Abstract
AIM: To investigate the association between GSTM1 and GSTT1 polymorphisms and the risk of hepatocellular carcinoma (HCC) in Chinese population.
METHODS: Literature databases including PubMed, ISI web of science and other databases were searched. Pooled odds ratio (OR) and 95% CI were calculated using random- or fixed- effects model. Subgroup analysis and sensitivity analysis were also performed.
RESULTS: Nineteen studies of GSTM1 (2660 cases and 4017 controls) and 16 studies of GSTT1 (2410 cases and 3669 controls) were included. The GSTM1/GSTT1 null genotypes were associated with increased risk of HCC in Chinese population (for GSTM1, OR = 1.487, 95% CI: 1.159 to 1.908, P = 0.002; for GSTT1, OR = 1.510, 95% CI: 1.236 to 1.845, P = 0.000). No publication bias was detected. In subgroup analysis, glutathione S-transferases polymorphisms were significantly associated with HCC risk among the subjects living in high-incidence areas, but not among the subjects living in low-incidence areas.
CONCLUSION: The present meta-analysis suggests that GSTM1/GSTT1 null genotypes are associated with increased risk of HCC in Chinese population.
Keywords: GSTM1, GSTT1, Polymorphism, Hepatocellular carcinoma, Liver cancer
INTRODUCTION
Liver cancer is one of the most common types of cancer and one of the most common causes of cancer-related death[1]. The death rates have increased for both men and women with liver cancer over the past two decades[2]. The incidence and mortality rates of liver cancer vary considerably among racial and ethnic groups[3]. Asians, particularly Chinese, have a high risk of developing liver cancer[4]. About 80%-90% of all cases of primary liver cancer are hepatocellular carcinoma (HCC).
The pathogenesis of HCC may have a genetic and environmental basis[5,6]. Epidemiological studies have shown that HCC is associated with many environmental factors, including alcoholism, chronic infection with hepatitis B virus (HBV), hepatitis C virus (HCV), and dietary exposure to aflatoxin B1 (AFB1)[7]. These hepatocarcinogens result in increased generation of reactive oxygen species and free radicals that cause liver damage and repair. Thus the accumulated multistage genetic mutations may lead to liver carcinogenesis[8-11].
During liver carcinogenesis, cellular defense mechanisms can alleviate the effects of oxidative stress and exogenous toxins. One of the essential antioxidant is the reducing compound glutathione. Reduced glutathione can be conjugated to various xenobiotics and endobiotics by glutathione S-transferases (GST), a superfamily of cytosolic soluble detoxification enzymes. In humans, cytosolic soluble GSTs are encoded by seven distinct genes: Alpha, Mu, Omega, Pi, Sigma, Theta and Zeta[12-14]. GSTs play an important role in cellular protection against oxidative stress and exogenous toxins. Homozygous deletion of GST genes (null genotype) results in decreased enzyme activity, which will impede detoxification and may ultimately increase the risk of many diseases, including HCC[13-16]. GSTM1 and GSTT1 have been the most extensively studied GST genes. While many studies have investigated the relationship between GSTM1 and GSTT1 polymorphisms and HCC risk, so far the results have been inconsistent.
Recently, a meta-analysis result did not suggest a statistically significant increased risk of HCC with the GST null genotypes[17]. However, a large number of studies were not reported in that meta-analysis (which included only 9 studies of GSTM1 and 8 studies of GSTT1 in Chinese population)[17]. Since the ethnic background and the environmental exposures may vary greatly across populations in different geographic regions, the conclusion could not be drawn[17]. During the preparation of this paper, another meta-analysis was published showing that the null genotypes of GSTM1 and GSTT1 are both associated with an increased HCC risk[18]. However, these investigators omitted some important data and introduced some incorrect information[18]. For example, Indians are mainly of Indo-European and Dravidian ancestries, which should be distinguished from East Asian population[18]. Many studies, including our previous studies[16,19-22], have reported on the effects of ethnic differences on genetic predisposition to human diseases. In addition, it is important to note that the allele frequencies and the genotype distributions for GST genes differed significantly across ethnic groups[23]. For example, the frequency of GSTM1 null genotype is about 0.53 in Caucasians/Asians and about 0.28 in Africans; and the frequency of GSTT1 null genotype is about 0.20 in Caucasians and about 0.52 in Asians[23]. Therefore, heterogeneity was introduced in that meta-analysis, which may not accurately assess the effects of GSTM1 and GSTT1 null genotypes on the risk of HCC[18]. In this study, we reinvestigated the relationship between GSTM1/GSTT1 polymorphisms and the risk of HCC. We focused on the association between GST polymorphisms and the risk of HCC in Chinese population, because Chinese are at a much greater risk of developing HCC compared to other ethnic groups. A total of 19 studies of GSTM1 (2660 cases and 4017 controls) and 16 studies of GSTT1 (2410 cases and 3669 controls) were included. This meta-analysis has a much greater number of subjects and thus a much greater statistical power; therefore, it may define the effects of GST gene polymorphisms on the risk of HCC more precisely.
MATERIALS AND METHODS
Literature and search strategy
We searched the literature databases including PubMed (1950 to 2010), ISI web of science (1975 to 2010), Embase (1966 to 2010), Chinese Biomedical Database (1978 to 2010), China National Knowledge Infrastructure (1979 to 2010, in Chinese) and Wanfang Data (1982 to 2010, in Chinese).
The search strategy to identify all possible studies involved use of combinations of the following key words: (“glutathione S-transferase” or “GST” or “GSTM1” or “GSTT1”) and (“hepatocellular carcinoma” or “liver cancer” or “HCC”) and (“China” or “Chinese”). The reference lists of reviews and retrieved articles were also searched. Supplementary data were searched for missing data points. If more than one article were published using the same case series, only the study with largest sample size was selected. The literature search was updated on Oct. 20th, 2010.
Inclusion criteria and data extraction
The studies included in the meta-analysis must meet all the following inclusion criteria: (1) evaluating the association between GSTM1 or GSTT1 null genotypes and HCC risk; (2) case-control design; (3) in Chinese population; and (4) sufficient data for calculation of odds ratio (OR) with CI. The following information was extracted from each study: (1) name of the first author; (2) year of publication; (3) language of publication; (4) residence area of the subjects; (5) source of control subjects; (6) numbers of cases and controls; (7) numbers of null genotypes for GSTM1 and GSTT1 in cases and controls; and (8) OR and 95% CI. The authors independently assessed the articles for compliance with the inclusion/exclusion criteria, resolved disagreements and reached a consistent decision.
