Summary
Asymmetric cell division requires the establishment of cortical cell polarity and the orientation of the mitotic spindle along the axis of cell polarity. Evidence from invertebrates demonstrates that the Par3/Par6/aPKC and NuMA/LGN/Gαi complexes, which are thought to be physically linked by the adapter protein mInscuteable (mInsc), play indispensable roles in this process. However, the molecular basis for the binding of LGN to NuMA and mInsc is poorly understood. The high resolution structures of the LGN/NuMA and LGN/mInsc complexes presented here provide mechanistic insights into the distinct and highly specific interactions of the LGN TPR repeats with mInsc and NuMA. Structural comparisons, together with biochemical and cell biology studies, demonstrate that the interactions of NuMA and mInsc with LGN are mutually exclusive, with mInsc binding preferentially. Our results suggest that the Par3/mInsc/LGN and NuMA/LGN/Gαi complexes play sequential and partially overlapping roles in asymmetric cell division.
Introduction
Asymmetric cell division (ACD), the process by which a mother cell gives rise to two distinct daughter cells, is a fundamental process widely used to regulate stem cell function and generate cellular diversity during development in metazoa (Cowan and Hyman, 2004; Morrison and Kimble, 2006; Neumuller and Knoblich, 2009; Siller and Doe, 2009). This process is governed by two mechanisms (Horvitz and Herskowitz, 1992). External cues, such as niche-derived signals or external polarity, surrounding mother cells can lead to asymmetric cell division (Lin, 2002; Morrison and Spradling, 2008; Zigman et al., 2005). Alternatively, asymmetric partitioning of cell fate determinants within mother cells (i.e. via the “intrinsic” mechanism that is independent of surrounding cells) can also cause cells to divide asymmetrically (Gonczy, 2008; Knoblich, 2008; Neumuller and Knoblich, 2009).
Drosophila neuroblasts (NB) provide an excellent model system for studying ACD. ACD generally involves three steps: the establishment of mother cell polarity, the orientation of mitotic spindles and the segregation of cell fate determinants. NBs inherit apical-basal polarity cues from the neuroepithelium, which contains the Par complex, an evolutionarily conserved tripartite complex composed of atypical protein kinase C (aPKC) (Wodarz et al., 2000), Par6 (Petronczki and Knoblich, 2001) and Bazooka (Baz, a Drosophila homologue of Par3) (Kuchinke et al., 1998). The Par complex is localized in a crescent at the apical cell cortex right below the overlaying epithelium (Kuchinke et al., 1998; Petronczki and Knoblich, 2001; Wodarz et al., 2000). Temporally, the Par proteins are the first molecules to localize to the apical cortices of cells. During late interphase and early prophase, Baz recruits the adaptor protein Inscuteable (Insc) (Kraut and Campos-Ortega, 1996; Kraut et al., 1996) which in turn recruits Partner of Inscuteable (Pins; its mammalian counterpart is LGN) to the apical cortex, as Insc can simultaneously bind to Baz (Schober et al., 1999; Wodarz et al., 1999) and Pins (Parmentier et al., 2000; Yu et al., 2000). The apical Pins then serves as a molecular linker to build up another evolutionarily conserved tripartite complex, Mud/Pins/Gαi (NuMA/LGN/Gαi in mammals), which functions in a receptor-independent G-protein pathway to orient mitotic spindles along the apical-basal axes of cells (Bowman et al., 2006; Izumi et al., 2006; Schaefer et al., 2001; Siller et al., 2006), likely via dynein-mediated pulling forces on astral microtubules (Siller and Doe, 2009), ensuring that the mitotic cleavage plane is perpendicular to the apical-basal axis.
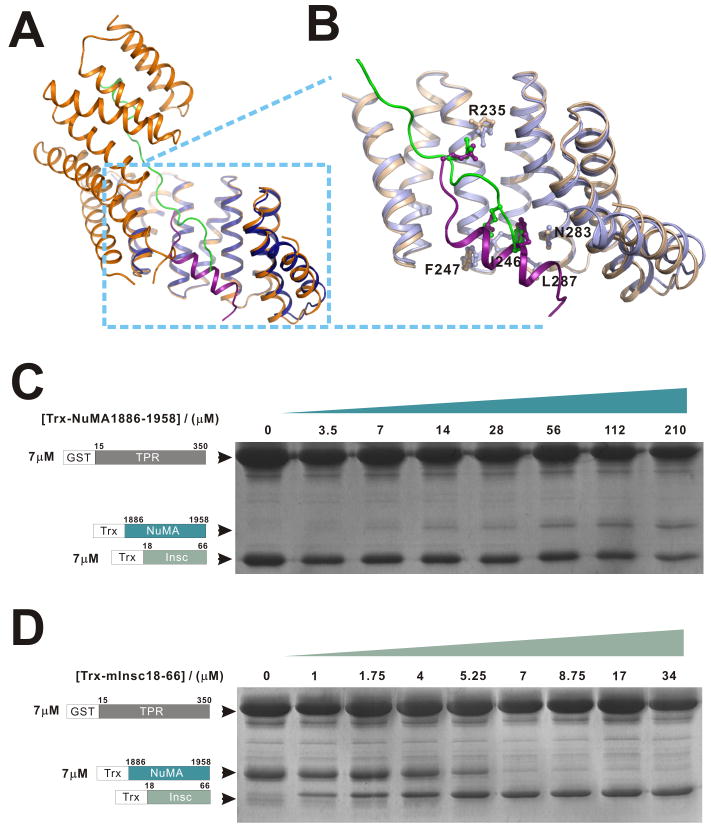
Pins associates with GDP-bound Gαi via the three GoLoco motifs at its C-terminus (Parmentier et al., 2000; Schaefer et al., 2001; Schaefer et al., 2000). Through the seven tetratricopeptide repeats (TPR) at its N-terminus, Pins localizes apically with the Par complex by binding to Insc (Yu et al., 2000). Using the same TPR repeats, Pins recruits Mud to the apical cortex, forming the Mud/Pins/Gαi complex to direct spindle orientation (Bowman et al., 2006; Izumi et al., 2006; Siller et al., 2006). The vertebrate NuMA/LGN/Gαi complex appear to function in a similar manner in orchestrating ACD (Du et al., 2001; Du et al., 2002; Poulson and Lechler, 2010; Williams et al., 2011; Zigman et al., 2005). Recently, ectopically expressed mInsc has been shown to co-localize with LGN in the developing epidermis to regulate ACD of the epidermis (Poulson and Lechler, 2010; Williams et al., 2011) and retina (Zigman et al., 2005). However, it is still unclear whether mInsc functions in the same way as the Drosophila counterpart. Additionally, the molecular basis of LGN-mediated ACD protein complex assembly is largely unclear. Detailed biochemical and structural studies are urgently needed to understand the molecular mechanisms by which LGN connects the Par/mInsc and NuMA/Gαi complexes to coordinate cell polarization and spindle orientation during ACD.
