Abstract
Alcoholism is a complex disease caused by a confluence of environmental and genetic factors influencing multiple brain pathways to produce a variety of behavioral sequelae, including addiction. Genetic factors contribute to over 50% of the risk for alcoholism and recent evidence points to a large number of genes with small effect sizes as the likely molecular basis for this disease. Recent progress in genomics (microarrays or RNA-Seq) and genetics has led to the identification of a large number of potential candidate genes influencing ethanol behaviors or alcoholism itself. To organize this complex information, investigators have begun to focus on the contribution of gene networks, rather than individual genes, for various ethanol-induced behaviors in animal models or behavioral endophenotypes comprising alcoholism. This chapter reviews some of the methods used for constructing gene networks from genomic data and some of the recent progress made in applying such approaches to the study of the neurobiology of ethanol. We show that rapid technology development in gathering genomic data, together with sophisticated experimental design and a growing collection of sophisticated tools are producing novel insights for understanding the molecular basis of alcoholism and that such approaches promise new opportunities for therapeutic development.
Introduction
Alcoholism is a prevalent and serious behavioral disease characterized by progression from intermittent social use of ethanol to abusive and uncontrolled consumption. As with other drug abuse disorders, the underlying neurobiological etiology of alcohol abuse and alcoholism is thought to involve long-lasting aberrant molecular plasticity in the central nervous system. Although multiple biological and environmental factors may converge en route to the manifestation and sustainability of this disease, altered function or expression of genes and gene networks are considered major factors contributing to long-lasting CNS changes causing the behavioral phenotype of alcoholism. The advent of high-throughput, unbiased approaches to studying genomic structure and expression, such as proteomics, DNA microarrays, whole genome SNP analysis and Next-Gen sequencing technologies, are providing new insights into gene sets involved in complex diseases. However, long lists of genes do not in themselves provide improved understanding of diseases such as alcoholism. New experimental approaches, combined with advanced statistical and bioinformatics support, have recently allowed organization of results from whole-genome studies into novel functional networks of genes related to the trait under study. Rather than focusing on an individual gene, the investigator can now simultaneously probe the entire genome to assess the interaction among individual genes. Provided with enough information a causal network may be constructed to predict functional mechanisms related to complex phenotypes (Zhu et al. 2008).
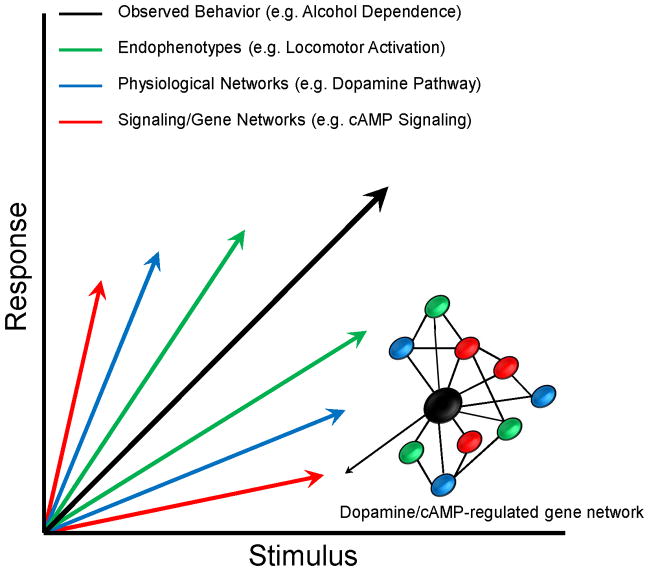
The overall framework leading to the full onset of alcohol dependence involves the progression from initial acute exposure toward compulsive drug use with frequent intermixed reoccurring bouts of tolerance and withdrawal (Koob and Volkow 2010). The disease involves reward seeking, compulsivity and habit formation, aversive stimuli (e.g. withdrawal) and many other behavioral facets. There is likely no single causative factor in alcoholism, and thus each facet of this disorder may provide an important area of scientific inquiry. For example, interpreting the gene network structure of an organism undergoing withdrawal may impart novel mechanistic information contributing to the neuroadaptations driving relapse behavior. The overall phenotype of alcoholism could thus be considered a “behavioral vector” that is made up of multiple component vectors subserving endophenotypes as mentioned above (Figure 1). Vectors of interacting neuronal/glial networks across multiple brain regions, in turn, likely control each of these endophenotype vectors. Drilling down yet further, these neural networks are ultimately controlled by regulation/function of multiple gene networks expressed within individual neurons or glial cells. As depicted in Figure 1, this hierarchy of nested response vectors, extending from the molecular to the behavioral, likely explains the tremendous difficulty encountered in studying mechanisms of complex traits such as alcoholism. This degree of complexity also explains why efforts to correlate function/expression of single genes to a complex disease are exceedingly difficult. When considered in this light, it becomes apparent that progress in studying the mechanisms of complex disease requires a combined distillation of traits into endophenotypes and the amalgamation of brain regional gene expression/function into networks relevant to the trait vectors. Once we have mapped the network structure of these varying endophenotypes, we may be able to identify major genetic hubs for developing more rational pharmacotherapies in the treatment of alcoholism. This manuscript will review the process of using whole genome expression analysis to define gene networks that contribute to the complex nature of alcoholism.
Figure 1. Diagram of multiple potential subcomponent vectors contributing to the observed behavioral phenomenon of alcohol dependence.
The figure illustrates the premise that observed behavioral phenotypes studied in humans or animal models are actually made up of multiple behavioral subcomponents (e.g. endophenotypes) that are in turn controlled by neural networks. The gene networks derived by genomic studies are a syncytium of gene regulation events occurring within multiple cells comprising a given neural network.
