Abstract
Purpose
To identify differentially expressed microRNAs (miRNAs) and expression patterns of specific miRNAs during meiosis in human oocytes.
Materials and methods
To identify differentially expressed miRNAs, GV oocytes and MII oocytes matured at conventional FSH levels (5.5 ng/ml) were analyzed by miRNA microarray. Real-time RT-PCR was used to confirm the changed miRNAs. To validate the dynamic changes of miRNAs from GV to MII stages, oocytes were divided into four groups (#1–4), corresponding to GV oocytes, MI oocytes, MII oocytes matured in conventional FSH level and MII oocytes matured in high FSH level (2,000 ng/ml) respectively.
Results
Compared with GV oocytes, MII oocytes exhibited up-regulation of 4 miRNAs (hsa-miR-193a-5p, hsa-miR-297, hsa-miR-625 and hsa-miR-602), and down-regulation of 11 miRNAs (hsa-miR-888*, hsa-miR-212, hsa-miR-662, hsa-miR-299-5p, hsa-miR-339-5p, hsa-miR-20a, hsa-miR-486-5p, hsa-miR-141*, hsa-miR-768-5p, hsa-miR-376a and hsa-miR-15a). RT-PCR analysis of hsa-miR-15a and hsa-miR-20a expression revealed concordant dynamic changes in oocytes from group 1 to group 4.
Conclusion(s)
Specific miRNAs in human oocytes had dynamic changes during meiosis. High-concentration FSH in IVM medium led to reverse effect on the expression of hsa-miR-15a and hsa-miR-20a.
Keywords: miRNA, Oocytes, In vitro maturation, qRT-PCR
Oocyte growth and early development requires large amounts of maternally-derived transcripts which are subjected to massive destruction as oocytes mature. Of the estimated 85 pg of polyadenylated mRNAs present in a germinal vesicle (GV)-stage mouse oocyte, 50 pg of mRNAs are degraded during oocyte maturation [1]. Furthermore, transcript degradation is highly selective, primarily affecting genes involved in processes associated with meiotic arrest at the GV-stage and progression of oocyte maturation, such as oxidative phosphorylation, energy production, protein synthesis and metabolism [2]. On the other hand, up-regulation of a number of transcripts during oocyte maturation was also observed in mice, cattle, and humans in recent years [3–5].
miRNAs are a family of small non-coding RNAs that play important regulatory roles in gene expression. Specifically, miRNA-mediated translational regulation involves cleavage of messenger RNAs or repression of mRNA translation [6]. It has been estimated that 30% or more of human mRNAs are regulated by miRNAs [7]. Likewise, miRNAs may also play an important role in modulating gene expression in oocytes [8].
Dynamic changes in miRNA expression during oogenesis were first revealed by a real-time PCR-based miRNA expression profiling method in single mouse oocytes [8]. A complex population of miRNAs, with member of the miR-30, miR-16, and let-7 families being detected at the highest levels in mouse GV oocytes, raised the possibility of a role for miRNAs in controlling the pattern of mRNAs in maturing oocytes [9].
The impact of miRNAs on mRNAs in oocytes was further studied in mice lacking Dicer. Dicer is a conserved ribonuclease whose function is known to be responsible for converting the miRNA precursors into mature miRNAs. The relative amount of Dicer mRNA in oocytes is the highest when compared with other cell types [10]. Abnormal gene expression and spindle configuration was exhibited in the Dicer knockout oocytes, indicating the critical roles for Dicer in the female germline [9].
The microinjection of Dicer siRNA to knockdown the transcripts of Dicer was studied by Liu et al. [11]. Dicer siRNA significantly reduced the transcripts of Dicer and miRNAs in mouse oocytes, and also the transcriptions of genes related to spindle formation proteins and spindle checkpoint regulation, leading to MI arrest, misaligned chromosomes and abnormal meiotic spindle assembly.
However, the role of miRNAs is still controversial. In a recent study, oocytes deficient in Dgcr8, a gene encodes a RNA-binding protein specifically required for miRNA processing [12] were examined. Although miRNA levels with Dgcr8 deletion were reduced to the similar levels as Dicer-deficient mice, meiosis was not affected. However, the Dgcr8 knockout mice suffered from unexpected low fecundity. Thus, the exact role of miRNAs in the oocytes remains elusive.
Very little information is available regarding the miRNA function during human oogenesis. Previously, we showed that more than 60% human oocytes became aneuploidy when matured at 2,000 ng/ml follicle-stimulating hormone (FSH) [13]. To investigate miRNA expression during human oocyte meiosis, we herein examined the miRNA expression profiles in human oocytes from the GV stage to the MII stage using miRNA microarrays, and three differentially expressed miRNAs were verified by quantitative RT-PCR. Moreover, expression of specific miRNAs in human oocytes matured at high FSH level was also compared with that under conventional FSH condition.
Materials and methods
Patient selection and treatment
This study was approved by the ethical committee of the First Affiliated Hospital of Sun Yat-sen University. From August 2008 to February 2010, we collected a total of 392 oocytes at germinal vesical stage and 43 oocytes at metaphase I stage during 251 intracytoplasmic sperm injection (ICSI) cycles in the Reproductive Medical Center of the Hospital.
