Abstract
Two lineages of enterohemorrhagic Escherichia coli O157:H7 (EDL933, Stx1+ and Stx2+) and 86-24 (Stx2+) were investigated to determine the genetic basis of biofilm formation on abiotic surfaces. Strain EDL933 formed a robust biofilm while strain 86-24 formed almost no biofilm on either polystyrene plates or polyethylene tubes. Whole-transcriptome profiles of EDL933 vs. 86-24 revealed that in the strong biofilm-forming strain, genes involved in curli biosynthesis (csgBAC and csgDEFG) and cellulose production (adrA) were significantly induced, whereas genes involved in indole signaling (trpLED and mtr) were most repressed. Additionally, 49 phage genes were highly induced and repressed between the two strains. Curli assays using Congo red plates and scanning electron microscopy corroborated the microarray data as the EDL933 strain produced a large amount of curli, while strain 86-24 formed much less curli. Moreover, EDL933 produced 19-fold more cellulose than 86-24, and indole production in EDL933 was two times lower than that of the strain 86-24. Therefore, it appears E. coli O157:H7 EDL933 produces more biofilm because of its increased curli and cellulose production and reduced indole production.
Keywords: biofilm, curli, indole, Escherichia coli O157:H7
1. Introduction
Enterohemorrhagic Escherichia coli serotype O157:H7 is the most common human pathogen responsible for outbreaks of hemorrhagic colitis causing bloody diarrhea that can lead to the life-threatening hemolytic-uremic syndrome that affects mostly children around the world (Boyce et al., 1995). Understanding E. coli O157:H7 infections is important given that there are approximately 73,000 infections annually in the U.S., directly leading to 2,000 hospitalizations, 60 deaths, and an economic cost up to $405 million (in 2003 dollars) (Frenzen et al., 2005).
Depending on the specific strain, E. coli serotype O157:H7 can produce both Shiga toxin 1 (Stx1) and 2 (Stx2) that are responsible for hemorrhagic colitis (Nataro & Kaper 1998). It has a very low infectious dose (as low as 50 CFU in one outbreak) and colonizes the intestinal epithelial cells, where it causes attaching and effacing lesions (Nataro & Kaper 1998). E. coli serotype O157:H7 strain EDL933 was implicated in two outbreaks of hemorrhagic colitis in the USA during 1982 (Wells et al., 1983) and produces both Stx1 and Stx2 toxins (Strockbine et al., 1986). E. coli O157:H7 strain 86-24, caused a hemorrhagic colitis outbreak in the USA during 1986 (Griffin et al., 1988) and produces only Stx2 (Jarvis & Kaper 1996). Epidemiological data suggest that Stx2 is more important than Stx1 in the development of hemolytic-uremic syndrome (Griffin 1995), although this result is not conclusive because, unlike Stx1, Stx2 has many variants (Nataro & Kaper 1998).
Bacterial biofilms are ubiquitous in natural, medical, and engineering environments (Potera 1999). Biofilms have been associated with many chronic infections such as prostatitis, biliary tract infections, and urinary catheter cystitis by pathogenic E. coli due to their high resistance to antimicrobial agents (Costerton et al., 1999). Food-borne microorganisms, such as E. coli O157:H7, can readily attach to and form biofilms on various surfaces, such as stainless steel, glass, and polystyrene (Ryu & Beuchat 2005, Rivas et al., 2007). The genetic mechanism of biofilm formation of E. coli O157:H7 is a complex process and is now beginning to be unveiled. The production of curli fimbriae (Ryu & Beuchat 2005, Uhlich et al., 2006, Saldaña et al., 2009) is the most common contributor to the biofilm formation in E. coli O157:H7. Diverse proteins also play an important role in the biofilm formation of E. coli O157:H7 (Wells et al., 2008, Puttamreddy et al., 2010, Lee et al., 2008b). Additionally, intercellular signal molecules, such as autoinducer-2 (Yoon & Sofos 2008, Bansal et al., 2008) and indole (Lee et al., 2007, Lee & Lee 2010), are involved in biofilm formation of E. coli O157:H7.
In this study, we initially observed a significant difference in the biofilm formation of the two E. coli O157:H7 strains, EDL933 and 86-24. DNA microarrays were utilized to identify the genetic basis for this difference in biofilm formation. Global gene expression from the microarray data was corroborated by phenotypic assays including those for curli, cellulose, and indole. It was found that EHEC biofilm formation depends chiefly on enhanced curli and cellulose production along with reduced indole production.
2. Materials and Methods
Bacterial strains, materials, and growth rate measurements
Two pathogenic strains of enterohemorrhagic E. coli O157:H7, strain EDL933 (ATCC43895) (Strockbine et al., 1986)) and strain 86-24 (kindly provided by Dr. Arul Jayaraman of Texas A&M University) (Griffin et al., 1988) were used. EDL933 was sequenced (Perna et al., 2001), whereas the strain 86-24 has not been sequenced. Luria-Bertani medium (LB) (Sambrook et al., 1989) was used for growth. All chemicals (Congo red, Coomasie brilliant blue, indole, calcofluor, crystal violet, sodium phosphate, and β-mercapto ethanol) were purchased from Sigma-Aldrich Co. (Missouri, USA). Glutaraldehyde, formaldehyde, acetonitrile, amyl alcohol, ethyl alcohol, hydrochloric acid, OsO4, and ρ-dimethylamino-benzaldehyde were purchased from Junsei Chemical Co. (Tokyo, Japan) or Duksan Pure Chemical Co. (Ansan, Korea). All experiments were performed with LB medium at 37°C (human body temperature). The strains were initially streaked from −80°C glycerol stocks and a fresh single colony was inoculated 25 ml LB medium in 250 ml flasks and cultured at 250 rpm. Overnight cultures were diluted 1:100 using LB medium. For cell growth measurements, the turbidity was measured at 600 nm (OD600) with a spectrophotometer (UV-160, Shimadzu, Japan). When the value of OD600 was above 0.7, the culture sample was diluted into the linear range of 0.2 to 0.7. Each experiment was performed with at least two independent cultures.