Methods of Gene Expression Network Analysis
a. Experimental design
Although this topic cannot be discussed in detail here, a variety of platforms exist for detecting differential gene expression. Under certain circumstances array platforms with a limited or more focused set of genes (e.g. arrays targeted against cytokine mRNA or protein) may be advantageous. However, the construction of gene networks as discussed in this chapter, generally requires a more inclusive approach that utilizes unbiased whole-genome arrays. In any case, the construction and analysis of the aforementioned gene networks relevant to complex phenotypes such as alcoholism cannot occur without a proper experimental design permitting higher order regression analysis and methods for obtaining robust measures of gene expression. Two major factors have to be considered in experimental design. The first concerns whether there is a balance between a well-controlled experiment, and having enough phenotypic variance and power to allow meaningful gene-gene and gene-phenotype correlations. Secondly, technical design of the experiment must avoid systematic environmental factors (such as batch effects) causing false positive correlation structures in the data (Chesler et al. 2002).
Performing a simple control versus treatment comparison in one mouse strain will not allow significant use of the expression data itself in defining networks relevant to ethanol/alcoholism. As described below, differential expression across two conditions can be superimposed upon gene networks derived by other methods but this does not take advantage of the biological specificity contained within the experimental expression data. Thus, investigators have chosen experimental designs combining larger numbers of conditions such as varying animal strains, drug treatments across responsive vs. unresponsive strains, dose responses and time courses.
Perhaps the most extensive example of such experimental designs comes in the use of “genetical genomics” where whole genome expression profiling is done across many genetically defined strains. By simultaneously analyzing genetic marker data, phenotypic measures (such as ethanol drinking behavior) and gene expression across large model system genetic panels or human subject populations, robust gene-gene and gene-phenotype correlation networks can be derived (Jansen and Nap 2001; Schadt et al. 2003; Chesler et al. 2005). Additionally, the genetic marker information allows mapping of chromosomal loci contributing to the variation in both gene expression and phenotypic traits. Such loci are termed quantitative trait loci (QTL) for phenotypic measures and eQTL for gene expression traits. These eQTL can either be “cis” or “trans” to the chromosomal location showing linkage to the expression of a particular gene. Cis-eQTL are eQTL located at the same location as the gene itself while trans-eQTL are located elsewhere. In some instances, there can be many genes showing trans-eQTL at the same location (Schadt et al. 2003; Chesler et al. 2005).
The identification of trans-eQTL provides a novel mechanism for defining gene expression networks, based upon their presumed common regulation by a gene or genes at a given trans genetic location. Furthermore, correlation of such trans-eQTL loci with behavioral traits such as alcoholism provides a powerful approach to defining and organizing the multiple gene networks contributing to complex traits (Chesler and Williams 2004; Chesler et al. 2005). An excellent example for this approach is the recent dissection by Williams and colleagues of a complex locus on the distal end of mouse Chr 1, where multiple behavioral traits relevant to alcohol and drug abuse have been mapped (Mozhui et al. 2008). This analysis identified the genes Rgs7 and Fmn2 as strong candidates for Chr 1 genes controlling the expression of large gene transcription networks and influencing the genetic variance of multiple behavioral phenotypes relevant to ethanol and drugs of abuse.
As alluded to above, the caveat to using genetical genomic approaches for identifying gene networks in complex traits concerns the possibility of multiple technical factors producing false correlation structures. In addition to the possibility of introducing batch effects from random environmental or technical factors during the analysis of large numbers of RNA samples and microarrays, the design of the microarrays themselves can be a problem when studying expression across different genotypes. All microarray designs using short oligonucleotides as probes can suffer from hybridization kinetic effects caused by polymorphisms or insertion/deletions occurring in the region of a gene interrogated by the microarray probe. Thus, when studying expression across the BXD panel of recombinant inbred mice or across human subjects, any sample with a genotype that destabilizes probe performance will give the appearance of reduced hybridization for that probe – with a false-positive decrease in expression for that gene. Since this would occur across all similar genotypes, the result would be a false-positive cis-eQTL (Walter et al. 2007). However, there are statistical approaches for detecting or minimizing false cis-eQTL caused by SNPs (Chen et al. 2009). Additionally, newer methods such as high throughput sequencing (RNA-Seq) for quantitation of transcript abundance avoid SNP artifacts altogether and offer the advantage of detecting alternative transcripts that might further enhance the genetical genomics analysis (Liu et al. 2010).
Although some limitations exist, whole genome expression profiling approaches offer a strong starting point for shaping the framework governing gene network analysis. Sensitivity and specificity issues of microarray hybridization analysis have been extensively discussed. High-density oligonuceleotide arrays are capable of detecting expression with frequencies between 1:300 and 1:300,000, with 1:300,000 representing 1 to 3 copies of mRNA per cell (Lockhart et al. 1996). Recent publications suggest that RNA-Seq analysis may extend sensitivity to both lower and higher abundance transcripts (Bottomly et al. 2011). This degree of sensitivity for detecting low abundance genes within a heterogenous environment such as the CNS has the potential to identify novel candidates for complex phenotypes. When viewed in the framework of a network such candidates or hub genes can be systematically tested for variation in human populations or experimentally validated in model organisms (Schadt et al. 2005; Zhu et al. 2008; Yang et al. 2009).
The resilience of identifying causal disease related gene networks depends on the ability to predict the impact of a gene network on a phenotype. In some instances protein expression and gene expression may not directly correlate and genome-wide analysis of protein abundance would seem a more accurate method for connecting gene networks to phenotypes. However, coordinate changes in the mRNA expression of a gene network can infer that the abundance and/or functional interactions of the cognate proteins are altered. Furthermore, whole genome expression profiling of RNA transcripts with microarrays or RNA-Seq is currently a more technically feasible approach than whole genome proteomic studies. If available, protein-protein interaction data as well as other types of molecular information can added to improve predictive validity of a gene expression network (Zhu et al. 2008). As public databases of genomic and proteomic data continue to expand, the structure of gene expression networks can be further evaluated based upon multiple cellular systems extending from mRNA/protein expression to transcription factor binding events, micro-RNA processing, and the epigenetic regulation.
b. Construction of gene networks based upon expression studies
Once genes differentially expressed across different conditions or animal strains relative to the disease model have been identified or robust expression data has been derived across a large panel of phenotyped/genotyped subjects, a relational network needs to be established across individual genes. A gene network could be considered either a set of genes that interact functionally (e.g. metabolite processing, protein-protein interaction, or protein modification) with one-another to accommodate the needs of the cellular environment or a set of genes that share a common regulatory mechanism and show highly correlated expression patterns. Obviously, in many cases these two factors may overlap within a given gene network.