All the patients received the routine long protocol. Gonadotropin administration with the dosage of 150–300 IU/day FSH (Gonal-F; Merck-Serono, Switzerland) was initiated from cycle days 3–5. Oocyte retrieval was performed 34–36 h after 10,000 IU hCG was administered. Germinal vesicle (GV) or metaphase I (MI) stage oocytes were collected with patients’ written consent of donation for research. Our study excluded patients older than 40, or with more than three treatment cycles.
In vitro maturation
Cumulus—oocyte complexes (COCs) were observed under an inverted microscope (×40) following a 20 s exposure in the presence of hyaluronidase. GV oocytes were surrounded by tight and dense granulosa cells. Tissue culture medium (TCM 199; Sigma, USA) was supplemented with 10% serum protein substitute (SPS; Sage, USA), 50,000 IU/L penicillin, 50 mg/L streptomycin and 25 mmol/L sodium pyruvate. GV oocytes (207 and 59) were dispensed into IVM medium with 5.5 ng/ml FSH or 2,000 ng/ml Gonal-F (300 IU; Merck-Serono, Switzerland), resulting in 133 and 40 MII oocytes respectively.
miRNA microarray analysis
Denuded oocytes were stored in Trizol at -80°C prior to RNA purification. Pooled RNAs from 20 oocytes were measured at 300–400 ng, exceeding the minimum requirement of 250 ng RNA for microarrary analysis. Subsequently, pooled RNAs from GV and MII oocytes matured in vitro with 5.5 ng/ml FSH, 3 samples each, were separately analyzed on 6 microarrays. A total of 60 GV oocytes and 60 MII oocytes were used. The average age of patients corresponding to the GV and MII oocytes was 30.65 ± 3.87 and 32.15 ± 4.63, respectively. No apparent bias was detected between these two groups.
MicroRNA expression was surveyed by using the miRCURY LNA™microarray platform (Exiqon, Denmark). All procedures were carried out according to the manufacturer’s protocol. Briefly, miRNAs were enriched from extracted total RNAs using RNasey Mini Kit (Qiagen p/n 74104) and labeled with miRCURYTM Array Power Labeling kit (Exiqon, Denmark). Labeled miRNAs were used for hybridization on a miRCURY LNA™ microarray (v.10.0—hsa, mmu & rno array) containing 722 human miRNAs corresponding to all microRNAs annotated in miRBase 10.0 as well as viral microRNAs (http://microrna.sanger.ac.uk).
Following hybridization, arrays were stained, washed, and scanned using Axon GenePix 4000B microarray scanner and GnenPix pro V6.0 software. Four replicate spots of each probe on the same slide were averaged. After normalization, the statistical significance of differential miRNA expression was analyzed by T-test. Unsupervised hierarchical clustering and Correlation analysis was performed with the miRNA data. Differentially expressed miRNAs were defined as those with expression in the MII oocytes differing by no less than 1.5 fold from that in the GV oocytes (P < 0.05).
Quantitative RT-PCR
GV and MII oocytes were collected in the validation tests of differentially expressed miRNAs. Three differentially expressed miRNAs, hsa-miR-15a, hsa-miR-20a and hsa-miR-602, were identified by RT-PCR. The numbers of GV oocytes in the identification of hsa-miR-15a, hsa-miR-20a and hsa-miR-602 were 12, 12, and 14 respectively. The numbers of MII oocytes in the identification of hsa-miR-15a, hsa-miR-20a and hsa-miR-602 were 14, 13 and 13 respectively.
To further study the dynamic changes of miRNAs during meiosis, oocytes were divided into 4 maturity groups. Group 1 to group 4 contained 27 GV oocytes, 43 MI oocytes, 34 MII oocytes matured at conventional FSH levels and 40 MII oocytes matured at high FSH levels respectively.
All oocytes were tested individually because it was difficult to obtain more than one human oocyte at the same maturity stage in the same patient. In addition, it was technically challenging to normalize the reaction volume and composition if oocytes were transferred into the same reaction tube at different times.
Briefly, individual oocytes were incubated with 25 μL lysis solution containing DNase I for 8 min at room temperature. Subsequently, 2.5 μL of stop solution was added directly into each lysis reaction. Each RT reaction mixture contained 1.5 μL 10X RT Buffer, 0.15 μL dNTP Mix, 0.19 μL RNase Inhibitor, 1 μL MultiScribe (TM) RT, 4.16 μL Nuclease-free Water, 3 μL RT primer, and 5 μL Cells-to-CT sample lysate. Unless otherwise indicated, all reagents used were purchased from ABI (Applied Biosystems). RT was performed on a thermal cycler (geneAMP PCR system 9700) as follows: 30 min at 16°C, 30 min at 42°C, 5 min at 85°C, and held at 4°C.
