufd2 and Rad23 is cell division cycle protein 48 (Cdc48), which acts on distinct and overlapping sets of substrates (1, 11). Cdc48 belongs to the family of ATPase associated with proteins with diverse cellular activities and believed to function as chaperones and to regulate cell-cycle progression, membrane fusion, and proteolysis (17, 18). Cdc48 is important for a subset of proteolytic pathways, including a protein quality-control process termed “endoplasmic reticulum-associated protein degradation” (ERAD), which selects malfolded secretory proteins for destruction (19). Cdc48 has two copies of the ATPase domain, each containing the consensus Walker A and B motifs that are responsible for ATP binding and hydrolysis. After ATP binding, Cdc48 undergoes structural changes, which could generate a pulling force to disassemble a protein complex (17). Indeed, with the help of two cofactors, ubiquitin fusion degradation-1 (Ufd1) and Npl4, the major function of Cdc48 in ERAD is to catalyze, by an ATP-driven mechanism, the disassembly of a protein complex and to extract a ubiquitylated substrate out of the endoplasmic reticulum (ER), facilitating the substrate’s delivery to and degradation by the proteasome (17, 19).
Cdc48 was shown previously to bind Ufd2 (16) and participates in the Ufd2/Rad23-dependent proteolytic process termed the “ubiquitin fusion degradation” (UFD) pathway that targets an artificially designed substrate UbV76–V–β-gal for proteolysis (20, 21). However, the specific roles of Cdc48 in the UFD pathway remain unclear. It has been proposed that Cdc48 brings Ufd2 to its substrate and thereby facilitates the ubiquitylation reaction by Ufd2 (13), based mainly on data from coimmunoprecipitation assays which demonstrate that the Ufd2–substrate interaction is markedly attenuated in cells with compromised activity of Cdc48 or its cofactors Npl4 and Ufd1 (13). In addition to its function in preubiquitylation, Cdc48 also may regulate a postubiquitylation step. Earlier studies suggest that Cdc48 likely acts downstream of the ubiquitylation reaction by Ufd2 in vivo, because UFD substrates are stabilized and fully ubiquitylated in cdc48 and ufd1 mutants (16, 21). It is possible that Cdc48 has multiple roles in the UFD pathway, because Cdc48 is known to regulate multiple events in ERAD through its association with diverse cofactors (17).
By analogy to its role in ERAD processing, we reasoned that Cdc48 mediates the disassembly of the complex through its association with Ufd2. A key to unraveling the specific function of Cdc48 in the UFD pathway then is to determine the significance and function of the Ufd2–Cdc48 interaction. In this work, rather than using a ufd2 deletion strain and temperature-sensitive mutants of Cdc48 or Npl4 that have multiple known and unknown defects, we isolated alleles of UFD2 (i.e., G274D, C385Y) that are specifically defective in Cdc48 binding. We found that the Ufd2–Cdc48 interaction is pivotal for substrate degradation in vivo. Moreover, Cdc48 destabilizes the Ufd2–Rad23 interaction in an ATP- and Ufd2-binding–dependent manner. We also demonstrate that Ufd2 requires the binding of Cdc48 for its function in ERAD. Our results provide one biochemical mechanism by which Cdc48 may modulate substrate proteolysis and further suggest that various protein–protein interactions are key to the transfer of ubiquitylated substrates to the proteasome.
Results
Mutations in Ufd2 Specifically Disrupt Its in Vivo Binding to Cdc48.
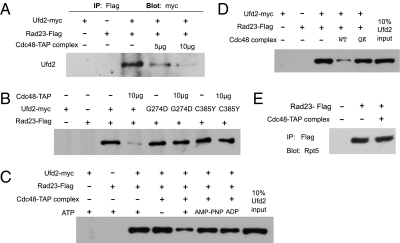
Ufd2 was shown to bind Cdc48 and Rad23 (13, 15, 16) (Fig. 1 and SI Appendix, Fig. S1A). We have shown previously that the Ufd2–Rad23 interaction is pivotal for substrate degradation (15). To establish the importance of the Ufd2–Cdc48 association in regulated proteolysis, we isolated mutants that abolish Ufd2–Cdc48 interaction. Ufd2 is an E4 enzyme involved in specific substrate degradation (16), whereas Cdc48 regulates many cellular processes essential for cell growth and survival (17). Therefore, we performed a “negative two-hybrid” screen to isolate alleles of UFD2 that are defective in Cdc48 binding. The yeast two-hybrid vector bearing Ufd2 fused to the Gal4 DNA-binding domain was mutagenized randomly with hydroxylamine and then transformed into yeast. The mutant Ufd2 proteins generated were analyzed subsequently by two-hybrid assay for their ability to interact with Rad23 and Cdc48. We found that the G274D and C385Y mutations in Ufd2 disrupted binding to Cdc48 in vivo but did not impair interaction with Rad23 (Fig. 1A).
Fig. 1.