Several related experimental analysis approaches have been used for constructing gene networks. All of these approaches have variants that utilize differing algorithms for calculating network membership or topography but such details are beyond the scope of this review. In one commonly used approach, a statistically filtered gene set is superimposed upon prior organized networks or lists of genes. This “over-representation analysis” depends upon first defining a statistically filtered (or ranked) gene list of interest (e.g. ethanol treated vs. control animals) and then interrogating this list for over-representation in existing gene networks or lists that have been constructed from prior experimental data such as protein-protein binding or regulation, transcription factor binding, or other associations defined in the biomedical literature (e.g. two genes appearing together in the same abstract). For example, statistically filtered gene lists can be interrogated for enrichment in previously defined signaling pathways such as those within the Kyoto Encyclopedia of Genes and Genomes (KEGG) (Kanehisa et al. 2008) or gene ontology groups. These interconnected systems are again derived from the existing biomedical literature, and defined by their shared functional involvement such as protein-protein interactions, gene-gene interactions, or signaling systems. Gene ontology groups are nested lists of defined gene categories grouped by cellular component, biological process, or molecular function (www.geneontology.org). The Database for Annotation, Visualization and Integrated Discovery (DAVID; http://david.abcc.ncifcrf.gov/) is one commonly used web-based resource for performing gene ontology over-representation analyses.
Several commercial and open-source applications exist for performing gene network over-representation analysis. These include those produced by Ingenuity Pathway Analysis (www.ingenuity.com) or GeneGo (www.genego.com). The Cytoscape suite of programs is an open-source approach to such network analyses (http://www.cytoscape.org/) and has many plug-in applications that integrate a wide variety of approaches for analyzing gene networks. A potential drawback of over-representation analyses for interpreting the biological significance within a given dataset is the inclusion of larger more widely studied categories, rather than more novel or empiric datasets better suited for a particular line of scientific inquiry. Therefore, caution should be exercised when interpreting the functional relevance of a gene set based on predetermined categories.
As a second approach for gene network analysis, gene-gene relations are defined de novo based upon expression/expression, expression/genotype or expression/phenotype correlations. This method depends on the inherent correlation structure generated by datasets such as microarray expression studies. Genes with highly correlated expression profiles across a large number of different experimental conditions are hypothesized to share biologically relevant relationships. Such “cluster analysis” can provide information about the function of uncharacterized genes or the biological mechanisms underlying a given drug action or phenotypic trait (Eisen et al. 1998; Hughes et al. 2000). However, without sufficient specific biological variance in the expression of individual genes, correlation-based network approaches can produce statistically parsimonious relationships between genes that might have little biological relevance. Furthermore, environmental or technical factors can produce artifactual correlations. A cluster or principal component analysis on “treatments or samples” (vs. genes) should reveal groupings relevant to the biology, rather than factors such as processing order, reagent lot or personnel handling of the samples.
A large number of methodologies have been developed to classify co-expression networks in complex phenotypes (Butte et al. 2000; Baldwin et al. 2005; Chesler et al. 2005; Zhang and Horvath 2005). For example, weighted gene co-expression network analysis (WGCNA) of human brain tissue was able to identify cell-type specific networks shared among distinct anatomical brain regions, assign cell-type classification to a protein of unknown function, and distinguish between cell-type specific subpopulations (Oldham et al. 2008). Given the complex heterogeneous environment of the human brain, this study successfully demonstrated the strength of a network approach for providing novel insight into the brain transcriptome.
An evolving approach to increase the biological information content and functional specificity of gene expression correlation networks involves essentially overlaying additional sources of biological gene-gene connectivity onto the expression correlates. Integrated meta-analysis of genes and proteins from multiple informative resources leverages biological function onto dense genomic expression profiling studies to help define disease associated molecular networks. STRING (Snel et al. 2000) and ToppGene Suite (Chen et al. 2009) are two examples of tools that integrate multiple bioinformatic resources to create functional networks based on a user-defined gene list.
STRING (Search Tool for the Retrieval of Interacting Genes/Proteins) is an in-depth meta-resource tool for illustrating a functional network of proteins across 630 organisms (Jensen et al. 2009). Networks are based upon a composite of known functional associations and protein-protein interactions exisiting within the biomedical literature, as well as predicted relationships in order to potentially provide a more comprehensive view of the system. The predicted functional relationship amongst given gene sets are determined through an array of experimentally derived conditions from phylogenetic and co-expression profiles. Additionally, predicted associations are incorporated into STRING based upon text-mining using natural language processing (Saric et al. 2006) and conserved organismic transfer of protein interactions (von Mering et al. 2005). Resulting networks can be filtered for confidence scores, and used as a prominent exploratory tool for investigating all of the possible biological relationships among a potentially more limited user-defined gene set.
ToppGene Suite is one example of an assimilated resource that can mine gene networks for enrichment of ontologies and phenotypes, as well as rank candidate genes within a network for a targeted validation of genes that may be critical to the phenotype under question. The ranking or prioritizing can be assigned based upon functional annotation and phenotype information (Chen et al. 2007) or protein-protein interaction networks (Chen et al. 2009). Such network based prioritization methods are important for connecting and validating genes in the context of molecular networks.
These higher-order organizational frameworks comprised of biological relationships, physical interactions, expression correlation structures and predicted associations encompass some of the current network approaches designed for understanding the molecular basis of disease in systems biology. The adoption of network analyses for ascertaining disrupted molecular networks associated with ethanol abuse and dependence is still in its infancy. However, expression profiling of brain tissue from humans and animals is opening the door to the discovery of fundamental networks inherent in alcoholism.