Statistical analysis
The association between GSTM1/GSTT1 polymorphisms and the risk of HCC was estimated by calculating pooled OR and 95% CI. The significance of the pooled OR was determined by Z test with P < 0.05 considered statistically significant. Q test was performed to evaluate whether the variation was due to heterogeneity or by chance. A random- (DerSimonian-Laird method[24]) or fixed- (Mantel-Haenszel method[25]) effects model was used to calculate pooled effect estimates in the presence (P ≤ 0.10) or absence (P > 0.10) of heterogeneity, respectively. Begg’s funnel plot, a scatter plot of effect against a measure of study size, was generated as a visual aid to detecting bias or systematic heterogeneity[26]. An asymmetric funnel plot indicates a relationship between effect and study size, which suggests the possibility of either publication bias or a systematic difference between smaller and larger studies (“small study effects”). Publication bias was assessed by Begg’s test and Egger’s test[27] with P < 0.05 considered statistically significant. Subgroup analyses were performed to examine the effect of heterogeneity on meta-analysis results. The following subgroup comparisons were analyzed: residence area of the subjects (high-incidence area vs low-incidence area), number of cases (< 100 vs ≥ 100), and source of controls (population-based vs hospital-based). To evaluate the stability of results, sensitivity analysis was performed by removing one study at a time and calculating the overall homogeneity and effect size. Data analysis was performed using STATA version 10 (StataCorp LP, College Station, Texas, USA).
RESULTS
Characteristics of the studies
The literature search identified a total of 137 potential relevant papers. The full text articles were retrieved and carefully reviewed to assess the eligibility according to the inclusion criteria. Forty-three papers met the inclusion criteria[28-70]. However, 23 papers were excluded because they were earlier or smaller reports from the same groups[29,30,32-37,43-46,48-50,53,55-58,61,62,64]. Nineteen studies of GSTM1 (2660 cases and 4017 controls) and 16 studies of GSTT1 (2410 cases and 3669 controls) were included in the meta-analysis, respectively. Most of the cases and controls included in this meta-analysis were HBV carriers. The characteristics of the included studies are listed in Tables 1 and 2.
Table 1.
Characteristics of studies included in the meta-analysis of GSTM1
| Ref. | Language of publication | Residence area of subjects |
Case |
Control |
Source of control | OR (95% CI) | Earlier or smaller reports | ||||
| Null | Positive | Total | Null | Positive | Total | ||||||
| Dong et al[28], 1997 | Chinese | Jiangsu, Guangxi and Hebei | 62 | 48 | 110 | 50 | 62 | 112 | Population | 1.602 (0.943-2.721) | [29,30] |
| Yu et al[31], 1999 | English | Taiwan | 42 | 42 | 84 | 216 | 159 | 375 | Hospital | 0.736 (0.458-1.183) | [32-37] |
| Wu et al[38], 2000 | Chinese | Hu’nan | 38 | 16 | 54 | 62 | 74 | 136 | Population | 2.835 (1.444-5.565) | NA |
| Sun et al[39], 2001 | English | Taiwan | 26 | 43 | 69 | 77 | 51 | 128 | Population | 0.400 (0.219-0.731) | NA |
| Zhu et al[40], 2001 | Chinese | Guangdong | 34 | 18 | 52 | 41 | 59 | 100 | Hospital | 2.718 (1.354-5.455) | NA |
| Chen et al[41], 2002 | English | Taiwan | 60 | 41 | 101 | 19 | 16 | 35 | Hospital | 1.232 (0.568-2.674) | NA |
| Liu et al[42], 2002 | Chinese | Jiangsu | 56 | 28 | 84 | 69 | 75 | 144 | Population | 2.174 (1.243-3.803) | [43-46] |
| McGlynn et al[47], 2003 | English | Jiangsu | NA | NA | 231 | NA | NA | 256 | Population | 0.830 (0.570-1.210) | [48-50] |
| Li et al[51], 2004 | Chinese | Jiangsu | 122 | 85 | 207 | 118 | 89 | 207 | Population | 1.083 (0.733-1.600) | NA |
| Chen et al[52], 2005 | English | Taiwan | 322 | 255 | 577 | 231 | 158 | 389 | Population | 0.864 (0.666-1.121) | [53] |
| Deng et al[54], 2005 | English | Guangxi | 117 | 64 | 181 | 172 | 188 | 360 | Hospital | 1.998 (1.383-2.888) | [55-58] |
| Guo et al[59], 2005 | Chinese | Henan | 67 | 28 | 95 | 52 | 51 | 103 | Population | 2.347 (1.306-4.218) | NA |
| He et al[60], 2005 | Chinese | Guangxi | 68 | 37 | 105 | 77 | 74 | 151 | Hospital | 1.766 (1.059-2.947) | [61,62] |
| Long et al[63], 2005 | Chinese | Guangxi | 92 | 48 | 140 | 254 | 282 | 536 | Hospital | 2.128 (1.444-3.137) | [64] |
| Ma et al[65], 2005 | Chinese | Guangxi | 37 | 25 | 62 | 29 | 44 | 73 | Population | 2.246 (1.125-4.481) | NA |
| Zhang et al[66], 2005 | Chinese | Hubei | 37 | 23 | 60 | 28 | 45 | 73 | Hospital | 2.585 (1.281-5.219) | NA |
| Zhu et al[67], 2005 | Chinese | Zhejiang | 56 | 35 | 91 | 61 | 69 | 130 | Hospital | 1.810 (1.049-3.121) | NA |
| Long et al[68], 2006 | English | Guangxi | 179 | 78 | 257 | 312 | 337 | 649 | Hospital | 2.479 (1.823-3.370) | NA |
| Yang et al[69], 2009 | Chinese | Guangxi | 59 | 41 | 100 | 41 | 19 | 60 | Hospital | 0.667 (0.340-1.309) | NA |
NA: Not available; OR: Odds ratio.
Table 2.
