Abstract
Introduction
Studies from selected candidate genes suggest that single nucleotide polymorphisms (SNP) involved in glutathione metabolism, DNA repair, or inflammatory responses may affect overall survival (OS) in stages I-II or low stage non-small cell lung cancer (LS-NSCLC); however, results are inconclusive. In this study, we took a systematic pathway-based approach to simultaneously evaluate the impact of genetic variation from these three pathways on OS following LS-NSCLC diagnosis.
Methods
DNA from 647 patients with LS-NSCLC was genotyped for 480 SNPs (tagSNPs) tagging 57 genes from the three candidate pathways. Associations of tagSNPs with OS were assessed at the individual SNP and whole gene levels, adjusting for age, tumor stage, surgery type, and adjuvant therapy. The genotype combinations of the SNPs associated with OS was also estimated.
Results
Among the 412 tagSNPs that were successfully genotyped and passed multi-step quality assessments, 28 showed association with OS (p<0.05). Two of the 28 were estimated to have less than a 20% chance of being false positives (rs3768490 in GSTM4 gene: p=1.32×10-4, q=0.06; rs1729786 in ABCC4 gene: p=9.25×10-4, q=0.20). Gene-based analysis suggested that, in addition to GSTM4 and ABCC4, variation in two other genes, PTGS2 and GSTA2, was also associated with OS.
Conclusions
We describe further evidence that variations in genes involved in the glutathione and inflammatory response pathways are associated with OS in patients with LS-NSCLC. Further studies are warranted to verify our findings and elucidate their functional mechanisms and clinical utility leading to improved survival for lung cancer patients.
Keywords: glutathione metabolism, DNA repair, inflammation response, genetic polymorphisms, non-small-cell lung cancer, survival analysis
Introduction
Lung cancer is the leading cause of cancer death of men and women combined worldwide(1), and approximately 80% of lung cancer cases are classified histologically as non-small-cell lung cancer (NSCLC). For patients with low stage (stage I-II) NSCLC (LS-NSCLC), surgical resection is the standard primary treatment, and a definitive role for adjuvant therapy has yet to be established(2). The 5-year survival rates following surgical resection of LS-NSCLC range from 30-70%(3); the use of adjuvant therapy (pre- or post-surgery) or during cancer recurrence may improve survival, particularly for those who responded well to the initial treatment. To our best knowledge, however, no such prognostic factors or biomarkers are available to accurately stratify the patients to optimal treatment plans. Evidence has shown that genetic factors, such as variants of the genes involved in the glutathione metabolism, DNA repair and inflammatory response pathways, may impact the prognosis of patients with NSCLC(4-10), which requires more systematic confirmation and mechanistic investigation.
The glutathione pathway is comprised of enzymes responsible for glutathione synthesis, redox, glutathione conjugation, and transporters that remove glutathione conjugates from cells. Studies have shown that genes in this system are highly polymorphic and that many are correlated with enzyme activities of the pathway and can alter responses in individuals exposed to certain environmental hazards(11). The DNA repair system has a critical role protecting the genome from insults caused by carcinogenic agents(12). Polymorphisms which affect protein activity in DNA repair genes may alter the efficiency of these processes and lead to genetic instability. As a double-edged sword, they may affect the effectiveness of chemotherapy agents targeting DNA(13, 14). Inflammatory responses to environmental exposures, such as tobacco smoke and cytotoxic agents, may play a role in lung carcinogenesis and prognosis following disease onset(10). Mutations or genetic variants of the genes involved in this pathway may perturb the balance between tumor growth and host response and thereby influence disease progression. A number of studies in recent years have demonstrated the significant role of genetic variation in the glutathione metabolism, DNA repair, and inflammatory response pathways in the overall survival of NSCLC patients(10, 12, 14-22), but controversies remain. The majority of these studies focused on advanced-stage NSCLC. Knowledge for LS-NSCLC is very limited.
The primary aim of our study was to systematically evaluate the potential effects of individual genetic variants in selected genes from the glutathione metabolism, DNA repair, and inflammatory response pathways on overall survival (OS) in LS- NSCLC. A secondary aim was to use the identified risk variants to construct an OS prediction model.
Materials and Methods
Patient Cohort
Lung cancer patients with LS-NSCLC diagnosed and treated with surgical resection at Mayo Clinic from 1997 to 2007 were included in this study. Detailed descriptions of patient identification, enrollment, blood collection, and follow-up have been published previously(3). Briefly, each subject was identified through the Mayo Clinic pathologic diagnostic (Co-Path) system, and patient medical records were abstracted by a trained nurse to obtain sex, age at diagnosis, history of tobacco exposure use, lung cancer stage and histological cell type, surgery type, adjuvant therapies, and the presence of comorbidities at diagnosis. The abstracted comorbid conditions included a range of major illnesses, such as other cancers, cardiovascular diseases, diabetes, hypertension, chronic bronchitis, and chronic obstructive pulmonary disease (COPD). For purposes of our analyses, we classified patients according to whether or not any comorbidity was present at diagnosis. Never smokers were defined as individuals who smoked fewer than 100 cigarettes during their lifetimes; former smokers were those who had quit smoking for six months or more before their lung cancer diagnosis; and current smokers were those who were smoking at the time of diagnosis. A blood sample was collected from all participants after informed consent was obtained. Survival status and cause of death were determined by reviewing the Mayo Clinic registration database and medical records, obtaining correspondence from patients’ next-of-kin, and by monitoring a variety of other sources, including death certificates, obituary documents, the Mayo Clinic Tumor Registry, and the Social Security Death Index website. Eight percent of non-Caucasian patients were excluded from this study to minimize population heterogeneity. This study was reviewed and approved by the Mayo Clinic Institutional Review Board.