In this work, we characterize the interactions of LGN with mInsc and NuMA in detail. The structures of the LGN/mInsc and LGN/NuMA complexes solved here reveal that LGN binds to the two target proteins with distinct mechanisms. We further demonstrate that the interactions of mInsc and NuMA with LGN are mutually exclusive. Interestingly, NuMA cannot bind to LGN in the presence of mInsc, although NuMA and mInsc display comparable affinities for LGN. The competitive binding of mInsc and NuMA to LGN is consistent with recent findings showing that the Par/Insc interaction pathway and the NuMA/LGN/Gαi pathway may function independently in ACDs.
Results
The Interaction between LGN and NuMA
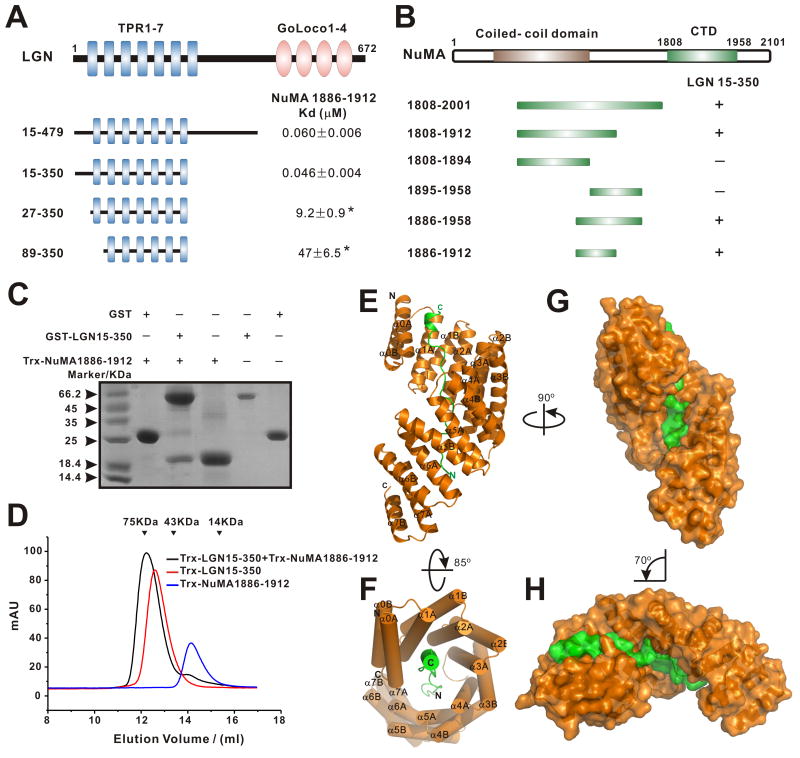
Recent studies have shown that Drosophila Mud binds robustly to the Pins TPR1-7 repeats (Bowman et al., 2006; Izumi et al., 2006; Siller et al., 2006). In mammals, LGN recruits NuMA to the cell cortex by binding to the C-terminal tail of NuMA (aa 1878-1910) through its seven TPR repeats, and this interaction regulates spindle orientation during mitosis (Du and Macara, 2004; Du et al., 2002; Zheng et al., 2010). We first confirmed the LGN/NuMA interaction by showing that a LGN fragments (aa 15-479) which contains a part of the N-terminus (aa 15-49), TPR1-7 (aa 50-350), and the linker between the TPR repeats and the Goloco motifs binds to a C-terminal fragment of NuMA with a Kd ∼60 nM (Fig. 1A). Deletion of the LGN linker (aa 351-479) did not have any observable impact on the binding of LGN to NuMA (Fig. 1A). Interestingly, a slightly shorter fragment of LGN (aa 27-350), which still contains all 7 TPR repeats (Figs. S1&3A), showed a ∼200 fold decrease in NuMA binding (Fig. 1A and Figs. S2A&B). These data indicate that an extension sequence N-terminal to TPR1-7 in LGN plays an important role in the interaction between LGN and NuMA. As expected, the removal of TPR1 further decreased the binding of LGN to NuMA (Fig. 1A). Using a similar truncation-based approach, we mapped the minimal LGN-binding region of NuMA to a 27-residue fragment (aa 1886-1912; Figs. 1B&C). We further showed that LGN and NuMA form a stable 1:1 stoichiometric complex (Fig. 1D and Fig. S2).