Interactions between Ufd2 and Rad23 or Cdc48. (A) Two-hybrid analysis of the interaction between Ufd2 derivatives and Rad23 or Cdc48. Yeast cells were cotransformed with plasmids encoding the Gal4 DNA-binding domain fused to Ufd2 or its mutant alleles as indicated and the Gal4 activation domain fused to Rad23 or Cdc48. Fourfold dilutions were spotted on the indicated plates. (Right) Growth on the SD-Leu-Trp–His plate is indicative of protein–protein interaction. (Left) Control plate (SD-Leu-Trp) indicates that equal amounts of cells were plated. (B) Purification of Ufd2, Rad23, and Cdc48 from E. coli. A Coomassie blue-stained gel shows purified RGS/His6-tagged Cdc48, Flag-tagged Rad23, and myc-tagged Ufd2 alleles. (C and D) Coimmunoprecipitation analysis of associations between Ufd2 and Cdc48 (C) or Ufd2 and Rad23 (D). Purified Ufd2-myc (2 μg) and RGS/His6-Cdc48 (2 μg) or Flag-Rad23 (2 μg) were mixed in various combinations as indicated in 150 μL binding buffer [50 mM Na-Hepes (pH 7.5), 150 mM NaCl, 5 mM EDTA, 2% Triton X-100, 0.2 mg/mL BSA] containing 1× protease inhibitor mix (Roche) and were incubated with 10 μL (bed volume) of beads coated with various antibodies for 2 h at 4 °C. The beads were washed four times with the binding buffer, followed by SDS/PAGE of the retained proteins and immunoblotting with the antibody indicated.
Ufd2 has been shown to interact directly with Cdc48 or Rad23. To ensure that the mutations directly affect these Ufd2 interactions, we set up in vitro binding assays with these Ufd2 alleles. Specifically, Flag-tagged Rad23, His6-tagged Cdc48, and wild-type and mutant alleles of Myc-tagged Ufd2 were expressed in Escherichia coli and purified separately (Fig. 1B). These purified proteins were incubated in various combinations, and immunoprecipitations were carried out. We found that these Ufd2 mutants are specifically defective for Cdc48 binding but retain normal Rad23 binding in vitro (Fig. 1 C and D).
Cdc48 Binding Is Essential for Ufd2 Function in the UFD Pathway.
To address the functional significance of the Ufd2–Cdc48 interaction, the mutations described above were introduced into UFD2 under the regulation of its own promoter on a low-copy plasmid to avoid potential complications associated with overexpression. Mutations in UFD2 did not affect its expression levels in vivo (Fig. 2A). These plasmids carrying UFD2 derivatives were cotransformed with a plasmid expressing the UFD substrate UbV76–V–β-gal into ufd2Δ cells. We first used the LacZ assay to assess the effect of the UFD2 mutations on the concentration of the UbV76–V–β-gal, and found that the expression of wild-type but not mutant UFD2 in the ufd2Δ cells restores low levels of β-gal activity (Fig. 2B), suggesting that UFD substrate degradation is impaired in Ufd2 mutants defective for Cdc48 interaction. We also performed an expression–shut-off assay to measure the stability of the UFD substrate in these cells. As expected, wild-type but not mutant ufd2 alleles rendered UbV76–V–β-gal unstable (Fig. 2 C and D). These results indicate that Cdc48 binding is essential for the function of Ufd2 in UFD substrate turnover.
Fig. 2.
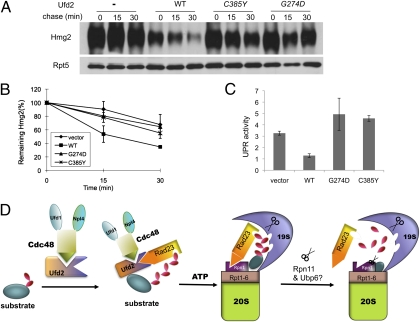
UFD2 mutants impair substrate degradation but not ubiquitylation. (A) Expression of Ufd2 derivatives in yeast. A low-copy plasmid with or without expressing myc-tagged Ufd2 derivatives from the UFD2 promoter was transformed into ufd2Δ cells. Ufd2 was precipitated with myc beads and analyzed by immunoblotting. The amount of Rpt5 in the yeast extracts serves as the loading control. (B) Levels of β-gal activity in ufd2Δ cells carried a plasmid bearing UbV76–V–β-gal and a low-copy plasmid with or without expressing UFD2 derivatives from the UFD2 promoter as indicated. The experiments were done more than three times, and average values with SD are shown. (C) UFD substrate UbV76–V–β-gal degradation is compromised in ufd2 mutant cells. First, yeast cells containing indicated Ufd2 alleles and a GAL1 promoter-regulated UbV76–V–β-gal were grown in raffinose-containing medium. Expression of UbV76–V–β-gal was induced by the addition of galactose for 1 h. Samples were taken after promoter shutoff at the time points indicated and analyzed by anti–β-gal Western blot. (D) Quantitation of the data in C for UbV76–V–β-gal. SDs are shown. (E) Ufd2 mutants maintain substrate ubiquitylation. UbV76–V–β-gal was cotransformed with plasmid p81 bearing Ha-tagged Ub into wild-type or ufd2Δ cells expressing various Ufd2 derivatives. UbV76–V–β-gal was isolated with β-gal antibody-coated beads and probed with anti-Ha antibody to detect ubiquitylated UbV76–V–β-gal species. Rpt5 serves as a loading control. The UFD2 alleles on the plasmids are labeled, and the UFD2 status in the strains is indicated at the top.
While our work was in progress, the C-terminal fragment of Cdc48 was shown to be responsible for its binding to Ufd2 and ubiquitin fusion degradation-3 (Ufd3), a Ub-binding protein involved in Ub homeostasis, histone ubiquitylation, protein trafficking, and autophagy (22, 23). Mutations in Cdc48 were used to demonstrate the functional significance of its binding to Ufd2 and Ufd3 in proteolysis (22). However, these Cdc48 mutations impaired interaction with both Ufd2 and Ufd3, not allowing a clear separation of two distinct functions of Cdc48. Cdc48 regulates a variety of important cellular processes, including cell-cycle progression, autophagy, and proteolysis. Its functional diversity is achieved through different binding partners (e.g., Ufd1–3, Ubx1–7) (17, 18). Instead of using mutations of Cdc48 that have multiple known and unknown defects, we characterized point mutations in Ufd2 that specifically alter its binding to Cdc48. In addition, our study reveals a more specific role of Cdc48 in the Ufd2 pathway (see below).