Reactions for qRT-PCR including negative controls were performed in duplicates. Hybridization probe assay specific for real-time PCR detection of microRNAs was optimized according to the recommended criteria using the 2× TaqMan Mastermix and TaqMan MicroRNA assay. To evaluate the reproducibility of the RT-PCR at the level of the single cell, cDNA reaction mixtures after RT from individual cells were repeated three times. Each PCR reaction mixture contained 10 μL 2× TaqMan Mastermix, 1 μL TaqMan MicroRNA assay, 7.67 μL Nuclease-free water, 1.33 μL RT products. Reactions were run in a thermal cycler (ABI 7500 real-time PCR) as follows: 95°C for 10 min, followed by 40 cycles of 95°C for 10 s and 60°C for 60 s.
For relative quantification at the single cell level, it is problematic to choose a housekeeping gene that shows constant expression levels between individual samples. Therefore, we used relative quantification method with normalization to the cell number in this study. ΔCt was calculated using the equation: ΔCt = Ct(calibrator)-Ct (test). The Ct value of the first sample was set as default calibrator value. The fold change was calculated using the equation: Ratio(test/calibrator) = 2 Ct(calibrator)-Ct (test). Unless otherwise indicated, all relative quantification was analyzed by this method.
Secondly, we used relative quantification method by 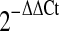 in the study of dynamic change of hsa-miR-15a. Specifically, the Ct values of the test miRNAs were normalized to the Ct values of the endogenous control (U6 RNA). The fold change was calculated using the equation
in the study of dynamic change of hsa-miR-15a. Specifically, the Ct values of the test miRNAs were normalized to the Ct values of the endogenous control (U6 RNA). The fold change was calculated using the equation 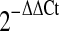 . The human miR-15a/miR-16 cluster is located at chromosome 13q14. miR-16 is considered as a “housekeeping” miRNA that appears to express in all the tissues tested at similarly abundant levels [14]. Therefore, miR-16 was also used as an alternative internal control.
. The human miR-15a/miR-16 cluster is located at chromosome 13q14. miR-16 is considered as a “housekeeping” miRNA that appears to express in all the tissues tested at similarly abundant levels [14]. Therefore, miR-16 was also used as an alternative internal control.
Significance was defined according to P-values gained from two-tailed t-test analysis, or for single-cell ΔCT comparisons, one-tailed t-test analysis.
Bioinformatic analysis
Putative mRNA targets were predicted by three algorithms: DIANA, TargetScan 4.0 and PicTar. Only the targets identified by all of these algorithms were analyzed further.
Results
miRNA microarray analysis
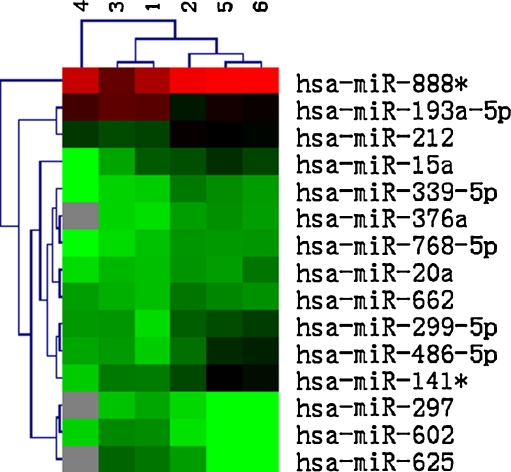
Hierachical clustering analyses of miRNA expression showed significant changes (>1.5-fold difference) in expression levels for 15 out of 722 human miRNAs (2.08%) between GV and MII oocytes (Fig. 1). Compared with the immature GV oocytes, 4 miRNAs were up-regulated (hsa-miR-193a-5p, hsa-miR-297, hsa-miR-625 and hsa-miR-602), and 11 miRNAs were down-regulated (hsa-miR-888*, hsa-miR-212, hsa-miR-662, hsa-miR-299-5p, hsa-miR-339-5p, hsa-miR-20a, hsa-miR-486-5p, hsa-miR-141*, hsa-miR-768-5p, hsa-miR-376a and hsa-miR-15a) in oocytes matured in vitro.
Fig. 1.
Heat map displaying 15 miRNAs with 1.5-fold or more differential expression between the GV stage (sample 1, 3, and 4) and the MII stage (sample 2, 5 and 6). The scale bar on top indicates relative levels, with red corresponding to high expression and green to low expression
Validation of miRNA expression
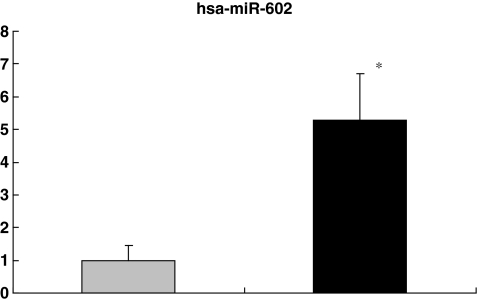
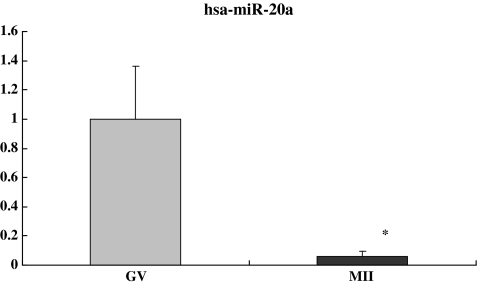
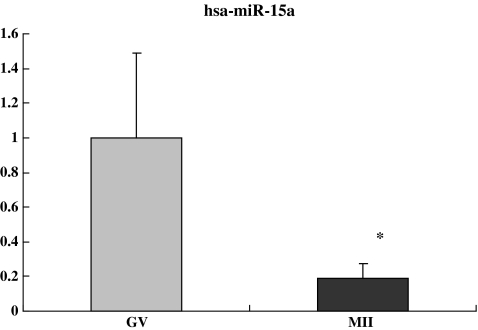
Compared with GV oocytes, the relative fold changes of hsa-miR-602, hsa-miR-15a and hsa-miR-20a in the MII oocytes were 5.28, 0.19, and 0.06 respectively (P < 0.05), consistent with the observed up- and down-regulation of these miRNAs in microarray analysis (5.01, 0.31, and 0.47 respectively) (Figs. 2, 3 and 4). Although individual variation was observed from cell to cell, our results were consistent, confirming the reliability of using real time PCR to quantify miRNAs levels in the single oocytes.