Crystal-violet biofilm assay
A static biofilm formation assay was performed in 96-well polystyrene plates (SPL life sciences, Korea) or 14 mL polyethylene test tubes (SPL life sciences) as previously reported (Pratt & Kolter 1998). Briefly, cells were inoculated at an initial turbidity at 600 nm of 0.05 for 24 h without shaking. Cell growth and total biofilm were measured using crystal violet staining. Each data point was averaged from at least twelve replicate wells (six wells from each of at least two independent cultures).
Total RNA isolation
For the microarray experiments, planktonic cells of EDL933 and 86-24 were cultured to a turbidity of 4.0 at 600 nm. Due to the very low biofilm formation of 86-24, planktonic cells were used to study the whole transcriptome profiles. Cells were immediately chilled with dry ice and 95% ethanol (to prevent RNA degradation) for 30 sec before centrifugation at 13,000 g for 2 min; cell pellets were frozen immediately with dry ice and stored −80°C. RNA was isolated using Qiagen RNeasy mini Kit described previously (Ren et al., 2004).
DNA Microarray analysis
The E. coli GeneChip Genome 2.0 Array (Affymetrix, P/N 900551, Santa Clara, USA) was used to study the differential gene expression profile for EDL933 vs. 86-24 as described in the Gene Expression Technical Manual. Ten μg of total RNA from each sample was converted to cDNA using random primers. The purified cDNA was fragmented using 0.6 U/μg of DNAse I and end-labeled by terminal transferase reaction incorporating a biotinylated dideoxynucleotide. Hybridization was performed for 16 hours at 45°C and 60 rpm as described in the GeneChip Expression Analysis Technical Manual (Affymetrix). After hybridization, the chips were stained and washed in a GeneChip Fluidics Station 450 (Affymetrix) and scanned by using a GeneChip Array scanner 3000 7G (Affymetrix), and the total cell intensity was scaled automatically in the software to an average value of 500. The probe array images were inspected for any image artifact. Background values, noise values, and scaling factors of both arrays were examined and were comparable. The intensities of the polyadenosine RNA controls of Bacillus subtilis (lys, phe, thr, and dap) at different concentrations were used to monitor the labeling and scanning process. A gene was considered differentially expressed when the p-value for comparing two chips was lower than 0.05 (to assure that the change in gene expression was statistically significant and that false positives arise less than 5%) and when the expression ratio was higher (3-fold) than the standard deviations for whole microarray (2.6-fold). Gene functions were obtained from the Affymetrix–NetAffx Analysis Center (https://www.affymetrix.com/analysis/netaffx/index.affx).
Quantitative real-time reverse transcription polymerase chain reaction (qRT-PCR)
To corroborate the DNA microarray data and curli assay and to confirm the induction of indole production, qRT-PCR was used to investigate the transcription level of csgA (encoding a curlin major subunit) and tnaA (encoding indole-synthesizing tryptophanase) using two independent RNA samples. Two primers for csgA (forward primer 5′-AGATGTTGGTCAGGGCTCAG-3′ and reverse primer 5′-CGTTGTTACCAAAGCCAACC-3′) and two primers for tnaA (forward primer 5′-TACACCATTCCGACTCACCA-3′ and reverse primer 5′-CCGTATCGAAGGCTTCTTTG-3′) were used. The expression level of the housekeeping gene rrsG (16S rRNA, forward primer 5′-TATTGCACAATGGGCGCAAG-3′ and reverse primer 5′-ACTTAACAAACCGCCTGCGT-3′) was used to normalize the expression data of interesting genes. The method of qRT-PCR was adapted (Lee et al., 2008a). qRT-PCR was performed with SYBR Green master mix (Applied Biosystems, Foster City, USA) and ABI 7500 Real-Time PCR System (Applied Biosystems).
Curli assay
To measure curli production, Congo red plates (LB agar medium containing 20 μg/ml Congo red and 10 μg/ml Coomassie brilliant blue, and 15 g/L agar) were used as described previously to visualize curli production after 24 h incubation (Reisner et al., 2006). In order to corroborate the plate assay, scanning electron microscopy (SEM) was used by modifying the protocol (Hossain et al, 1995). Briefly, E. coli cells were cultured until a turbidity of 2.0 at 600 nm, and cells were directly fixed by adding glutaraldehyde (2.5% final) and formaldehyde (2% final) and incubated at 4°C overnight. Fixed cells were collected by filtering with a 0.45 μm nylon filter (Nalgene, New York, USA) with vacuum. The filter containing cells was cut into 0.5 × 0.5 mm squares and washed with 0.2 M sodium phosphate buffer before fixating for 90 min with osmium solution (containing 1.5 mL of sodium phosphate buffer 0.2 M, 3 mL of 2% OsO4 and 3 mL deionized water). Then, samples were washed and dehydrated by successive 10 min incubations in 50% ethanol, 70% ethanol, 80% ethanol, 90% ethanol, and 95% ethanol followed by two successive incubations for 20 min in 100% ethanol. After dehydrating, the filters with cells were incubated in isoamyl acetate for 20 min and dried with critical-point dryer (HCP-2, Hitachi, Japan). The nylon filters were affixed to SEM stubs and coated with white gold for 200 seconds using ion sputtering (E-1030, Hitachi). The specimens were examined using the SEM S-4100 (Hitachi, Tokyo, Japan). The voltage was set at 15 kV and viewed at a magnification from x 2,000 to x15,000.
