Highlights
► Xylulose kinase (XKS1) is a key enzyme for xylose utilization in Saccharomyces cerevisiae. ► XKS1 was recombinantly produced in E. coli, and native and tagged forms of the enzyme were isolated. ► XKS1 was highly unstable, losing its activity rapidly during purification or storage. ► Isolated XKS1 was shown by MS to be structurally intact. ► XKS1 harboring a C-terminal Strep-tag II was purified with retention of activity and characterized.
Keywords: Saccharomyces cerevisiae xylulose kinase, XKS1, Protein expression/purification, Stability/stabilization, Protein–protein interactions, Pentose fermentation
Abstract
The Saccharomyces cerevisiae gene encoding xylulose kinase (XKS1) was over-expressed to an abundance of ⩾10% intracellular protein in Escherichia coli. Instability of XKS1, not pointed out in previous reports of the enzyme, prevented isolation of active enzyme in native or “tagged” form under a wide range of purification conditions. A fusion protein haboring C-terminal Strep-tag II (XKS1-Strep) displayed activity (∼20 U/mg) as isolated. However, the half-life time of purified XKS1-Strep was only ∼1.5 h at 4 °C and could not be enhanced substantially by an assortment of extrinsic stabilizers (osmolytes, protein, substrates). Peptide mass mapping and N-terminal sequencing showed that the recombinant protein was structurally intact, ruling out proteolytic processing and chemical modifications as possible factors to compromise the stability of the enzyme as isolated. Partial functional complementation of a largely inactive XKS1 preparation by the high-molecular mass fraction (⩾10 kDa) of cell extract prepared from an E. coli BL21 (DE3) expression host suggests a possible role for heterotropic protein-XKS1 interactions in conferring activity/stability to the enzyme. Michaelis–Menten constants of XKS1-Strep were determined: d-xylulose (210 ± 40 μM) and Mg2+-ATP (1.70 ± 0.10 mM).
Introduction
Production of fuel ethanol from lignocellulose hydrolyzates involves microbial fermentation of hexose and pentose sugars [1,2], often using engineered Saccharomyces cerevisiae [3–8]. The major pentoses, d-xylose and l-arabinose, are converted in most of the reported yeast strains [9–11] through metabolic pathways that pass through the level of d-xylulose. Xylulose kinase catalyzes phosphorylation of d-xylulose by ATP (Eq. (1)), and the resulting d-xylulose 5-phosphate is further converted via the non-oxidative part of the pentose phosphate shunt.
| (1) |
The open reading frame YGR194c encodes a xylulose kinase (XKS1)1 in S. cerevisiae [12,13]. YGR194c is essential for growth on d-xylulose [14] although its expression in the native yeast is generally low. Moderately enhanced expression of YGR194c improved the ethanol yield in xylose-fermenting strains of S. cerevisiae [4–7,15,16]. However, some “toxic effects” resulting from uncontrolled over-expression of YGR194c were also noted [6,13,17]. One possible explanation is that XK reacts with substrates other than d-xylulose and the phosphorylated products obtained are toxic. Another is that balance of intracellular ATP is perturbed by XK present in excess. Work of Jeffries and colleagues show that toxicity resulting from XK over-expression can be removed under certain conditions and that it is not relevant when glucose is used instead of xylose [18–20].
Little is currently known about the properties of XKS1 and generally, yeast XK enzymes [21]. The mode of action of the XKS1 and the performance of the enzyme under physiological boundary conditions are not well understood. This presents a serious limitation to targeted and rational optimization of the catabolic pathways used for pentose fermentation in industrial strains of S. cerevisiae in which XKS1 has a key role. Richard et al. succeeded in partially purifying XKS1 from a S. cerevisiae over-expressing YGR194c. These authors measured the substrate specificity of the enzyme and obtained a limited set of kinetic parameters [17]. A His-tagged form of XKS1 was recently derived from an Escherichia coli production strain. The purified enzyme preparation was used without any further characterization in an assay of transketolase activity [22].
Considering the reported heterologous production of XKS1 [22], we felt strongly encouraged to perform a molecular and kinetic characterization of the enzyme, using a recombinant preparation derived from an E. coli expression host. As mentioned above, this important information was not available for XKS1, hence promoting this study. Unfortunately, the published protocol [22] was not useful in our hands due to the extreme lability of XKS1 activity. We therefore needed to examine various alternative routes of protein production (e.g. considering the “N-end rule”; [23]) and isolation that we wish to communicate herein, and were eventually successful in obtaining a purified preparation of active enzyme that could be characterized. Evidence from applying an assortment of analytical techniques (e.g. peptide mass mapping, N-terminal Edman sequencing, functional complementation of inactive enzyme) to the study of XKS1 revealed that recombinant production in E. coli had yielded a structurally intact protein, both in native or C-terminally tagged form. It also indicated that activity and stability of XKS1 were stimulated by the presence of one or more components contained in ⩾10 kDa subfraction prepared from the cell extract of the E. coli expression host. Purified XKS1 was therefore largely inactive. However, XKS1 harboring Strep-Tag II was isolated with retention of activity and characterized kinetically. A possible role for heterotropic protein-XKS1 interactions in conferring activity/stability to the enzyme is suggested.
Materials and methods
Chemicals and enzymes
Enzymes for molecular cloning were obtained from MBI Fermentas (St. Leon-Rot, Germany) except Pfu DNA polymerase, which was from Promega (Madison, WI, USA). pTrcHisA was purchased from Invitrogen (Carlsbad, CA, USA). The plasmids pTrc99A and pGEX-6P-2 were from Amersham Biosciences (GE-Healthcare, Piscataway, NJ, USA) and pET-26(b+) was from Novagen (Nottingham, UK). d-Xylulose (⩾ 95% purity) was produced by microbial oxidation of d-arabitol following a published protocol [24], Phosphoenolpyruvate, pyruvate kinase (PK, 986 U/mL), lactate dehydrogenase (LDH, 1230 U/mL), ATP (99% purity) and the protease inhibitor cocktail were from Sigma Aldrich (St. Louis, MO, USA). NADH (⩾98%) was from Roth (Karlsruhe, Germany). All other chemicals were reagent-grade and obtained from Sigma Aldrich or Roth (Karlsruhe, Germany).
Cloning and expression of the YGR194c gene in E. coli
The YGR194c gene was amplified from the previously described plasmid vector pRS416GPD-XKS1 [25] using Pfu DNA polymerase. Forward and reverse oligonucleotide primers introducing suitable restriction sites for sub-cloning and primers for the deletion of the first amino acid (Leu) and replacement of Leu by Val are listed in Table 1. Plasmid vectors were transformed into different electro-competent strains of E. coli (see Table 1), and those selected by colony PCR were sequenced (MWG-Biotech, Martinsried, Germany).