Figure 1. Interaction between LGN and NuMA and the structure of the LGN/NuMA complex.
(A) Fluorescence- and ITC-based measurements of the bindings between the LGN TPR repeats and a C-terminal fragment of NuMA (aa 1886-1912). The binding affinities between LGN TPR0-7 and NuMA were obtained by titrating LGN TPR repeats to a FITC-labeled NuMA peptide. Binding affinities obtained by ITC-based assay are indicated by asterisks. (B) Analytical gel-filtration-based analysis of the bindings of NuMA fragments to LGN TPR0-7. (C) Pull-down assays showing that GST-LGN(15-350) robustly binds to Trx-NuMA(1886-1912). (D) Analytical gel-filtration analysis showing that Trx-LGN(15-350) and Trx-NuMA(1886-1912) formed a 1:1 stoichiometric complex. The elution volumes of the molecular mass standards are indicated at the top of the panel. (E) Ribbon diagram representation of the LGN (orange)/NuMA (green) complex viewed from the side. (F) Cylinder representation of the LGN/NuMA complex structure viewed from the top. (G)&(H) Surface representations of the LGN/NuMA complex with their orientations corresponding to those shown in panels (A)& (B), respectively. Also see Figs. S1-3.
Crystal structure of the LGN/NuMA complex
To understand the molecular mechanism underlying the LGN/NuMA interaction, we solved the crystal structure of the LGN (aa 15-350)/NuMA peptide (aa 1885-1912) complex at 2.3 Å resolution (Table 1). The structure of LGN(15-350) is composed of sixteen α-helices arranged into eight sequential helix-turn-helix repeats (Figs. 1E&F). The entire length of LGN(15-350) is well resolved except for the four residues in the loop connecting αA and αB of TPR3 (aa 58-61) and six residues at the C terminus. Most of the residues of the NuMA peptide are also clearly defined. The NuMA peptide adopts an extended conformation occupying essentially the entire concave channel formed by the 7 TPR repeats as well as a pseudo-TPR repeat (referred to as TPR0) formed by the N-terminal extension of LGN, burying a total surface area of 4896 Å2 (Figs. 1E&F). The C- and N-terminal ends of the NuMA peptide make contact with the N- and C-terminal TPR repeats, respectively.
Table 1. Crystallographic Data and Refinement Statistics.
| Data collection | ||
| Data sets | LGN/mInsc | LGN/NuMA |
| Space group | C2 | P6122 |
| Unit cell (Å) | a=88.196, b=75.606, c=35.346 |
a=91.305, b=91.305, c=178.376 |
| Resolution range (Å) | 50.00-1.10 (1.12-1.10) | 50.00-2.30 (2.34-2.30) |
| No. of unique reflections | 84934 (2703) | 20308 (984) |
| Rmerge (%) | 4.8 (51.1) | 8.7 (65.6) |
| I/s | 29.7(2.4) | 35.5 (4.9) |
| Redundancy | 5.3 (3.2) | 17.6 (16.6) |
| Completeness (%) | 94.4 (60.6) | 99.7 (99.9) |
|
| ||
| Refinement | ||
| Resolution (Å) | 20.00-1.10 (1.16-1.10) | 36.1-2.30 (2.36-2.30) |
| Rcryst/Rfree (%) | 12.0 (21.1)/15.0 | 21.5(22.9)/27.1(30.3) |
| No. of atoms | ||
| Proteins | 1536 | 2671 |
| Water | 316 | 103 |
| Other atoms | 14 | 18 |
| No. of reflections | ||
| Working set | 80648 | 18723 |
| Test set | 4259 | 1585 |
| Mean B factor of protein/peptide | ||
| Main chain | 11.0/14.5 | 49.8/90.4 |
| Side chain | 15.1/22.9 | 51.8/93.7 |
| R.m.s. deviations | ||
| Bond length (Å) | 0.014 | 0.008 |
| Bond angles (°) | 0.030 | 1.00 |
| Ramachandran plot (%) | ||
| Most favored | 98.63 | 97.08 |
| Additionally allowed | 1.37 | 2.34 |
| generously allowed | 0 | 0.58 |
Numbers in parentheses represent the value for the highest resolution shell.
Rmerge = SUM (ABS(I - <I>)) / SUM (I), where I is the intensity of measured reflection and <I> is the mean intensity of all symmetry-related reflections.
Rcryst= Σ|Fcalc − Fobs|/ΣFobs,where Fobs and Fcalc are observed and calculated structure factors.
Rfree= ΣT|Fcalc − Fobs|/ΣFobs, where T is a test data set of about 5% of the total unique reflections randomly chosen and set aside prior to refinement.
B factors are caculated by combining the residual B-factor and TLS parameters using TLSANL program in CCP4.