Cellulose assay
A cellulose assay using calcofluor (Ma & Wood 2009) was used. Cells (2 ml) at a turbidity of 4 were centrifuged and re-suspended in 1 ml of 1% tryptone with 16 mg/ml calcofluor and incubated for 2 h at 250 rpm. Bacterial bound calcofluor was removed by centrifugation for 5 min at 17 000 g, and the amount of calcofluor unbound to cellulose was determined by measuring the absorbance of the supernatant at 350 nm.
Indole assay
To measure the concentration of extracellular indole, EDL933 and 86-24 were grown to a turbidity of 4.0 at 600 nm. The turbidity of 4.0 represents the time point of the stationary phase at which the DNA microarray samples were taken. Indole concentrations were measured with the previous protocol using the Kovac’s reagent (Kawamura-Sato et al., 1999). Briefly, Kovac’s reagent (0.4 mL) was mixed with supernatants (1 mL) of bacterial cultures. The reaction mixture was diluted 1:10 in HCl-amyl alcohol solution (30 mL of HCl and 90 mL of amyl alcohol), and the absorbance of the mixture was measured at 540 nm with a spectrophotometer (Shimadzu UV-160). In addition, the spectrophotometric indole assay was corroborated with reverse-phase HPLC using a 100 × 4.6 mm Chromolith Performance RP-18e column (Merck KGaA, Darmstadt, Germany) and elution with H2O-0.1% (v/v) trifluoroacetic acid and acetonitrile as the mobile phases at a flow rate of 0.5 mL/min (50:50). Under these conditions, the retention time and the absorbance maximum was 5.1 min/271 nm for indole.
3. Results
The main goal of this research was to investigate the genetic mechanism of biofilm formation of the two E. coli O157:H7 strains, EDL933 (good biofilm former) and 86-24 (poor biofilm former). DNA microarrays were initially utilized to predict the phenotypic changes, and phenotypic assays for curli formation, cellulose, and indole production were performed to corroborate the whole transcriptome data.
Biofilm formation
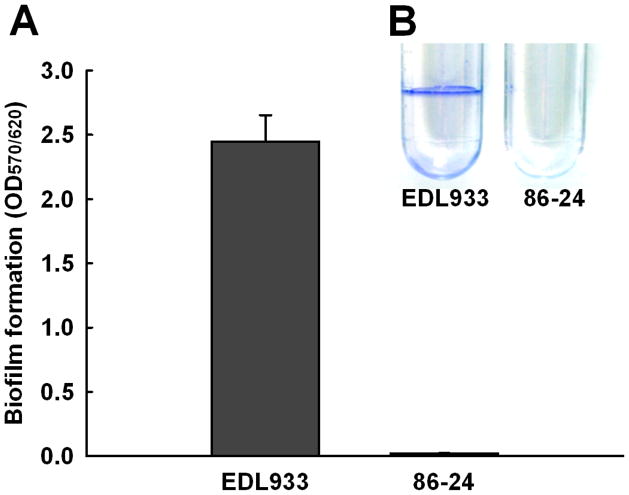
Biofilm formation of the two E. coli O157:H7 strains was tested using 96-well polystyrene plates and polyethylene test tubes. While EDL933 formed robust biofilms, 86-24 formed almost no biofilm on both polystyrene plates (Fig. 1A) and polyethylene tubes (Fig. 1B). The specific growth rates of the two E. coli O157:H7 strains were almost identical (1.38 ± 0.07/h for EDL933 and 1.40 ± 0.08/h for 86-24); therefore, the changes in biofilm formation were not due to differences in growth rates.
Fig. 1.
Biofilm formation of the two E. coli O157:H7 strains, EDL933 and 86-24, in 96-well polystyrene plates (A) and in polyethylene tubes (B) in LB medium without shaking at 37°C after 24 h. At least four independent experiments were conducted (a total 12 wells), and error bars indicate one standard deviation.
Differential gene expression of EDL933 vs. 86-24
To investigate the global genetic basis for the difference in biofilm formation, DNA microarrays were used. For EDL933 vs. 86-24, 440 genes were differentially expressed more than 3-fold; 325 genes were induced and 115 genes were repressed. Partial lists of genes most induced and repressed are shown in the Table 1 and Table 2, and the whole set of expression data for the two strains has been deposited in the NCBI GENE Expression Omnibus and are accessible through accession number GSE 19953. As expected, 86-24 showed almost no signal for the stx1B and stx1A genes (Table 1) because strain 86-24 lacks Stx1 (Jarvis & Kaper 1996). Among the differentially expressed genes, many prophage genes of CP-933M, BP-933W, CP-933O, and CP-933V were highly induced in EDL933 (Table 1), whereas prophage genes of CP-933T were highly repressed in EDL933 (Table 2).
Table 1.
Genes induced more than 6-fold for planktonic cells of EDL933 vs. 86-24. Cells were grown in LB medium at 37 °C to a turbidity of 4.0.