Transcriptional Networks of Alcohol Abuse and Alcoholism
Genetic predisposition contributes an underlying vulnerability to the risk of developing alcohol dependence (Goodwin et al. 1974; Prescott and Kendler 1999) as well as other substance abuse disorders. However, limited success has been achieved in the identification of candidate genes that contribute to the variable occurrence of alcohol dependence through linkage studies, single gene association or, more recently, genome-wide association studies (GWAS) (Johnson et al. 2006). This difficulty is likely due to the occurrence of one or more rare polymorphisms of small effect size in a large number of genes being causal elements in complex traits such as alcoholism. Moreover, even in cases where candidate genes for alcoholism have been implicated, the detection of regulatory network-wide systems is usually beyond the power of these approaches.
Alterations in mRNA transcript abundance evoked by ethanol or substance abuse are proposed as mechanisms underlying enduring neuro-adaptations leading to abuse and addiction (Miles 1995; Nestler and Aghajanian 1997). Disrupted homeostatic control of gene networks is also a possible mechanism underlying CNS toxicity from compulsive ethanol or drug use. Furthermore, genetic differences in gene expression responses to ethanol are also thought to be an important mechanism underlying a predisposition to alcoholism or other complex traits (Schadt et al. 2003; Chesler et al. 2005). Thus, studies in animal models or humans on gene expression networks associated with alcoholism or ethanol-related behaviors have been an area of intense study.
Using genetic animal models, a recent meta-analysis by Mulligan et al. identified ~3,800 differentially expressed unique genes in whole brain homogenates of ethanol-naïve mice across strains markedly divergent for ethanol drinking behavior (Mulligan et al. 2006). An additional meta-analysis of selectively bred ethanol-naïve mice, recombinant inbred mice (BXD mice), and a large panel of inbred mice identified >8,000 transcripts related to ethanol preference (Tabakoff et al. 2008). The findings for both of these studies were filtered for effect size, positional overlap with known behavioral quantitative trait loci (QTLs), and/or anatomical expression patterns to identify putative quantitative trait genes (QTGs) for ethanol preference. For example, in the Mulligan study, the most statistically significant genes from the meta-analysis (Q<0.01) showed over-representation for MAPK signaling and several transcription factor pathways. Based on both of these inquiries it is fairly clear that a large number of differentially expressed genes are cooperating within each of these datasets, and the genetic predisposition component of alcohol preference is not limited to a single factor.
Both Mulligan et al. and Tabakoff et al. narrowed their focus to putative QTGs, such as the sodium channel 4β subunit (Scn4b) that was common to both data sets. The large number of differentially expressed transcripts that correlated to alcohol preference may contribute to functionally distinct gene networks with major genetic hubs such as Scn4b. Unveiling the role of these genetic networks for this specific endophenotype may significantly impact our knowledge of the molecular basis for predisposition to risk for the development of alcohol dependence.
Differences in ethanol-induced signaling events are another dynamic aspect influencing the maturation of alcohol dependency. Divergent sensitivity to an initial acute exposure to ethanol is correlated with an individual s chances of developing alcoholism (Schuckit 1994), suggesting that acute ethanol-induced signaling events are an important “risk factor” for developing dependence. Alterations in gene expression can occur at biologically relevant concentrations of ethanol within 4 hours after an initial exposure and persist for extended periods of time (Miles et al. 1991). Early genomic work from our laboratory utilizing expression profiling in neuroblastoma cells identified some of the major gene targets of ethanol (Thibault et al. 2000). These expression profiles pointed to catecholamine metabolism (dopamine β-hydroxylase), cellular survival and oxidative stress, and cyclic-AMP (cAMP) signaling mechanisms. These results further implicated the cAMP system as a key regulator of acute and chronic ethanol action; however, it also exemplified that ethanol has unique signaling mechanisms as not all of the same genes were regulated solely by cAMP (Thibault et al. 2000; Hassan et al. 2003; Thibault et al. 2005). Characterizing these differences in gene expression both validated the use of microarrays by uncovering genes known to contribute to ethanol responses (i.e. cAMP signaling genes), and demonstrated the utility of genomic approaches to identify novel genes unique to an ethanol-induced response.
Genomic studies on acute or chronic ethanol exposure have also been done in vivo using rodent and human (autopsy) brain tissue (Lewohl et al. 2000; Daniels and Buck 2002; Mayfield et al. 2002; Treadwell and Singh 2004; Kerns et al. 2005; Liu et al. 2006). Kerns et al. profiled acute responses to ethanol (4 hours, 2 g/kg i.p.) across areas of the mesolimbocortical dopamine pathway – medial prefrontal cortex, nucleus accumbens and ventral tegmental area (Kerns et al. 2005). By performing these studies across the DBA/2J (D2) and C57BL/6J (B6) mouse strains, which are known to have widely divergent behavioral responses to acute ethanol and ethanol consumption behavior, these studies implicated both basal and ethanol-responsive expression networks in ethanol-related behaviors.
Kerns et al. showed multiple clusters of acute ethanol regulated genes within the nucleus accumbens, including a coordinately expressed cluster related to brain derived neurotrophic factor (Bdnf). Bdnf expression was strongly correlated with T-box brain 1 (Tbr1), forkhead box P1 (Foxp1), and pituitary adenylate cyclase-activating polypeptide (Pacap). The polypeptide Pacap, also known as Adcyap1, regulates the expression of Bdnf and activity of the NMDA receptor via its activation of the cAMP pathway (Yaka et al. 2003). Such coordinately expressed networks may have behavioral consequences as Bdnf is located within the support interval of a previously identified QTL for acute ethanol locomotor response (Actre3), and mice with altered expression of Pacap possess altered locomotor activity (Hashimoto et al. 2001; Tappe and Kuner 2006). Thus the differential expression of a Bdnf gene network evoked by acute ethanol, rather than just Bdnf itself, may be an important quantitative trait gene network (QTGN) for acute ethanol locomotor activation. Given the in vitro genomic evidence for ethanol action on cAMP signalling mentioned above, it is relevant that cAMP-response element binding protein (CREB) is known to regulate expression of Bdnf and other genes previously implicated in alcoholism or behavioral responses to ethanol, including neuropeptide Y (Npy), and corticotropin releasing factor (Crh). This may suggest that multiple genes relevant to behavioral and neurobiological actions of ethanol share common signaling mechanisms, affecting entire networks of genes.