Characteristics of studies included in the meta-analysis of GSTT1
| Ref. | Language of publication | Residence area of subjects |
Case |
Control |
Source of control | OR (95% CI) | Earlier or smaller reports | ||||
| Null | Positive | Total | Null | Positive | Total | ||||||
| Dong et al[28], 1997 | Chinese | Jiangsu, Guangxi and Hebei | 63 | 47 | 110 | 42 | 70 | 112 | Population | 2.234 (1.305-3.825) | [29,30] |
| Yu et al[31], 1999 | English | Taiwan | 41 | 42 | 83 | 181 | 194 | 375 | Hospital | 1.046 (0.650-1.683) | [32-37] |
| Sun et al[39], 2001 | English | Taiwan | 30 | 37 | 67 | 77 | 51 | 128 | Population | 0.537 (0.295-0.976) | NA |
| Liu et al[42], 2002 | Chinese | Jiangsu | 34 | 50 | 84 | 36 | 108 | 144 | Population | 2.040 (1.146-3.630) | [43-46] |
| McGlynn et al[47], 2003 | English | Jiangsu | NA | NA | 231 | NA | NA | 256 | Population | 0.880 (0.590-1.310) | [48-50] |
| Liu et al[70], 2003 | Chinese | Guangxi | 28 | 23 | 51 | 18 | 35 | 53 | Population | 2.367 (1.072-5.227) | NA |
| Li et al[51], 2004 | Chinese | Jiangsu | 108 | 99 | 207 | 97 | 110 | 207 | Population | 1.237 (0.841-1.820) | NA |
| Chen et al[52], 2005 | English | Taiwan | 298 | 279 | 577 | 199 | 190 | 389 | Population | 1.020 (0.788-1.319) | [53] |
| Deng et al[54], 2005 | English | Guangxi | 108 | 73 | 181 | 154 | 206 | 360 | Hospital | 1.979 (1.377-2.845) | [55-58] |
| Guo et al[59], 2005 | Chinese | Henan | 58 | 37 | 95 | 45 | 58 | 103 | Population | 2.020 (1.146-3.562) | NA |
| He et al[60], 2005 | Chinese | Guangxi | 43 | 62 | 105 | 50 | 101 | 151 | Hospital | 1.401 (0.836-2.347) | [61,62] |
| Long et al[63], 2005 | Chinese | Guangxi | 82 | 58 | 140 | 234 | 302 | 536 | Hospital | 1.825 (1.251-2.660) | [64] |
| Ma et al[65], 2005 | Chinese | Guangxi | 35 | 27 | 62 | 21 | 52 | 73 | Population | 3.210 (1.573-6.551) | NA |
| Zhang et al[66], 2005 | Chinese | Hubei | 38 | 22 | 60 | 34 | 39 | 73 | Hospital | 1.981 (0.986-3.982) | NA |
| Long et al[68], 2006 | English | Guangxi | 146 | 111 | 257 | 297 | 352 | 649 | Hospital | 1.559 (1.165-2.086) | NA |
| Yang et al[69], 2009 | Chinese | Guangxi | 33 | 67 | 100 | 11 | 49 | 60 | Hospital | 2.194 (1.010-4.765) | NA |
NA: Not available; OR: Odds ratio.
Meta-analysis results of the association between GSTM1 polymorphisms and HCC
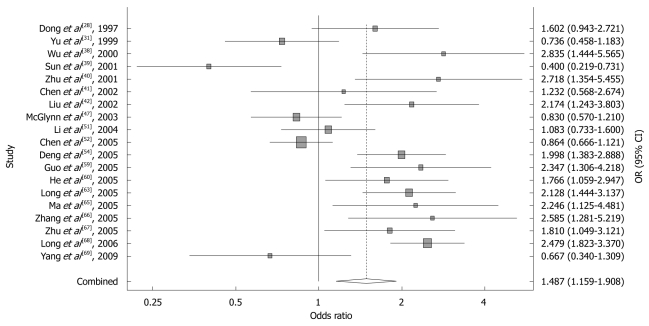
Nineteen studies of GSTM1, including 2660 cases and 4017 controls, were included in the meta-analysis. The relative frequency of GSTM1 null genotype among control groups ranged from 0.384 to 0.683 in Chinese population. Using a random-effects model, the overall meta-analysis result showed that there was a statistically significant association between GSTM1 null genotype and HCC risk in Chinese population (OR = 1.487, 95% CI: 1.159 to 1.908, P = 0.002). The forest plot is shown in Figure 1.
Figure 1.
Forest plot of the meta-analysis of the association between GSTM1 polymorphism and hepatocellular carcinoma risk.
Meta-analysis results of the association between GSTT1 polymorphisms and HCC
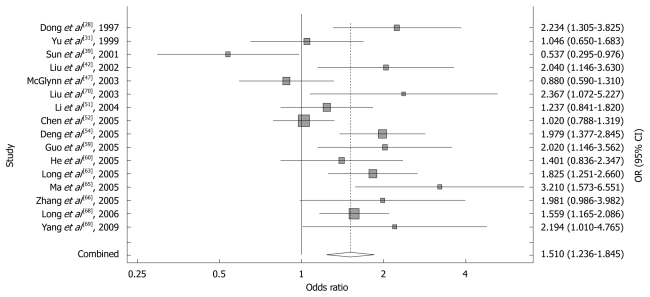
Sixteen studies of GSTT1, including 2410 cases and 3669 controls, were included in the meta-analysis. The relative frequency of GSTT1 null genotype among control groups ranged from 0.183 to 0.602 in Chinese population. Using a random-effects model, the overall meta-analysis result showed that there was a statistically significant association between GSTT1 null genotype and HCC risk in Chinese population (OR = 1.510, 95% CI: 1.236 to 1.845, P = 0.000). The forest plot is shown in Figure 2.
Figure 2.
Forest plot of the meta-analysis of the association between GSTT1 polymorphism and hepatocellular carcinoma risk.
Subgroup analysis
To examine the effect of heterogeneity between studies on meta-analysis results, we conducted subgroup analyses stratified by the following: residence area of the subjects (high-incidence area vs low-incidence area), number of cases (< 100 vs ≥ 100), and source of controls (population-based vs hospital-based). GST polymorphisms were significantly associated with HCC risk among the subjects living in high-incidence areas (Jiangsu, Zhejiang, Guangxi and Guangdong provinces), but not among the studies living in low-incidence areas. The result of subgroup analysis is shown in Tables 3 and 4.
Table 3.