The TPR repeats of LGN exhibit a number of features distinct from those of other TPR proteins with known structures. Most of the α helices in LGN-TPR are composed of 18 residues, four residues longer than those of canonical TPR motifs. The αA and αB helices of LGN TPR3 are composed of 20 and 26 residues, respectively, and an extended 14-residue linker connects the two helices (Fig. S3A). Like other TPR domains (Das et al., 1998; Jinek et al., 2004; Scheufler et al., 2000; Wang et al., 2009; Zhang et al., 2010), the eight TPR units of LGN are arranged in parallel with adjacent α-helices antiparallel to each other, crossing at an angle of ∼20° to create a right-handed superhelix (Table S1). The LGN TPR superhelix is ∼80 Å long and 35 Å wide, with a pitch height of 50 Å (Figs. S3B&D). One complete superhelical turn in LGN is comprised of six TPR repeats, whereas one superhelical turn in the O-linked GlcNAc transferase (OGT) contains seven TPR repeats (Fig. 1F, and Fig. S3B-D) (Jinek et al., 2004). This is because the inter-helix angles between the TPR repeat of LGN are significantly larger than those of conventional TPR repeats (Table S1). The formation of the superhelix creates an elongated target binding channel along the concave inner surface of the LGN TPR repeats (Fig. 1E-H). The full-length Cdc16 (or human APC6) has 14 TPR repeats and interaction between its C-terminal 8 TPR repeats with a peptide fragment of Cdc26 observed in the structures of the Cdc16/Cdc26 complex (Zhang et al., 2010; Wang et al., 2009) shares some similarities with that between LGN and NuMA reported in this study (Fig. S3E-G). Firstly, in both complexes, the targets of TPR repeats adopt an extended conformation binding to the inner groove of the TPR superhelix. Secondly, analogous to the NuMA peptide, the C-terminal end of Cdc26 also forms an α-helix. Interestingly, the Cdc26 helix pairs with the orphan helix 15A of Cdc16 TPR, as if this helix complements helix 15A forming an additional TPR motif (Fig. S3G). The differences between the two complexes are nonetheless obvious. For example, the Cdc16 TPR superhelix is even shorter (five repeats per superhelical turn; Fig. S3E). Additionally, NuMA binds to the LGN TPR superhelix in an antiparallel manner, whereas Cdc26 binds to Cdc16 in parallel (Fig. S3F-G).
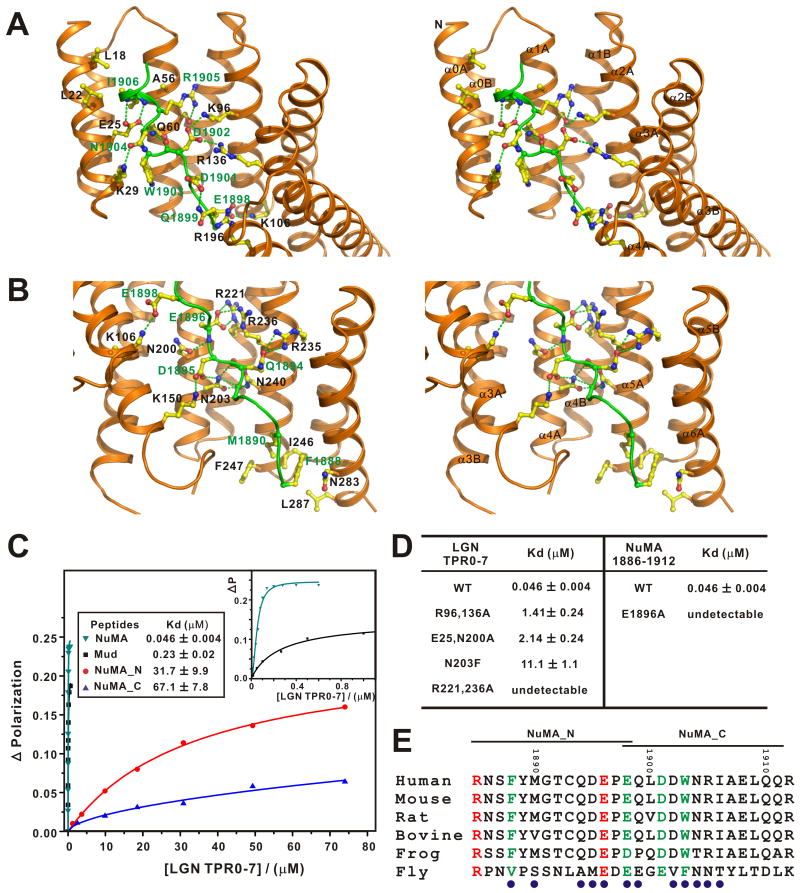
The LGN/NuMA interface can be divided into two regions based on the organization of the TPR superhelix (Figs. 2A&B): (I) TPR0-3 and the C-terminal half of the NuMA peptide (Fig. 2A), and (II) TPR4-7 and the N-terminal half of the NuMA peptide (Fig. 2B). The structure clearly shows that the pseudo-TPR0 makes contacts with the C-terminus of NuMA peptide at several points (e.g. Lys29 and Glu25, form hydrogen bonds with the side chain of Asn1904NuMA and the backbone amide nitrogen of Ala1907NuMA, respectively), explaining why TPR0 is critical for the interaction between LGN and NuMA. The binding of LGN to NuMA is mainly mediated by an extensive network of hydrogen bonds and electrostatic interactions. Consistent with the interactions observed in the complex structure, mutations that disrupt these polar interactions weaken the LGN/NuMA interaction (Fig. 2D). For example, substitution of E1896 NuMA with Ala or replacement of R221&236 in LGN with Ala both lead to the complete disruption of the LGN/NuMA complex formation (Fig. 2D). A significant decrease in LGN/NuMA binding was observed when Asn203 of LGN (corresponding to Asn226 of Drosophila Pins) was substituted with Phe (Fig. 2D). This is in agreement with previous findings showing that N226F-Pins has compromised binding to Mud, and the mutant Pins fails to align mitotic spindles along the apical-basal axes of fly S2 cells (Johnston et al., 2009). To evaluate the contributions of the two packing regions to LGN/NuMA complex formation, we divided the NuMA peptide into its “N” (NuMA_N, 1885-1898) and “C” parts (NuMA_C, 1898-1912). The binding affinities of NuMA_N and NuMA-C to LGN were 31.7 and 67.1 μM, respectively (Fig. 2C), both much lower than the affinity of the full length peptide. These structure and mutagenesis-based data suggest that the strong LGN/NuMA interaction results from a combination of numerous relatively weak charge-charge and hydrogen bonding interactions interactions along the LGN TPR repeats and the NuMA peptide. Therefore, the LGN/NuMA interaction is highly specific. However, this also implies that a ligand that binds strongly to a smaller section of LGN can displace NuMA despite having an overall LGN affinity similar to that of NuMA – this turns out to be how mInsc displaces NuMA from LGN, as we show below.