To delineate the specific proteolytic function of the Ufd2–Cdc48 association, we examined the effect of Ufd2 mutations on the ubiquitylation pattern of UbV76–V–β-gal. Consistent with earlier results, levels of multiubiquitylated UbV76–V–β-gal species were reduced in ufd2Δ cells. The expression of wild-type UFD2 restored the appearance of UbV76–V–β-gal multiubiquitylation products (Fig. 2E). Interestingly, ufd2 mutants also rescued the ubiquitylation deficiency in ufd2Δ cells, suggesting that, although these mutants maintain their ubiquitylation activity, the proteolytic defects observed lie in compromised postubiquitylation function.
Cdc48 Promotes Disassembly of the Ufd2–Rad23 Complex.
We next set out to elucidate the function of Cdc48 in the Ufd2 pathway. Previous studies have shown that Cdc48 uses its ATPase activity to disassemble protein complexes (17, 18). Therefore, it is likely that Cdc48 functions similarly in the UFD pathway by mediating the disassembly of protein complexes through its association with Ufd2. However, the identity of proteins liberated by the Cdc48Ufd1/Npl4 complex remains unknown. Because the UBL motif of Rad23 binds separately to Ufd2 and Rpn1 (15), two proteins with distinct functions, the Ufd2–Rad23 complex presents an obvious candidate target of Cdc48.
To address directly the role of Cdc48 in the Ufd2–Rad23 association, we set up in vitro binding assays with various Ufd2 mutant controls to eliminate artifacts. Cdc48 alone has little effect on the Ufd2–Rad23 interaction (SI Appendix, Fig. S1B). Similar to its involvement in ERAD, Cdc48 probably would require various cofactors (e.g., Ufd1, Npl4, Shp1) for its function in the UFD pathway. Following a previously established procedure for TAP purification (24), we purified the Cdc48-containing complex from yeast cells containing endogenous Cdc48 appended with a tandem affinity purification (TAP) tag. Consistent with earlier studies (17, 18), Ufd1 protein was present in the Cdc48 immunoprecipitates (SI Appendix, Fig. S1C). Interestingly, the Ufd2–Rad23 interaction was destabilized by increasing amounts of the Cdc48-containing complex (Fig. 3A). Furthermore, Ufd2 mutants defective for Cdc48 binding retained their affinity for Rad23 (Fig. 3B), suggesting that Ufd2–Cdc48 binding is crucial for modulation of the stability of the Ufd2–Rad23 complex.
Fig. 3.
Cdc48 promotes the disassembly of the Ufd2–Rad23 complex. (A) Increasing the amount of Cdc48-containing complex destabilizes the Ufd2–Rad23 interaction. The Cdc48-containing complex was purified from Cdc48-TAP strain following a previously described protocol for TAP purification (24). Purified Ufd2-myc (2 μg) and Rad23-Flag (2 μg) (Fig. 1B) were mixed as indicated in 150 μL binding buffer containing ATP and MgCl2 (36) and were incubated with 10 μL Flag-agarose (Sigma) for 2 h at 4 °C. After the beads were washed two times with the binding buffer, increasing amounts of the Cdc48-containing fraction (5 μg or 10 μg) isolated from the Cdc48-TAP strain were added and incubated for an additional hour (36). The beads were washed three times, followed by SDS/PAGE and immunoblotting with a monoclonal anti-myc antibody. (B and C) The effect of Cdc48 on the Ufd2–Rad23 association requires ATP and Ufd2 binding. The experiments were preformed as described above, except that Ufd2 mutants or ATP analogs (i.e., AMP-PNP, ADP) were used or ATP was omitted in the reaction. Ten micrograms of the Cdc48-containing fraction derived from the Cdc48–TAP complex was used in the experiment. (D) The Cdc48 E588Q mutant defective for ATP hydrolysis is ineffective in promoting Ufd2–Rad23 dissociation. The binding assay was carried out as described above. Wild-type and E588Q Cdc48 tagged with the His6 epitope were purified as described in Fig. 1B. These purified proteins were incubated with yeast extracts to bring down other Cdc48 cofactors in the presence of Ni-NTA beads. The Cdc48-containing complex was eluted with 250 mM imidazole and used in the Ufd2–Rad23 disassembly experiment as in A. Ten micrograms of the Cdc48-containing complex was used in the experiment. (E) Cdc48 does not affect the Rad23–proteasome interaction. Purified Flag-Rad23 was mixed with yeast extracts to bring down the associated proteasome and then was incubated with or without 10 μg of the Cdc48-containing fraction derived from the Cdc48–TAP complex. The Rad23-associated proteasome was detected by anti-Rpt5 antibody.
ATP is essential to the functioning of Cdc48 (17, 18, 25, 26). Importantly, we found that the Ufd2–Rad23 interaction was unaltered by the Cdc48-containing complex in the absence of ATP (Fig. 3C). In addition, two ATP analogs, β-γ-imidoadenosine 5′-phosphate (AMP-PNP) and ADP, were inefficient in promoting the disassembly of the Ufd2–Rad23 complex (Fig. 3C). Because AMP-PNP has a nonhydrolysable bond, the data suggest that ATP hydrolysis is important for Cdc48-mediated complex disassembly. To ascertain the requirement of ATP hydrolysis in this reaction, we used a mutation E588Q in the Walker B motif of Cdc48, which was shown to inactivate its ATP hydrolysis and impair proteolysis (17, 27). The E588Q mutation does not affect its binding to Ufd1 (SI Appendix, Fig. S1D). Interestingly, the E588Q mutant did not affect the Ufd2–Rad23 association significantly (Fig. 3D), supporting the significance of ATP hydrolysis. Combined, our results suggest that Cdc48 likely facilitates the disassembly of the Ufd2–Rad23 complex in both an ATP- and Ufd2-binding–dependent manner.