| gene | fold change | Descriptions |
|---|---|---|
| Shiga-like toxin genes | ||
| stx1B | 5405 | shiga-like toxin 1 subunit B |
| stx1A | 955 | shiga-like toxin 1 subunit A |
| Curli and cellulose genes | ||
| csgA | 49 | curlin major subunit, coiled surface structures |
| csgB | 79 | minor curlin subunit precursor, similar to CsgA |
| csgC | 84 | putative curli production protein |
| csgD | 17 | putative 2-component transcriptional regulator for curli operon |
| csgE | 24 | curli production assembly transport component, |
| csgF | 28 | curli production assembly transport component |
| csgG | 24 | curli production assembly transport component |
| adrA | 8 | regulated by CsgD involved in cellulose biosynthesis |
| Prophage genes and unknown genes | ||
| z1215 | 6 | unknown protein |
| z1218 | 8 | hypothetical protein |
| z1353 | 6 | putative antirepressor protein of cryptic prophage CP-933M |
| z1354 | 9 | putative endopeptidase of cryptic prophage CP-933M |
| z1362 | 6 | unknown protein encoded by cryptic prophage CP-933M |
| z1364 | 6 | unknown protein encoded by cryptic prophage CP-933M |
| z1373 | 23 | unknown protein encoded by cryptic prophage CP-933M |
| z1443 | 362 | unknown protein encoded by bacteriophage BP-933W |
| z1444 | 1911 | putative serinethreonine kinase encoded by bacteriophage BP-933W |
| z1445 | 10,809 | unknown protein encoded by bacteriophage BP-933W |
| z1446 | 1261 | unknown protein encoded by bacteriophage BP-933W |
| z1448 | 256 | regulatory protein Cro of bacteriophage BP-933W |
| z1469 | 158 | putative lysozyme protein R of bacteriophage BP-933W |
| z1663 | 6 | unknown protein |
| z1811 | 137 | putative tail component encoded by prophage CP-933N |
| z1921 | 6 | unknown protein encoded by prophage CP-933X |
| z1957 | 832 | transposase for IS629 |
| z2112 | 37 | putative ClpP-like protease encoded within prophage CP-933O |
| z2113 | 32 | unknown protein encoded within prophage CP-933O |
| z2114 | 891 | unknown protein encoded within prophage CP-933O |
| z2115 | 16 | unknown protein encoded within prophage CP-933O |
| z2116 | 832 | unknown protein encoded within prophage CP-933O |
| z2117 | 30 | unknown protein encoded within prophage CP-933O |
| z3083 | 69 | putative tail fiber component M of prophage CP-933U |
| z3341 | 147 | unknown protein encoded within prophage CP-933V |
| z3342 | 69 | unknown protein encoded within prophage CP-933V |
| z3347 | 147 | unknown protein encoded within prophage CP-933V |
| z3348 | 169 | unknown protein encoded within prophage CP-933V |
| z3357 | 549 | putative regulatory protein CII of prophage CP-933V |
| z3388 | 208 | unknown protein |
| z5148 | 6 | unknown protein |
| z5878 | 69 | putative integrase |
| z5881 | 338 | unknown protein |
| z5882 | 42 | unknown protein |
| z5883 | 7 | unknown protein |
| z5884 | 91 | unknown protein |
| z5885 | 13 | putative resolvase |
| z5886 | 2195 | unknown protein |
| z5887 | 16 | unknown protein |
| z5888 | 119 | unknown protein |
| z5889 | 119 | unknown protein |
| ybcK | 6 | unknown protein |
| ybgS | 8 | unknown protein |
| yccT | 13 | unknown protein |
| yhiM | 9 | unknown protein |
| yibH | 8 | unknown protein |
| yohC | 6 | unknown protein |
| Other genes | ||
| aidB | 6 | putative acyl coenzyme A |
| appB | 12 | probable third cytochrome oxidase, subunit I |
| appC | 12 | probable third cytochrome oxidase, subunit I |
| prpB | 23 | putative phosphonomutase 2 |
| prpC | 17 | putative citrate synthase; propionate metabolism |
| prpD | 21 | hypothetical protein |
| prpE | 18 | putative propionyl-CoA synthetase |
| rpoS | 11 | RNA polymerase, sigma S (sigma38) factor |
| ssuA | 28 | iphatic sulphonate binding protein precursor |
| tfaR | 9 | Rac prophage |
Table 2.
Genes repressed more than 6-fold for planktonic cells of EDL933 vs. 86-24. Cells were grown in LB medium at 37 °C to a turbidity of 4.0.
| Gene | fold change | Descriptions |
|---|---|---|
| Prophage genes and unknown genes | ||
| intT | −91 | IntT, integrase for prophage CP-933T |
| z2967 | −128 | unknown protein encoded by prophage CP-933T |
| z2968 | −5405 | unknown protein encoded by prophage CP-933T |
| z2969 | −1261 | unknown protein encoded by prophage CP-933T |
| z2970 | −169 | putative regulator for prophage CP-933T |
| z2971 | −8 | unknown protein encoded by prophage CP-933T |
| z2972 | −8 | unknown protein encoded by prophage CP-933T |
| z2973 | −64 | unknown protein encoded by prophage CP-933T |
| z2974 | −23 | unknown protein encoded by prophage CP-933T |
| z2976 | −79 | unknown protein encoded by prophage CP-933T |
| z2977 | −7 | unknown protein encoded by prophage CP-933T |
| z2979 | −12 | putative stabilitypartitioning protein encoded within prophage CP-933T |
| z2980 | −17 | putative stabilitypartitioning protein encoded within CP-933T |
| z2983 | −15 | putative tail fiber assembly protein of prophage CP-933T |
| z2984 | −74 | putative serine acetlyltransferase of prophage CP-933T |
| z2986 | −37 | putative tail fiber protein of prophage CP-933T |
| z2987 | −34 | putative tail fiber component of prophage CP-933T |
| z2988 | −9 | putative tail fiber protein component of prophage CP-933T |
| z2989 | −11 | unknown protein encoded by prophage CP-933T |
| z2990 | −39 | putative tail fiber component of prophage CP-933T |
| z2991 | −39 | putative tail sheath protein of prophage CP-933T |
| z2992 | −34 | putative tail assembly protein of prophage CP-933T |
| z2994 | −315 | unknown protein encoded by prophage CP-933T |
| z3271 | −8 | unknown protein |
| z5102 | −6 | unknown protein |
| z5111 | −6 | unknown protein |
| ydfV | −23 | unknown protein, Qin prophage |
| Indole related genes | ||
| trpD | −12 | anthranilate synthase component II |
| trpE | −15 | anthranilate synthase component I |
| trpL | −11 | trp operon leader peptide |
| mtr | −13 | tryptophan-specific transport protein |
| Other genes | ||
| dxs | −9 | deoxy-xylulose-P synthase |
| nlpA | −28 | lipoprotein-28 |
Among the most induced genes in EDL933 were seven curli genes (csgBAC and csgDEFG) (17- to 84-fold) (Table 1). Among the most repressed genes in EDL933 were, trpLED, involved in tryptophan synthesis (a substrate required for indole synthesis), and mtr, involved in indole-and tryptophan-uptake (Table 2).