Figure 2. The interaction interface of the LGN/NuMA complex.
The LGN/NuMA interface is divided into two parts corresponding to LGN TPR0-3/NuMA_C (A) and LGN TPR4-7/NuMA_N (B). The interaction details between LGN and NuMA in the two parts are shown in stereo-view. The sidechains of the residues involved in the inter-domain interactions are drawn in the stick model. Charge-charge and hydrogen bonding interactions are highlighted by dashed lines in green. (C) Fluorescence polarization-based measurement of the binding affinities of LGN TPR0-7 to various NuMA and Mud peptides (sequences shown in panel E). The insert shows the expanded view of the binding curves of the NuMA and Mud peptides to LGN TPR0-7. (D) Summary of the bindings of LGN TPR0-7 and its mutants with the NuMA peptides. (E) Sequence alignment of the NuMA peptide showing that the residues involved in making contact with LGN are evolutionary conserved. The residues involved in the LGN interaction are indicated with blue circles.
Amino acid sequence alignment analysis reveals that most of the key residues involved in the LGN/NuMA interaction are evolutionary conserved, indicating that the main features of the LGN/NuMA interaction mode observed here can be extended to corresponding interactions in other species (Fig. 2E and Fig. S1). Supporting this notion, we found that a corresponding Mud peptide can bind to LGN with an affinity Kd ∼ 0.23 μM, similar to the affinity between the NuMA peptide and LGN (Fig. 2C).
NuMA targets LGN to mitotic spindles
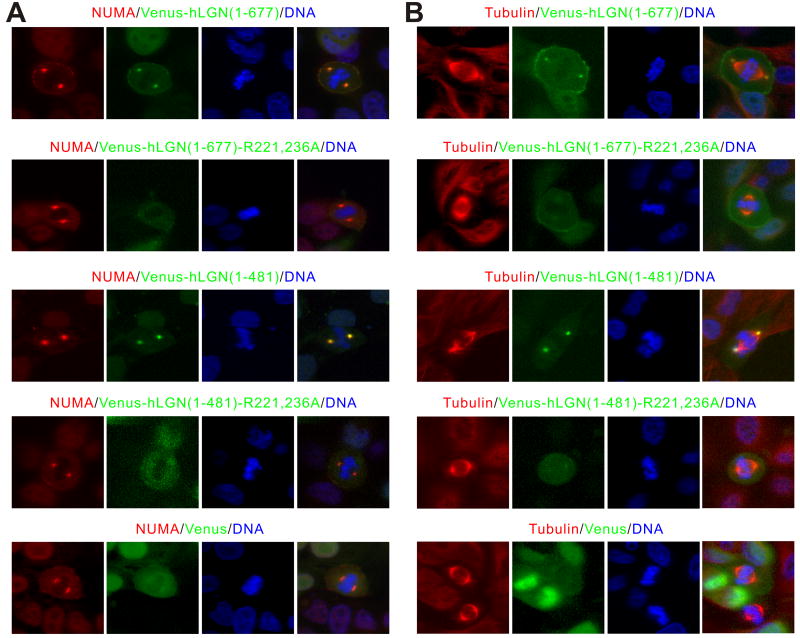
The structure of the LGN/NuMA complex obtained here allows us to design point mutations to specifically disrupt the LGN/NuMA interaction and to assess the impact of these mutations on mitotic spindle formation. We chose to use the human LGN-R221,236A (hLGN-R221,236A) mutant to test this, as its mouse counterpart has been shown to have no detectable binding to NuMA (Fig. 2D) although the mutant can still bind to Insc in vitro (data not shown). In MDCK cells transfected with Venus-hLGN(1-677), LGN colocalizes with endogenous NuMA at spindle poles as well as cell cortices in mitotic cells (Fig. 3A) as shown earlier (Du and Macara, 2004). In contrast, the Venus-hLGN(1-677)-R221,236A mutant failed to localize at spindle poles, but the spindle pole localization of NuMA was unaltered (Fig. 3A). This finding reveals that the interaction between TPR and NuMA is required for the spindle pole localization of LGN. Interestingly, the cortical localization of NuMA was noticeably diminished in Venus-hLGN(1-677)-R221,236A expressing cells (Fig. 3A), in agreement with earlier findings showing that LGN, most likely via its C-terminal GoLoco repeats, tethers NuMA to cell cortices (Du and Macara, 2004). Consistent with our findings in full-length LGN, the wild type Venus-tagged N-terminal half of hLGN(1-481) colocalizes well with NuMA at spindle poles, whereas Venus-hLGN(1-481)-R221,236A failed to do so (Fig. 3A). Importantly, endogenous NuMA failed to localize to cell cortices in cells expressing the wild type Venus-hLGN(1-481), presumably due to the dominant sequestration effect caused by the TRP repeats of LGN. Notably, weak cortical localization of NuMA can still be observed in cells expressing Venus-hLGN(1-481)-R221,236A (Fig. 3A), which can be attributed to the endogenous LGN/NuMA interaction.