Subgroup analysis of the association between GSTM1 polymorphism and hepatocellular carcinoma risk
| Group | No. of studies (cases/controls) | Statistical method | OR (95% CI) | P |
| All studies | 19 (2660/4017) | Random | 1.487 (1.159-1.908) | 0.002 |
| Residence area of the subjects | ||||
| High-incidence area | 11 (1510/2666) | Random | 1.659 (1.264-2.177) | 0.000 |
| Low-incidence area | 7 (1040/1239) | Random | 1.235 (0.753-2.026) | 0.402 |
| Mixed areas | 1 (110/112) | - | 1.602 (0.943-2.721) | 0.081 |
| No. of cases | ||||
| < 100 | 9 (651/1262) | Random | 1.676 (1.061-2.649) | 0.027 |
| ≥ 100 | 10 (2009/2755) | Random | 1.365 (1.005-1.853) | 0.046 |
| Source of controls | ||||
| Population-based | 9 (1489/1548) | Random | 1.316 (0.915-1.892) | 0.139 |
| Hospital-based | 10 (1171/2469) | Random | 1.675 (1.251-2.243) | 0.001 |
OR: Odds ratio.
Table 4.
Subgroup analysis of the association between GSTT1 polymorphism and hepatocellular carcinoma risk
| Group | No. of studies (cases/controls) | Statistical method | OR (95% CI) | P |
| All studies | 16 (2410/3669) | Random | 1.510 (1.236-1.845) | 0.000 |
| Residence area of the subjects | ||||
| High-incidence area | 10 (1418/2489) | Random | 1.641 (1.328-2.027) | 0.000 |
| Low-incidence area | 5 (882/1068) | Random | 1.152 (0.777-1.707) | 0.483 |
| Mixed areas | 1 (110/112) | - | 2.234 (1.305-3.825) | 0.003 |
| Number of cases | ||||
| < 100 | 7 (502/949) | Random | 1.617 (1.035-2.528) | 0.035 |
| ≥ 100 | 9 (1908/2720) | Random | 1.457 (1.173-1.810) | 0.001 |
| Source of controls | ||||
| Population-based | 9 (1484/1465) | Random | 1.441 (1.039-1.997) | 0.028 |
| Hospital-based | 7 (926/2204) | Fixed | 1.635 (1.391-1.921) | 0.000 |
OR: Odds ratio.
Sensitivity analysis
Sensitivity analysis was performed by excluding each study at a time. The analysis confirmed the stability of the association between GSTM1 and GSTT1 polymorphisms and HCC risk (data not shown).
Potential publication bias
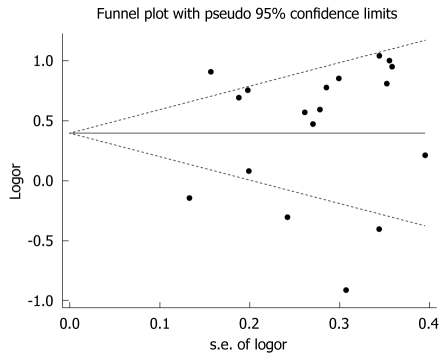
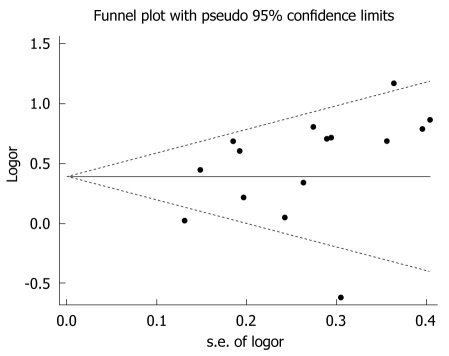
Begg’s funnel plots were generated to assess potential publication bias (Figure 3 for GSTM1 and Figure 4 for GSTT1). No publication bias was detected (Egger’s test, P = 0.542 for GSTM1 and P = 0.136 for GSTT1; Begg’s test, P = 0.677 for GSTM1 and P = 0.299 for GSTT1).
Figure 3.
Funnel plot of the meta-analysis of the association between GSTM1 polymorphism and hepatocellular carcinoma risk.
Figure 4.
Funnel plot of the meta-analysis of the association between GSTT1 polymorphism and hepatocellular carcinoma risk.
DISCUSSION
GST polymorphisms are implicated in the development of HCC. In the present study, we investigated the relationship between GSTM1 and GSTT1 polymorphisms and the risk of HCC in Chinese population. To minimize language bias, all available studies published in both English and Chinese languages were assessed, including a total of 19 studies of GSTM1 (2660 cases and 4017 controls) and 16 studies of GSTT1 (2410 cases and 3669 controls). The present meta-analysis has a much greater number of subjects and thus a much greater statistical power; therefore, it can define the effect of GST genes polymorphisms on HCC risk more precisely. The pooled analysis of two genes produced similar risk estimates (for GSTM1, OR = 1.487, 95% CI: 1.159 to 1.908, P = 0.002; for GSTT1, OR = 1.510, 95% CI: 1.236 to 1.845, P = 0.000), suggesting that both GSTM1 and GSTT1 null genotypes are associated with an increased risk of HCC in Chinese population.
The difference between our meta-analysis and the previous metaanalysis may be due to language bias introduced in the previous metaanalysis, which was based primarily on reports in English[17]. Therefore, all available studies published in both English and Chinese languages were assessed in the current meta-analysis. Another recent meta-analysis suggested that null genotypes of GSTM1 and GSTT1 were both associated with increased risk of HCC[18]. However, these investigators omitted some data and included some studies with incorrect information[52,53,59,65,69]. Therefore, the effects of GSTM1 and GSTT1 null genotypes on HCC was not assessed accurately[18]. Considering that Chinese people are at a much greater risk of developing HCC, we focused on the relationship between GST polymorphisms and HCC risk in Chinese population, thereby minimizing ethnic/racial differences. In the subgroup analysis, GST polymorphisms were significantly associated with HCC risk among the subjects living in high-incidence areas, but not among the subjects living in low-incidence areas. This result suggests that the effect of GST polymorphisms on HCC risk may be enhanced by environmental risk factors, such as dietary exposure to AFB1. Since sample size could influence the results, we also performed subgroup analysis stratified by sample size of cases. The results showed that the studies with either large (number of cases ≥ 100) or small sample size (number of cases < 100) had similar risk estimates, indicating that small study effects may not exist in this meta-analysis. In addition, we generated Begg’s funnel plots and found no publication bias among the studies included in this meta-analysis (Begg’s test and Egger’s test, P > 0.1).