Additional ethanol-responsive genes identified by Kerns et al. included a group of genes related to myelin structure or regulation. The prefrontal cortex showed both basal and ethanol-response expression differences for this myelin gene network. While B6 mice showed increased basal myelin gene expression, D2 mice showed much greater ethanol induction of these genes. Furthermore, this work also suggested a role for the Creb1 gene in regulation of the myelin gene network (Kerns et al. 2005).
The regulation of a myelin gene network by acute ethanol in mice has substantially greater importance since similar observations have been made with genomic studies on human prefrontal cortex gene expression in autopsy material from studies on schizophrenia and alcoholism. Harris, Mayfield and colleagues have used microarray studies in human brain postmortem tissue to identify gene sets and networks that show alterations with alcoholism (Lewohl et al. 2000; Mayfield et al. 2002; Lewohl et al. 2005; Liu et al. 2006). Strikingly, among the most reproducible findings in these microarray studies on brain tissue from alcoholics is the coordinate down-regulation of many myelin-related genes, particularly in prefrontal cortex. Thus, at a functional group or gene network level, these genomic studies across humans and mice strongly point to myelin gene network as an important molecular mechanism possibly contributing either to ethanol neurotoxicity, neural plasticity related to ethanol-evoked behaviors, or alcohol dependence.
Cross species studies, as illustrated above for the myelin genes, have been used to parse the most relevant genes or gene networks out of genomic and genetic studies on ethanol or alcoholism. For example, convergent functional genomics (CFG) is a translational approach for connecting genes between human and animal models in complex disorders (Bertsch et al. 2005). Although it is not a network approach per se it does constrict copious amounts of gene expression and genetic linkage data into experimentally derived groups with shared biological relationships. When applied to alcoholism research, this approach has identified candidate genes within multiple signaling systems, with some degree of association to other neuropsychiatric disorders such as schizophrenia (Rodd et al. 2007).
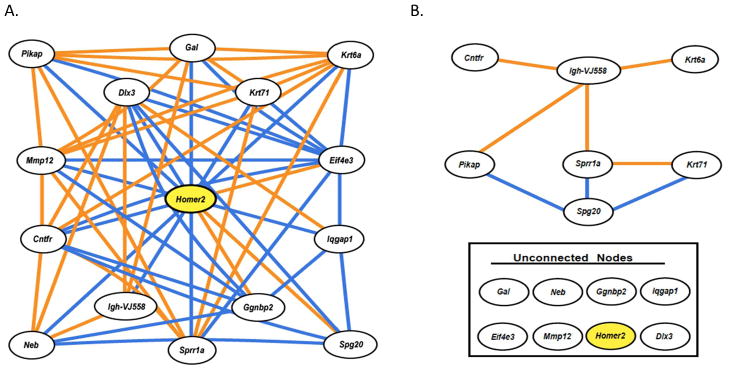
Ethanol exposure may also disrupt homeostatic control of gene networks. Expression profiling of medial prefrontal cortex in the BXD recombinant inbred panel, as well as B6 and D2 progenitors, provides an illustration of disrupted homeostatic control in a Homer2-associated gene network (Figure 2). Homer2 is a glutamatergic-related scaffolding protein implicated in functional neuroplasticity of ethanol abuse (Szumlinski et al. 2005), whose baseline correlation structure (Fig. 2A) is distinctly different 4 hours post acute ethanol (1.8 g/kg) (Fig. 2B). Under baseline conditions Homer2 is inversely correlated to galanin (Gal) but this connectivity is lost following acute ethanol treatment. Galanin is known to attenuate drug reinforcement (Narasimhaiah et al. 2009). Such network dysregulation of Homer2 and Gal, as well as other motifs or networks not mentioned, may have profound impact on subsequent behavioral responses to drug or alcohol exposure.
Figure 2. Acute ethanol disruption of baseline Homer2-associated gene network in BXD recombinant inbred mice.
(A) Homer2 expression correlation network in prefrontal cortex under baseline conditions (B) Homer2 baseline correlation network 4 hours after acute ethanol (1.8 g/kg) treatment. The highly coherent network seen across control BXD strains was severely rearranged by acute ethanol treatment. This suggests that elements of the Homer 2 expression network are targets of acute ethanol action. Orange lines represent positive Pearson correlation coefficients ≥ 0.60; Blue lines represent negative Pearson correlation coefficients ≤ −0.60 (n = 29).
Conclusion and Closing Remarks
Neuropsychiatric conditions including alcoholism are multifaceted diseases of complex origin. Extensive efforts across multiple fields of scientific research have actively sought to explain the origins of alcoholism; however, no solitary molecular mechanism has yet been established. Emerging evidence from a trove of genome-wide association and differential gene expression studies have illustrated that variants and expression differences in multiple genes can account for the manifestation of complex diseases. In order to grasp the functional importance of these individual genes, visualizing the shared interconnection among gene products using network-based approaches is essential.
Functional genomics is rapidly producing prodigious amounts of data, outlining the systems biology of alcohol abuse and alcoholism. Defining the multitude of sub-networks within this framework is awaiting continued investigation. However, existing datasets across multiple species are helping identify conserved regulatory elements and major genetic hubs of network activity, pushing us beyond distinct candidate genes towards quantitative trait gene networks (QTGNs).
Networks can be derived from multiple sources of information and theoretical frameworks. The precise determination of how to build such networks is open to interpretation, but undoubtedly will rely on some type of an organizing framework such as correlation structures or functional associations. We have suggested here that the most informative network strategies, those most likely to predict functional gene/phenotype associations, are likely to be constructed from multiple types of data from both human and animal models. Coupling large scale phenotypic, genetic and genomic investigations across temporal and phenotypic space will allow productive use of approaches such as structural equation modeling (Moore et al. 2007) in better defining network centrality and generating testable hypotheses. These informative lines of interpretation will critically depend on the continued development of large-scale bioinformatics resources that can help define temporal and spatial patterns of gene expression or other gene-gene interactions. This is particularly important given the complex organization and degree of regulation, both cellular and molecular, within the nervous system. Indeed, this review has not even touched upon the topic of defining genomic expression networks at a cellular level. Thus, many of the existing genomic data sets and network constructions are very likely composites across multiple cell types. Efforts such as the Allen Brain Atlas (Lein et al. 2007), which is constructing tools to define cellular expression patterns on a genomic scale, are but a modest beginning at taking gene network efforts to the level of resolution needed to understand neural network dysregulation as occurs in alcoholism.