Figure 3. NuMA targets LGN to spindle poles via binding to the TPR repeats.
(A) Over-expressed Venus-hLGN(1-677) and Venus-hLGN(1-481) co-localized with the endogenous NuMA at spindle poles in mitotic MDCK cells. In contrast, no NuMA co-localization could be detected in cells expressing Venus-hLGN(1-677)-R221,236A or Venus-hLGN(1-481)-R221,236A. Cells were fixed and stained using anti-NuMA antibody 24 hrs after transfection. DNA was stained with Hoechst 33342. (B) Over-expression of Venus-hLGN(1-677) or Venus-hLGN(1-481), but not the corresponding NuMA-binding defective LGN mutants, led to separation of spindle poles from centrosomes and misalignment of chromosomes in mitotic MDCK cells. Cells were fixed and stained using anti-α-tubulin antibody 24 hrs after transfection.
We also co-stained MDCK cells expressing various forms of LGN with anti-α-tubulin antibody (Fig. 3B). It is noteworthy that over-expression of the wild type Venus-hLGN(1-677) or Venus-hLGN(1-481) causes the separation of centrosomes from spindle poles and the misalignment of chromosomes (Fig. 3B), as has been previously observed in LGN-overexpressing mitotic cells (Du et al., 2001). These phenotypes, however, were not observed in cells expressing Venus-hLGN(1-677)-R221,236A or Venus-hLGN (1-481)-R221,236A (Fig. 3B), suggesting that they are caused by the binding of over-expressed LGN to endogenous NuMA, which may interfere with the association between NuMA and microtubules as we proposed previously (Du et al. 2002).
The Interaction between LGN and mInsc
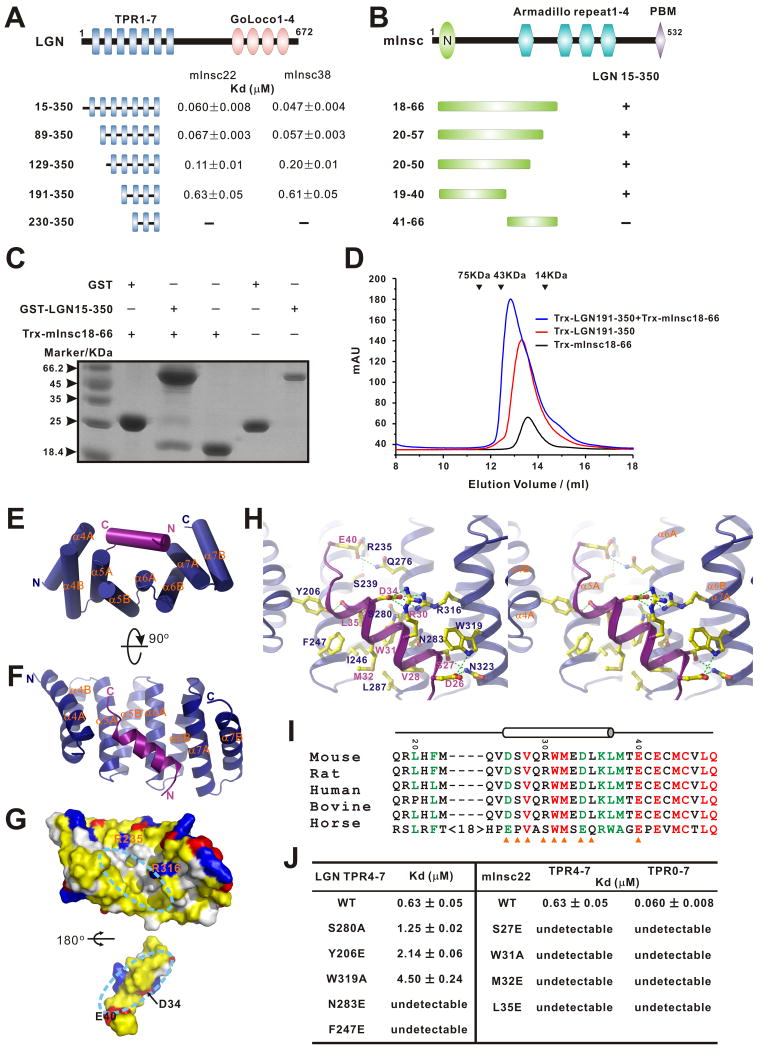
We next studied the interaction between LGN and mInsc in detail. To map the LGN/mInsc interaction regions more precisely, we made a series of truncation mutants of the LGN TPR repeats and mouse Insc. A direct binding experiment showed that TPR0 and TPR1 do not contribute to LGN/mInsc binding, whereas deletions up to the N-terminal four TPR repeats (TPR0-3) decreased binding affinity by ∼10 fold. Further deletion of TPR4 completely disrupts binding between LGN and mInsc (Fig. 4A-D). Thus, we conclude that TPR2-7 is sufficient for mInsc binding, and the last four TPR repeats of LGN (TPR4-7, aa 191-350) contain most of the mInsc binding region.