The current meta-analysis has vital advantages compared to other studies; however, it does have some limitations. First, the present meta-analysis was based on unadjusted effect estimates and confidence intervals due to insufficient data available for most of the studies. Although the cases and controls were matched on age, sex and residence in all studies, these confounding factors might slightly modify the effect estimates. Second, the effect of gene-environment interactions was not studied in this meta-analysis. Alcoholism, HBV/HCV infections, and dietary exposure to AFB1 may be environmental risk factors that modify the effect estimates. Third, although most primary liver cancer cases are HCC, some of the included studies did not state whether the primary liver cancer patients were histologically confirmed to be HCC. Fourth, the heterogeneity between studies was not well addressed by subgroup analysis, suggesting there were other potential confounding factors in the included studies. Fifth, the results of subgroup analysis should be interpreted with caution because of limited statistical power. We anticipate these issues will be addressed in future studies.
In summary, our research suggests that GSTM1/GSTT1 null genotypes are associated with increased risk of HCC in Chinese population. Considering the increasing prevalence of HCC in China and other countries worldwide, our finding may have important clinical and public health implications. More epidemiological and mechanistic studies are needed to further elucidate the role of GST polymorphisms in HCC and other liver cancers.
COMMENTS
Background
Asians, particularly Chinese, have a high risk of developing liver cancer. About 80%-90% of all cases of primary liver cancer diagnosed are hepatocellular carcinoma (HCC). Previous studies suggest that glutathione S-transferase (GST) polymorphisms (GSTM1 and GSTT1) may be risk factors for HCC. However, recent findings have been inconsistent.
Research frontiers
Meta-analysis was performed to assess the association between GSTM1 and GSTT1 polymorphisms and HCC risk in Chinese population.
Innovations and breakthroughs
The meta-analysis provided new evidence for the association between GSTM1 and GSTT1 polymorphisms and HCC risk in Chinese population. The results of this meta-analysis show that the GSTM1 and GSTT1 null genotypes are both associated with increased risk of HCC in Chinese population, suggesting that GSTM1/GSTT1 null genotype carriers have 1.5 fold higher risk of developing HCC.
Applications
Since GSTM1/GSTT1 polymorphisms are implicated in the pathogenesis of HCC, population-based genetic screening in future may help to identify the individuals at high risk of developing liver cancers.
Terminology
Meta-analysis, which combines the results of several studies that address a set of related research hypotheses, is an important component of a systematic review procedure.
Peer review
This meta-analysis provided new insights into liver cancer research.
Footnotes
Supported by (partially) The Heilongjiang Provincial Health Department, No. 2009-201; and the Administration of Traditional Chinese Medicine of Heilongjiang Province, No. ZHY10-293
Peer reviewer: Wen Zhuang, Professor, Department of Gastrointestinal Surgery, West China Hospital/West China Medical Center, Sichuan University, Chengdu 610041, Sichuan Province, China
S- Editor Sun H L- Editor Wang XL E- Editor Zheng XM
References
- 1.Jemal A, Siegel R, Xu J, Ward E. Cancer statistics, 2010. CA Cancer J Clin. 2010;60:277–300. doi: 10.3322/caac.20073. [DOI] [PubMed] [Google Scholar]
- 2.Jemal A, Center MM, DeSantis C, Ward EM. Global patterns of cancer incidence and mortality rates and trends. Cancer Epidemiol Biomarkers Prev. 2010;19:1893–1907. doi: 10.1158/1055-9965.EPI-10-0437. [DOI] [PubMed] [Google Scholar]
- 3.Ward E, Jemal A, Cokkinides V, Singh GK, Cardinez C, Ghafoor A, Thun M. Cancer disparities by race/ethnicity and socioeconomic status. CA Cancer J Clin. 2004;54:78–93. doi: 10.3322/canjclin.54.2.78. [DOI] [PubMed] [Google Scholar]
- 4.McCracken M, Olsen M, Chen MS, Jemal A, Thun M, Cokkinides V, Deapen D, Ward E. Cancer incidence, mortality, and associated risk factors among Asian Americans of Chinese, Filipino, Vietnamese, Korean, and Japanese ethnicities. CA Cancer J Clin. 2007;57:190–205. doi: 10.3322/canjclin.57.4.190. [DOI] [PubMed] [Google Scholar]
- 5.Brechot C, Kremsdorf D, Soussan P, Pineau P, Dejean A, Paterlini-Brechot P, Tiollais P. Hepatitis B virus (HBV)-related hepatocellular carcinoma (HCC): molecular mechanisms and novel paradigms. Pathol Biol (Paris) 2010;58:278–287. doi: 10.1016/j.patbio.2010.05.001. [DOI] [PubMed] [Google Scholar]
- 6.Wild CP, Law GR, Roman E. Molecular epidemiology and cancer: promising areas for future research in the post-genomic era. Mutat Res. 2002;499:3–12. doi: 10.1016/s0027-5107(01)00290-1. [DOI] [PubMed] [Google Scholar]
- 7.Schütte K, Bornschein J, Malfertheiner P. Hepatocellular carcinoma--epidemiological trends and risk factors. Dig Dis. 2009;27:80–92. doi: 10.1159/000218339. [DOI] [PubMed] [Google Scholar]