Additional limitations are imposed upon network-based gene discovery approaches due to inadequate resources. For example, a whole genome association study may lack sufficient sample size to detect low abundance genes or rare SNPs hypothesized to contribute to the occurrence of complex traits. Efforts such as Next-Gen sequencing of the “exome” and RNA-Seq analyses of splice variation and non-coding RNA regulation are now technically possible but the financial, statistical and computational resources needed for such efforts are largely vestigial at this time. Overcoming these inherent limitations, and making network analyses capable of predicting causal factors in disease, will require innovative strategies for resource sharing, the continuance of high-throughput molecular studies, and the development of novel methods of data exploration and interpretation.
Acknowledgments
The authors would like to thank Nate Bruce, Paul Vorster, and Alexander Putman for their contributions in generating some of the discussed BXD data, and Robert Williams at University of Tennessee Health Science Center for collaboration in the use of GeneNetwork. This work was supported in part by grants from the National Institute on Alcohol Abuse and Alcoholism (F31AA018615 to SPF; U01AA016662, U01AA016667, RO1AA014717, and P20AA017828 to MFM).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
V. References
- Baldwin NE, Chesler EJ, Kirov S, Langston MA, Snoddy JR, Williams RW, et al. Computational, integrative, and comparative methods for the elucidation of genetic coexpression networks. J Biomed Biotechnol. 2005;2005(2):172–80. doi: 10.1155/JBB.2005.172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bertsch B, Ogden CA, Sidhu K, Le-Niculescu H, Kuczenski R, Niculescu AB. Convergent functional genomics: a Bayesian candidate gene identification approach for complex disorders. Methods. 2005;37(3):274–9. doi: 10.1016/j.ymeth.2005.03.012. [DOI] [PubMed] [Google Scholar]
- Bottomly D, Walter NA, Hunter JE, Darakjian P, Kawane S, Buck KJ, et al. Evaluating Gene Expression in C57BL/6J and DBA/2J Mouse Striatum Using RNA-Seq and Microarrays. PloS one. 2011;6(3):e17820. doi: 10.1371/journal.pone.0017820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Butte AJ, Tamayo P, Slonim D, Golub TR, Kohane IS. Discovering functional relationships between RNA expression and chemotherapeutic susceptibility using relevance networks. Proc Natl Acad Sci U S A. 2000;97(22):12182–6. doi: 10.1073/pnas.220392197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen J, Aronow BJ, Jegga AG. Disease candidate gene identification and prioritization using protein interaction networks. BMC Bioinformatics. 2009;10:73. doi: 10.1186/1471-2105-10-73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen J, Bardes EE, Aronow BJ, Jegga AG. ToppGene Suite for gene list enrichment analysis and candidate gene prioritization. Nucleic Acids Res. 2009;37(Web Server issue):W305–11. doi: 10.1093/nar/gkp427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen J, Xu H, Aronow BJ, Jegga AG. Improved human disease candidate gene prioritization using mouse phenotype. BMC Bioinformatics. 2007;8:392. doi: 10.1186/1471-2105-8-392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen L, Page GP, Mehta T, Feng R, Cui X. Single nucleotide polymorphisms affect both cis- and trans-eQTLs. Genomics. 2009;93(6):501–8. doi: 10.1016/j.ygeno.2009.01.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chesler EJ, Lu L, Shou S, Qu Y, Gu J, Wang J, et al. Complex trait analysis of gene expression uncovers polygenic and pleiotropic networks that modulate nervous system function. Nat Genet. 2005;37(3):233–42. doi: 10.1038/ng1518. [DOI] [PubMed] [Google Scholar]
- Chesler EJ, Williams RW. Brain gene expression: genomics and genetics. Int Rev Neurobiol. 2004;60:59–95. doi: 10.1016/S0074-7742(04)60003-1. [DOI] [PubMed] [Google Scholar]
- Chesler EJ, Wilson SG, Lariviere WR, Rodriguez-Zas SL, Mogil JS. Identification and ranking of genetic and laboratory environment factors influencing a behavioral trait, thermal nociception, via computational analysis of a large data archive. Neurosci Biobehav Rev. 2002;26(8):907–23. doi: 10.1016/s0149-7634(02)00103-3. [DOI] [PubMed] [Google Scholar]
- Daniels GM, Buck KJ. Expression profiling identifies strain-specific changes associated with ethanol withdrawal in mice. Genes Brain Behav. 2002;1(1):35–45. doi: 10.1046/j.1601-1848.2001.00008.x. [DOI] [PubMed] [Google Scholar]
- Eisen MB, Spellman PT, Brown PO, Botstein D. Cluster analysis and display of genome-wide expression patterns. Proc Natl Acad Sci U S A. 1998;95(25):14863–8. doi: 10.1073/pnas.95.25.14863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goodwin DW, Schulsinger F, Moller N, Hermansen L, Winokur G, Guze SB. Drinking problems in adopted and nonadopted sons of alcoholics. Arch Gen Psychiatry. 1974;31(2):164–9. doi: 10.1001/archpsyc.1974.01760140022003. [DOI] [PubMed] [Google Scholar]
- Hashimoto H, Shintani N, Tanaka K, Mori W, Hirose M, Matsuda T, et al. Altered psychomotor behaviors in mice lacking pituitary adenylate cyclase-activating polypeptide (PACAP) Proc Natl Acad Sci U S A. 2001;98(23):13355–60. doi: 10.1073/pnas.231094498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hassan S, Duong B, Kim KS, Miles MF. Pharmacogenomic analysis of mechanisms mediating ethanol regulation of dopamine beta-hydroxylase. J Biol Chem. 2003;278(40):38860–9. doi: 10.1074/jbc.M305040200. [DOI] [PubMed] [Google Scholar]