Curli formation
Curli formation is important for biofilm formation for E. coli O157:H7 (Saldaña et al., 2009, Puttamreddy et al., 2010, Ryu & Beuchat 2005, Uhlich et al., 2006). Since the microarray data showed that seven curli genes (csgBAC and csgDEFG) were highly induced (Table 1), curli production was measured using Congo red plates and SEM. Congo red plates were used to observe the curli production with colonies grown under static conditions, while SEM images were used to examine the curli production from planktonic cells that were used for the DNA microarrays.
EDL933 produced a large amount of curli, whereas 86-24 produced almost no curli on the Congo red plate (Fig. 2A) and with SEM (Fig. 2B). For SEM, polymerized curli appeared as 4- to 7-nm wide fibers of varying lengths, and a large amount of curli appeared as a tangled and amorphous matrix surrounding bacterial cells (Chapman et al., 2002). EDL933 obviously produced a large amount of curli, while 86-24 produced few curli (Fig. 2B). These results corroborate well the microarray data (Table 1). Therefore, these results indicate that curli production is an important genetic mechanism of the different biofilm formation of the two E. coli O157:H7 strains. In addition, it is interesting that E. coli O157:H7 EDL933 is capable of making copious amounts of curli at 37°C whereas E. coli K-12 only makes curli at low temperatures(less than 32°C)(Gualdi et al., 2008).
Fig. 2.

Curli formation of the two E. coli O157:H7 strains, EDL933 and 86-24. Curli production at 37°C after 24 h was observed using Congo red plates (A) and SEM (B). For SEM analysis, planktonic cells were grown in LB medium to turbidity of 2.0. The inset shows the magnified amorphous curli/cellulose matrix.
Cellulose production
The curli protein CsgD also stimulates cellulose production directly by activating transcription of adrA (Zogaj et al., 2001) and also curli and cellulose play a synergistic role in biofilm formation (Saldaña et al., 2009). Since adrA was induced 8-fold in EDL933 (Table 1), cellulose production was measured for the two strains using cellulose specific calcofluor (Ma & Wood 2009). Indeed, EDL933 produced 19-fold more cellulose than 86-24 (0.57 ± 0.08 μg cellulose bound calcofluor/ml cells for EDL933 and 0.03 ± 0.06 μg cellulose bound calcofluor/ml cells for 86-24). Therefore, EDL933 produces much more of both curli and cellulose than 86-24.
Indole production
We discovered that extracellular indole inhibits the biofilm formation of E. coli O157:H7 (Lee et al., 2007, Bansal et al., 2007). Since the whole-transcriptome analysis showed that indole-related genes (trpLED and mtr) were significantly repressed in EDL933 (Table 2), extracellular indole concentrations were measured for the two strains. Corroborating the microarray data, EDL933 produced 2-fold less extracellular indole than 86-24 (210 ± 10 μM for EDL933 vs. 430 ± 10 μM for 86-24). This result partially explains the difference in biofilm formation for these two strains.
Confirmation of the microarray data
qRT-PCR was used to investigate the transcription of csgA (encoding a curlin major subunit) and tnaA (encoding indole-synthesizing tryptophanase). The qRT-PCR results were comparable with the DNA microarray data for csgA (10.2 ± 0.1-fold induction via qRT-PCR vs. 49-fold in the microarrays). Additionally, qRT-PCR showed 2.6 ± 0.4-fold lower of tnaA expression with EDL933 than 86-24, which corroborated the enhanced indole production in the EDL933.
4. Discussion
DNA microarrays have been utilized extensively to obtain whole-transcriptome profiling. For example, two separate lineages of E. coli O157:H7 were compared (EDL933 strains LI and LII) to determine if there are differences in expression of virulence-related genes (Dowd & Ishizaki 2006). They found differential expression of stx2b, ureD, curli (csgAFEG), stress-related genes (hslJ, cspG, ibpB, ibpA), type III secretion apparatus, LPS, and flagella under anaerobic conditions (Dowd & Ishizaki 2006). Here, we also used DNA microarrays to understand the genetic mechanisms of the biofilm formation of the two E. coli O157:H7 strains, EDL933 (ATCC43895) and 86-24.
In the current study, about 12% (440 genes) of the total genome (5416 genes, (Perna et al., 2001)) was differentially expressed (more than 3-fold). Among the highly induced and repressed genes (more than 6-fold), 49 prophage genes were identified (Table 1 and Table 2), which indicates the two tested strains have evolved differently. The antibiotic norfloxacin induces many prophage genes in the BP-933W prophage genome (Herold et al., 2005) including z1443 through z1469 (Table 1), and prophage genes play a role in E. coli K-12 biofilm formation (Wang et al., 2009). Also, the deletion of the stx2 or the entire Stx2-encoding phage genes in the strain 86-24 did not affect E. coli O157:H7 colonization in sheep (Cornick et al., 2007). Therefore, it would be interesting to investigate whether the EDL933 specific prophage genes (Table 1) affect the biofilm formation and the antibiotic resistance of E. coli O157:H7.
The whole transcriptomic profiling showed that curli genes (csgBAC and csgDEFG), a cellulose gene (adrA), and indole-related genes (trpLED and mtr) are the most differentially expressed loci between the two strains (Table 1). The curli fibers produced by E. coli have many of the same biochemical and biophysical properties in common with human amyloid that is associated with Alzheimer’s and prion diseases (Chapman et al., 2002), and curli formation is important for biofilm formation of pathogenic E. coli O157:H7 (Ryu & Beuchat 2005, Uhlich et al., 2006) as well as non-pathogenic E. coli (Prigent-Combaret et al., 2000, Landini 2009). Using a Congo red assay and SEM, this study confirmed that curli formation is an important positive factor for the biofilm formation of E. coli O157:H7 (Fig. 2AB). In agreement with this result, a study using transposon mutagenesis also showed that curli genes play a role in biofilm development in E. coli O157:H7 (Puttamreddy et al. 2010).