Fig. 2.
Confirmation of microarray data by quantitative real-time PCR. The relative fold change of hsa-miR-602 in the MII oocytes was 5.28. The average Ct values of hsa-miR-602 were 36.09 ± 0.74 and 33.69 ± 0.33 in GV and MII oocytes respectively. * P < 0.05 compared to GV oocytes
Fig. 3.
The relative fold change of hsa-miR-20a in the MII oocytes was 0.06. The average Ct values of hsa-miR-20a were 29.48 ± 0.11 and 33.46 ± 0.22 in GV and MII oocytes respectively. * P < 0.05 compared to GV oocytes
Fig. 4.
The relative fold change of hsa-miR-15a in the MII oocytes was 0.19. The average Ct values of hsa-miR-15a were 30.52 ± 0.23 and 32.95 ± 0.70 in GV and MII oocytes respectively. * P < 0.05 compared to GV oocytes
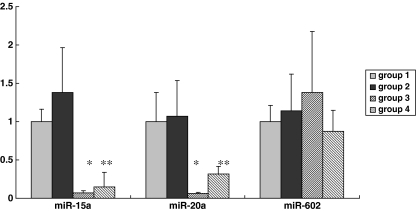
Dynamic changes of hsa-miR-15a, hsa-miR-20a and hsa-miR-602
Expression of hsa-miR-15a
Dynamic changes of hsa-miR-15a during meiosis were showed in this study. The numbers of oocytes from group 1 to group 4 were 7, 10, 10 and 11 respectively, when relative quantification was calculated with normalization to the cell number. The average Ct values of hsa-miR-15a were 31.00 ± 0.43, 30.53 ± 0.30, 34.75 ± 0.30 and 33.69 ± 0.34 in group 1–4 respectively. Compared with the GV oocytes (group 1), the relative fold changes of hsa-miR-15a expression were 1.38, 0.07 and 0.15 respectively, for groups 2, 3, and 4 (Fig. 5). No significance was detected between MI and GV oocytes. However, the expression levels of both group 3 and group 4 were significantly lower than those in both MI and GV oocytes (P < 0.001). Group 4 with oocytes matured in high FSH level showed higher expression compared with group 3, with a fold change of 2.08 (P < 0.05).
Fig. 5.
Dynamic expression of hsa-miRNA-15a, hsa-miRNA-20a and hsa-miRNA-602. Group 1–4 corresponds to GV, MI, MII oocytes matured at normal FSH level, and MII oocytes matured at high FSH level, respectively. The average Ct values of hsa-miR-15a were 31.00 ± 0.43, 30.53 ± 0.30, 34.75 ± 0.30 and 33.69 ± 0.34 in group 1–4 respectively. The average Ct values of hsa-miR-20a were 29.21 ± 0.39, 29.10 ± 0.27, 33.27 ± 0.63 and 30.85 ± 0.41 in group 1–4 respectively. The average Ct values of hsa-miR-602 were 34.10 ± 0.67, 34.16 ± 0.35, 34.05 ± 0.46 and 35.04 ± 0.45 in group 1–4 respectively. * P < 0.001 compared to group 1 and group 2, ** P < 0.05 compared to group 3
When U6 was selected as an internal control, the numbers of oocytes from group 1 to group 4 were 5, 5, 5 and 4 respectively. The relative fold changes of hsa-miR-15a expression were 1.88, 0.09 and 0.12, respectively, for groups 2, 3, and 4. The mean Ct values of hsa-miR-15a in group 3 and 4 were 34.16 ± 1.68 and 33.77 ± 1.42 respectively, yet both significantly higher than 30.73 ± 1.42 in group 1 (P < 0.01).
The expression level of miR-16 appeared to remain constant in both MII and GV oocytes in our unpublished study. As an additional validation of the dynamic changes in hsa-miR-15a expression, miR-16 was used as an additional internal control in qRT-PCR analysis. The numbers of oocytes from group 1 to group 4 were 4, 4, 4 and 5 respectively. The relative fold changes of hsa-miR-15a expression for groups 2, 3, and 4 were 1.65, 0.08 and 0.10 respectively, in good accordance with the above results with U6 as the internal control. The mean Ct values in groups 3 and 4 were 34.72 ± 0.39 and 34.45 ± 1.14 respectively, significantly higher than 31.17 ± 1.97 in group 1 (P < 0.01).