- Hughes TR, Marton MJ, Jones AR, Roberts CJ, Stoughton R, Armour CD, et al. Functional discovery via a compendium of expression profiles. Cell. 2000;102(1):109–26. doi: 10.1016/s0092-8674(00)00015-5. [DOI] [PubMed] [Google Scholar]
- Jansen RC, Nap JP. Genetical genomics: the added value from segregation. Trends Genet. 2001;17(7):388–91. doi: 10.1016/s0168-9525(01)02310-1. [DOI] [PubMed] [Google Scholar]
- Jensen LJ, Kuhn M, Stark M, Chaffron S, Creevey C, Muller J, et al. STRING 8--a global view on proteins and their functional interactions in 630 organisms. Nucleic Acids Res. 2009;37(Database issue):D412–6. doi: 10.1093/nar/gkn760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson C, Drgon T, Liu QR, Walther D, Edenberg H, Rice J, et al. Pooled association genome scanning for alcohol dependence using 104,268 SNPs: validation and use to identify alcoholism vulnerability loci in unrelated individuals from the collaborative study on the genetics of alcoholism. Am J Med Genet B Neuropsychiatr Genet. 2006;141B(8):844–53. doi: 10.1002/ajmg.b.30346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanehisa M, Araki M, Goto S, Hattori M, Hirakawa M, Itoh M, et al. KEGG for linking genomes to life and the environment. Nucleic Acids Res. 2008;36(Database issue):D480–4. doi: 10.1093/nar/gkm882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kerns RT, Ravindranathan A, Hassan S, Cage MP, York T, Sikela JM, et al. Ethanol-responsive brain region expression networks: implications for behavioral responses to acute ethanol in DBA/2J versus C57BL/6J mice. J Neurosci. 2005;25(9):2255–66. doi: 10.1523/JNEUROSCI.4372-04.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koob GF, Volkow ND. Neurocircuitry of addiction. Neuropsychopharmacology. 2010;35(1):217–38. doi: 10.1038/npp.2009.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lein ES, Hawrylycz MJ, Ao N, Ayres M, Bensinger A, Bernard A, et al. Genome-wide atlas of gene expression in the adult mouse brain. Nature. 2007;445(7124):168–76. doi: 10.1038/nature05453. [DOI] [PubMed] [Google Scholar]
- Lewohl JM, Wang L, Miles MF, Zhang L, Dodd PR, Harris RA. Gene expression in human alcoholism: microarray analysis of frontal cortex. Alcohol Clin Exp Res. 2000;24(12):1873–82. [PubMed] [Google Scholar]
- Lewohl JM, Wixey J, Harper CG, Dodd PR. Expression of MBP, PLP, MAG, CNP, and GFAP in the Human Alcoholic Brain. Alcohol Clin Exp Res. 2005;29(9):1698–705. doi: 10.1097/01.alc.0000179406.98868.59. [DOI] [PubMed] [Google Scholar]
- Liu J, Lewohl JM, Harris RA, Iyer VR, Dodd PR, Randall PK, et al. Patterns of gene expression in the frontal cortex discriminate alcoholic from nonalcoholic individuals. Neuropsychopharmacology. 2006;31(7):1574–82. doi: 10.1038/sj.npp.1300947. [DOI] [PubMed] [Google Scholar]
- Liu S, Lin L, Jiang P, Wang D, Xing Y. A comparison of RNA-Seq and high-density exon array for detecting differential gene expression between closely related species. Nucleic Acids Res. 2010 doi: 10.1093/nar/gkq817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lockhart DJ, Dong H, Byrne MC, Follettie MT, Gallo MV, Chee MS, et al. Expression monitoring by hybridization to high-density oligonucleotide arrays. Nature Biotechnology. 1996;14:1675–80. doi: 10.1038/nbt1296-1675. [DOI] [PubMed] [Google Scholar]
- Mayfield RD, Lewohl JM, Dodd PR, Herlihy A, Liu J, Harris RA. Patterns of gene expression are altered in the frontal and motor cortices of human alcoholics. J Neurochem. 2002;81(4):802–13. doi: 10.1046/j.1471-4159.2002.00860.x. [DOI] [PubMed] [Google Scholar]
- Miles MF. Alcohol's effects on gene expression. Alcohol Health Res World. 1995;(19):237–43. [PMC free article] [PubMed] [Google Scholar]
- Miles MF, Diaz JE, DeGuzman VS. Mechanisms of neuronal adaptation to ethanol. Ethanol induces Hsc70 gene transcription in NG108-15 neuroblastoma x glioma cells. J Biol Chem. 1991;266(4):2409–14. [PubMed] [Google Scholar]
- Moore DF, Gelderman MP, Ferreira PA, Fuhrmann SR, Yi H, Elkahloun A, et al. Genomic abnormalities of the murine model of Fabry disease after disease-related perturbation, a systems biology approach. Proc Natl Acad Sci U S A. 2007;104(19):8065–70. doi: 10.1073/pnas.0701991104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mozhui K, Ciobanu DC, Schikorski T, Wang X, Lu L, Williams RW. Dissection of a QTL hotspot on mouse distal chromosome 1 that modulates neurobehavioral phenotypes and gene expression. PLoS Genet. 2008;4(11):e1000260. doi: 10.1371/journal.pgen.1000260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mulligan MK, Ponomarev I, Hitzemann RJ, Belknap JK, Tabakoff B, Harris RA, et al. Toward understanding the genetics of alcohol drinking through transcriptome meta-analysis. Proc Natl Acad Sci U S A. 2006;103(16):6368–73. doi: 10.1073/pnas.0510188103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Narasimhaiah R, Kamens HM, Picciotto MR. Effects of galanin on cocaine-mediated conditioned place preference and ERK signaling in mice. Psychopharmacology (Berl) 2009;204(1):95–102. doi: 10.1007/s00213-008-1438-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nestler EJ, Aghajanian GK. Molecular and cellular basis of addiction. Science. 1997;278(5335):58–63. doi: 10.1126/science.278.5335.58. [DOI] [PubMed] [Google Scholar]