Corroborating the high expression of the cellulose gene adrA (Table 1), EDL933 produces 19-fold more cellulose than 86-24 although the expression of bacterial cellulose biosynthesis cluster (bcsABZC) was not changed in this transcriptome study. Cellulose production increases the binding of E. coli O157:H7 to sprouts (Matthysse et al., 2008) and increases biofilm formation in E. coli K-12 on hydrophilic surfaces, such as glass (Ma & Wood 2009). Also, cellulose production as well as curli production positively influence the biofilm formation of E. coli O157:H7 (Saldaña et al., 2009).
In addition, intercellular signal molecules, such as autoinducer-2 (Yoon & Sofos 2008) and indole (Lee et al., 2007, Bansal et al., 2007) are also be involved in biofilm formation of E. coli O157:H7. Since 86-24 produced more indole than EDL933 and formed almost no biofilm (Fig. 1), this result supports that elevated indole concentrations are partially responsible for the lack of biofilm formation by 86-24. Further support of this is that addition of 1 mM indole to EDL933 reduces its biofilm formation dramatically (Lee et al., 2007).
In the previous transcriptome study of the strong biofilm-forming strain EDL933, biofilm cells of EDL933 formed on glass wool repressed 20 pathogenic genes from the LEE island compared to planktonic cells of EDL933 which indicated that the virulence genes were not utilized in biofilm cells (Lee et al., 2007). The current study also could not identify any positive relationship between virulence gene expression and biofilm formation for the two E. coli O157:H7 strains. Hence, it is possible that environmental cues are missing that induce virulence genes under laboratory conditions within a single species.
To date, no effective therapy for E. coli O157:H7 serotypes has been found (Tarr et al., 2005, Boyce et al., 1995) because antibiotics, anti-motility agents, narcotics, and non-steroidal anti-inflammatory drugs are not usually provided as they increase the risk of developing hemolytic-uremic syndrome, a major cause of acute renal failure in children (Tarr et al., 2005); these agents induce an SOS response in E. coli O157:H7 which induces the prophage-based Shiga toxins (Kimmitt et al., 2000). Hence, controlling biofilm formation of E. coli O157:H7 is important in medicine as well as in food industry. In addition to antimicrobial therapy, anti-virulence therapies (Cegelski et al., 2008, Lesic et al., 2007) have been proposed because unlike antimicrobials, anti-virulence compounds do not affect growth and so there is less chance of developing resistance (Hentzer et al., 2002, Lesic et al., 2007). Since the two tested E. coli O157:H7 strains showed same growth rates but showed distinctive biofilm formation due to mainly curli and cellulose formation, controlling either curli formation or cellulose formation are possible ways to prevent biofilm formation of E. coli O157:H7.
Acknowledgments
This research was supported by the Yeungnam University research grant (to J. Lee). J-H. Lee and Y-G Kim were supported by the Brain Korea 21 Project from the Ministry of Education and Human Resources, Korea. T. Wood is the T. Michael O’Connor endowed chair and is also supported by the NIH (R01 GM089999).
References
- Bansal T, Englert D, Lee J, Hegde M, Wood TK, Jayaraman A. Differential effects of epinephrine, norepinephrine, and indole on Escherichia coli O157:H7 chemotaxis, colonization, and gene expression. Infect Immun. 2007;75:4597–4607. doi: 10.1128/IAI.00630-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bansal T, Jesudhasan P, Pillai S, Wood TK, Jayaraman A. Temporal regulation of enterohemorrhagic Escherichia coli virulence mediated by autoinducer-2. Appl Microbiol Biotechnol. 2008;78:811–819. doi: 10.1007/s00253-008-1359-8. [DOI] [PubMed] [Google Scholar]
- Boyce TG, Swerdlow DL, Griffin PM. Escherichia coli O157:H7 and the hemolytic-uremic syndrome. N Engl J Med. 1995;333:364–368. doi: 10.1056/NEJM199508103330608. [DOI] [PubMed] [Google Scholar]
- Cegelski L, Marshall GR, Eldridge GR, Hultgren SJ. The biology and future prospects of antivirulence therapies. Nat Rev Microbiol. 2008;6:17–27. doi: 10.1038/nrmicro1818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chapman MR, Robinson LS, Pinkner JS, Roth R, Heuser J, Hammar M, Normark S, Hultgren SJ. Role of Escherichia coli curli operons in directing amyloid fiber formation. Science. 2002;295:851–855. doi: 10.1126/science.1067484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cornick NA, Helgerson AF, Sharma V. Shiga toxin and Shiga toxin-encoding phage do not facilitate Escherichia coli O157:H7 colonization in sheep. Appl Environ Microbiol. 2007;73:344–346. doi: 10.1128/AEM.01328-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Costerton JW, Stewart PS, Greenberg EP. Bacterial biofilms: a common cause of persistent infections. Science. 1999;284:1318–1322. doi: 10.1126/science.284.5418.1318. [DOI] [PubMed] [Google Scholar]