SNP selection and Genotyping Procedure
We selected 29 genes from the glutathione metabolism pathway, as described previously(4). Additionally, 20 genes from the DNA repair pathway and 8 key genes from the inflammatory response pathway were included in this study, following a review of the literature for evidence of genetic associations with treatment response or survival in lung or other cancers. TagSNPs within these genes were selected based on HapMap data (http://www.hapmap.org/) of 60 unrelated Caucasian (CEU) subjects (Release 22/Phase II on NCBI B36) via Haploview (http://www.broad.mit.edu/mpg/haploview/). Candidate SNPs meeting the quality thresholds of having ≥75% of individuals with genotypes, Hardy-Weinberg p-values >0.001, and minor allele frequencies >0.001 were identified from the candidate genes. TagSNPs for each gene were selected using an r2 threshold of 0.8(23), ignoring SNPs >500 kb apart.
Four hundred seventy tagSNPs, 267 from the glutathione metabolism pathway, 152 from the DNA repair pathway, and 51 from the inflammatory response pathway were genotyped using a custom-designed Illumina GoldenGate panel. Quality control was implemented in multiple steps, as described previously(24). In brief, SNPs were excluded from further analysis if they had call rates less than 95% or Hardy-Weinberg equilibrium p-values less than 10-5 or minor allele frequencies less than 0.01 in this study population.
Statistical Analysis
The clinical characteristics of the patients were summarized by person-years of follow-up, and five-year survival was estimated, with corresponding 95% confidence intervals (95% CI) using the Kaplan-Meier approach. Demographic and clinical variables were assessed for associations with OS in univariate and multivariate Cox proportional hazards regression models. These models were assessed to verify that the data conformed to assumptions of regression approach. In order to identify the subset of demographic and clinical factors as adjustment variables (covariates) in genetic association tests, we performed a stepwise selection process using Cox proportional hazards regression and retained variables whose p-values were less than 0.05.
For each SNP that passed quality control thresholds, we used a Cox regression model to assess the associations between the SNP genotypes and OS, while adjusting for covariates. A genetic model-free approach was utilized, where the heterozygote and rare homozygote genotypes were simultaneously compared to the reference genotype (common homozygote). We extracted hazard ratios (HR), 95% confidence intervals, and 2 degrees of freedom p-values, and computed q-values corresponding to the p-values from each of the candidate SNPs to assess the false discovery rates(25).
Gene-based association tests were also used to obtain additional information about the potential effects of the selected genes in glutathione metabolism, DNA repair, and inflammatory response pathways on OS. A principal components analysis was performed on the tagSNP genotypes from each gene to identify uncorrelated linear combinations of tagSNP data that captured at least 80% of the variability present in the tagSNPs within the gene. The primary genetic coding was based on a log-additive genetic model in which SNP genotypes were coded to reflect the number of minor alleles carried by each individual; parallel analyses were performed for dominant and recessive genetic models. The identified principal components were used to perform an omnibus test of significance for the association between each of the candidate genes and OS in multivariable Cox proportional hazards regression models. P-values for the global tests were obtained, along with outcome summaries of the principal components analysis(26, 27).
We tested the combined effects of the tagSNPs with false discovery rates (q-values) no larger than 20%. We first formed all possible pairs of genotypes of the multiple significant tagSNPs and compared all genotype combinations to the reference combination of the major allele homozygotes of the component SNPs. We also identified the at-risk genotypes and formed a score that reflected the ordinal number of at-risk genotypes carried by each subject. HRs and 95% confidence intervals associated with these at-risk genotypes combinations were estimated, both individually and as an ordinal trend. All analyses were carried out using SAS (SAS Institute, Inc., Cary, NC) and R (http://www.r-project.org/) software systems.
Results
Demographic and Clinical Characteristics
Demographic and clinical characteristics of 647 stage I and II NSCLC patients are shown in Table 1. The average age of all patients was 66.6 years, with slightly more men (53.8%) than women (46.2%), and 82% were stage I at the time of diagnosis. By the time of analysis, more than one-half of the subjects were still living, with a median follow-up time of 7.06 years. The 5-year OS in this cohort was 73.6% (95% CI=70.0%-76.9%). In univariate assessments, five variables were significantly associated with OS (p-values<0.05); while in our multivariate analysis, four variables were simultaneously associated with OS: age at diagnosis, surgery subtype, adjuvant therapy, and stage; they were considered to be potential confounders and were included as adjustment variables in the SNP- and gene-based analyses. Assessments of the multivariable models did not identify large deviations from the assumptions required by the Cox proportional hazards regression model.
Table 1.