- 8.Wang JS, Groopman JD. DNA damage by mycotoxins. Mutat Res. 1999;424:167–181. doi: 10.1016/s0027-5107(99)00017-2. [DOI] [PubMed] [Google Scholar]
- 9.Chen PJ, Chen DS. Hepatitis B virus infection and hepatocellular carcinoma: molecular genetics and clinical perspectives. Semin Liver Dis. 1999;19:253–262. doi: 10.1055/s-2007-1007115. [DOI] [PubMed] [Google Scholar]
- 10.Kew MC. Synergistic interaction between aflatoxin B1 and hepatitis B virus in hepatocarcinogenesis. Liver Int. 2003;23:405–409. doi: 10.1111/j.1478-3231.2003.00869.x. [DOI] [PubMed] [Google Scholar]
- 11.Seitz HK, Stickel F. Risk factors and mechanisms of hepatocarcinogenesis with special emphasis on alcohol and oxidative stress. Biol Chem. 2006;387:349–360. doi: 10.1515/BC.2006.047. [DOI] [PubMed] [Google Scholar]
- 12.Strange RC, Spiteri MA, Ramachandran S, Fryer AA. Glutathione-S-transferase family of enzymes. Mutat Res. 2001;482:21–26. doi: 10.1016/s0027-5107(01)00206-8. [DOI] [PubMed] [Google Scholar]
- 13.Parl FF. Glutathione S-transferase genotypes and cancer risk. Cancer Lett. 2005;221:123–129. doi: 10.1016/j.canlet.2004.06.016. [DOI] [PubMed] [Google Scholar]
- 14.McIlwain CC, Townsend DM, Tew KD. Glutathione S-transferase polymorphisms: cancer incidence and therapy. Oncogene. 2006;25:1639–1648. doi: 10.1038/sj.onc.1209373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hayes JD, Strange RC. Glutathione S-transferase polymorphisms and their biological consequences. Pharmacology. 2000;61:154–166. doi: 10.1159/000028396. [DOI] [PubMed] [Google Scholar]
- 16.Sun L, Xi B, Yu L, Gao XC, Shi DJ, Yan YK, Xu DJ, Han Q, Wang C. Association of glutathione S-transferases polymorphisms (GSTM1 and GSTT1) with senile cataract: a meta-analysis. Invest Ophthalmol Vis Sci. 2010;51:6381–6386. doi: 10.1167/iovs.10-5815. [DOI] [PubMed] [Google Scholar]
- 17.White DL, Li D, Nurgalieva Z, El-Serag HB. Genetic variants of glutathione S-transferase as possible risk factors for hepatocellular carcinoma: a HuGE systematic review and meta-analysis. Am J Epidemiol. 2008;167:377–389. doi: 10.1093/aje/kwm315. [DOI] [PubMed] [Google Scholar]
- 18.Wang B, Huang G, Wang D, Li A, Xu Z, Dong R, Zhang D, Zhou W. Null genotypes of GSTM1 and GSTT1 contribute to hepatocellular carcinoma risk: evidence from an updated meta-analysis. J Hepatol. 2010;53:508–518. doi: 10.1016/j.jhep.2010.03.026. [DOI] [PubMed] [Google Scholar]
- 19.Liu L, Zhuang W, Wang C, Chen Z, Wu XT, Zhou Y. Interleukin-8 -251 A/T gene polymorphism and gastric cancer susceptibility: a meta-analysis of epidemiological studies. Cytokine. 2010;50:328–334. doi: 10.1016/j.cyto.2010.03.008. [DOI] [PubMed] [Google Scholar]
- 20.Wang R, Zhong B, Liu Y, Wang C. Association between alpha-adducin gene polymorphism (Gly460Trp) and genetic predisposition to salt sensitivity: a meta-analysis. J Appl Genet. 2010;51:87–94. doi: 10.1007/BF03195715. [DOI] [PubMed] [Google Scholar]
- 21.Xi B, Wang C, Wang R, Huang Y. FTO gene polymorphisms are associated with obesity and type 2 diabetes in East Asian Populations: an update. Obesity. 2011;19:236–237. doi: 10.1038/oby.2010.139. [DOI] [PubMed] [Google Scholar]
- 22.Zhang S, Wang C, Xi B, Li X. Association between the tumour necrosis factor-α-308G/A polymorphism and chronic obstructive pulmonary disease: an update. Respirology. 2011;16:107–115. doi: 10.1111/j.1440-1843.2010.01879.x. [DOI] [PubMed] [Google Scholar]
- 23.Garte S, Gaspari L, Alexandrie AK, Ambrosone C, Autrup H, Autrup JL, Baranova H, Bathum L, Benhamou S, Boffetta P, et al. Metabolic gene polymorphism frequencies in control populations. Cancer Epidemiol Biomarkers Prev. 2001;10:1239–1248. [PubMed] [Google Scholar]
- 24.DerSimonian R, Laird N. Meta-analysis in clinical trials. Control Clin Trials. 1986;7:177–188. doi: 10.1016/0197-2456(86)90046-2. [DOI] [PubMed] [Google Scholar]
- 25.MANTEL N, HAENSZEL W. Statistical aspects of the analysis of data from retrospective studies of disease. J Natl Cancer Inst. 1959;22:719–748. [PubMed] [Google Scholar]
- 26.Begg CB, Mazumdar M. Operating characteristics of a rank correlation test for publication bias. Biometrics. 1994;50:1088–1101. [PubMed] [Google Scholar]
- 27.Egger M, Davey Smith G, Schneider M, Minder C. Bias in meta-analysis detected by a simple, graphical test. BMJ. 1997;315:629–634. doi: 10.1136/bmj.315.7109.629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Dong CH, Yu SZ, Chen GC, Zhao DM, Fu YP. Polymorphisms of GSTT1 and M1 genotypes and their effects on elevated aflatoxin exposure and increased risk of hepatocellular carcinoma. Zhongliu Fangzhi Yanjiu. 1997;24:327–329. [Google Scholar]
- 29.Dong CH, Yu SZ, Chen GC, Zhao DM, Hu Y. Association of polymorphisms of glutathione S-transferase M1 and T1 genotypes with elevated aflatoxin and increased risk of primary liver cancer. Shijie Huaren Xiaohua Zazhi. 1998;6:463–466. [Google Scholar]
- 30.Dong CH, Zi XL, Yu SZ, Han JS. Relationship between deletion of Glutathion S-transferase gene and susceptibility to primary hepatocellular carcinoma. Zhongguo Gonggong Weisheng Xuebao. 1997;16:141–142. [Google Scholar]