- Dowd SE, Ishizaki H. Microarray based comparison of two Escherichia coli O157:H7 lineages. BMC Microbiol. 2006;6:30. doi: 10.1186/1471-2180-6-30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frenzen PD, Drake A, Angulo FJ. Economic cost of illness due to Escherichia coli O157 infections in the United States. J Food Prot. 2005;68:2623–2630. doi: 10.4315/0362-028x-68.12.2623. [DOI] [PubMed] [Google Scholar]
- Griffin PM. Escherichia coli O157:H7 and other enterohemorrhagic Escherichia coli. In: Blaser PDSMJ, Ravdin JI, Greenberg HB, Guerrant RL, editors. Infections of the gastrointestinal tract. New York: Raven Press; 1995. pp. 739–761. [Google Scholar]
- Griffin PM, Ostroff SM, Tauxe RV, Greene KD, Wells JG, Lewis JH, Blake PA. Illnesses associated with Escherichia coli O157:H7 infections. A broad clinical spectrum. Ann Intern Med. 1988;109:705–712. doi: 10.7326/0003-4819-109-9-705. [DOI] [PubMed] [Google Scholar]
- Gualdi L, Tagliabue L, Bertagnoli S, Ierano T, De Castro C, Landini P. Cellulose modulates biofilm formation by counteracting curli-mediated colonization of solid surfaces in Escherichia coli. Microbiology. 2008;154:2017–2024. doi: 10.1099/mic.0.2008/018093-0. [DOI] [PubMed] [Google Scholar]
- Hentzer M, Riedel K, Rasmussen TB, Heydorn A, Andersen JB, Parsek MR, Rice SA, Eberl L, Molin S, Høiby N, Kjelleberg S, Givskov M. Inhibition of quorum sensing in Pseudomonas aeruginosa biofilm bacteria by a halogenated furanone compound. Microbiology. 2002;148:87–102. doi: 10.1099/00221287-148-1-87. [DOI] [PubMed] [Google Scholar]
- Herold S, Siebert J, Huber A, Schmidt H. Global expression of prophage genes in Escherichia coli O157:H7 strain EDL933 in response to norfloxacin. Antimicrob Agents Chemother. 2005;49:931–944. doi: 10.1128/AAC.49.3.931-944.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jarvis KG, Kaper JB. Secretion of extracellular proteins by enterohemorrhagic Escherichia coli via a putative type III secretion system. Infect Immun. 1996;64:4826–4829. doi: 10.1128/iai.64.11.4826-4829.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawamura-Sato K, Shibayama K, Horii T, Iimuma Y, Arakawa Y, Ohta M. Role of multiple efflux pumps in Escherichia coli in indole expulsion. FEMS Microbiol Lett. 1999;179:345–352. doi: 10.1111/j.1574-6968.1999.tb08748.x. [DOI] [PubMed] [Google Scholar]
- Kimmitt PT, Harwood CR, Barer MR. Toxin gene expression by shiga toxin-producing Escherichia coli: the role of antibiotics and the bacterial SOS response. Emerg Infect Dis. 2000;6:458–465. doi: 10.3201/eid0605.000503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Landini P. Cross-talk mechanisms in biofilm formation and responses to environmental and physiological stress in Escherichia coli. Res Microbiol. 2009;160:259–266. doi: 10.1016/j.resmic.2009.03.001. [DOI] [PubMed] [Google Scholar]
- Lee JH, Lee J. Indole as an Intercellular Signal in Microbial Community. FEMS Microbiol rev. 2010;34:426–444. doi: 10.1111/j.1574-6976.2009.00204.x. [DOI] [PubMed] [Google Scholar]
- Lee J, Bansal T, Jayaraman A, Bentley WE, Wood TK. Enterohemorrhagic Escherichia coli biofilms are inhibited by 7-hydroxyindole and stimulated by isatin. Appl Environ Microbiol. 2007;73:4100–4109. doi: 10.1128/AEM.00360-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee J, Zhang XS, Hegde M, Bentley WE, Jayaraman A, Wood TK. Indole cell signaling occurs primarily at low temperatures in Escherichia coli. ISME J. 2008a;2:1007–1023. doi: 10.1038/ismej.2008.54. [DOI] [PubMed] [Google Scholar]
- Lee Y, Kim Y, Yeom S, Kim S, Park S, Jeon CO, Park W. The role of disulfide bond isomerase A (DsbA) of Escherichia coli O157: H7 in biofilm formation and virulence. Fems Microbiology Letters. 2008b;278:213–222. doi: 10.1111/j.1574-6968.2007.00993.x. [DOI] [PubMed] [Google Scholar]
- Lesic B, Lépine F, Déziel E, Zhang J, Zhang Q, Padfield K, Castonguay MH, Milot S, Stachel S, Tzika AA, Tompkins RG, Rahme LG. Inhibitors of pathogen intercellular signals as selective anti-infective compounds. PLoS Pathog. 2007;3:1229–1239. doi: 10.1371/journal.ppat.0030126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma Q, Wood TK. OmpA influences Escherichia coli biofilm formation by repressing cellulose production through the CpxRA two-component system. Environ Microbiol. 2009;11:2735–2746. doi: 10.1111/j.1462-2920.2009.02000.x. [DOI] [PubMed] [Google Scholar]
- Matthysse AG, Deora R, Mishra M, Torres AG. Polysaccharides cellulose, poly-beta-1,6-N-acetyl-D-glucosamine, and colanic acid are required for optimal binding of Escherichia coli O157:H7 strains to alfalfa sprouts and K-12 strains to plastic but not for binding to epithelial cells. Appl Environ Microbiol. 2008;74:2384–2390. doi: 10.1128/AEM.01854-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nataro JP, Kaper JB. Diarrheagenic Escherichia coli. Clin Microbiol Rev. 1998;11:142–201. doi: 10.1128/cmr.11.1.142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perna NT, Plunkett G, 3rd, Burland V, Mau B, Glasner JD, Rose DJ, Mayhew GF, Evans PS, Gregor J, Kirkpatrick HA, Posfai G, Hackett J, Klink S, Boutin A, Shao Y, Miller L, Grotbeck EJ, Davis NW, Lim A, Dimalanta ET, Potamousis KD, Apodaca J, Anantharaman TS, Lin J, Yen G, Schwartz DC, Welch RA, Blattner FR. Genome sequence of enterohaemorrhagic Escherichia coli O157:H7. Nature. 2001;409:529–533. doi: 10.1038/35054089. [DOI] [PubMed] [Google Scholar]