Univariate and Multivariate Analysis for Survival by Demographic and Clinical Characteristics
| Characteristics | Total (%) N=647 | Person-years of Follow-up | Dead N=279 | Univariate Analysis | Multivariate Analysis | ||
|---|---|---|---|---|---|---|---|
| HR (95%CI) | p-value | HR (95%CI) | p-value | ||||
| Sex | |||||||
| Female | 299 (46.2) | 1764.1 | 117 | 1.00 (reference) | 1.00 (reference) | ||
| Male | 348 (53.8) | 1980.1 | 162 | 1.22 (0.96-1.55) | 0.11 | 1.08 (0.83-1.39) | 5.66×10-1 |
| Age at Diagnosis (Mean±SD) | 66.6±8.8 | 68.4±8.8 | 1.03 (1.02-1.05) | 1.96×10-5 | 1.04 (1.03-1.06) | 7.55×10-7 | |
| Smoking Status | |||||||
| Never Smokers | 102 (15.8) | 589.4 | 37 | 1.00 (reference) | 1.00 (reference) | ||
| Former Smokers | 350 (54.1) | 2051.0 | 162 | 1.20 (0.84-1.71) | 3.27×10-1 | 0.89 (0.60-1.31) | 5.49×10-1 |
| Current Smokers | 195 (30.1) | 1105.7 | 80 | 1.14 (0.77-1.68) | 5.10×10-1 | 0.95 (0.63-1.44) | 8.24×10-1 |
| Surgery Subtype | |||||||
| Lobectomy | 435 (67.2) | 2614.4 | 168 | 1.00 (reference) | 1.00 (reference) | ||
| More extensive resection | 110 (17.0) | 607.9 | 58 | 1.48 (1.10-2.00) | 1.05×10-2 | 1.14 (0.81-1.60) | 4.59×10-1 |
| Others | 102 (15.8) | 524.3 | 53 | 1.61 (1.18-2.20) | 2.46×10-3 | 1.51 (1.10-2.07) | 1.17×10-2 |
| Adjuvant Therapy | |||||||
| Surgery Alone | 455 (70.3) | 2837.4 | 160 | 1.00 (reference) | 1.00 (reference) | ||
| Surgery and Chemotherapy | 99 (15.3) | 452.1 | 55 | 2.37 (1.74-3.23) | 4.95×10-8 | 2.87 (1.91-3.65) | 3.65×10-9 |
| Surgery and Radiation | 28 (4.3) | 143.5 | 18 | 2.30 (1.41-3.76) | 8.07×10-4 | 1.19 (1.14-3.09) | 1.33×10-2 |
| Surgery and Chemo+Radiation | 65 (10.0) | 313.3 | 46 | 2.72 (1.96-3.79) | 2.43×10-9 | 12.3 (2.03-4.05) | 1.84×10-9 |
| Lung Cancer Subtype | |||||||
| Adenocarcinoma | 383 (59.2) | 2320.5 | 146 | 1.00 (reference) | 1.00 (reference) | ||
| Squamous | 184 (28.4) | 1026.4 | 94 | 1.46 (1.12-1.89) | 4.62×10-3 | 1.19 (0.90-1.59) | 2.21×10-1 |
| Others | 80 (12.4) | 420.3 | 39 | 1.57 (1.10-2.24) | 1.26×10-2 | 1.23 (0.85-1.79) | 2.62×10-1 |
| Lung Cancer Stage | |||||||
| Stage IA | 309 (47.8) | 1898.4 | 106 | 1.00 (reference) | 1.00 (reference) | ||
| Stage IB | 221 (34.2) | 1246.4 | 112 | 1.57 (1.21-2.05) | 8.51×10-4 | 1.34 (1.01-1.76) | 4.00×10-2 |
| Stage IIA | 29 (4.5) | 170.4 | 12 | 1.24 (0.68-2.25) | 4.88×10-1 | 1.35 (0.74-2.46) | 3.12×10-1 |
| Stage IIB | 88 (13.6) | 431.2 | 49 | 2.07 (1.48-2.91) | 2.55×10-5 | 1.67 (1.13-2.48) | 1.01×10-2 |
| Comorbidity at Diagnosis | |||||||
| No | 141 (21.8) | 824.9 | 54 | 1.00 (reference) | 1.00 (reference) | ||
| Yes | 506 (78.2) | 2919.6 | 225 | 1.15 (0.86-1.55) | 3.50×10-1 | 1.09 (0.81-1.48) | 5.71×10-1 |
Note: Values are presented as a number (percent) unless otherwise indicated; age at diagnosis, surgery subtype, adjuvant therapy, and lung cancer stage, were identified as adjustment variables for genetic association tests.
Single SNP Analysis
From 57 candidate genes in the three pathways of interest, we genotyped 480 tagSNPs. Thirty-two of the 480 tagSNPs were failed by the genotyping center; 9 had a minor allele frequency <0.01; 11 were monomorphic; and 16 were not in Hardy-Weinberg equilibrium (9 had copy number variant issues). This resulted in a total of 412 tagSNPs for analysis. Supplementary Table 1 contains a listing of all of the SNPs that were genotyped in this study. The average call rate for these SNPs was 99.5%, and the concordance between control DNA samples was 100%. No individual sample failed genotyping for this panel of SNPs.
Twenty-eight SNPs, distributed among 20 genes, had p-values less than 0.05 for the genetic model-free tests. After estimating q-values to evaluate the probability of false positive findings, two of the 28 had q-values no larger than 0.20: rs3768490 (p-value=1.32×10-4, q-value=0.06) in the GSTM5 gene and rs1729786 (p-value=9.25×10-4, q-value=0.20) in the ABCC4 gene. The results of the SNP-based association tests are shown in Supplementary Table 1.
Gene-based Analysis
Gene-based association results are presented in Supplementary Table 2. A total of 51 genes were tested using the principal components approach described in the methods section. The two genes containing the individually significant SNPs, GSTM5 and ABCC4, were both confirmed to be significantly associated with OS (p-values=2.22×10-3 and 1.46×10-2 for GSTM5 and ABCC4, respectively). In the whole-gene tests, both PTGS2 and GSTA2 were also suggestively associated with OS (p-values=2.11×10-2 and 4.32×10-2 for PTGS2 and GSTA2, respectively).