- 31.Yu MW, Chiu YH, Chiang YC, Chen CH, Lee TH, Santella RM, Chern HD, Liaw YF, Chen CJ. Plasma carotenoids, glutathione S-transferase M1 and T1 genetic polymorphisms, and risk of hepatocellular carcinoma: independent and interactive effects. Am J Epidemiol. 1999;149:621–629. doi: 10.1093/oxfordjournals.aje.a009862. [DOI] [PubMed] [Google Scholar]
- 32.Chen CJ, Yu MW, Liaw YF, Wang LW, Chiamprasert S, Matin F, Hirvonen A, Bell DA, Santella RM. Chronic hepatitis B carriers with null genotypes of glutathione S-transferase M1 and T1 polymorphisms who are exposed to aflatoxin are at increased risk of hepatocellular carcinoma. Am J Hum Genet. 1996;59:128–134. [PMC free article] [PubMed] [Google Scholar]
- 33.Hsieh LL, Huang RC, Yu MW, Chen CJ, Liaw YF. L-myc, GST M1 genetic polymorphism and hepatocellular carcinoma risk among chronic hepatitis B carriers. Cancer Lett. 1996;103:171–176. doi: 10.1016/0304-3835(96)04209-7. [DOI] [PubMed] [Google Scholar]
- 34.Yu MW, Chiu YH, Yang SY, Santella RM, Chern HD, Liaw YF, Chen CJ. Cytochrome P450 1A1 genetic polymorphisms and risk of hepatocellular carcinoma among chronic hepatitis B carriers. Br J Cancer. 1999;80:598–603. doi: 10.1038/sj.bjc.6690397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Yu MW, Gladek-Yarborough A, Chiamprasert S, Santella RM, Liaw YF, Chen CJ. Cytochrome P450 2E1 and glutathione S-transferase M1 polymorphisms and susceptibility to hepatocellular carcinoma. Gastroenterology. 1995;109:1266–1273. doi: 10.1016/0016-5085(95)90587-1. [DOI] [PubMed] [Google Scholar]
- 36.Yu MW, Lien JP, Chiu YH, Santella RM, Liaw YF, Chen CJ. Effect of aflatoxin metabolism and DNA adduct formation on hepatocellular carcinoma among chronic hepatitis B carriers in Taiwan. J Hepatol. 1997;27:320–330. doi: 10.1016/s0168-8278(97)80178-x. [DOI] [PubMed] [Google Scholar]
- 37.Yu MW, Yang SY, Chiu YH, Chiang YC, Liaw YF, Chen CJ. A p53 genetic polymorphism as a modulator of hepatocellular carcinoma risk in relation to chronic liver disease, familial tendency, and cigarette smoking in hepatitis B carriers. Hepatology. 1999;29:697–702. doi: 10.1002/hep.510290330. [DOI] [PubMed] [Google Scholar]
- 38.Wu HL, Chen MN, Liu PX, Zhang RN. Relationship between GSTM1 gene polymorphism and genetic susceptibility to primary hepatocellular carcinoma. Shiyong Aizheng Zazhi. 2000;15:463–465. [Google Scholar]
- 39.Sun CA, Wang LY, Chen CJ, Lu SN, You SL, Wang LW, Wang Q, Wu DM, Santella RM. Genetic polymorphisms of glutathione S-transferases M1 and T1 associated with susceptibility to aflatoxin-related hepatocarcinogenesis among chronic hepatitis B carriers: a nested case-control study in Taiwan. Carcinogenesis. 2001;22:1289–1294. doi: 10.1093/carcin/22.8.1289. [DOI] [PubMed] [Google Scholar]
- 40.Zhu WC, Chen Q, Luo CL, Chu XW, Wu M. Relationship study between gene polymorphism of CYP1A1, GSTM1 and genetic susceptibility of primary hepatocellular carcinoma. Zhongliu Fangzhi Zazhi. 2001;8:572–574. [Google Scholar]
- 41.Chen SY, Wang LY, Lunn RM, Tsai WY, Lee PH, Lee CS, Ahsan H, Zhang YJ, Chen CJ, Santella RM. Polycyclic aromatic hydrocarbon-DNA adducts in liver tissues of hepatocellular carcinoma patients and controls. Int J Cancer. 2002;99:14–21. doi: 10.1002/ijc.10291. [DOI] [PubMed] [Google Scholar]
- 42.Liu CZ, Bian JC, Jiang F, Shen FM. Genetic polymorphism of glutathion S-transferase M1, T1, P1 on susceptibility hepatocellular carcinoma. Zhongguo Gonggong Weisheng. 2002;18:935–936. [Google Scholar]
- 43.Bian J, Shen F, Wang J, Chen G, Zhang B, Wu Y. [The mutation of deletion for glutathione S-transferase M1 gene in the tissue of hepatocellular carcinoma] Zhonghua Yixue Yichuanxue Zazhi. 1999;16:171–173. [PubMed] [Google Scholar]
- 44.Hu Y, Shen FM. Association between GSTM1 gene polymorphism of primary hepatocellular carcinoma and mutation of p53 codon 249. Zhonghua Yixue Yichuanxue Zazhi. 1997;14:76–78. [Google Scholar]
- 45.Bian JC, Shen FM, Shen L, Wang TR, Wang XH, Jiang F, Lu M, Liu CZ, Chen GD, Wang JB. Susceptibility to hepatocelluar carcinoma is associated with the null genotypes of GSTM1 and GSTT1. Zhonghua Xiaohua Zazhi. 1999;19:87–90. [Google Scholar]
- 46.Bian JC, Shen FM, Shen L, Wang TR, Wang XH, Chen GC, Wang JB. Susceptibility to hepatocellular carcinoma associated with null genotypes of GSTM1 and GSTT1. World J Gastroenterol. 2000;6:228–230. doi: 10.3748/wjg.v6.i2.228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.McGlynn KA, Hunter K, LeVoyer T, Roush J, Wise P, Michielli RA, Shen FM, Evans AA, London WT, Buetow KH. Susceptibility to aflatoxin B1-related primary hepatocellular carcinoma in mice and humans. Cancer Res. 2003;63:4594–4601. [PubMed] [Google Scholar]
- 48.London WT, Evans AA, Buetow K, Litwin S, McGlynn K, Zhou T, Clapper M, Ross E, Wild C, Shen FM. Molecular and genetic epidemiology of hepatocellular carcinoma: studies in China and Senegal. Princess Takamatsu Symp. 1995;25:51–60. [PubMed] [Google Scholar]
- 49.London WT, Evans AA, McGlynn K, Buetow K, An P, Gao L, Lustbader E, Ross E, Chen G, Shen F. Viral, host and environmental risk factors for hepatocellular carcinoma: a prospective study in Haimen City, China. Intervirology. 1995;38:155–161. doi: 10.1159/000150426. [DOI] [PubMed] [Google Scholar]