Table 1.
Cloning strategy for production of functional XKS1 in E. coli.
| Primer (restriction sites in bold; mismatched bases in italic; tags underlined) | Target plasmid | Restriction sites | Resulting plasmid | Resulting strain/XK activity |
|---|---|---|---|---|
| Fw: 5′-CGCGGATCCTTGTGTT | ||||
| CAGTAATTCAGAGACAGACAAG-3′ | ||||
| Rev: 5′-CGGGAATTCTTAGATGA | ||||
| GAGTCTTTTCCAGTTCGCTTAAG-3′ | ||||
| Annealing temperature: 59 °C (30 cycles) | pTrcHisA | BamH1/EcoR1 | pTrcHisA[XKS1]a | E. coli Top 10 [pTrcHisAXKS1]/– U/mg |
| Fw: 5′CCGGAATTCTTGTGTTC | ||||
| AGTAATTCAGAGACAGACAAG3′ | ||||
| Rev: 5′TTCTGCAGTTAGATGAG | ||||
| AGTCTTTTCCAGTTCGCT3′ | ||||
| Annealing temperature: 59 °C (30 cycles) | pTrc99A | EcoR1/Pst1 | pTrc99A[XKS1]b | E. coli XL1-Blue [pTrc99AXKS1]/4.5 U/mg |
| Fw: 5′CCGGAATTCTTGTGTTCA | ||||
| GTAATTCAGAGACAGACAAG3′ | ||||
| Rev: 5′-TTCTGCAGTTAATGATGAT | ||||
| GATGATGATGGATGAGAGTCTTTTC | ||||
| CAG TTCGCT-3′ | ||||
| Annealing temperature: 62 °C (30 cycles) | pTrc99A | EcoR1/Pst1 | pTrc99A[XKS1His]c | E. coli XL1-Blue pTrc99A[XKS1His]/3 U/mg |
| Fw: 5′-CCGGAATTCTTGTGTTCAG | ||||
| TAATTCAGAGACAGACAAG3′ | ||||
| Rev: 5′-TTCTGCAGTTATTTTTCGA | ||||
| ACTGCGGGTGGCTCCAAGCGCTG | ||||
| ATGAGAGTCTTTTCCAGTTCGCT3′ | ||||
| Annealing temperature: 65 °C (30 cycles) | pTrc99A | EcoR1/Pst1 | pTrc99A[XKS1Strep]d | E. coli XL1-Blue pTrc99A[XKS1Strep]/2.3 U/mg |
| Fw: 5′-CGCGGATCCTTGTGTTCAG | ||||
| TAATTCAGAGACAGACAAG-3′ | ||||
| Rev: 5′-CGGGAATTCTTAGATGAGA | ||||
| GTCTTTTCCAGTTCGCTTAAG-3′ | ||||
| Annealing temperature: 59 °C (30 cycles) | pGEX-6P-2 | BamH1/EcoR1 | pGEX[XKS1]e | E. coli BL21 (DE3) pGEX[XKS1]/1.7 U/mg |
| Fw: 5′-ATACATATGTGTTCAGTAATTC | ||||
| AGAGA-3′ | ||||
| Rev: 5′-GAAGGATCCTTAATGATGATGA | ||||
| TGATGGATGAGAGTCTTTTCCAGTTCGCT-3′ | ||||
| Annealing temperature: 60 °C (30 cycles) | pET26b(+) | NdeI/BamHI | pET[XKS1ΔL-His]f | E. coli BL21 (DE3) pET26b(+)[XKS1ΔL] 5.6 U/mg |
| Fw: 5′ATACATATGGTTTGTTCA | ||||
| GTAATTCAGAGA-3′ | ||||
| Rev: 5′GAAGGATCCTTAATGATGATGAT | ||||
| GATGGATGAGAGTCTTTTCCAGTTCGCT-3′ | ||||
| Annealing temperature: 62 °C (30 cycles) | pET26b(+) | NdeI/BamHI | pET[XKS1V-His]g | E. coli BL21 (DE3) pET26b(+)[XKS1V-His] 3.4 U/mg |
N-terminal His-Tag.
Native XKS1.
C-terminal His-Tag.
C-terminal Strep-Tag.
N-terminal GST-Tag.
Leu1 deletion mutant; C-terminal His-Tag.
Mutant, where Leu1 was replaced by Val; C-terminal His-Tag.
Standard conditions were used for enzyme production in 1-L baffled shake flasks (250 mL LB medium, 115 μg/mL ampicillin or 50 μg/mL kanamycin; 110 rpm; 37 °C). Isopropyl β-d-thiogalactoside (IPTG; 0.5 mM) was used for induction of exponentially growing cells (20 °C, 18 h). Crude cell extract was prepared by passing suspended E. coli biomass (50 mM Tris/HCl, pH 7.8) twice through a French Press (1500 psi). The temperature of the cells was kept low by incubation on ice for 5 min in between the two passages. Following ultra-centrifugation using a T-865 fixed-angle titanium rotor (90000g; 45 min, 4 °C; angle: 23.5°, Thermo Scientific, Bonn, Germany) and 25 mL centrifugation tubes (diameter 24 mm) the enzyme preparation was stored on ice. If not mentioned otherwise, the above Tris/HCl buffer was used.
Expression of the YGR194c gene in S. cerevisiae
S. cerevisiae CEN.PK 113–5D harboring the plasmid [pRs416GPDXKS1] was cultured using conditions reported elsewhere [25]. Preparation of a crude cell extract was exactly as described above for E. coli.
Assay for XKS1 activity
A continuous initial-rate assay was used, where XKS1-catalyzed formation of ADP (Eq. (1)) was coupled to oxidation of NADH via the reactions of PK and LDH. Consumption of NADH was measured at 340 nm (εNADH = 6220 M−1 cm−1) using a Beckman DU-800 spectrophotometer. Unless otherwise stated, the assay was performed at 30 °C in 50 mM HEPES buffer (pH 7.4), containing 10 mM MgCl2, 50 mM KCl, 0.30 mM NADH, 1.00 mM phophoenolpyruvate, 3 U/mL of PK, 5.4 U/mL of LDH, 5.00 mM d-xylulose, 5.00 mM ATP and 1.0 g/L bovine serum albumin (BSA). In kinetic studies, the concentration of d-xylulose (0.01–2.00 mM) or ATP (0.10–5.00 mM) was varied at a constant concentration of ATP (5.00 mM) and d-xylulose (5.00 mM), respectively. Reactions were started by adding 10 μL of XKS1 solution to 490 μL of reaction mixture, appropriately pre-incubated for about 2 min to allow conversion of traces of ADP present in the commercial ATP preparation. ATPase activity was measured using the above reaction mixture lacking d-xylulose. NADH oxidase activity was obtained under conditions in which the assay mixture lacked both d-xylulose and ATP. Reported initial rates for XKS1 are corrected for ATPase.