Cumulative risk assessment
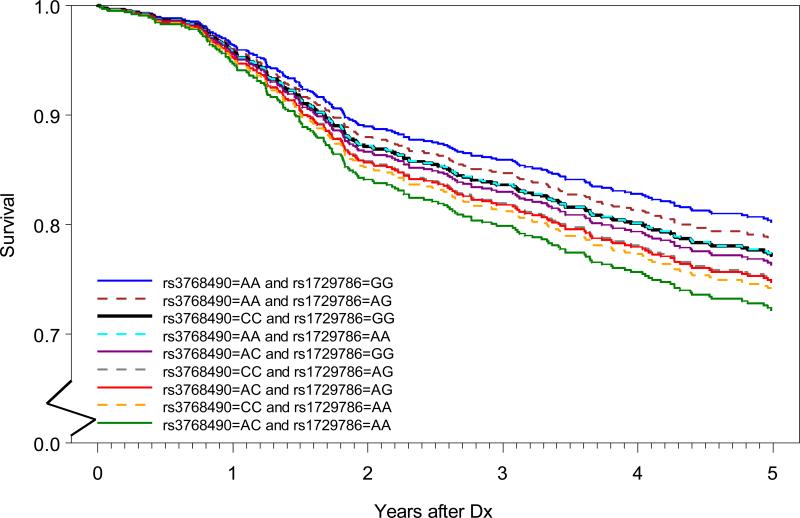
The risk associations arising from combining data from the two independent high-risk tagSNPs, that were identified in the SNP-based analyses and confirmed by gene-based analysis, are presented in Tables 3 and 4. First, all combinations of the two genotypes were formed, and specific HR estimates were obtained; the combination of the common homozygote of the two SNPs was the reference group; age at diagnosis, lung cancer stage, surgery subtype, and adjuvant therapy were the adjustment variables. The HR estimates ranged from 0.53 to 2.06 (Table 3): the lowest HR was observed in patients with the A/A genotype for rs3768490 and G/G genotype for rs1729786; whereas, the highest HR was observed in patients with the A/C genotype for rs3768490 and A/A genotype for rs1729786. When comparing this saturated model to the model that only included the main effects of the SNPs, no significant interaction was detected between the two SNPs (p=0.90). Figure 1 depicts the adjusted survival curves for the patients with different combinations of the top two candidate SNPs. There are nine survival curves, one for each of the nine genotype combinations between rs3768490 and rs1729786. Compared to the reference group (CC/GG), five genotype combination groups showed significant difference, 4 with worse and 1 with better survival (Table 3 and Figure 1). For comparison, we also provided the unadjusted Kaplan-Meier survival curves for the nine combinations in Supplemental Figure 1.
Table 3.
The Effects of Combined Genotypes on Survival in LS- Non-Small Cell Lung Cancer
| rs3768490 | rs1729786 | Total (%) N=646* | Person-years Of Follow-up | Dead N=279 | HR(95%CI) | Global P Value |
|---|---|---|---|---|---|---|
| CC | GG | 88(13.62) | 511.4 | 36 | 1.00(reference) | 9.00×10-4 |
| CC | AG | 136(21.05) | 794.2 | 58 | 1.36(1.03-1.80) | |
| CC | AA | 48(7.43) | 286.1 | 24 | 1.73(1.22-2.47) | |
| AC | GG | 85(13.16) | 517.0 | 34 | 1.19(0.92-1.53) | |
| AC | AG | 145(22.45) | 760.7 | 75 | 1.62(1.12-2.35) | |
| AC | AA | 50(7.74) | 275.2 | 28 | 2.06(1.35-3.14) | |
| AA | GG | 44(6.81) | 295.6 | 9 | 0.53(0.34-0.83) | |
| AA | AG | 42(6.50) | 261.0 | 13 | 0.73(0.43-1.25) | |
| AA | AA | 8(1.24) | 46.5 | 2 | 0.93(0.52-1.65) |
N=646 (1 with missing genotype in RS1729786)
Table 4.
Genotype Score-Based Prediction Model for Survival in LS- Non-Small Cell Lung Cancer
| Genotype Risk Score | Total (%) N=646* | Person-years of Follow-up | Dead N=279 | HR(95%CI) | P Value |
|---|---|---|---|---|---|
| 0 | 44(6.81) | 295.6 | 9 | 1.00(reference) | |
| 1 | 42(6.50) | 261.0 | 13 | 1.63(0.69-3.84) | 2.67×10-1 |
| 2 | 181(28.02) | 1074.9 | 72 | 2.09(1.04-4.21) | 3.80×10-2 |
| 3 | 281(43.50) | 1554.9 | 133 | 2.89(1.47-5.70) | 2.20×10-3 |
| 4 | 98(15.17) | 561.3 | 52 | 3.83(1.87-7.81) | 2.00×10-4 |
| Per-score (Trend Test) | 1.37(1.21-1.56) | < 0.0001 | |||
N=646 (1 with missing genotype in RS1729786)
Figure 1. Adjusted survival curves for the nine combinations of the top two SNP genotypes.
Each line represents one of the nine genotype combinations between rs3768490 and rs1729786. Average method was used for the multivariate adjusted curves. The black line (CC/GG of rs3768490 and rs1729786) was the reference group used for statistical testing of significance. Survival probability from 0 to 0.5 on y-axis was cut for better readability of the curves and survival time was truncated at 5 years post diagnosis.
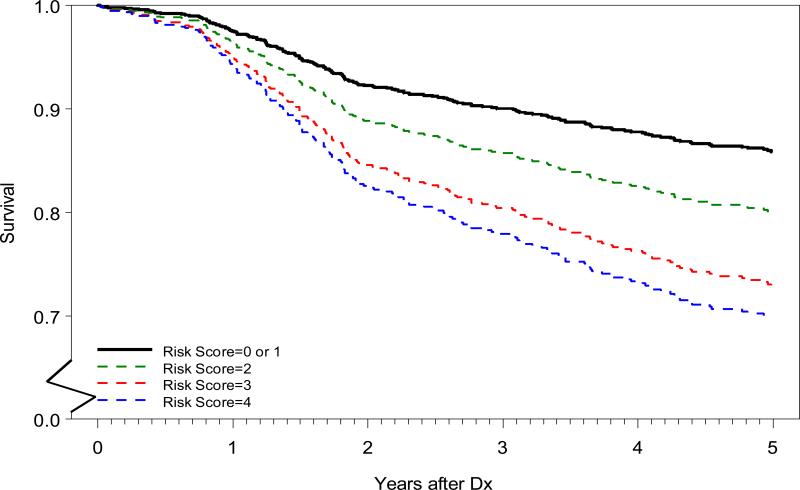
In addition to examining the simultaneous inclusion of both SNPs in a single model, we also created a risk score based on the number of high-risk alleles. The results suggested that rs3768490 acted in a recessive mode, while rs1729786 followed an ordinal pattern. Therefore, corresponding to rs3768490, individuals with zero and one copy of the minor allele received a score of 0, while individuals carrying 2 copies received a score of 2; the risk score corresponding to rs1729786 was the number of the minor alleles carried by the patient; and the final risk score was the sum of component risk scores from both SNPs (Table 4). These risk scores ranged from 0 to 4, with increasing values of the risk score being associated with increased risk of death; specifically, each increment of 1 unit in risk score suggests a 1.37-fold higher risk of dying post LS-NSCLC diagnosis with adjustment of 4 potential confounders. Figure 2 illustrates the adjusted survival curves for patients with the different risk scores (Supplementary Figure 2 for the unadjusted Kaplan Meier survival curves). Patients with a risk score of 1 were similar to those with a score of zero, both had a mortality rate lower than those with higher risk scores (>=2); patients with a genotype risk score of 4 had a 3.83-fold (95% CI: 1.87-7.81) higher risk of dying than those with a score of 0 (Table 4).