Expression of hsa-miR-20a
The expression of hsa-miR-20a had similar dynamic changes from group 1 to group 4 (Fig. 5), when relative quantification was calculated with normalization to the cell number. The numbers of oocytes from group 1 to group 4 were 5, 12, 7 and 10 respectively. The average Ct values of hsa-miR-20a were 29.21 ± 0.39, 29.10 ± 0.27, 33.27 ± 0.63 and 30.85 ± 0.41 in group 1–4 respectively. Compared with the GV oocytes, the relative fold changes of hsa-miR-20a expression were 1.07, 0.06 and 0.32 respectively, for groups 2, 3, and 4. No significance was detected between MI and GV oocytes. However, the expression levels of both group 3 and group 4 were significantly lower than those in both MI and GV oocytes (P < 0.001). Group 4 with oocytes matured in high FSH concentration showed higher expression level compared with group 3, with a fold change of 5.36 (P = 0.003).
Expression of hsa-miR-602
The expression of hsa-miR-602 exhibited the trend of moderate yet consistent up-regulation during oocyte maturation when relative quantification was calculated with normalization to the cell number (Fig. 5). The numbers of oocytes from group 1 to group 4 were 6, 12, 8 and 10 respectively. The average Ct values of hsa-miR-602 were 34.10 ± 0.67, 34.16 ± 0.35, 34.05 ± 0.46 and 35.04 ± 0.45 in group 1–4 respectively. The relative fold changes of hsa-miR-602 expression in groups 2, 3, and 4 were 1.14, 1.38 and 0.87, respectively. The relative expression level in group 4 was not significantly different from that of group 3.
Putative miRNA target prediction
Putative mRNA targets of the 15 differentially expressed miRNAs identified above were predicted by three algorithms: DIANA, TargetScan 4.0 and PicTar. Only the targets identified by all three algorithms were further analyzed. The transcript annotation results suggested that targets of miR-15a are predominantly involved in the regulation of cell division and cell growth, including BCL-2 family members and CDC25A. However, targets of the other 14 miRNAs are poorly defined.
Discussion
During mammalian oocyte development, oocytes remain arrested at the dictyate stage of prophase I until the resumption of meiosis. Despite massive transcript turnover, it is well accepted that meiotic maturation of oocytes is transcriptionally quiescent. Fertilization and early embryo development depends on maternal transcripts accumulated during oocyte growth. The maternal-to-zygotic transition in mouse eggs initiates in the one-cell embryo during mid-S phase [15]. In human embryos, genome activation occurs at the six- to eight-cell stage [16].
In recent years, there are conflicting reports on the occurrence of transcription following the resumption of oocyte meiotic maturation. Up-regulation of a number of transcripts during oocyte maturation in mice, in cattle, and in humans was observed by comparison between immature (GV stage) and in vivo matured oocytes [3–5]. In bovine oocytes, 2117 out of 8489 transcripts were found to be differentially expressed between immature and IVM oocytes, corresponding to 1528 transcripts that were significantly lower and 589 that were significantly higher in abundance in IVM oocytes compared to their immature counterparts [4]. Among 8728 genes detected by oligonucleotide microarrays in human MII oocytes, 803 genes were underexpressed and 444 genes were overexpressed [5]. The overexpression may be explained by a specific expression patterns related to the near completion of meiosis.
As key regulators of gene expression at the post-transcriptional level, miRNAs may play important roles during meiosis and early development. It was reported that miRNAs directly or indirectly control more than one third of the maternal genes expressed in oocytes [8]. High expressions of Dicer and miRNAs insured normal oocyte maturation and maintained meiotic spindle integrity which is necessary for normal meiotic maturation [10].
Previously, dynamic changes in miRNA expression during oogenesis have been found in growing mouse oocytes obtained from females 15–16 d after birth (postnatal days 15–16 [P15-P16]), at P20–P21, and mature oocytes from adult females [8]. Several miRNAs, such as miR-103, let-7 d, miR-16, miR-30b and miR-30c, were suggested to play fundamental roles in the meiosis of mouse GV oocytes [9].
However, recent studies on the Dgcr8-deficient oocytes demonstrated miRNA function is globally suppressed in mouse oocytes and early embryos [12, 17]. Although miRNA levels of mouse oocytes with Dgcr8 deletion were reduced to the similar levels as Dicer-deficient mice, meiosis was not affected as in the oocytes from Dicer knockout mice. However, the exact role of miRNAs in the oocytes remains elusive, since the Dgcr8 knockout mice suffered from unexpected low fecundity.
In the current study, only 2.08% miRNAs from 722 human miRNAs displayed differential expression in GV and MII oocytes. Such limited miRNA changes in human oocytes during meiosis supports the notion that miRNAs may play specific, rather than global, regulatory functions in gene expression during oocyte maturation [12, 17].