Purification of XKS1
Purification was monitored by SDS PAGE as well as by determination of total protein and activity. A detailed description of the methods used is provided in the Supplementary Material. The following procedures were used: Immobilized metal affinity chromatography (IMAC), anion exchange chromatography (AEX), hydrophobic interaction chromatography (HIC), glutathione-S-transferase tag (GST-tag) purification, and Strep-tag purification.
Structural characterization of XKS1
The procedures used for peptide mass mapping, N-terminal sequencing, and mass spectrometric analysis of XKS1 as isolated are described in the Supplementary material.
Restoration of XKS1 activity
A site-directed mutant of C-terminally His-tagged XKS1 (XKS1-His) was used in which Leu-1 was substituted by Val (XKS1V-His). A 100 μL of solution containing isolated XKS1V-His (2.0 mg/mL; specific activity: 0.1 U/mg) were mixed with 100 μL of cell extract prepared from E. coli BL21 (DE3) (final protein content: 10, 20, 40 or 160 mg/mL) or S. cerevisiae CEN.PK 113-5D (40 mg/mL) and incubated on ice for 400 min. Note that neither cell extract contained recombinant protein. Samples were taken at certain times and the kinase activity was measured. Appropriate controls where the activity was obtained under conditions in which the assay mixture lacked both d-xylulose and ATP were also recorded and subtracted from XKS1 activity. In a second experiment, the E. coli cell extract was fractionated using a Vivaspin concentrator tube (10 kDa cut-off; Sartorius, Goettingen, Germany) and 100 μL of the retentate (protein content 200 mg/mL) or flow-through (protein content 2 mg/mL), respectively, were used for incubation with XKS1V-His.
Results and discussion
Functional expression of XKS1 in E. coli
Plasmid vectors and strains used for production of wild-type and variant forms of XKS1 are listed in Table 1. Induced expression of the different constructs was generally accompanied by formation of inclusion bodies, inferred from visual or microscopic inspection of E. coli cells prior to their disruption in the French press (data not shown). In agreement with literature [22], no enzyme activity was measurable in cell extracts of the strain producing His-XKS1. All other strains, however, contained functional XK, the amount of specific XK activity produced being in the range 1.7–5.6 U/mg (Table 1). Non-induced strains lacked XK activity above the level of the ATPase background (∼0.2 U/mg). A reference XL1-Blue strain that did not carry a plasmid vector was likewise devoid of a measurable amount of XK activity. An SDS polyacrylamide gel in which crude extracts of the different E. coli strains containing XKS1 activity (Table 1) were compared, showed a prominent band migrating to a position expected from the molecular mass of recombinant XKS1 (67 kDa). Using a value of ∼20 U/mg for purified XKS1 (see later in this work), we can estimate that recombinant enzyme accounted for about 10–20% of total soluble E. coli protein, consistent with results of a semi-quantitative densitometric analysis of the SDS gel (data not shown). Akinterinwa and Cirino [25] expressed the Pichia stipitis gene encoding XK in E. coli and reached a specific activity for the recombinant enzyme (3 U/mg) in the cell extract that is well comparable to the specific activities of XKS1 in Table 1. However, isolation of the recombinant P. stipitis XK was not reported. The native P. stipitis XK was purified before [21].
The XKS1 activity in E. coli cell extracts was not stable. It was lost completely after a single freeze-and-thaw cycle at −20 °C. Fast inactivation (i.e. within a few hours at room temperature) could be prevented when cell extracts were stored on ice. The half-life of the XKS1 activity was ⩽1 week under these conditions. The stabilities of “native” and tagged forms of the enzyme were identical under these conditions. Addition of a commercial mix of protease inhibitors (Protease Inhibitor Cocktail 5 mL, P8465 Sigma Aldrich, St. Louis, MO, USA) to the cell extract (1 mL/20 mL of sample) did not improve enzyme stability. Note: P. stipitis XK was also reported to be “relatively unstable” and lose all activity after 24 h at 4 °C [21].
We therefore considered production of XKS1 in S. cerevisiae. However, the specific XKS1 activity synthesized by the different E. coli systems (Table 1) was about 1 order of magnitude higher than the corresponding specific XKS1 activity (∼0.3 U/mg) produced by a S. cerevisiae strain harboring the previously described plasmid vector [pRs416GPDXKS1] [25]. Moreover, the XKS1 activity in the yeast cell extract was extremely unstable, undergoing partial inactivation even by dilution into the assay buffer. The level of XKS1 activity in S. cerevisiae was therefore not well defined, as also found by Krahulec et al. [26]. Addition of glycerol (30%, by vol.) or BSA (1 g/L) to all buffers did not improve the stability of XKS1, and the enzyme produced in the yeast was therefore not further pursued.
Richard et al. [17] reported a value of 3.5 U/mg for the specific activity of XKS1 produced in S. cerevisiae and for reasons unknown to us, these authors appear to not have had similar problems of enzyme stability as we had. When over-expressing the XKS1 gene in S. cerevisiae, Johansson et al. [6] considered the so-called “N-end rule” [23], according to which the N-terminal amino acid has an effect upon the in vivo stability of yeast and bacterial proteins. Because the “N-end rule” predicts Leu-1 of XKS1 to be destabilizing, the leucine was replaced by valine. However, consequences of the site-directed substitution with respect to stability of XKS1 were not reported by Johansson et al. [6]. We prepared a L1V variant of XKS1-His and show in Table 1 that the specific kinase activity in the E. coli cell extract (3.4 U/mg) was not affected by the amino acid substitution. However, loss of activity in crude enzyme preparation (cell extract) occurred at identical rate within limits of error for the mutated XKS1 as compared to the corresponding wild-type enzyme.