Figure 2. Adjusted survival curves by composite risk scores.
Adjusted survival curves were created for the patients with risk scores of 0/1, 2, 3, and 4 using average method. The adjusted variables included age at diagnosis, lung cancer stage, surgery subtype, and adjuvant therapy. Patients with a score of 0 were not statistically different from those with a score of 1 and the two groups were therefore combined and used as reference (black line). Survival probability from 0 to 0.5 on y-axis was cut for better readability of the curves and survival time was truncated at 5 years post diagnosis. .
Comparing the adjusted survival curves (Figures 1 and 2) with unadjusted Kaplan Meier survival curves (Supplementary Figures 1 and 2), we noticed some consistencies, but also some differences, suggesting a need to closely look at the model fit for the Cox proportional hazards analysis. Our assessments demonstrated that the assumptions of the multivariable regression models were reasonably well approximated by the data; therefore, we believe that our adjusted estimates provided an appropriate reflection of associations between the chosen SNPs and OS following diagnosis of NSCLC.
Discussion
Almost all LS-NSCLC patients are treated with surgical resection, yet outcomes vary statistically from one individual to another. The relatively homogeneous treatment modality coupled with the highly variable outcomes suggests the effects of host variations on surgery induced outcomes. In this study, we took a systematic approach and genotyped tagSNPs within 57 common genes from the glutathione metabolism, DNA repair, and inflammatory response pathways. Our purpose was to test the roles of the key genes in these pathways in OS of LS-NSCLC after surgical resection, and to measure the magnitude of any identified effects. Our results showed that 2 tagSNPs, rs3768490 and rs1729786, from the GSTM4 and ABCC4 genes were significantly associated with survival of LS-NSCLC after adjusting for potential confounding variables. Through gene-based analyses, the survival predictive effects of GSTM4 and ABCC4 genes were confirmed. Two additional genes, PTGS2 and GSTA2, were identified to be significantly associated with OS of LS-NSCLC. The fact that a significant association was observed only from a gene-based test suggests that either combinations of genotyped alleles or individual SNPs that were not genotyped may better explain the genetic associations than the individual tagSNPs that were genotyped. To understand the cumulative effects of multiple variants in different genes on survival, we constructed a cumulative risk prediction model for OS based on the combination of the two tagSNPs, which could better segregate high and low risk patients.
Glutathione S-transferase mu 4(GSTM4) belongs to the family of the Mu class of glutathione-S-transferases (GSTs) widely expressed in lung tissue. GSTs catalyze a number of reactions between glutathione (GSH) and lipophilic compounds with electrophilic centres. When the reaction forms a covalent bond, the resultant more water-soluble GSH conjugate is usually no longer toxic and may be excreted(28). The GST Mu class of GST genes consists of five members, GSTM1, GSTM2, GSTM3, GSTM4, and GSTM5, which are arranged in tandem spanning a 97-kb region on chromosome 1p13(29-31). GSTM4 is a less well-studied family member and has been demonstrated to recognize the same substrates as GSTM1, GSTM2, and GSTM3 but has lower specific activity(32). Comstock et al(33) reported the complete gene sequence and cDNA sequence of the gene, of which the deduced amino acid sequence was highly identical to GSTM1, GSTM2, and GSTM3. The same study has demonstrated the presence of GSTM4 mRNA in a variety of human tissues, including lung. The GSTM4 gene structure is very similar to that of GSTM1b with a different length of intron 7(34). To our knowledge, associations between GSTM4 polymorphism and lung cancer survival have not been reported. The tagSNP rs3768490 that showed a significant association with OS in this study is intronic, and its functional significance is not yet understood; it remains possible that the significance arises only through its linkage disequilibrium with other functional SNPs.
ATP-binding cassette sub-family C member 4 (ABCC4) is a member of the multidrug resistance protein family, which transports glutathione conjugates across the cell membrane(35). ABCC4 is localized in the apical luminal membrane of polarized epithelial cells of several excretion organs (e.g., liver, intestine, and kidney). Evidence shows that ABCC4 has very important roles in the pharmacogenetics of cancer(36); for example, in childhood acute lymphoblastic leukemia (T-1393C and A934C (Lys304Asn) polymorphisms)(37) and with cyclophosphamide-induced adverse drug reactions in breast cancer patients (rs9561778)(38). Among the 63 tagSNPs analyzed from ABCC4 in our study, rs1729786 was found to be significantly associated with OS of LS- NSCLC; like rs3768490, rs1729786 is intronic, and its functional significance needs to be further elucidated.