Figure 4. Interaction between LGN and mInsc and the structure of the LGN/mInsc complex.
(A) Fluorescence polarization-based measurements of the bindings between various LGN TPR fragments and the two mInsc peptides (mInsc22 and mInsc38). (B) Analytical gel-filtration-based mapping of the minimal LGN binding region of mInsc. (C) Pull-down assays showing that GST-LGN(15-350) robustly binds to an N-terminal fragment of mInsc containing residues of 18-66. (D) Analytical gel-filtration analysis showing that Trx-LGN(191-350) forms a 1:1 stoichiometric complex with mInsc(18-66). (E&F) Cylinder ribbon diagram representations of the LGN TPR4-7 (blue)/mInsc22 (magenta) complex viewed from two angles. (G) Open-book view of the LGN TPR4-7/mInsc22 complex showing the surface complementation between the TPR repeats and mInsc22. In this drawing, the hydrophobic residues are in yellow, the positively charged residues in blue, the negatively charged residues in red, and the rest of the amino acids in grey. The orientation of TPR4-7 is the same as in panel F. (H) Stereo views showing the interaction details between LGN TPR4-7 and mInsc22. The charge-charge and hydrogen bonding interactions are highlighted by dashed lines in green. (I) Sequence alignment of the LGN binding domain of Insc. In this alignment, the absolutely conserved amino acids are highlighted in red, and the highly conserved residues are in green. The residues involved in the LGN interaction are indicated with orange triangles. (J) Summary of the bindings between various LGN TPR repeats and different mInsc22 peptides derived from fluorescence-based assays. Also see Fig. S4.
Using a similar truncation-based approach, we found that a 38-residue peptide fragment in the N-terminal end of mInsc (aa 20-57, referred to as mInsc38) is sufficient for LGN binding. The minimal binding region was further narrowed down to a 22-residue fragment (aa 19-40, referred to as mInsc22; Figs. 4A&B). We also demonstrated that LGN and mInsc form a 1:1 stoichiometric complex with a dissociation constant of 47 nM (Figs. 4A&D).
Crystal structure of the LGN/mInsc complex
To understand how LGN and mInsc bind to each other, we tried to determine the crystal structure of LGN TPR0-7 in complex with the 38-residue fragment of mInsc but our efforts failed. However, we succeeded in obtaining crystals of the TPR4-7/mInsc22 peptide complex, and the structure was solved to a resolution of 1.1 Å (Table 1). All of the amino acid residues in the TPR4-7/mInsc22 complex have clearly defined electron densities. The structure of LGN TPR4-7 is composed of eight α-helices arranged into four sequential TPR repeats (Figs. 4E&F). Unlike the extended structure observed in the LGN-bound NuMA peptide, the mInsc22 peptide forms an α-helix that sits snugly at the center of the concave inner surface of the LGN TPR4-7 repeats. The mInsc helix make contact with all four of the TPR repeats, with its N- and C-termini pointing to TPR7 and TPR4 of LGN, respectively (Figs. 4E&F). The LGN/mInsc packing interface is dominated by extensive hydrophobic interactions, burying a total of ∼2665 Å2 surface area (Fig. 4G). Specifically, the side chains of L35mInsc and M32mInsc make contact with the side chains of I246LGN and F247LGN; the side chain of W31mInsc interacts with I246LGN and L287LGN, and V28mInsc interacts with both T286LGN and L287LGN (Fig. 4H). In addition to these hydrophobic interactions, several pairs of hydrogen bonding and charge-charge interactions further stabilize the LGN/mInsc complex (see Fig. 4H for details). Importantly, most of the residues involved in the formation of the interface between Pins and Insc are highly conserved from flies to humans (Fig. 4I and Fig. S1), implying that the LGN/mInsc interaction observed in the current study also occurs in corresponding proteins in other species.
We performed a series of mutagenesis studies to validate the interactions observed in the TPR4-7/mInsc22 complex structure. To probe the extensive hydrophobic interactions, we substituted Asn283 (a polar residue completely buried in the hydrophobic interface of the complex), Phe247 from LGN, and Met32, Leu35 from mInsc individually with Glu; every one of these substitution mutations abolished the LGN/mInsc interaction (Fig. 4J). Disruption of the hydrogen bonds or the salt bridge networks in the LGN/mInsc interface, such as the S280A, Y206E, W319A mutations from LGN, and S27E, W31A mutations from mInsc, led to decreased or abolished binding between TPR4-7 and mInsc22 (Fig. 4J). Since the mInsc38 and mInsc22 peptides display indistinguishable binding affinities to various LGN TPR repeats (Fig. 4A) and TPR4-7 binds to mInsc22 with only a slightly weaker affinity than TPR0-7 to mInsc38, the TPR4-7/mInsc22 peptide complex obtained here is likely to be a good representation of the interaction between the two proteins in their full-length forms. Consistent with this notion, various mInsc22 mutant peptides showed identical binding profiles towards LGN TPR4-7 and TPR0-7 (Fig. 4J). Nonetheless, it is formally possible that the interaction between the two proteins in their full length forms may be more complicated than shown here.