Isolation of an active preparation of XKS1
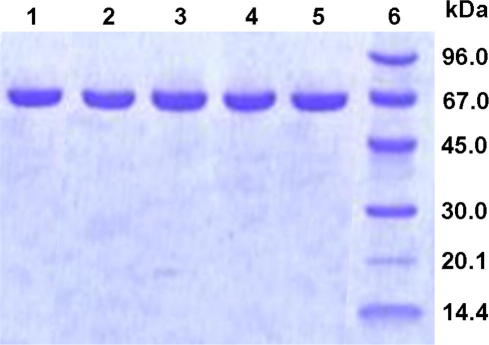
XKS1-His, XKS1ΔL-His (Leu-1 deleted) and XKS1V-His were purified to apparent electrophoretic homogeneity (Fig. 1) using Ni2+ or Cu2+ chelate chromatography. Elution of the enzymes was independent of the used metal ion. However, XKS1-His, XKS1ΔL-His and XKS1V-His isolated by either procedure were completely inactive, in contrast to findings of Lee et al. [22]. It was confirmed that enzyme activity was not lost to other proteins eluting from the column (data not shown). A low amount of activity (∼0.8 U/mg) could be restored in the purified XKS1 protein using dialysis against 50 mM Tris/HCl buffer, pH 7.8, supplemented with 30% (by vol.) glycerol. The dialysis had to be done in two steps where first, 200 mM MgCl2 were added to the buffer. Unfortunately, the “rescued” activity was by no means stable when incubated in a protein solution of 1.5 mg/mL, decaying with a half-life of ∼9 h on ice and being lost completely upon freezing. We determined that imidazole brought about a severe destabilization of XKS1-His in the crude cell extract, causing irreversible loss of enzyme activity with a half-life of ∼18 h. Despite addition of various stabilizers to the sample and the elution buffers (see the Supplementary material), IMAC gave in our hands just inactive protein.
Fig. 1.
SDS polyacrylamide gel showing purified preparation of XKS1. Lane 1, XKS1-Strep; lane 2, XKS1-His; lane 3, GST-XKS1; lane 4, XKS1V-His; lane 5, XKS1ΔL-His; lane 6, low molecular mass standard proteins (GE Healthcare). Approximately 10 μg of each XKS1 preparation were loaded on the gel. Protein bands were visualized by staining with Coomassie Blue.
The problem of enzyme inactivation during chromatography persisted while purifying GST-XKS1 and native XKS1. Isolated GST-XKS1 (Fig. 1) was inactive and could not be “rescued” by dialysis. Native XKS1 lost most of its activity upon dilution for AEX or addition of ammonium sulfate for HIC. Table 2 shows that from a series of potential stabilizers tested, glycerol and PEG 4000 were most effective in preserving the activity of native XKS1 in the cell extract. Similar results were obtained using the different C-terminally tagged forms of XKS1, including XKS1-Strep (data not shown). Despite the extra stability conferred by glycerol, most of the applied native XKS1 activity was lost during AEX and HIC, rendering either method unsuitable for purification of the native enzyme.
Table 2.
Stability of XKS1 in a crude E. coli cell extract after 24 h on ice.a
| Stabilizing additive | Retention of activity (%)b |
|---|---|
| None | 18 |
| 30% (v/v) glycerol | 99 |
| 30% (w/v) PEG 4000 | 98 |
| 200 mM MgCl2 | 20 |
| 300 mM trehalose | 82 |
| 300 mM mannitol | 80 |
| 300 mM sorbitol | 65 |
Protein concentration was 3 mg/mL.
100% refers to the XK activity in the undiluted E. coli cell extract containing the native XKS1 (12 U/mL; 4 U/mg).
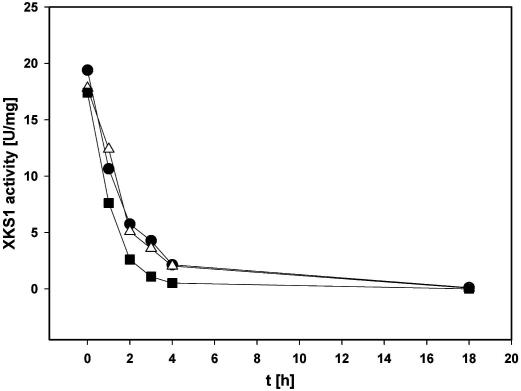
XKS1-Strep was obtained as pure protein (Fig. 1) and displayed XK activity (∼18 U/mg). This specific activity agrees with previous findings for XKS1 [17,22] and is in accordance with the specific activity for P. stipitis XK [21]. The overall yield of XKS1-Strep was only 6% based on the total amount of activity applied on and recovered from the Strep-Tactin column. Isolated XKS1-Strep was not stable on ice (Fig. 2), even in the presence of compounds that were best stabilizing the activity in the cell extract (see Table 2). Considering results to be presented later in the section Restoration of activity in XKS1V-His, it is important to note that the activity of XKS1-Strep was much more stable, roughly by an order of magnitude in buffer without any supplement, when the enzyme was present in the crude E. coli cell extract as compared to the purified protein. Inactivation of purified XKS1-Strep was not accompanied by a visible precipitation of the protein, suggesting that insoluble aggregates are not formed. The addition of BSA which was shown to stabilize XK from E. coli to some extent [27] did not help with XKS1-Strep. Instability of XKS1 is consistent with findings for P. stipitis XK [21], showing that the Pichia enzyme lost its activity at 4 °C within a day. However, other XK enzymes such as those from calf liver, Aerobacter aerogenes and Lactobacillus pentosus (reviewed in Ref. [21]) appear to be relatively stable when stored cold or frozen.
Fig. 2.
Stability of purified XKS1-Strep. Incubations were carried out on ice in 50 mM Tris/HCl buffer (pH 7.4), containing 30% glycerol (●), 30% PEG 4000 (■) or 5 mM d-xylulose (Δ). The protein concentration was 0.18 mg/mL.
Primary structure analysis for XKS1
Fig. 1 shows that the molecular mass of isolated XKS1-Strep corresponds to the size of the full-length protein expected from the coding gene (∼67 kDa). However, the different observations with respect to XKS1 stability made herein (Table 2; Fig. 2) and in previous reports [17,22] might be explained if the recombinant protein was chemically modified during enzyme production and work-up. Proteolysis and oxidation are two possible types of “chemical denaturation”. We therefore sequenced peptides of purified XKS1-Strep, based on proteolysis of the target protein with seven different enzymes, and summarize the results in Table 3. Using LC-MS/MS analysis of the respective peptide mixtures, we were able to identify 170 distinct peptides and thus obtained an overall sequence coverage of 92%. Using sequence similarity, Asp28 and Asp299 are positional homologs of the major catalytic residues, Asp6 and Asp233, in E. coli XK [27] and other members of the FGGY carbohydrate kinase family [28]. The peptides containing Asp28 and Asp299 were clearly identified by peptide sequencing of XKS1-Strep. Interestingly, however, despite the high sequence coverage, the N-terminal peptide of XKS1-Strep was not identified. The MS data therefore suggested that XKS1 may have been produced as an N-terminally degraded protein, and we considered the possibility that absence of the authentic N-terminus could have an effect upon the stability of the isolated enzyme.