The prostaglandin-endoperoxide synthase 2 gene (PTGS2), encoding the COX-2 enzyme, has been found to be over-expressed in many tumor types, including lung cancer(39), and polymorphisms in PTGS2 were also reported to be associated with lung cancer(40). The glutathione S-transferase alpha 2 gene (GSTA2) is highly polymorphic, and its function is less investigated. GSTA2 and GSTA1 are co-expressed and form heterodimers in most human tissues(41). Our gene-based analysis results implied that multiple independent polymorphic markers with weak effects, or individual SNPs that were not genotyped, in the PTGS2 and GSTA2 genes may affect OS in LS-NSCLC.
There are several advantages in our study design, results from which contributed comprehensive and robust knowledge to our understanding of survival after the diagnosis of NSCLC. First, we recruited a relatively large and homogeneous cohort of patients, thus improved statistical power. Second, we focused on low stage (stage I and II) NSCLC patients with Caucasian ethnic background, which reduced the possibility of false discovery caused by disease heterogeneity and population stratification. Disease heterogeneity and population stratification are key reasons why findings from genetic association studies cannot be replicated easily. Third, we conducted both SNP- and gene-based association tests, which improved the opportunities to detect the true association signals for a given gene. Fourth, relatively stringent control of the false discovery rate via the estimation of q-values improved the robustness of the positive findings. Fifth, we generated a cumulative risk prediction model to bridge the gap between genetic information and potential clinical application, which points to the clinical utility of these genomic markers in patient management.
A few points need to be addressed in further investigations. First, it is important to replicate these findings in additional collections of patients, both within Caucasians as a validation, as well as in other ethnic populations as an assessment of whether our findings have population-specific effects. Second, it is important to carry out functional studies to uncover the biological mechanisms by which these genetic variants influence patient survival. Third, it is important to understand whether there are differential SNP effects by specific cytotoxic treatment agent. In our current study, with only 25% of the patients receiving chemotherapy, and with over 70% of those receiving platinum and taxane doublets, analyses of specific drug combinations was not adequately informative. In other words, we would not have adequate power to evaluate the interaction effects between genetic variations and various drug combinations.
In summary, our results show that genetic variants in the glutathione metabolic and inflammatory response pathways may influence OS following NSCLC diagnosis in a Caucasian population. Specifically, we have found association signals for GSTM5, ABCC4, PTGS2, and GSTA2 from SNP-, and/or gene-based analyses. The high-risk tagSNPs in the GSTM5 and ABCC4 genes may be good combinatorial predictors of mortality following NSCLC diagnosis. Further validation of these SNP associations with functional characterization of their roles in lung cancer outcomes is warranted.
Supplementary Material
Supplementary Tables:
Supplementary Table 1. All genotyped and tested SNPs from the genes in the glutathione, DNA repair, and inflammation response pathways, along with their associations with overall survival in low stage NSCLC.
Supplementary Table 2. Genes tested with multi-SNP tests of association, and the results of these tests.
Supplementary Figures:
Supplementary Figure 1. Kaplan-Meier survival curves by 9 combinations of the top two SNP genotypes.
Supplementary Figure 2. Kaplan-Meier survival curves by composite risk scores derived from the top two SNP genotypes.
Table 2.
Significant SNPs after Adjusting for Clinical Variables
| Characteristics | Total (%) N=647 | Person-years of Follow-up | Dead N=279 | Univariate Analysis | Multivariate Analysis1 | ||
|---|---|---|---|---|---|---|---|
| HR (95%CI) | p-value | HR (95%CI) | p-value | ||||
| RS3768490 (GSTM5) | |||||||
| CC | 272 (42.0) | 1591.2 | 118 | 1.00 (reference) | 2.00×10-4 | 1.00 (reference) | 1.32×10-4 |
| AC | 281 (43.4) | 1555.0 | 137 | 1.21 (0.94-1.55) | 1.19 (0.92-1.53) | ||
| AA | 94 (14.5) | 603.0 | 24 | 0.51 (0.33-0.80) | 0.53 (0.34-0.83) | ||
| RS1729786 (ABCC4)* | |||||||
| GG | 217 (33.6) | 1324.1 | 79 | 1.00 (reference) | 1.90×10-3 | 1.00 (reference) | 9.25×10-4 |
| AG | 323 (50.0) | 1815.3 | 146 | 1.43 (1.08-1.89) | 1.36 (1.03-1.81) | ||
| AA | 106 (16.4) | 607.8 | 54 | 1.85 (1.30-2.63) | 1.73 (1.22-1.47) | ||
Note: Values are presented as a number (percent) unless otherwise indicated.
Age at diagnosis, surgery subtype, adjuvant therapy, and lung cancer stage were selected as adjustment variables in genetic association tests.
N=646 (1 with missing genotype)
Acknowledgements
The authors thank Susan Ernst, M.A., for her technical assistance with the manuscript development.
Funding Sources: This work was supported by the U.S. National Cancer Institute grants (RO3 CA77118, RO1 CA80127, and RO1 CA84354) and Mayo Foundation Funds.