- 50.McGlynn KA, Rosvold EA, Lustbader ED, Hu Y, Clapper ML, Zhou T, Wild CP, Xia XL, Baffoe-Bonnie A, Ofori-Adjei D. Susceptibility to hepatocellular carcinoma is associated with genetic variation in the enzymatic detoxification of aflatoxin B1. Proc Natl Acad Sci USA. 1995;92:2384–2387. doi: 10.1073/pnas.92.6.2384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Li SP, Wu JZ, Ding JH, Gao CM, Cao HX, Zhou XF. Impact of genetic polymorphisms of glutathione S-transferaseT1, M1 on the risk of primary hepatocellular carcinoma in alchohol drinkers. Shiyong Aizheng Zazhi. 2004;19:229–232. [Google Scholar]
- 52.Chen CC, Yang SY, Liu CJ, Lin CL, Liaw YF, Lin SM, Lee SD, Chen PJ, Chen CJ, Yu MW. Association of cytokine and DNA repair gene polymorphisms with hepatitis B-related hepatocellular carcinoma. Int J Epidemiol. 2005;34:1310–1318. doi: 10.1093/ije/dyi191. [DOI] [PubMed] [Google Scholar]
- 53.Yu MW, Yang SY, Pan IJ, Lin CL, Liu CJ, Liaw YF, Lin SM, Chen PJ, Lee SD, Chen CJ. Polymorphisms in XRCC1 and glutathione S-transferase genes and hepatitis B-related hepatocellular carcinoma. J Natl Cancer Inst. 2003;95:1485–1488. doi: 10.1093/jnci/djg051. [DOI] [PubMed] [Google Scholar]
- 54.Deng ZL, Wei YP, Ma Y. Polymorphism of glutathione S-transferase mu 1 and theta 1 genes and hepatocellular carcinoma in southern Guangxi, China. World J Gastroenterol. 2005;11:272–274. doi: 10.3748/wjg.v11.i2.272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Ma Y, Deng ZL, Wei YP. GSTM1 gene polymorphisms of hepatocellular carcinoma from aflatoxin B1 high contaminated area in Guangxi, China. Aizheng. 2000;19:868–870. [Google Scholar]
- 56.Deng Z, Wei Y, Ma Y. [Glutathione-S-transferase M1 genotype in patients with hepatocellular carcinoma] Zhonghua Zhongliu Zazhi. 2001;23:477–479. [PubMed] [Google Scholar]
- 57.Wei YP, Ma Y, Deng ZL. Genetic polymorphisms of Glutathione S-transferase M1 and T1 and the risk of hepatocellular carcinoma. Zhongliu. 2003;23:464–466. [Google Scholar]
- 58.Deng ZL, Wei YP, Ma Y. Genetic deletion of GSTM1 and GSTT1 detoxicated enzymes in relation to hepatocellular carcinoma in Guangxi. Guangxi Kexue. 2005;12:55–57. [Google Scholar]
- 59.Guo HY, Bian JC, Jiang F, Wang QM, Zhang ZM, Fan WW, Wang QJ, Zhu X, Tang BM. The null genotypes of GSTM1 and GSTT1 and the genetic susceptibility of primary liver cancer in Luoyang, China. Zhongliu. 2005;25:58–61. [Google Scholar]
- 60.He SJ, Qin JR, Gu YY, Zhong WG, Su SG. Analysis of GSTM1, GSTT1 polymorphisms in liver cancer patients. Guangxi Yike Daxue Xuebao. 2005;22:875–877. [Google Scholar]
- 61.He SJ, Qin JR, Gu YY, Zhong WG, Su SG. Relationship between polymorphisms of phase II metabolic genes and the susceptibility to hepatocellular carcinoma in Guangxi. Shiyong Aizheng Zazhi. 2004;19:460–462, 473. [Google Scholar]
- 62.He SJ, Gu YY, Liao ZH. The relationship between the susceptibility to primary liver cancer and GSTM1 polymorphism, cigarette smoking and alcohol drinking. Guangxi Yike Daxue Xuebao. 2008;25:567–568. [Google Scholar]
- 63.Long XD, Ma Y, Wei YP, Deng ZL. [Study on the detoxication gene gstM1-gstT1-null and susceptibility to aflatoxin B1 related hepatocellular carcinoma in Guangxi] Zhonghua Liuxingbingxue Zazhi. 2005;26:777–781. [PubMed] [Google Scholar]
- 64.Long XD, Ma Y, Wei YP, Deng ZL. [A study about the association of detoxication gene GSTM1 polymorphism and the susceptibility to aflatoxin B1-related hepatocellular carcinoma] Zhonghua Ganzangbing Zazhi. 2005;13:668–670. [PubMed] [Google Scholar]
- 65.Ma DL, Chen YX, Li Y, Zhao HT, Xie XM. Glutathione-S-transferase M1 and T1 polymorphisms (deficiency) and susceptibility to liver cancer in hepatitis B surface antigen positive (HBsAg positive) population. Guangxi Yixue. 2005;27:656–657. [Google Scholar]
- 66.Zhang YC, Deng CS, Zhu YQ. Study on genetic polymorphisms of xenobiotica metabolizing enzymes in hepatitis B virus-associated hepatic diseases. Wenzhou Yixueyuan Xuebao. 2005;35:464–467. [Google Scholar]
- 67.Zhu MH, Chen XH, Zhou LF. [Association of genetic polymorphisms in glutathione S-transferases M1 with hepatitis beta-related hepatocellular carcinoma] Zhejiang Da Xue Xue Bao Yi Xue Ban. 2005;34:126–130. doi: 10.3785/j.issn.1008-9292.2005.02.007. [DOI] [PubMed] [Google Scholar]
- 68.Long XD, Ma Y, Wei YP, Deng ZL. The polymorphisms of GSTM1, GSTT1, HYL1*2, and XRCC1, and aflatoxin B1-related hepatocellular carcinoma in Guangxi population, China. Hepatol Res. 2006;36:48–55. doi: 10.1016/j.hepres.2006.06.004. [DOI] [PubMed] [Google Scholar]
- 69.Yang ZG, Xie YA, Kuang ZP, Luo XL, Zhang WM, Leng CH. Relationship between genetic polymorphisms of glutathione-S-transferase M1, T1 genes and susceptibility to hepatocellular carcinoma in population of Fusui District of Guangxi Zhuang Autonomous Region. Zhonghua Zhongliu Fangzhi Zazhi. 2009;16:970–975. [Google Scholar]
- 70.Liu ZG, Wei YP, Ma Y, Deng ZL. Population with GSTT1 gene deletion and the relationship to hepatocellular carcinoma from Guangxi. Guangxi Yike Daxue Xuebao. 2003;20:161–163. [Google Scholar]