Table 3.
Distinct peptides identified by LC-MSMS analysis of S. cerevisiae Xylulokinase XKS1 (NCBI Nr. 6321633). The two conserved catalytic aspartate residues D28 and D299 are indicated in bold. The sequence coverage was 92%.
| Peptide sequence | Experimental parent mass (Da) | Mass error (Da) | Start residue |
|---|---|---|---|
| (M)SLDSYYLGFD28LSTQQLK(C) | 1977.98 | 0.0015 | 18 |
| (K)CLAINQDLK(I) | 1074.561 | −0.0003 | 35 |
| (C)LAINQDLKIV(H) | 1126.683 | 0.0003 | 36 |
| CK)IVHSETVEFEK(D) | 1317.669 | −0.0002 | 44 |
| CK)DLPHYHTKKGVYIHG(D) | 1764.918 | 0.0009 | 55 |
| (K)KGVYIHGDTIECPVAMWLEALDLVLSK(Y) | 3057.579 | 0.002 | 63 |
| (K)VMAVSGSCQQHGSVYWSSQAESLLEQLNK(K) | 3223.515 | 0 | 100 |
| (E)KDLLHYVSSVAFARQTAPNWQDHSTAKQCQE(F) | 3615.74 | −0.0108 | 132 |
| (A)KQCQEFEECIGGPEKM(A) | 1969.845 | 0.0009 | 158 |
| (K)MAQLTGSRAHFRFTGPQILK(T) | 2259.218 | 0.0003 | 173 |
| (K)IAQLEPEAYEK(T) | 1290.658 | −0.0007 | 193 |
| (E)KTKTISLVSNFLTSILVGHLVE(L) | 2399.391 | 0.0034 | 203 |
| (K)TISLVSNFLTSILVGHLVELEEADACGMNLYDIRERK(F) | 4206.157 | 0.004 | 206 |
| (K)FSDELLHLIDSSSKDK(T) | 1833.923 | 0.0002 | 243 |
| (K)LMRAPMK(N) | 846.469 | −0.0003 | 264 |
| (K)NLIAGTICK(Y) | 989.545 | 0.0001 | 271 |
| (G)TICKYFIEKYGFNT(N) | 1783.872 | 0.002 | 276 |
| (K)VSPMTGD299NLATICSLPLRK(N) | 2073.083 | −0.0001 | 293 |
| (K)NDVLVSLGTSTTVLLVTDK(Y) | 1975.096 | 0 | 312 |
| (L)LVTDKYHPSPN(Y) | 1270.643 | −0.0001 | 326 |
| (N)YHLFIHPT(L) | 1027.536 | −0.0005 | 337 |
| (R)IRDELNKEREN(N) | 1415.724 | −0.0002 | 365 |
| (K)TNDWTLFNQAVLDDSESSENELGVYFPLGEIVPSVK(A) | 4012.934 | 0.0014 | 380 |
| (E)IWSVKAINKRVTFNPKTGMIERE(V) | 2739.57 | 0.0015 | 410 |
| (E)VAKFKDKRHDAKNIVE(S) | 1898.061 | 0.0003 | 434 |
| (H)DAKNIVESQALSCRVRISPLLS(D) | 2456.329 | 0.0023 | 443 |
| (R)ISPLLSDSNASSQQRLNEDTIVKFDYDESPLR(D) | 3637.798 | −0.002 | 459 |
| (R)DYLKKRPERTFFVGGASKN(D) | 2199.131 | −0.0013 | 491 |
| (D)AIVKKFAQVIGATKGNFRLE(T) | 2190.276 | 0.0005 | 511 |
| (K)GNFRLETPNSCALGGCYK(A) | 2043.938 | −0.001 | 525 |
| (K)AMWSLLYDSNK(I) | 1327.635 | −0.0006 | 543 |
| (I)AWFDKFLNDNFPWHV(M) | 1945.959 | −0.0003 | 555 |
| (K)FLNDNFPWHVMESISDVDNENWDRYNSKIVPLSELEK(T) | 4480.119 | 0.0115 | 561 |
Automated Edman sequencing of a purified, yet barely active preparation of XKS1V-His (Fig. 1; MALDI-MS mass: 68.873 kDa; calculated mass: 68.858 kDa) revealed the presence of the expected N-terminal amino acid Val-1. However, additional amino acids were found in the first degradation cycle, including Cys-2 and Ser-3. The relative abundance of Val, Cys and Ser suggested that about one-third of XKS1V-His was intact while the remainder protein was N-terminally degraded with about half of it lacking Val-1 and the rest lacking Val-1 and Cys-2. However, these results are at variance with evidence from MS analysis of XKS1V-His (see above) and XKS1ΔL-His (MALDI-MS mass: 68.739 kDa; calculated mass: 68.739 kDa) indicating that isolated enzyme preparations were actually unprocessed. We cannot exclude the possibility that the N-terminus of XKS1V-His is chemically labile and undergoes degradation during sample preparation for Edman degradation. However, the N-terminal sequencing result confirming the presence of Val-1 in (at least) 30% of XKS1V-His as isolated provides good evidence indicating that proteolytic truncation of the N-terminus is not the prime reason for the observed instability of XKS1, native or “tagged”. If it were, one would expect stability in about one-third of the protein preparation, contrary to experimental observations for XKS1V-His. We therefore did not pursue N-terminal sequencing of other forms of the enzyme such as XKS1-Strep, for example.
Flanagan and Waites [21] reported that P. stipitis XK has a native molecular mass of 120–130 kDa. Considering a molecular mass of 70 kDa for the protein subunit, it was concluded that the Pichia XK is a functional homodimer. By way of comparison, E. coli XK is also a homodimer [27]. We examined purified XKS1-Strep (∼18 U/mg) by gel filtration analysis. Protein was eluted from the Superdex 200 column (see Supporting Information) in no discrete peak and all of the applied activity was lost during the analysis. The molecular mass of functional XKS1 remains elusive.