The restricted availability of human oocytes allowed qRT-PCR validation of only 3 out of 15 microRNAs with differential expression in microarray analysis. Relative quantification method by normalization with a housekeeping gene is the most common method in RT-PCR. However, it is technically problematic to choose a housekeeping gene that shows constant expression levels between individual samples at the single cell level. It was believed that the occurrence of stochastic production of mRNA in bursts renders the use of constantly expressed reference genes for sample comparison invalid at the level of single cells [18]. Currently, single cell data are routinely analyzed as absolute mRNA/cDNA copies or as relative quantities without any normalization except for the number of cells (one) [19].
Therefore, we used relative quantification method with normalization to the cell number. The relative fold changes of hsa-miR-15a, hsa-miR-20a and hsa-miR-602 expression in the MII oocytes were 0.19, 0.06 and 5.28 respectively, consistent with the observed up- and down-regulation of these miRNAs in microarray analysis (0.31, 0.47 and 5.01 respectively).
We further selected both U6 and miR-16 to verify the reproducibility of relative quantification method with normalization to the cell number. The human miR-15a/miR-16 cluster is located at chromosome 13q14. miR-16 is considered as a “housekeeping” miRNA that appears to express in all the tissues tested at similarly abundance. qRT-PCR analysis of hsa-miR-15a expression using two different internal controls showed highly consistent down-regulation concordant with the microarray result.
Dynamic changes of hsa-miR-15a and hsa-miR-20a during meiosis were observed in our study. No significant changes of both miRNAs were observed in MI oocytes, compared with GV oocytes. Our results are consistent with the results of the human cumulus-oocyte complex gene-expression profile. Very few genes over or under expressed between GV and MI oocytes, indicating a very similar expression profile, as opposed to MII oocytes. Fifty-two genes were identified with progressively increasing expression during oocyte maturation in humans [5].
Species difference may account for the distinct temporal miRNA expression patterns during oocytes development [20]. In a previous study on genes involved in miRNA processing, oocyte expression of DICER, GEMIN, TNRC6B, RANGAP1 and RNASEN mRNAs appeared to be regulated differently in mouse and rhesus monkey.
The expression of hsa-miR-602 in the analysis of dynamic changes from group 1 to group 4 only exhibited the trend of moderate yet consistent up-regulation. The relative fold changes were lower than the results of both microarray analysis and qRT-PCR in the identification of differentially expressed microRNAs. The reason may be due to the small sampling size and individual variability in single cell qRT-PCR.
Computational predictions of miRNA target genes may provide a global view of the gene regulation network by miRNAs. The stringent prediction criterion used in the current study revealed a possible role of miR-15a in the regulation of cell growth and division [21]. Previously, miR-15a and miR-16-1 expression was found to inversely correlate with Bcl2 expression in chronic lymphocytic leukemia (CLL); furthermore, both miRNAs appeared to negatively regulate Bcl2 at a post-transcriptional level [20]. BCL2L10, a BCL2 family member, has been shown to be exclusively expressed in oocytes, with a stage-specific redistribution along the pericortical regulatory ooplasm [22, 23]. No temporal expression pattern of BCL-2 family mRNAs has been observed in human oocyte; in contrast, the expression of BCL2L10 in rhesus monkey oocytes increased three fold between the GV stage and the MII stage. The observed down-regulation of miR-15a in the current study may be corresponding to the latter finding, suggesting a possible role of BCL2L10 in oocyte maturation [24].
CDC25A was among the 52 genes whose expression was found to gradually increase during human oocyte maturation [5]. The CDC25A phosphatase is a key cell cycle regulator in vertebrates, promoting cell cycle progression by dephosphorylating and consequently activating specific cyclin-dependent kinases (Cdks) [25]. Expression of exogenous CDC25A overcomes cAMP-mediated maintenance of meiotic arrest, whereas reduction of CDC25A results in fewer oocytes resuming meiosis and consequently reaching MII [26]. Future experimental studies are required to confirm the possible regulation of CDC25A by miR-15a. On the other hand, prediction analysis did not yield much insight on the other 14 miRNAs, perhaps due to the scarcity of information on MII-stage oocytes with possible tissue- and developmental-specific expression patterns.
FSH is a crucial gondadotrophin for follicle recruitment. Accumulating evidence in animal studies indicates that elevated FSH levels may be an underlying cause of aneuploidy [27]. In our previous study, the percentage of aneuploid MII oocytes at 0 ng/ml, 5.5 ng/ml, 22 ng/ml, 100 ng/ml and 2,000 ng/ml of FSH was 26.7%, 23.3%, 36.75%, 46.7% and 63.3%, respectively. No difference in spindle morphology was detected between 2,000 ng/ml and 5.5 ng/ml FSH groups (45.0% versus 50.0%) [13]. The exact mechanism by which FSH induces aneuploidy remains unclear, although several possibilities have been proposed, including oocyte susceptibility to environmental factors during the first meiosis [28–30], effects of FSH on microtubules [31, 32], and enhanced meiotic resumption of abnormal oocytes which may be otherwise destined for atresia.