- Oldham MC, Konopka G, Iwamoto K, Langfelder P, Kato T, Horvath S, et al. Functional organization of the transcriptome in human brain. Nat Neurosci. 2008;11(11):1271–82. doi: 10.1038/nn.2207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prescott CA, Kendler KS. Genetic and environmental contributions to alcohol abuse and dependence in a population-based sample of male twins. Am J Psychiatry. 1999;156(1):34–40. doi: 10.1176/ajp.156.1.34. [DOI] [PubMed] [Google Scholar]
- Rodd ZA, Bertsch BA, Strother WN, Le-Niculescu H, Balaraman Y, Hayden E, et al. Candidate genes, pathways and mechanisms for alcoholism: an expanded convergent functional genomics approach. Pharmacogenomics J. 2007;7(4):222–56. doi: 10.1038/sj.tpj.6500420. [DOI] [PubMed] [Google Scholar]
- Saric J, Jensen LJ, Ouzounova R, Rojas I, Bork P. Extraction of regulatory gene/protein networks from Medline. Bioinformatics. 2006;22(6):645–50. doi: 10.1093/bioinformatics/bti597. [DOI] [PubMed] [Google Scholar]
- Schadt EE, Lamb J, Yang X, Zhu J, Edwards S, Guhathakurta D, et al. An integrative genomics approach to infer causal associations between gene expression and disease. Nat Genet. 2005;37(7):710–7. doi: 10.1038/ng1589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schadt EE, Monks SA, Drake TA, Lusis AJ, Che N, Colinayo V, et al. Genetics of gene expression surveyed in maize, mouse and man. Nature. 2003;422(6929):297–302. doi: 10.1038/nature01434. [DOI] [PubMed] [Google Scholar]
- Schuckit MA. Low level of response to alcohol as a predictor of future alcoholism. Am J Psychiatry. 1994;151(2):184–9. doi: 10.1176/ajp.151.2.184. [DOI] [PubMed] [Google Scholar]
- Snel B, Lehmann G, Bork P, Huynen MA. STRING: a web-server to retrieve and display the repeatedly occurring neighbourhood of a gene. Nucleic Acids Res. 2000;28(18):3442–4. doi: 10.1093/nar/28.18.3442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szumlinski KK, Lominac KD, Oleson EB, Walker JK, Mason A, Dehoff MH, et al. Homer2 is necessary for EtOH-induced neuroplasticity. J Neurosci. 2005;25(30):7054–61. doi: 10.1523/JNEUROSCI.1529-05.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tabakoff B, Saba L, Kechris K, Hu W, Bhave SV, Finn DA, et al. The genomic determinants of alcohol preference in mice. Mamm Genome. 2008;19(5):352–65. doi: 10.1007/s00335-008-9115-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tappe A, Kuner R. Regulation of motor performance and striatal function by synaptic scaffolding proteins of the Homer1 family. Proc Natl Acad Sci U S A. 2006;103(3):774–9. doi: 10.1073/pnas.0505900103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thibault C, Hassan S, Miles MF. Using in vitro models for expression profiling studies on ethanol and drugs of abuse. Addiction Biology. 2005;10(1):53–62. doi: 10.1080/13556210412331308949. [DOI] [PubMed] [Google Scholar]
- Thibault C, Lai C, Wilke N, Duong B, Olive MF, Rahman S, et al. Expression profiling of neural cells reveals specific patterns of ethanol-responsive gene expression. Mol Pharmacol. 2000;58(6):1593–600. doi: 10.1124/mol.58.6.1593. [DOI] [PubMed] [Google Scholar]
- Thibault C, Lai C, Wilke N, Duong B, Olive MF, Rahman S, et al. Expression profiling of neural cells reveals specific patterns of ethanol-responsive gene expression. Mol Pharmacol. 2000;58(6):1593–600. doi: 10.1124/mol.58.6.1593. [DOI] [PubMed] [Google Scholar]
- Treadwell JA, Singh SM. Microarray analysis of mouse brain gene expression following acute ethanol treatment. Neurochem Res. 2004;29(2):357–69. doi: 10.1023/b:nere.0000013738.06437.a6. [DOI] [PubMed] [Google Scholar]
- von Mering C, Jensen LJ, Snel B, Hooper SD, Krupp M, Foglierini M, et al. STRING: known and predicted protein-protein associations, integrated and transferred across organisms. Nucleic Acids Res. 2005;33(Database issue):D433–7. doi: 10.1093/nar/gki005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walter NA, McWeeney SK, Peters ST, Belknap JK, Hitzemann R, Buck KJ. SNPs matter: impact on detection of differential expression. Nat Methods. 2007;4(9):679–80. doi: 10.1038/nmeth0907-679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yaka R, He DY, Phamluong K, Ron D. Pituitary adenylate cyclase-activating polypeptide (PACAP(1-38)) enhances N-methyl-D-aspartate receptor function and brain-derived neurotrophic factor expression via RACK1. J Biol Chem. 2003;278(11):9630–8. doi: 10.1074/jbc.M209141200. [DOI] [PubMed] [Google Scholar]
- Yang X, Deignan JL, Qi H, Zhu J, Qian S, Zhong J, et al. Validation of candidate causal genes for obesity that affect shared metabolic pathways and networks. Nature genetics. 2009;41(4):415–23. doi: 10.1038/ng.325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang B, Horvath S. A general framework for weighted gene co-expression network analysis. Stat Appl Genet Mol Biol. 2005;4:Article17. doi: 10.2202/1544-6115.1128. [DOI] [PubMed] [Google Scholar]
- Zhu J, Zhang B, Smith EN, Drees B, Brem RB, Kruglyak L, et al. Integrating large-scale functional genomic data to dissect the complexity of yeast regulatory networks. Nature genetics. 2008;40(7):854–61. doi: 10.1038/ng.167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu J, Zhang B, Smith EN, Drees B, Brem RB, Kruglyak L, et al. Integrating large-scale functional genomic data to dissect the complexity of yeast regulatory networks. Nat Genet. 2008;40(7):854–61. doi: 10.1038/ng.167. [DOI] [PMC free article] [PubMed] [Google Scholar]