Restoration of activity in XKS1V-His
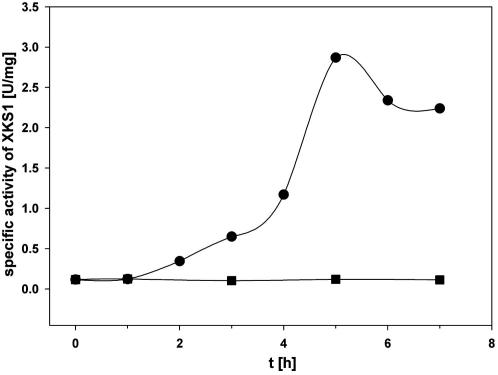
Observation that XKS1 (native or “tagged”) was generally much more stable in the E. coli cell extract (Table 2) as compared to the isolated enzyme preparation (XKS1-Strep; Fig. 2) inspired an experiment in which isolated XKS1V-His (0.1 U/mg) was mixed with cell extract (to a final protein concentration of 40 mg/mL) and kinase activity was measured in dependence of the incubation time. Fig. 3 shows that enzyme activity was enhanced to a level of about 2.3 U/mg when cell extract was present, reflecting a 20-fold functional complementation of the original XKS1 preparation. The recovered activity was fully stable for at least 48 h when the enzyme was stored on ice. This result is consistent with enhanced stability of the enzyme activity in crude cell extract as compared to the isolated protein (XKS1-Strep).
Fig. 3.
Restoration of XKS1 activity. A 100 μL of purified XKS1 (specific activity: 0.1 U/mg; protein concentration: 2 mg/mL) were incubated with 100 μL of E. coli BL21 DE3 crude extract (protein concentration: 40 mg/mL) for 400 min at 25 °C (●). The same experiment was performed using 100 μL of S. cerevisiae CEN.PK-113-5D crude extract (protein concentration: 40 mg/mL) for 400 min at 25 °C (■). The specific activity of XKS1 was determined at defined points using the standard XKS1 activity assay (see Materials and Methods).
Restoration of XKS1 activity was dependent on the concentration of E. coli protein charged to the assay. The concentration of XKS1 in the assay was constant. Use of diluted cell extract (10 and 20 mg/mL) failed to “activate” XKS1V-His to a measurable extent while a concentrated preparation (160 mg/mL) was about as effective as the originally used E. coli cell extract (40 mg/mL; Fig. 3). It was surprising that addition of cell extract obtained from S. cerevisiae CEN.PK 113-5D in a protein concentration of 40 mg/mL did not complement the activity of XKS1V-His (Fig. 3). BSA in the same concentration range was likewise inactive. Mixtures of cell extracts or cell extract and BSA were not applied.
The E. coli cell extract was fractionated using ultrafiltration with a molecular mass cut-off of 10 kDa. The low-molecular-mass permeate was inactive towards functional complementation of XKS1V-His. The macromolecular retentate (200 mg/mL) gave reactivation of XKS1V-His to an extent expected from results obtained with crude cell extract. This result supports the notion that heterotropic protein–protein interactions might be involved in determining XKS1 stability and/or activity. A more general role of macromolecular crowding on the stability of XKS1 was ruled out because in that case one expects that BSA or the S. cerevisiae cell extract should likewise be stabilizing. It is clear that further research will be needed to identify the macromolecular factor(s) required for stability of XKS1. This, however, was considered to be beyond the scope of the present paper.
Little is known about relationships between structure and stability in XKS1 and other members of the FGGY family of carbohydrate kinases. XKS1 is only 16% identical in primary structure to XK from E. coli, implying that properties of the eukaryotic enzyme cannot be directly inferred from the bacterial counterpart enzyme that has been characterized crystallographically [27]. However, low stability is not uncommon among XK enzymes [21] and more generally, members of the FGGY carbohydrate kinase family as demonstrated for the glycerol kinase from E. coli [29] and Enterococcus casseliflavus [30] which required the addition of extrinsic stabilizers (e.g. glycerol) during purification. However, no generalization is possible as the example of glucose kinases shows where the enzyme from Streptomyces peuceticus var. caesius required added glucose to prevent dissociative denaturation of the native tetramer while the also tetrameric enzyme from the closely related Streptomyces ceolicolor was stable in the absence of glucose [31,32]. Inactivation during IMAC is not a general feature of FGGY-type carbohydrate kinases because l-fucokinase from Arabidopsis thaliana was successfully isolated [33]. Heterotropic protein–protein interactions that may be needed for stability of XKS1 are not without precedence among members of the FGGY carbohydrate kinase family. E. coli glycerol kinase is inhibited allosterically by binding of the phosphotransferase system phosphocarrier protein IIAGlc [33].
Kinetic characterization of XKS1-Strep
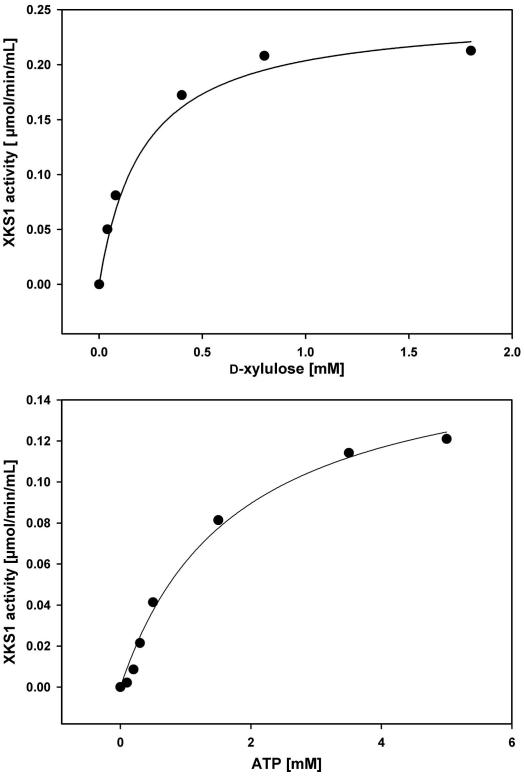
Isolated XKS1-Strep was characterized using steady-state kinetic analysis. Fig. 4 shows the results. Fits of a hyperbola to the data gave Km values for d-xylulose and ATP of 210 ± 40 μM and 1.70 ± 0.10 mM (see Fig. 4). These data are in good agreement with kinetic parameters for native XKS1 partially purified from an YGR194c over-expressing strain of S. cerevisiae [17]. Km values for XKS1 are similar to the Km values reported for P. stipitis XK [21].
Fig. 4.
Michaelis–Menten plots for ATP-dependent phosphorylation of d-xylulose by purified XKS1-Strep. Initial rates were obtained using a concentration of 20 μg/mL purified XKS1-Strep. Solid lines show fits of a hyperbola to the data. Extrapolated to saturating substrate concentration, both plots give a maximum specific rate of about 18 μmol/min/mg.