In the present study, miRNA expression exhibited opposing changes from the MI stage to the MII stage in oocytes matured at high FSH levels compared with those at conventional FSH levels. Definitive evidence will require greater sample size in future studies.
Taken together, our data revealed differential expression of specific miRNAs during human oocyte development. Future functional and mechanistic studies on the dynamic changes of miRNA expression during meiosis may yield better understanding of the regulatory role of miRNAs in meiosis.
Footnotes
Yan-Wen Xu and Bin Wang contributed equally to this study.
This study was supported by National Basic Research Program of China (grant no. 2007CB948101), National Natural Science Foundation of China (grant no. 30700910 and 31071272).
Capsule
Differentially expressed miRNAs in the human oocytes between GV and MII stages were identified by miRNA microarrays. Dynamic changes of miR-15a, hsa-miR-20a, and miR-602 during meiosis were validated by qRT-PCR.
References
- 1.Paynton BV, Rempel R, Bachvarova R. Changes in state of adenylation and time course of degradation of maternal mRNAs during oocyte maturation and early embryonic development in the mouse. Dev Biol. 1988;129:304–314. doi: 10.1016/0012-1606(88)90377-6. [DOI] [PubMed] [Google Scholar]
- 2.Su YQ, Sugiura K, Woo Y, Wiggleworth K, Kamdar S, Affourtit J, Eppig JJ. Selective degradation of transcripts during meiotic maturation of mouse oocytes. Dev Biol. 2007;302(1):104–117. doi: 10.1016/j.ydbio.2006.09.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Cui XS, Li XY, Yin XJ, Kong IK, Kang JJ, Kim NH. Maternal gene transcription in mouse oocytes genes implicated in oocyte maturation and fertilization. J Reprod Dev. 2007;53(2):405–418. doi: 10.1262/jrd.18113. [DOI] [PubMed] [Google Scholar]
- 4.Mamo S, Carter F, Lonergan P, Leal C, Naib A, McGettigan P, et al. Sequential analysis of global gene expression profiles in immature and in vitro matured bovine oocytes: potential molecular markers of oocyte maturation. BMC Genomics. 2011;12:151. doi: 10.1186/1471-2164-12-151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Assou S, Anahory T, Pantesco V, Carrour T, Pellestor F, Klein B, et al. The human cumulus–oocyte complex gene-expression profile. Hum Reprod. 2006;21(7):1705–1719. doi: 10.1093/humrep/del065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bartel DP. MicroRNAs: fenomics, biogenesis, mechanism, and function. Cell. 2004;116:281–297. doi: 10.1016/S0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]
- 7.Lewis BP, Burge CB, Bartel DP. Conserved seed pairing, often flanked by adenosines, indicates that thousands of human genes are microRNA targets. Cell. 2005;120:15–20. doi: 10.1016/j.cell.2004.12.035. [DOI] [PubMed] [Google Scholar]
- 8.Tang F, Kaneda M, O’Carroll D, Hajkova P, Barton S, Sun YA, et al. Maternal microRNAs are essential for mouse zygotic development. Genes Dev. 2007;21(6):644–648. doi: 10.1101/gad.418707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Murchison EP, Stein P, Xuan Z, Pan H, Zhang MQ, Schultz RM, Hannon GJ. Critical roles for Dicer in the female germline. Genes Dev. 2007;21(6):682–693. doi: 10.1101/gad.1521307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Svoboda P, Stein P, Hayashi H, Schultz RM. Selective reduction of dormant maternal mRNAs in mouse oocytes by RNA interference. Development. 2000;127:4147–56. doi: 10.1242/dev.127.19.4147. [DOI] [PubMed] [Google Scholar]
- 11.Liu HC, Tang YX, He ZY, Rosenwaks Z. Dicer is a key player in oocyte maturation. J Assist Reprod Genet. 2010;27:571–850. doi: 10.1007/s10815-010-9456-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Suh N, Baehner L, Moltzahn F, Melton C, Shenoy A, Chen J, Blelloch R. MicroRNA function is globally suppressed in mouse oocytes and early embryos. Curr Biol. 2010;20:271–277. doi: 10.1016/j.cub.2009.12.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Xu YW, Peng YT, Wang B, Zeng YH, Zhuang GL, Zhou CQ. High follicle-stimulating hormone increases aneuploidy in human oocytes matured in vitro. Fertil & Steril. 2011;95(1):99–104. doi: 10.1016/j.fertnstert.2010.04.037. [DOI] [PubMed] [Google Scholar]
- 14.Ro S, Park C, Sanders KM, McCarrey JR, Yan W. Cloning and expression profiling of testis-expressed microRNAs. Dev Biol. 2007;311:592–602. doi: 10.1016/j.ydbio.2007.09.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Schultz RM, Davis W, Jr, Stein P, Svoboda P. Reprogramming of gene expression during preimplantation development. J Exp Zool. 1999;285(3):276–82. doi: 10.1002/(SICI)1097-010X(19991015)285:3<276::AID-JEZ11>3.0.CO;2-K. [DOI] [PubMed] [Google Scholar]