Footnotes
Conflicts of Interest
None of the authors have any conflicts of interest.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.International Agency for Research on Cancer GLOBALCAN 2008. Int J Cancer. 2010;127:2893–2917. [Google Scholar]
- 2.Hoffman PC, Mauer AM, Vokes EE. Lung Cancer. Lancet. 2000;355:479–485. doi: 10.1016/S0140-6736(00)82038-3. [DOI] [PubMed] [Google Scholar]
- 3.Yang P, Allen MS, Aubry MC, et al. Clinical Features of 5,628 Primary Lung Cancer Patients: Experience at Mayo Clinic from 1997-2003. Chest. 2005;128:452–462. doi: 10.1378/chest.128.1.452. [DOI] [PubMed] [Google Scholar]
- 4.Yang P, Ebbert JO, Sun Z, et al. Role of the glutathione metabolic pathway in lung cancer treatment and prognosis: a review. J Clin Oncol. 2006;24:1761–1769. doi: 10.1200/JCO.2005.02.7110. [DOI] [PubMed] [Google Scholar]
- 5.Moyer AM, Sun Z, Batzler AJ, et al. Glutathione pathway genetic polymorphisms and lung cancer survival after platinum-based chemotherapy. Cancer Epidemiol Biomarkers Prev. 2010;19:811–821. doi: 10.1158/1055-9965.EPI-09-0871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Grimminger PP, Stohlmacher J, Vallbohmer D, et al. Prognostic Significance and Clinicopathological Associations of COX-2 SNP in Patients with Nonsmall Cell Lung Cancer. Journal of oncology. 2009;2009:139590. doi: 10.1155/2009/139590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Yin Z, Zhou B, He Q, et al. Association between polymorphisms in DNA repair genes and survival of non-smoking female patients with lung adenocarcinoma. BMC Cancer. 2009;9:439. doi: 10.1186/1471-2407-9-439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Huang YT, Heist RS, Chirieac LR, et al. Genome-wide analysis of survival in early-stage non-small-cell lung cancer. J Clin Oncol. 2009;27:2660–2667. doi: 10.1200/JCO.2008.18.7906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Takenaka T, Yano T, Kiyohara C, et al. Effects of excision repair cross-complementation group 1 (ERCC1) single nucleotide polymorphisms on the prognosis of non-small cell lung cancer patients. Lung Cancer. 2010;67:101–107. doi: 10.1016/j.lungcan.2009.03.007. [DOI] [PubMed] [Google Scholar]
- 10.Matakidou A, el Galta R, Webb EL, et al. Genetic variation in the DNA repair genes is predictive of outcome in lung cancer. Hum Mol Genet. 2007;16:2333–2340. doi: 10.1093/hmg/ddm190. [DOI] [PubMed] [Google Scholar]
- 11.Reszka E, Wasowicz W, Gromadzinska J. Antioxidant defense markers modulated by glutathione S-transferase genetic polymorphism: results of lung cancer case-control study. Genes & nutrition. 2007;2:287–294. doi: 10.1007/s12263-007-0057-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kalikaki A, Kanaki M, Vassalou H, et al. DNA repair gene polymorphisms predict favorable clinical outcome in advanced non-small-cell lung cancer. Clin Lung Cancer. 2009;10:118–123. doi: 10.3816/CLC.2009.n.015. [DOI] [PubMed] [Google Scholar]
- 13.Wei Q, Frazier ML, Levin B. DNA repair: a double-edged sword. J Natl Cancer Inst. 2000;92:440–441. doi: 10.1093/jnci/92.6.440. [DOI] [PubMed] [Google Scholar]
- 14.Yu D, Zhang X, Liu J, et al. Characterization of functional excision repair cross-complementation group 1 variants and their association with lung cancer risk and prognosis. Clin Cancer Res. 2008;14:2878–2886. doi: 10.1158/1078-0432.CCR-07-1612. [DOI] [PubMed] [Google Scholar]
- 15.Dong S, Guo AL, Chen ZH, et al. RRM1 single nucleotide polymorphism -37C-->A correlates with progression-free survival in NSCLC patients after gemcitabine-based chemotherapy. Journal of hematology & oncology. 2010;3:10. doi: 10.1186/1756-8722-3-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zhou C, Ren S, Zhou S, et al. Predictive effects of ERCC1 and XRCC3 SNP on efficacy of platinum-based chemotherapy in advanced NSCLC patients. Jpn J Clin Oncol. 2010;40:954–960. doi: 10.1093/jjco/hyq071. [DOI] [PubMed] [Google Scholar]
- 17.Bi N, Yang M, Zhang L, et al. Cyclooxygenase-2 genetic variants are associated with survival in unresectable locally advanced non-small cell lung cancer. Clin Cancer Res. 2010;16:2383–2390. doi: 10.1158/1078-0432.CCR-09-2793. [DOI] [PubMed] [Google Scholar]
- 18.Muller PJ, Dally H, Klappenecker CN, et al. Polymorphisms in ABCG2, ABCC3 and CNT1 genes and their possible impact on chemotherapy outcome of lung cancer patients. Int J Cancer. 2009;124:1669–1674. doi: 10.1002/ijc.23956. [DOI] [PubMed] [Google Scholar]
- 19.Wu X, Lu C, Ye Y, et al. Germline genetic variations in drug action pathways predict clinical outcomes in advanced lung cancer treated with platinum-based chemotherapy. Pharmacogenetics and genomics. 2008;18:955–965. doi: 10.1097/FPC.0b013e32830efdd4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Tibaldi C, Giovannetti E, Vasile E, et al. Correlation of CDA, ERCC1, and XPD polymorphisms with response and survival in gemcitabine/cisplatin-treated advanced non-small cell lung cancer patients. Clin Cancer Res. 2008;14:1797–1803. doi: 10.1158/1078-0432.CCR-07-1364. [DOI] [PubMed] [Google Scholar]
- 21.Giachino DF, Ghio P, Regazzoni S, et al. Prospective assessment of XPD Lys751Gln and XRCC1 Arg399Gln single nucleotide polymorphisms in lung cancer. Clin Cancer Res. 2007;13:2876–2881. doi: 10.1158/1078-0432.CCR-06-2543. [DOI] [PubMed] [Google Scholar]
- 22.Park SY, Hong YC, Kim JH, et al. Effect of ERCC1 polymorphisms and the modification by smoking on the survival of non-small cell lung cancer patients. Med Oncol. 2006;23:489–498. doi: 10.1385/MO:23:4:489. [DOI] [PubMed] [Google Scholar]