The specific activity of XKS1-Strep under conditions in which both substrates are saturating was 18 U/mg. Using a molecular mass of 67 kDa, a turnover number (kcat) of 14 s−1 can be calculated. While the kinetic evidence for XKS1-Strep supports the notion of a functional enzyme, the proprotion of active kinase in the isolated protein cannot be determined with the methods used.
Conclusions
Summarizing, of the numerous approaches of recombinant production and isolation of XKS1 that needed to be pursued in this work, construction of a fusion protein in which Strep-Tag II was appended to the C-terminal Ile600 of XKS1 eventually enabled isolation of a functional enzyme preparation from an E. coli production system. While this result could open up the possibility to explore in more detail structure–function relationships for purified XKS1-Strep, enzyme instability that was not encountered or recognized in previous reports on XKS1 [17,22] remains an issue. In support of our claims and the requirement for this study, we have provided clear evidence that XK activity and loss thereof was associated with the unprocessed full-length XKS1 protein, which was isolated and characterized. Both, the observation that XKS1-Strep was less prone to inactivation in the bacterial cell extract than in isolated form and that purified, inactive enzyme (XKS1V-His) was partly reactivated by incubation with crude E. coli extract could indicate a possible role for naturally activating/stabilizing macromolecular component(s), arguably protein(s), in the function of XKS1. Their identification should be a relevant goal for future research on this key enzyme of microbial pentose utilization. Others may also wish to consider that measurement of XK activity from S. cerevisiae cell extract is potentially afflicted with large error due to fast inactivation of the enzyme, by dilution for example.
Declaration of interest
The authors report no conflicts of interest. The authors alone are responsible for the content and writing of the paper.
Acknowledgments
Financial support from the Austrian Science Funds (FWF project P18275 to BN) is gratefully acknowledged. The authors would like to thank Dr. Sigrid Egger, Dr. Mario Klimacek and Dr. Regina Kratzer from the Institute of Biotechnology and Biochemical Engineering for useful discussions.
Footnotes
Abbreviations used: XKS1, xylulose kinase; XKS1 harboring C-terminal Strep-tag II; IMAC, immobilized metal affinity chromatography; AEX, anion exchange chromatography; HIC, hydrophobic interaction chromatography (HIC); GST-tag, glutathione-S-transferase tag; XKS1-His, C-terminally His-tagged XKS1.
Supplementary data associated with this article can be found, in the online version, at doi:10.1016/j.pep.2011.05.018.
Appendix A. Supplementary material
Supplementary material.
References
- 1.Ingram L.O., Doran J.B. Conversion of cellulosic materials to ethanol. FEMS Microbiol. Rev. 1995;16:235–241. [Google Scholar]
- 2.Olsson L., Hahn-Hägerdal B. Fermentation of lignocellulosic hydrolysates for ethanol production. Enzyme Microb. Technol. 1995;18:312–331. [Google Scholar]
- 3.Hahn-Hägerdal B., Wahlbom C.F., Gardonyi M., van Zyl W.H., Cordero Otero R.R., Jonsson L.J. Metabolic engineering of Saccharomyces cerevisiae for xylose utilization. Adv. Biochem. Eng. Biotechnol. 2001;73:53–84. doi: 10.1007/3-540-45300-8_4. [DOI] [PubMed] [Google Scholar]
- 4.Ho N.W., Chen Z., Brainard A.P. Genetically engineered Saccharomyces yeast capable of effective cofermentation of glucose and xylose. Appl. Environ. Microbiol. 1998;64(5):1852–1859. doi: 10.1128/aem.64.5.1852-1859.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Jeffries T.W., Jin Y.S. Metabolic engineering for improved fermentation of pentoses by yeasts. Appl. Microbiol. Biotechnol. 2004;63(5):495–509. doi: 10.1007/s00253-003-1450-0. [DOI] [PubMed] [Google Scholar]
- 6.Johansson B., Christensson C., Hobley T., Hahn-Hägerdal B. Xylulokinase overexpression in two strains of Saccharomyces cerevisiae also expressing xylose reductase and xylitol dehydrogenase and its effect on fermentation of xylose and lignocellulosic hydrolysate. Appl. Environ. Microbiol. 2001;67(9):4249–4255. doi: 10.1128/AEM.67.9.4249-4255.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Matsushika A., Sawayama S. Efficient bioethanol production from xylose by recombinant Saccharomyces cerevisiae requires high activity of xylose reductase and moderate xylulokinase activity. J. Biosci. Bioeng. 2008;106(3):306–309. doi: 10.1263/jbb.106.306. [DOI] [PubMed] [Google Scholar]
- 8.Van Vleet J.H., Jeffries T.M. Yeast metabolic engineering for hemicellulosic ethanol production. Curr. Opin. Biotechnol. 2009;20(3):300–306. doi: 10.1016/j.copbio.2009.06.001. [DOI] [PubMed] [Google Scholar]
- 9.Verho R., Putkonen M., Londesborough J., Penttilä M., Richard P. A novel NADH-linked l-xylulose reductase in the l-arabinose catabolic pathway of yeast. J. Biol. Chem. 2004;279(15):14746–14751. doi: 10.1074/jbc.M312533200. [DOI] [PubMed] [Google Scholar]
- 10.Richard P., Verho R., Putkonen M., Londesborough J., Penttilä M. Production of ethanol from l-arabinose by Saccharomyces cerevisiae containing a fungal l-arabinose pathway. FEMS Yeast Res. 2003;3(2):185–189. doi: 10.1016/S1567-1356(02)00184-8. [DOI] [PubMed] [Google Scholar]
- 11.Karhumaa K., Wiedemann B., Hahn-Hägerdal B., Boles E., Gorwa-Grauslund M.F. Co-utilization of l-arabinose and d-xylose by laboratory and industrial Saccharomyces cerevisiae strains. Microb. Cell Fact. 2006;5:18. doi: 10.1186/1475-2859-5-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ho N.W.Y., Chang S.F. Cloning of yeast xylulokinase gene by complementation of Escherichia coli and yeast mutations. Enzyme Microb. Technol. 1989;11:417–421. [Google Scholar]