In contrast to the effect of Cdc48 on the Ufd2–Rad23 interaction (Fig. 3A), Rad23–proteasome binding was unaffected by the Cdc48 complex (Fig. 3E). Furthermore, the Ufd2–Cdc48 association was not altered by increasing amounts of Rad23 (SI Appendix, Fig. S1E), supporting a specific regulatory role of Cdc48 on the Ufd2–Rad23 binding. Although other possibilities may exist, our combined results favor the model that Cdc48 promotes the disassembly of the Ufd2–Rad23 complex. Given their association with Ufd2, we also examined whether Cdc48 and Rad23 can be detected in one complex. We analyzed the in vivo binding between Cdc48 and Rad23 in ufd2Δ cells by coimmunoprecipitation. Without Ufd2, we did not detect the Cdc48–Rad23 interaction (SI Appendix, Fig. S2A, lane 2). Interestingly, the Cdc48–Rad23 association was observed upon Ufd2 expression (SI Appendix, Fig. S2A), suggesting that Ufd2 may be the mediator of the Cdc48–Rad23 interaction. When Ufd2, Rad23, and Cdc48 are mixed together in vitro, both Cdc48 and Rad23 bind Ufd2 efficiently (SI Appendix, Fig. S2B, first row), but the interaction between Rad23 and Cdc48 is rather weak and probably is transient (SI Appendix, Fig. S2B, second row).
Ufd2 Mutants Exhibit Increased Association with Rad23 in Vivo.
In the absence of Cdc48, Rad23 exhibits a similar affinity toward wild-type and mutant Ufd2 proteins in vitro (Fig. 1D). If Cdc48 promotes the in vivo disassembly of the Ufd2–Rad23 complex, Rad23 may be trapped by Ufd2 alleles defective for Cdc48 binding. Indeed, Ufd2 mutants exhibit enhanced two-hybrid interaction with Rad23 relative to wild-type Ufd2 in vivo (Fig. 1A). We also used coimmunoprecipitation analysis to evaluate the in vivo association between Ufd2 alleles and Rad23 or Cdc48 (Fig. 4 A and B). Interestingly, consistent with yeast two-hybrid results, Rad23 has a stronger affinity for Ufd2 mutants than for wild-type Ufd2 (Fig. 4A), suggesting that the binding between Rad23 and Ufd2 mutants is more stable without Cdc48. Taken together, these results support the conjecture that Cdc48 modulates the Ufd2–Rad23 interaction in vivo.
Fig. 4.
Mutations in Ufd2 alter its binding to Rad23 and Cdc48 in vivo. (A) Coimmunoprecipitation analysis of interactions between Rad23 and Ufd2 derivatives. Proteins were extracted from cells expressing Flag-tagged Rad23 and myc-tagged Ufd2 alleles and immunoprecipitated with beads coupled to various antibodies as indicated. Immunoprecipitates were separated on SDS/PAGE, transferred to a PVDF membrane, and then probed with antibodies. Mutations in Ufd2 are indicated above the panels. The antibodies used for immunoprecipitation (IP) and Western blot (Blot) are indicated at the right of the panels. (B) Coimmunoprecipitation analysis of interactions between Cdc48 and Ufd2 derivatives.
Mutations in Ufd2 Impair Degradation of Hmg2.
Ufd2 was shown to be required for the efficient elimination of several malfolded ERAD substrates, including Hmg2 and Ste6* (13, 25, 26). We therefore assessed whether Ufd2–Cdc48 interaction is important for Hmg2 turnover. As reported previously, Hmg2 is stabilized in ufd2Δ cells (13). The expression of wild-type but neither of two mutant ufd2 alleles in ufd2Δ cells restored the rapid turnover of Hmg2 (Fig. 5 A and B), suggesting that the Ufd2–Cdc48 interaction is pivotal for the degradation function of UFD2 in ERAD.
Fig. 5.
Compromised ERAD and UPR activities in ufd2 mutants. (A) Mutations of UFD2 affect the degradation of the ER membrane protein Hmg2. The plasmid expressing wild-type or mutant UFD2 alleles was introduced into ufd2Δ cells harboring myc-Hmg2. Time points were taken after expression shut-off. Membrane proteins were fractioned from these cells and were immunoprecipitated with beads coupled to myc antibody to enrich myc-Hmg2; immunoprecipitates were resolved by SDS/PAGE and then were probed with anti-myc antibody. The stable protein Rpt5 was used as the loading control. (B) Quantitation of the data in A for Hmg2. (C) Elevated UPR activity in cells bearing UFD2 mutants. Yeast cells were transformed with a plasmid that contains the LACZ gene under the control of the promoter of KAR2, which encodes an ER chaperone. Levels of β-gal activity in yeast cells were derived from more than three independent experiments. The strain genotypes are indicated. Bars indicate SD. (D) A model for substrate transfer to the proteasome mediated by a protein–protein interaction network. Cdc48 could play dual roles to promote substrate degradation by assisting Ufd2-mediated ubiquitylation and Rad23-facilitated substrate delivery. The Ub-conjugation phase depicted is based on published evidence (1, 13, 15, 16). The 26S proteasome is drawn as described in ref. 37 with minor modifications. Through interactions with Ufd2, Cdc48, and Rpn1, Rad23 recognizes the substrate and facilitates substrate transfer to the proteasome. See text for details.