- 16.Tesarik J, Kopecny V, Plachot M, Mandelbaum J. Early morphological signs of embryonic genome expression in human preimplantation development as revealed by quantitative electron microscopy. Dev Biol. 1988;128:15–20. doi: 10.1016/0012-1606(88)90261-8. [DOI] [PubMed] [Google Scholar]
- 17.Ma J, Flemr M, Stein P, Berninger P, Malik R, Zavolan M, Svoboda P, Schultz RM. MicroRNA activity is suppressed in mouse oocytes. Curr Biol. 2010;20:265–270. doi: 10.1016/j.cub.2009.12.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Stáhlerg A, Bengtsson M, Hemberg M, Semb H. Quantitative Transcription factor analysis of undifferentiated single human embryonic stem cells. Clin Chem. 2009;55(12):2162–2170. doi: 10.1373/clinchem.2009.131433. [DOI] [PubMed] [Google Scholar]
- 19.Stáhlerg A, Bengtsson M. Single-cell gene expression profiling using reverse transcription quantitative real-time PCR. Methods. 2010;50:282–288. doi: 10.1016/j.ymeth.2010.01.002. [DOI] [PubMed] [Google Scholar]
- 20.Mtango NR, Potireddy S, Latham KE. Expression of microRNA processing machinery genes in rhesus monkey oocytes and embryos of different developmental potentials. Mol Reprod Dev. 2009;76(3):255–69. doi: 10.1002/mrd.20950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Linsley PS, Schelter J, Burchard J, Kibukawa M, Martin MM, Bartz SR, et al. Transcripts targeted by the microRNA-16 family cooperatively regulate cell cycle progression. Mol Cell Biol. 2007;27(6):2240–2252. doi: 10.1128/MCB.02005-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Cimmino A, Calin GA, Fabbri M, Lorio MV, Ferracin M, Shimizu M, et al. miR-15 and miR-16 induce apoptosis by targeting BCL2. Proc Natl Acad Sci U S A. 2005;102(39):13944–13949. doi: 10.1073/pnas.0506654102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Yoon SJ, Kim EY, Kim YS. Role of Bcl2-like 10 (Bcl2l10) in regulating mouse oocyte maturation. Biol Reprod. 2009;81(3):497–506. doi: 10.1095/biolreprod.108.073759. [DOI] [PubMed] [Google Scholar]
- 24.Guillemin Y, Lalle P, Gillet G. Oocytes and early embryos selectively express the survival factor BCL2L10. J Mol Med. 2009;87:923–940. doi: 10.1007/s00109-009-0495-7. [DOI] [PubMed] [Google Scholar]
- 25.Isoda M, Kanemori Y, Nakajo N, Uchida S, Yamashita K, Ueno H, Sagata N. The extracellular signal-regulated kinase-mitogen-activated protein kinase pathway phosphorylates and targets Cdc25A for SCFβ-TrCP-dependent degradation for cell cycle arrest. Mol Biol Cell. 2009;20(20):2186–2195. doi: 10.1091/mbc.E09-01-0008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Solc P, Saskova A, Baran V, Kubelka M, Schultz RM, Motlik J. CDC25A phosphatase controls meiosis I progression in mouse oocytes. Dev Biol. 2008;317(1):260–269. doi: 10.1016/j.ydbio.2008.02.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Dursun P, Gultekin M, Yuce K, Ayhan A. What is the underlying cause of aneuploidy associated with increasing maternal age? Is it associated with elevated levels of gnoadotropins? Medical Hypotheses. 2006;66:143–147. doi: 10.1016/j.mehy.2004.10.022. [DOI] [PubMed] [Google Scholar]
- 28.Eichenlaub-Ritter U, Winterscheidt U, Vogt E, Shen Y, Tinneberg H, Sorensen R. 2-Methoxyestradiol induces spindle aberrations, chromosome congression failure, and nondisjunction in mouse oocytes. Biol Reprod. 2007;76:784–93. doi: 10.1095/biolreprod.106.055111. [DOI] [PubMed] [Google Scholar]
- 29.Yin H, Baart E, Betzendahl I, Eichenlaub-Ritter U. Diazepam induces meiotic delay, aneuploidy and predivision of homologues and chromatids in mammalian oocytes. Mutagenesis. 1998;13:567–80. doi: 10.1093/mutage/13.6.567. [DOI] [PubMed] [Google Scholar]
- 30.Can A, Semiz O. Diethylstilbestrol (DES)-induced cell cycle delay and meiotic spindle disruption in mouse oocytes during in-vitro maturation. Mol Hum Reprod. 2000;6:154–62. doi: 10.1093/molehr/6.2.154. [DOI] [PubMed] [Google Scholar]
- 31.Wakefield JG, Stephens DJ, Tavare JM. A role for glycogen synthase kinase-3 in mitotic spindle dynamics and chromosome alignment. J Cell Sci. 2003;116:637–46. doi: 10.1242/jcs.00273. [DOI] [PubMed] [Google Scholar]
- 32.Wang X, Liu XT, Dunn R, Ohl DA, Smith GD. Glycogen synthase kinase-3 regulates mouse oocyte homologue segregation. Mol Reprod Dev. 2003;64:96–105. doi: 10.1002/mrd.10213. [DOI] [PubMed] [Google Scholar]