- 13.Rodriguez-Pena J.M., Cid V.J., Arroyo J., Nombela C. The YGR194c (XKS1) gene encodes the xylulokinase from the budding yeast Saccharomyces cerevisiae. FEMS Microbiol. Lett. 1998;162(1):155–160. doi: 10.1111/j.1574-6968.1998.tb12993.x. [DOI] [PubMed] [Google Scholar]
- 14.Deng X.X., Ho N.W. Xylulokinase activity in various yeasts including Saccharomyces cerevisiae containing the cloned xylulokinase gene. Scientific note. Appl. Biochem. Biotechnol. 1990;24–25:193–199. doi: 10.1007/BF02920245. [DOI] [PubMed] [Google Scholar]
- 15.Toivari M.H., Aristidou A., Ruohonen L., Penttilä M. Conversion of xylose to ethanol by recombinant Saccharomyces cerevisiae: importance of xylulokinase (XKS1) and oxygen availability. Metab. Eng. 2001;3(3):236–249. doi: 10.1006/mben.2000.0191. [DOI] [PubMed] [Google Scholar]
- 16.Jin Y.S., Ni H., Laplaza J.M., Jeffries T.W. Optimal growth and ethanol production from xylose by recombinant Saccharomyces cerevisiae require moderate d-xylulokinase activity. Appl. Environ. Microbiol. 2003;69(1):495–503. doi: 10.1128/AEM.69.1.495-503.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Richard P., Toivari M.H., Penttilä M. The role of xylulokinase in Saccharomyces cerevisiae xylulose catabolism. FEMS Microbiol. Lett. 2000;190(1):39–43. doi: 10.1111/j.1574-6968.2000.tb09259.x. [DOI] [PubMed] [Google Scholar]
- 18.Van Vleet J.H., Jeffries T.M., Olsson L. Deletion of the para-nitrophenyl phosphatase (pNPPase), PHO13, in recombinant Saccharomyces cerevisiae improves growth and ethanol production on d-xylose. Metab. Eng. 2008;10(6):360–369. doi: 10.1016/j.ymben.2007.12.002. [DOI] [PubMed] [Google Scholar]
- 19.Ni H., Laplaza J.M., Jefrries T.W. Transposon mutagenesis to improve the growth of recombinant Saccharomyces cerevisiae on d-xylose. Appl. Environ. Mircobiol. 2007;73(7):2061–2066. doi: 10.1128/AEM.02564-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Jin Y.S., Ni H., Laplaza J.M., Jeffries T.W. Saccharomyces cerevisiae engineered for xylose metabolism exhibits a respiratory response. Appl. Environ. Microbiol. 2004;70(11):6816–6825. doi: 10.1128/AEM.70.11.6816-6825.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Flanagan T., Waites M.J. Purification and characterization of d-xylulose kinase from the pentose-fermenting yeast Pichia stipitis NCYC 1541. Enzyme Microb. Technol. 1992;14(10):975–979. [Google Scholar]
- 22.Lee J.Y., Cheong D.E., Kim G.J. A novel assay system for the measurement of transketolase activity using xylulokinase from Saccharomyces cerevisiae. Biotechnol. Lett. 2008;30(5):899–904. doi: 10.1007/s10529-007-9616-y. [DOI] [PubMed] [Google Scholar]
- 23.Varshavsky A. The N-end rule pathway of protein degradation. Genes Cells. 1997;2(1):13–28. doi: 10.1046/j.1365-2443.1997.1020301.x. [DOI] [PubMed] [Google Scholar]
- 24.Petschacher B., Nidetzky B. Altering the coenzyme preference of xylose reductase to favor utilization of NADH enhances ethanol yield from xylose in a metabolically engineered strain of Saccharomyces cerevisiae. Microb. Cell Fact. 2008;7:9. doi: 10.1186/1475-2859-7-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Akinterinwa O., Cirino P.C. Heterologous expression of d-xylulokinase from Pichia stipitis enables high levels of xylitol production by engineered Escherichia coli growing on xylose. Metab. Eng. 2009;11(1):48–55. doi: 10.1016/j.ymben.2008.07.006. [DOI] [PubMed] [Google Scholar]
- 26.Krahulec S., Klimacek M., Nidetzky B. Engineering of a matched pair of xylose reductase and xylitol dehydrogenase for xylose fermentation by Saccharomyces cerevisiae. Biotechnol. J. 2009;4(5):684–694. doi: 10.1002/biot.200800334. [DOI] [PubMed] [Google Scholar]
- 27.Di Luccio E., Petschacher B., Voegtli J., Chou H.T., Stahlberg H., Nidetzky B., Wilson D.K. Structural and kinetic studies of induced fit in xylulose kinase from Escherichia coli. J. Mol. Biol. 2007;365(3):783–798. doi: 10.1016/j.jmb.2006.10.068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kotake T., Hojo S., Tajima N., Matsuoka K., Koyama T., Tsumuraya Y. A bifunctional enzyme with l-fucokinase and GDP-l-fucose pyrophosphorylase activities salvages free l-fucose in Arabidopsis. J. Biol. Chem. 2008;283(13):8125–8135. doi: 10.1074/jbc.M710078200. [DOI] [PubMed] [Google Scholar]
- 29.Hayashi S.I., Lin E.C. Purification and properties of glycerol kinase from Escherichia coli. J. Biol. Chem. 1967;242(5):1030–1035. [PubMed] [Google Scholar]
- 30.Yeh J.I., Kettering R., Saxl R., Bourand A., Darbon E., Joly N., Briozzo P., Deutscher J. Structural characterizations of glycerol kinase: unraveling phosphorylation-induced long-range activation. Biochemistry. 2009;48(2):346–356. doi: 10.1021/bi8009407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Imriskova I., Arreguin-Espinosa R., Guzman S., Rodriguez-Sanoja R., Langley E., Sanchez S. Biochemical characterization of the glucose kinase from Streptomyces coelicolor compared to Streptomyces peucetius var. caesius. Res. Microbiol. 2005;156(3):361–366. doi: 10.1016/j.resmic.2004.11.001. [DOI] [PubMed] [Google Scholar]
- 32.Mahr K., van Wezel G.P., Svensson C., Krengel U., Bibb M.J., Titgemeyer F. Glucose kinase of Streptomyces coelicolor A3(2): large-scale purification and biochemical analysis. Antonie Van Leeuwenhoek. 2000;78(3–4):253–261. doi: 10.1023/a:1010234916745. [DOI] [PubMed] [Google Scholar]
- 33.Yu P., Lasagna M., Pawlyk A.C., Reinhart G.D., Pettigrew D.W. IIAGlc Inhibition of glycerol kinase: a communications network tunes protein motions at the allosteric site. Biochemistry. 2007;46(43):12355–12365. doi: 10.1021/bi7010948. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary material.