Consistent with the involvement of Ufd2 in malfolded protein degradation, cells lacking UFD2 show constitutive activation of the unfolded protein response (UPR) (26). We found that higher UPR activity remained in ufd2Δ mutant cells expressing Ufd2 alleles defective for Cdc48 binding (Fig. 5C).
Discussion
One of the most challenging questions in the field of proteolysis is how the substrate is delivered to the proteasome. It now is widely accepted that Ub-binding proteins (e.g., Rad23, Cdc48, and Rpn10) play important roles in this process (1, 3). These proteins have distinct and sometimes overlapping substrates and cooperate functionally for substrate proteolysis (1, 11, 13). However, how any one of these proteins works in vivo remains poorly understood. Our study reveals the specific role of Cdc48 in assisting Rad23’s function via Ufd2 binding. Although the interaction of Ufd2 with Cdc48 first was demonstrated 12 y ago (16), the significance and specific role of this association remained elusive. We isolated two point mutations in UFD2 that specifically disrupt Ufd2 protein binding to Cdc48. Our in vivo analysis demonstrates that the Ufd2–Cdc48 interaction is pivotal for substrate degradation. Furthermore, our results suggest that mutations in Ufd2 impair proteolysis at a postubiquitylation but preproteasome step, because UFD substrates are stabilized but efficiently ubiquitylated in cells harboring these Ufd2 alleles (Fig. 2E), a phenotype that also was detected in cdc48 and ufd1 mutants (16, 21). Because Cdc48 fulfills its diverse cellular functions through its interaction with different cofactors (e.g., Ufd1–3, Ubx1–7) (17), our study also suggests that it is advantageous to isolate and characterize mutations in these cofactors rather than in Cdc48 to elucidate the separate functions of Cdc48.
Like its involvement in ERAD (17), Cdc48 may play multiple roles in the UFD pathway. Two mutually compatible roles have been proposed. Based mainly on the results derived from coimmunoprecipitation analyses, it was proposed that Cdc48 acts upstream of Ufd2 and brings Ufd2 to its substrate, which in turn facilitates the ubiquitylation by Ufd2 (13). For example, the in vivo Ufd2–substrate association is disrupted in temperature-sensitive cells bearing npl4 or ufd1 mutations (13). Because Cdc48-Npl4-Ufd1 are essential for cell viability with key regulatory roles in diverse cellular processes (17), a caveat is that some of the phenotypes observed in these temperature-sensitive mutant cells may be indirect or nonspecific. Note that nearly all protein–protein interactions (e.g., Ufd2–Cdc48, Cdc48–substrates, Ufd2–substrates, Ufd2–Rad23, Rad23–substrates) are reduced in cells harboring npl4 or ufd1 mutations (13). Because the Cdc48Ufd1/Npl4 complex regulates multiple cellular events, it also is possible that one dominant deficiency in cells with compromised Npl4, Cdc48, or Ufd1 activity may mask other defects associated with these cells, as witnessed with various cell-cycle mutants including cdc28.
Besides its involvement in Ufd2-mediated ubiquitylation (13), Cdc48 has another possible, not mutually exclusive, function in proteolysis. Earlier studies in vivo suggest that Cdc48 may act downstream of the ubiquitylation reaction by Ufd2, because stabilized UFD substrates are fully ubiquitylated in cdc48 and ufd1 mutants (16, 21). Our results reveal the significance and specific role of the Ufd2–Cdc48 interaction. In ERAD, Cdc48 appears to perform multiple functions, because it can bind to both nonubiquitylated and ubiquitylated proteins (27, 28).
One postubiquitylation event that may be under the control of Cdc48 is the association between Ufd2 and Rad23. The proteolytic function of Cdc48 is understood best in the ERAD pathway, where it binds Ufd1 and Npl4 (17, 19, 27). The Cdc48Ufd1/Npl4 complex uses its chaperone activity to extract ubiquitylated ER proteins associated with the ER translocon and to facilitate degradation by the cytosolic proteasome. The proteolytic roles of both Ufd1 and Cdc48 were revealed first by their requirement for efficient UFD substrate degradation. The specific function of the Cdc48Ufd1/Npl4 complex in the UFD pathway was not known previously. We found that the Cdc48-containing complex destabilizes Ufd2–Rad23 binding, the specificity of which was demonstrated further by its dependence on ATP and Ufd2 binding (Fig. 3).
Rad23 plays a key role in bringing ubiquitylated proteins to the proteasome, although the underlying mechanism remains elusive (1). Our previous study (15) suggests that Rad23 employs the same UBL domain to bind separately to Ufd2, a ubiquitylation enzyme, and Rpn1, a proteasome subunit, which define distinct steps in substrate degradation. A dilemma then is how Rad23 switches between its binding partners, Ufd2 and Rpn1. Our data suggest that Cdc48 may promote the disassembly of the Ufd2–Rad23 complex (Fig. 3) and that Rad23–Rpn1 binding probably is regulated by factors other than Cdc48 (Fig. 3E). Rad23 also could bind to the proteasome through its bindings with the proteasome subunit Rpn13 and/or Ub conjugates via UBA domains (1, 3). These multiple interactions promote the docking of Rad23 to the proteasome. The Rad23–Rpn1 interaction probably is crucial for substrate proteolysis (Fig. 2 and ref. 15).