- Potera C. Forging a link between biofilms and disease. Science. 1999;283:1837–1839. doi: 10.1126/science.283.5409.1837. [DOI] [PubMed] [Google Scholar]
- Pratt LA, Kolter R. Genetic analysis of Escherichia coli biofilm formation: roles of flagella, motility, chemotaxis and type I pili. Mol Microbiol. 1998;30:285–293. doi: 10.1046/j.1365-2958.1998.01061.x. [DOI] [PubMed] [Google Scholar]
- Prigent-Combaret C, Prensier G, Le Thi TT, Vidal O, Lejeune P, Dorel C. Developmental pathway for biofilm formation in curli-producing Escherichia coli strains: role of flagella, curli and colanic acid. Environ Microbiol. 2000;2:450–464. doi: 10.1046/j.1462-2920.2000.00128.x. [DOI] [PubMed] [Google Scholar]
- Puttamreddy S, Cornick NA, Minion FC. Genome-wide transposon mutagenesis reveals a role for pO157 genes in biofilm development in Escherichia coli O157:H7 EDL933. Infect Immun. 2010 doi: 10.1128/IAI.00156-10. In press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reisner A, Krogfelt KA, Klein BM, Zechner EL, Molin S. In vitro biofilm formation of commensal and pathogenic Escherichia coli strains: impact of environmental and genetic factors. J Bacteriol. 2006;188:3572–3581. doi: 10.1128/JB.188.10.3572-3581.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ren D, Bedzyk LA, Ye RW, Thomas SM, Wood TK. Differential gene expression shows natural brominated furanones interfere with the autoinducer-2 bacterial signaling system of Escherichia coli. Biotechnol Bioeng. 2004;88:630–642. doi: 10.1002/bit.20259. [DOI] [PubMed] [Google Scholar]
- Rivas L, Dykes GA, Fegan N. A comparative study of biofilm formation by Shiga toxigenic Escherichia coli using epifluorescence microscopy on stainless steel and a microtitre plate method. Journal of Microbiological Methods. 2007;69:44–51. doi: 10.1016/j.mimet.2006.11.014. [DOI] [PubMed] [Google Scholar]
- Ryu JH, Beuchat LR. Biofilm formation by Escherichia coli O157:H7 on stainless steel: effect of exopolysaccharide and Curli production on its resistance to chlorine. Appl Environ Microbiol. 2005;71:247–254. doi: 10.1128/AEM.71.1.247-254.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saldaña Z, Xicohtencatl-Cortes J, Avelino F, Phillips AD, Kaper JB, Puente JL, Girón JA. Synergistic role of curli and cellulose in cell adherence and biofilm formation of attaching and effacing Escherichia coli and identification of Fis as a negative regulator of curli. Environ Microbiol. 2009;11:992–1006. doi: 10.1111/j.1462-2920.2008.01824.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sambrook J, Fritsch EF, Maniatis T. Molecular Cloning: A Laboratory Manual. Cold Spring Harbor Laboratory Press; Cold Spring Harbor, NY: 1989. [Google Scholar]
- Strockbine NA, Marques LR, Newland JW, Smith HW, Holmes RK, O’Brien AD. Two toxin-converting phages from Escherichia coli O157:H7 strain 933 encode antigenically distinct toxins with similar biologic activities. Infect Immun. 1986;53:135–140. doi: 10.1128/iai.53.1.135-140.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tarr PI, Gordon CA, Chandler WL. Shiga-toxin-producing Escherichia coli and haemolytic uraemic syndrome. Lancet. 2005;365:1073–1086. doi: 10.1016/S0140-6736(05)71144-2. [DOI] [PubMed] [Google Scholar]
- Uhlich GA, Cooke PH, Solomon EB. Analyses of the red-dry-rough phenotype of an Escherichia coli O157:H7 strain and its role in biofilm formation and resistance to antibacterial agents. Appl Environ Microbiol. 2006;72:2564–2572. doi: 10.1128/AEM.72.4.2564-2572.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X, Kim Y, Wood TK. Control and benefits of CP4-57 prophage excision in Escherichia coli biofilms. ISME J. 2009;3:1164–1179. doi: 10.1038/ismej.2009.59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wells JG, Davis BR, Wachsmuth IK, Riley LW, Remis RS, Sokolow R, Morris GK. Laboratory investigation of hemorrhagic colitis outbreaks associated with a rare Escherichia coli serotype. J Clin Microbiol. 1983;18:512–520. doi: 10.1128/jcm.18.3.512-520.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wells TJ, Sherlock O, Rivas L, Mahajan A, Beatson SA, Torpdahl M, Webb RI, Allsopp LP, Gobius KS, Gally DL, Schembri MA. EhaA is a novel autotransporter protein of enterohemorrhagic Escherichia coli O157:H7 that contributes to adhesion and biofilm formation. Environ Microbiol. 2008;10:589–604. doi: 10.1111/j.1462-2920.2007.01479.x. [DOI] [PubMed] [Google Scholar]
- Yoon Y, Sofos JN. Autoinducer-2 activity of gram-negative foodborne pathogenic bacteria and its influence on biofilm formation. J Food Sci. 2008;73:M140–147. doi: 10.1111/j.1750-3841.2008.00697.x. [DOI] [PubMed] [Google Scholar]
- Zogaj X, Nimtz M, Rohde M, Bokranz W, Römling U. The multicellular morphotypes of Salmonella typhimurium and Escherichia coli produce cellulose as the second component of the extracellular matrix. Mol Microbiol. 2001;39:1452–1463. doi: 10.1046/j.1365-2958.2001.02337.x. [DOI] [PubMed] [Google Scholar]