- 23.Carlson CS, Eberle MA, Rieder MJ, et al. Selecting a maximally informative set of single-nucleotide polymorphisms for association analyses using linkage disequilbrium. Am J Hum Genet. 2004;74:106–120. doi: 10.1086/381000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sun Z, Chen J, Aakre J, et al. Genetic variation in glutathione metabolism and DNA repair genes predicts survival of small-cell lung cancer patients. Ann Oncol. 2010 doi: 10.1093/annonc/mdq212. doi:10.1093/annonc/mdq1212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Schaid DJ, Rowland CM, Tines DE, et al. Score tests for association between traits and haplotypes when linkage phase is ambiguous. Am J Hum Genet. 2002;70(2):425–434. doi: 10.1086/338688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wang K, Abbott D. A principal components regression approach to multilocus genetic association studies. Genet Epidemiol. 2008;32:108–118. doi: 10.1002/gepi.20266. [DOI] [PubMed] [Google Scholar]
- 27.Gauderman WJ, Murcray C, Gilliland F, et al. Testing association between disease and multiple SNPs in a candidate gene. Genet Epidemiol. 2007;31:383–395. doi: 10.1002/gepi.20219. [DOI] [PubMed] [Google Scholar]
- 28.Hengstler JG, Arand M, Herrero ME, et al. Polymorphisms of N-acetyltransferases, glutathione S-transferases, microsomal epoxide hydrolase and sulfotransferases: influence on cancer susceptibility. Recent Results Cancer Res. 1998;154:47–85. doi: 10.1007/978-3-642-46870-4_4. [DOI] [PubMed] [Google Scholar]
- 29.Ross VL, Board PG, Webb GC. Chromosomal mapping of the human Mu class glutathione S-transferases to 1p13. Genomics. 1993;18:87–91. doi: 10.1006/geno.1993.1429. [DOI] [PubMed] [Google Scholar]
- 30.Pearson WR, Vorachek WR, Xu SJ, et al. Identification of class-mu glutathione transferase genes GSTM1-GSTM5 on human chromosome 1p13. Am J Hum Genet. 1993;53:220–233. [PMC free article] [PubMed] [Google Scholar]
- 31.Xu S, Wang Y, Roe B, et al. Characterization of the human class Mu glutathione S-transferase gene cluster and the GSTM1 deletion. J Biol Chem. 1998;273:3517–3527. doi: 10.1074/jbc.273.6.3517. [DOI] [PubMed] [Google Scholar]
- 32.Comstock KE, Widersten M, Hao XY, et al. A comparison of the enzymatic and physicochemical properties of human glutathione transferase M4-4 and three other human Mu class enzymes. Arch Biochem Biophys. 1994;311:487–495. doi: 10.1006/abbi.1994.1266. [DOI] [PubMed] [Google Scholar]
- 33.Comstock KE, Johnson KJ, Rifenbery D, et al. Isolation and analysis of the gene and cDNA for a human Mu class glutathione S-transferase, GSTM4. J Biol Chem. 1993;268:16958–16965. [PubMed] [Google Scholar]
- 34.Zhong S, Spurr NK, Hayes JD, et al. Deduced amino acid sequence, gene structure and chromosomal location of a novel human class Mu glutathione S-transferase, GSTM4. The Biochemical journal. 1993;291(Pt 1):41–50. doi: 10.1042/bj2910041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Rius M, Hummel-Eisenbeiss J, Hofmann AF, et al. Substrate specificity of human ABCC4 (MRP4)-mediated cotransport of bile acids and reduced glutathione. Am J Physiol Gastrointest Liver Physiol. 2006;290:G640–649. doi: 10.1152/ajpgi.00354.2005. [DOI] [PubMed] [Google Scholar]
- 36.Gradhand U, Kim RB. Pharmacogenomics of MRP transporters (ABCC1-5) and BCRP (ABCG2). Drug Metab Rev. 2008;40:317–354. doi: 10.1080/03602530801952617. [DOI] [PubMed] [Google Scholar]
- 37.Low SK, Kiyotani K, Mushiroda T, et al. Association study of genetic polymorphism in ABCC4 with cyclophosphamide-induced adverse drug reactions in breast cancer patients. J Hum Genet. 2009;54:564–571. doi: 10.1038/jhg.2009.79. [DOI] [PubMed] [Google Scholar]
- 38.Ansari M, Sauty G, Labuda M, et al. Polymorphisms in multidrug resistance-associated protein gene 4 is associated with outcome in childhood acute lymphoblastic leukemia. Blood. 2009;114:1383–1386. doi: 10.1182/blood-2008-11-191098. [DOI] [PubMed] [Google Scholar]
- 39.Strazisar M, Mlakar V, Glavac D. The expression of COX-2, hTERT, MDM2, LATS2 and S100A2 in different types of non-small cell lung cancer (NSCLC). Cellular & molecular biology letters. 2009;14:442–456. doi: 10.2478/s11658-009-0011-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Campa D, Zienolddiny S, Maggini V, et al. Association of a common polymorphism in the cyclooxygenase 2 gene with risk of non-small cell lung cancer. Carcinogenesis. 2004;25:229–235. doi: 10.1093/carcin/bgh008. [DOI] [PubMed] [Google Scholar]
- 41.Ning B, Wang C, Morel F, et al. Human glutathione S-transferase A2 polymorphisms: variant expression, distribution in prostate cancer cases/controls and a novel form. Pharmacogenetics. 2004;14:35–44. doi: 10.1097/00008571-200401000-00004. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Tables:
Supplementary Table 1. All genotyped and tested SNPs from the genes in the glutathione, DNA repair, and inflammation response pathways, along with their associations with overall survival in low stage NSCLC.
Supplementary Table 2. Genes tested with multi-SNP tests of association, and the results of these tests.
Supplementary Figures:
Supplementary Figure 1. Kaplan-Meier survival curves by 9 combinations of the top two SNP genotypes.
Supplementary Figure 2. Kaplan-Meier survival curves by composite risk scores derived from the top two SNP genotypes.