Based on these studies, we propose a model (Fig. 5D) in which Cdc48 plays two functions in helping both Ufd2-mediated protein ubiquitylation and Rad23-facilitated substrate delivery to the proteasome through a network of protein–protein interactions involving the components of the ubiquitylation machinery (e.g., Ufd2) and proteasome subunits (e.g., Rpn1). Specifically, following the appendage of a few Ubs onto the UFD substrate mediated by Ufd4 (E3) and other required components (e.g., E1, E2), Ufd2 further extends the Ub chain (16); this extension may require Cdc48 to recognize or extract some substrates (13, 17, 19, 27). Rad23 is recruited to the ubiquitylation machinery via its binding to Ufd2, which in turn facilitates target recognition by the UBA domain of Rad23. Then the Cdc48 ATPase comes in to promote the release of the UBL element of Rad23 from Ufd2, because the UBL motif is needed to bind the proteasome subunit Rpn1. The Cdc48-facilitated sequential interactions of Rad23 would allow an orderly handoff of the substrate from the ubiquitylation machinery to the proteasome. The Rad23–Rpn1 interaction helps bring the ubiquitylated substrate to the proteasome. The final transfer of the substrate to the proteasome remains enigmatic (1). It is tempting to speculate that the dockings of Rad23 and the Ub chain onto the proteasome subunits (e.g., Rpn1, Rpn10, Rpn13) ensure the loading of the substrate onto the proper site of the proteasome for subsequent unfolding and degradation. Ub chains later are cleaved from the substrate by deubiquitylation enzymes (e.g., Ubp6, Rpn11) (1), which not only may facilitate the unloading of the substrate onto the proteasome for processing but also may promote the release of Rad23 from the proteasome (Fig. 5D).
Multiple pathways are used in bringing the ubiquitylated substrates to the proteasome (1). Thus, yeast Ufd2 substrates are regulated by Rad23 and Cdc48, but not all Rad23 and Cdc48 substrates require Ufd2 (e.g., Ubc6*, ricin A chain) (13, 26, 29). Recent studies of Ufd2 have revealed the involvement of Ufd2 in protein quality control, transcription regulation, muscle formation, and tumor suppression (13, 26, 30, 31). Knockout of mouse UFD2 leads to cardiac defects and neurodegeneration (32). The model proposed above probably is applicable to most, if not all, Ufd2 targets.
All three proteins, Rad23, Ufd2, and Cdc48, are highly conserved from yeast to human. Homologs of Ufd2 and Cdc48 regulate the stability of Ataxin-3, a polyQ protein involved in Machado–Joseph disease (33, 34). Rad23 binds to Ataxin-3 (34) and is present in cellular inclusions formed by Ataxin-3. Our studies of the functional relationship among Rad23, Ufd2, and Cdc48 in a genetically amenable system may shed light on their roles in Ataxin-3 degradation and human diseases.
Materials and Methods
Strains, Media, and Plasmids.
Yeast cultures were grown in rich (YPD) or synthetic media containing standard ingredients and 2% glucose (SD medium), 2% raffinose (SR medium), or 2% raffinose + 2% galactose (SRG medium). Saccharomyces cerevisiae PJ69-4A (MATa lys2-801 ura3-52 trp1-109 his3-Δ200 leu2-3,112 Gal4Δ gal80Δ GAL2-ADE2 LYS2::GAL1-HIS3 met::GAL7-lacZ) was used for yeast two-hybrid assays. Strains EJY130 (UFD2::LEU2) and isogenic wild-type strain EJ128 were obtained from Alex Varshavsky (California Institute of Technology, Pasadena, CA).
The plasmids expressing the proteasomal substrates UbV76–V–β-gal or Hmg2-myc and the protein stability assays have been described previously (35). Two Ufd2 mutations were incorporated into yeast expression vector containing wild-type Ufd2 tagged with the myc epitope under the regulation of its own promoter (∼4 kb), pRS314-Pufd2-Ufd2 myc, by site-directed mutagenesis. Cdc48 was amplified by PCR, tagged with RGS/His6 epitope, and cloned into pRS415G vector for yeast expression. For its expression in E. coli, His6-tagged Cdc48 was cloned into pGEX4T (Pharmacia Biotech). The plasmids for Rad23 and Ufd2 expression have been described previously (15). Anti-Ufd1 antibody was a kind gift of C. L. Moore (Tufts University, Boston, MA).
Isolation of Ufd2 Mutants Defective in Cdc48 Binding.
To isolate Ufd2 mutants defective in Cdc48 binding, yeast two-hybrid plasmid pGBT-Ufd2 containing wild-type Ufd2 was mutagenized with hydroxylamine. Mutagenized plasmids then were transformed into PJ69-4A containing pGAD-Cdc48. The two-hybrid assay was used to determine the interaction between Ufd2 alleles and Cdc48 on SD-Leu-Trp-His plates. Ufd2 alleles defective in Cdc48 binding then were tested for Rad23 interaction by the two-hybrid assay.
Purification of Ufd2, Rad23, and Cdc48 from E. coli.
GST fusions of Ufd2-myc and Flag-Rad23 were purified as described previously (15). GST-His6–tagged Cdc48 was expressed in the BL21 strain. Cdc48 expression was induced with 1 mM isopropylthio-β-d-1-thiogalactoside at 30 °C for 4 h. Cells were dissolved in lysis buffer (0.25 M NaCl, 2.7 mM KCl, 100 mM Na2HPO4, 2 mM KH2PO4, 10% glycerol) and lysed by sonication. The extracts then were subjected to centrifugation at 17,000 × g for 30 min. The supernatant was incubated with glutathione-agarose beads for 2 h. We eluted His6-tagged Cdc48 proteins free of GST fusion with the thrombin protease that cleaves after the GST portion. The eluent then was incubated with Ni-NTA beads for 1 h at 4 °C. His6-tagged Cdc48 was eluted with buffer containing 250 mM imidazole as described previously (15).
Coimmunoprecipitation/Immunoblotting Assay.
Yeast cells carrying either vectors or pRS314-Pufd2-Ufd2myc (expressing myc-tagged Ufd2) and pRS416G-Flag-Rad23 (bearing Flag-tagged Rad23) or pRS415G-RGS/His6-Cdc48 (expressing RGS/His6-tagged Cdc48), as indicated, were grown in synthetic medium to log phase, followed by preparation of extracts, were immunoprecipitated with beads coated with the specific antibodies indicated, and were resolved by SDS/PAGE and immunoblotting separately with anti-Flag (Sigma-Aldrich), anti-myc, or His6 (Covance).
Supplementary Material
Acknowledgments
We thank C. L. Moore, D. Ng, R. Hampton, and P. A. Silver for reagents and the members of the H.R. laboratory for discussion. H.R. is supported by Grant GM078085from the National Institutes of Health, Grant AQ-1747 from the Welch Foundation, and by a University of Texas Health Science Center Institutional Pilot Grant.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1104051108/-/DCSupplemental.
References
- 1.Finley D. Recognition and processing of ubiquitin-protein conjugates by the proteasome. Annu Rev Biochem. 2009;78:477–513. doi: 10.1146/annurev.biochem.78.081507.101607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Hochstrasser M. Introduction to intracellular protein degradation. Chem Rev. 2009;109:1479–1480. doi: 10.1021/cr900054t. [DOI] [PubMed] [Google Scholar]
- 3.Liu F, Walters KJ. Multitasking with ubiquitin through multivalent interactions. Trends Biochem Sci. 2010;35:352–360. doi: 10.1016/j.tibs.2010.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bertolaet BL, et al. UBA domains of DNA damage-inducible proteins interact with ubiquitin. Nat Struct Biol. 2001;8:417–422. doi: 10.1038/87575. [DOI] [PubMed] [Google Scholar]
- 5.Chen L, Madura K. Rad23 promotes the targeting of proteolytic substrates to the proteasome. Mol Cell Biol. 2002;22:4902–4913. doi: 10.1128/MCB.22.13.4902-4913.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Schauber C, et al. Rad23 links DNA repair to the ubiquitin/proteasome pathway. Nature. 1998;391:715–718. doi: 10.1038/35661. [DOI] [PubMed] [Google Scholar]
- 7.Elsasser S, et al. Proteasome subunit Rpn1 binds ubiquitin-like protein domains. Nat Cell Biol. 2002;4:725–730. doi: 10.1038/ncb845. [DOI] [PubMed] [Google Scholar]
- 8.Rao H, Sastry A. Recognition of specific ubiquitin conjugates is important for the proteolytic functions of the ubiquitin-associated domain proteins Dsk2 and Rad23. J Biol Chem. 2002;277:11691–11695. doi: 10.1074/jbc.M200245200. [DOI] [PubMed] [Google Scholar]
- 9.Wilkinson CRM, et al. Proteins containing the UBA domain are able to bind to multi-ubiquitin chains. Nat Cell Biol. 2001;3:939–943. doi: 10.1038/ncb1001-939. [DOI] [PubMed] [Google Scholar]
- 10.Lambertson D, Chen L, Madura K. Pleiotropic defects caused by loss of the proteasome-interacting factors Rad23 and Rpn10 of Saccharomyces cerevisiae. Genetics. 1999;153:69–79. doi: 10.1093/genetics/153.1.69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Verma R, Oania R, Graumann J, Deshaies RJ. Multiubiquitin chain receptors define a layer of substrate selectivity in the ubiquitin-proteasome system. Cell. 2004;118:99–110. doi: 10.1016/j.cell.2004.06.014. [DOI] [PubMed] [Google Scholar]
- 12.Medicherla B, Kostova Z, Schaefer A, Wolf DH. A genomic screen identifies Dsk2p and Rad23p as essential components of ER-associated degradation. EMBO Rep. 2004;5:692–697. doi: 10.1038/sj.embor.7400164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Richly H, et al. A series of ubiquitin binding factors connects CDC48/p97 to substrate multiubiquitylation and proteasomal targeting. Cell. 2005;120:73–84. doi: 10.1016/j.cell.2004.11.013. [DOI] [PubMed] [Google Scholar]
- 14.Glockzin S, Ogi FX, Hengstermann A, Scheffner M, Blattner C. Involvement of the DNA repair protein hHR23 in p53 degradation. Mol Cell Biol. 2003;23:8960–8969. doi: 10.1128/MCB.23.24.8960-8969.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kim I, Mi K, Rao H. Multiple interactions of rad23 suggest a mechanism for ubiquitylated substrate delivery important in proteolysis. Mol Biol Cell. 2004;15:3357–3365. doi: 10.1091/mbc.E03-11-0835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Koegl M, et al. A novel ubiquitination factor, E4, is involved in multiubiquitin chain assembly. Cell. 1999;96:635–644. doi: 10.1016/s0092-8674(00)80574-7. [DOI] [PubMed] [Google Scholar]
- 17.Buchberger A. Control of ubiquitin conjugation by cdc48 and its cofactors. Subcell Biochem. 2010;54:17–30. doi: 10.1007/978-1-4419-6676-6_2. [DOI] [PubMed] [Google Scholar]
- 18.Jentsch S, Rumpf S. Cdc48 (p97): A “molecular gearbox” in the ubiquitin pathway? Trends Biochem Sci. 2007;32:6–11. doi: 10.1016/j.tibs.2006.11.005. [DOI] [PubMed] [Google Scholar]
- 19.Vembar SS, Brodsky JL. One step at a time: Endoplasmic reticulum-associated degradation. Nat Rev Mol Cell Biol. 2008;9:944–957. doi: 10.1038/nrm2546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ghislain M, Dohmen RJ, Levy F, Varshavsky A. Cdc48p interacts with Ufd3p, a WD repeat protein required for ubiquitin-mediated proteolysis in Saccharomyces cerevisiae. EMBO J. 1996;15:4884–4899. [PMC free article] [PubMed] [Google Scholar]
- 21.Johnson ES, Ma PCM, Ota IM, Varshavsky A. A proteolytic pathway that recognizes ubiquitin as a degradation signal. J Biol Chem. 1995;270:17442–17456. doi: 10.1074/jbc.270.29.17442. [DOI] [PubMed] [Google Scholar]
- 22.Bohm S, Lamberti G, Fernandez-Saiz V, Stapf C, Buchberger A. Cellular functions of Ufd2 and Ufd3 in proteasomal protein degradation depend on Cdc48 binding. Mol Cell Biol. 2011;31:1528–1539. doi: 10.1128/MCB.00962-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zhao G, Li G, Schindelin H, Lennarz WJ. An Armadillo motif in Ufd3 interacts with Cdc48 and is involved in ubiquitin homeostasis and protein degradation. Proc Natl Acad Sci USA. 2009;106:16197–16202. doi: 10.1073/pnas.0908321106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Keogh MC, et al. The Saccharomyces cerevisiae histone H2A variant Htz1 is acetylated by NuA4. Genes Dev. 2006;20:660–665. doi: 10.1101/gad.1388106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Nakatsukasa K, Huyer G, Michaelis S, Brodsky JL. Dissecting the ER-associated degradation of a misfolded polytopic membrane protein. Cell. 2008;132:101–112. doi: 10.1016/j.cell.2007.11.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Liu C, et al. Ubiquitin chain elongation enzyme Ufd2 regulates a subset of Doa10 substrates. J Biol Chem. 2010;285:10265–10272. doi: 10.1074/jbc.M110.110551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ye Y, Meyer HH, Rapoport TA. Function of the p97-Ufd1-Npl4 complex in retrotranslocation from the ER to the cytosol: Dual recognition of nonubiquitinated polypeptide segments and polyubiquitin chains. J Cell Biol. 2003;162:71–84. doi: 10.1083/jcb.200302169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Elkabetz Y, Shapira I, Rabinovich E, Bar-Nun S. Distinct steps in dislocation of luminal endoplasmic reticulum-associated degradation substrates: Roles of endoplasmic reticulum-bound p97/Cdc48p and proteasome. J Biol Chem. 2004;279:3980–3989. doi: 10.1074/jbc.M309938200. [DOI] [PubMed] [Google Scholar]
- 29.Kim I, et al. The Png1-Rad23 complex regulates glycoprotein turnover. J Cell Biol. 2006;172:211–219. doi: 10.1083/jcb.200507149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Krona C, et al. Screening for gene mutations in a 500 kb neuroblastoma tumor suppressor candidate region in chromosome 1p; mutation and stage-specific expression in UBE4B/UFD2. Oncogene. 2003;22:2343–2351. doi: 10.1038/sj.onc.1206324. [DOI] [PubMed] [Google Scholar]
- 31.Hoppe T, et al. Regulation of the myosin-directed chaperone UNC-45 by a novel E3/E4-multiubiquitylation complex in C. elegans. Cell. 2004;118:337–349. doi: 10.1016/j.cell.2004.07.014. [DOI] [PubMed] [Google Scholar]
- 32.Kaneko-Oshikawa C, et al. Mammalian E4 is required for cardiac development and maintenance of the nervous system. Mol Cell Biol. 2005;25:10953–10964. doi: 10.1128/MCB.25.24.10953-10964.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Matsumoto M, et al. Molecular clearance of ataxin-3 is regulated by a mammalian E4. EMBO J. 2004;23:659–669. doi: 10.1038/sj.emboj.7600081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wang G, Sawai N, Kotliarova S, Kanazawa I, Nukina N. Ataxin-3, the MJD1 gene product, interacts with the two human homologs of yeast DNA repair protein RAD23, HHR23A and HHR23B. Hum Mol Genet. 2000;9:1795–1803. doi: 10.1093/hmg/9.12.1795. [DOI] [PubMed] [Google Scholar]
- 35.Liu C, Apodaca J, Davis LE, Rao H. Proteasome inhibition in wild-type yeast Saccharomyces cerevisiae cells. Biotechniques. 2007;42:158, 160–162. doi: 10.2144/000112389. [DOI] [PubMed] [Google Scholar]
- 36.Verma R, et al. Proteasomal proteomics: Identification of nucleotide-sensitive proteasome-interacting proteins by mass spectrometric analysis of affinity-purified proteasomes. Mol Biol Cell. 2000;11:3425–3439. doi: 10.1091/mbc.11.10.3425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Rosenzweig R, Osmulski PA, Gaczynska M, Glickman MH. The central unit within the 19S regulatory particle of the proteasome. Nat Struct Mol Biol. 2008;15:573–580. doi: 10.1038/nsmb.1427. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.