mInsc can displace NuMA from LGN
The biochemical and structural data above demonstrate that the TPR repeats of LGN can specifically bind to mInsc and NuMA with comparable macroscopic binding affinities. We next asked whether LGN can bind to mInsc and NuMA simultaneously. To address this question, we compared the structures of the LGN/mInsc and LGN/NuMA complexes. The conformations of TPR4-7 in the LGN/mInsc and LGN/NuMA complexes are highly similar (r.m.s. deviation of 0.57 Å between 127 equivalent Cα atoms, Fig. 5A). The binding sites of the N-terminal half of the NuMA peptide and the mInsc peptide on LGN TPR4-7 overlap with each other. A close-up view of the TPR4-7 regions of the two complexes reveals that several residues from LGN TPR4-7 (e.g. Arg235, I246, F247, Asn283, and L287) are involved in binding to both mInsc and NuMA (Fig. 5B). This structural analysis suggests that the LGN TPR repeats are not likely to be able to bind to mInsc and NuMA simultaneously, and this prediction is directly supported by our biochemical experiments. When LGN TPR0-7 was incubated with both mInsc and NuMA in a 1:1:1 molar ratio, only mInsc was found in complex with LGN (Fig. S4). In vitro competition experiments further showed that sub-stoichiometric amounts of mInsc can effectively compete with NuMA for binding to LGN, whereas excess amounts of NuMA (e.g. even at a 10:1 molar ratio of NuMA to mInsc) cannot compete with mInsc for binding to LGN (Figs. 5C&D). This result seems odd, as the macroscopic dissociation constants of the LGN/NuMA and LGN/mInsc complexes are comparable (Figs. 1A&4A). However, the structures of the two complexes nicely explain these apparently contradicting observations. In the LGN/mInsc complex, the mInsc peptide forms a single α-helix, and the interaction surface is concentrated within TPR4-7. In contrast, the NuMA peptide spans the entire TPR0-7 of LGN, and the interaction between NuMA and LGN can be viewed as a coupling of multiple discrete weak sites into a thermodynamically strong macroscopic interaction. Consistent with this analysis, fitting of the LGN/NuMA binding curve requires a Hill coefficient of ∼2.0, indicating that multiple binding sites are conformationally coupled to each other (Fig. 2C). In contrast, the LGN/mInsc binding reaction in Fig. 4A can be fitted perfectly with the simple one-site binding mode of the complex. Thus, upon addition of the mInsc peptide, the N-terminal half of the NuMA peptide is displaced from the LGN/NuMA complex. The interaction between the C-terminal half of the NuMA peptide and LGN is very weak, and the NuMA peptide can then dissociate completely from LGN (Fig. 2C). Consistent with the competitive binding mode shown in Fig. 5, two mInsc mutants, “W31A” and L35E”, both are defective in binding to LGN as shown in Fig. 4J, are incapable of competing with NuMA for LGN (Fig. S5).
Figure 5. mInsc can displace NuMA from LGN.
(A) Ribbon diagram representation showing the superposition of the LGN/mInsc and LGN/NuMA complexes with the same coloring schemes as in Figs. 1&4. (B) An enlarged view showing the comparison of the LGN/mInsc and LGN/NuMA interfaces. A selected set of amino acid residues, which are in the inter-domain interfaces and share similar positions in the two complexes, are drawn in the stick model. (C) NuMA cannot compete with mInsc in binding to LGN TPR0-7. In this experiment, the total amounts of GST-LGN TPR0-7 and Trx-mInsc were fixed (7 μM), and the concentrations of Trx-NuMA were increased to as high as 210 μM. (D) Sub-stoichiometric amount of mInsc can displace NuMA from LGN. In this experiment, the amounts of GST-LGN TPR0-7 and Trx-NuMA in each assay were fixed at 7 μM, and the concentrations of Trx-mInsc were gradually increased. GSH-Sepharose beads pull-down experiment was used to assay the formations of the LGN/mInsc or LGN/NuMA complexes. Also see Fig. S5 and Fig. S7.
mInsc can block the association of LGN with NuMA in MDCK cells to cause spindle mis-orientation and defective cystogenesis
Our biochemical and structural data above clearly show that NuMA and mInsc are mutually exclusive in binding to LGN and that mInsc can displace NuMA from LGN. We used the MDCK cystogenesis as a model to verify this observation in vivo and to explore its functional implications. During cystogenesis, LGN is restricted to the lateral cell cortices of mitotic cells and functions to direct spindle orientation perpendicular to the apical-basal axis, which is critical for normal cystogenesis (Hao et al., 2010; Zheng et al., 2010). Firstly, we tested the effects of ectopic expression of full length mInsc on cystogenesis in MDCK cells. Interestingly, unlike in Drosophila epithelial cells, ectopic Venus-mInsc shows diffused cytoplasmic distribution and does not accumulate at the apical cortices of MDCK cells during cystogenesis (Fig. S6). This is probably due to the differential localization of endogenous Par3 in the two systems. In Drosophila epithelial cells, Baz localizes at the entire apical membranes. In contrast, in MDCK cells, Par-3 is restricted to the tight junctions (Bryant et al., 2010). Concomitantly, we did not observe apical accumulation of endogenous LGN in cells expressing Venus-mInsc, although the cortical localization of LGN appears weaker (Fig. S6A). Ectopic expression of Venus-mInsc, however, results in cystogenesis defects. The portion of cysts that could not form a single normal lumen significantly increased in Venus-mInsc cysts (44±3.0%), compared with control cysts (19±5.0%) (Fig. S6B). Spindle orientation is also more randomized in Venus-mInsc expressing cells during cystogenesis (Fig. S6C).
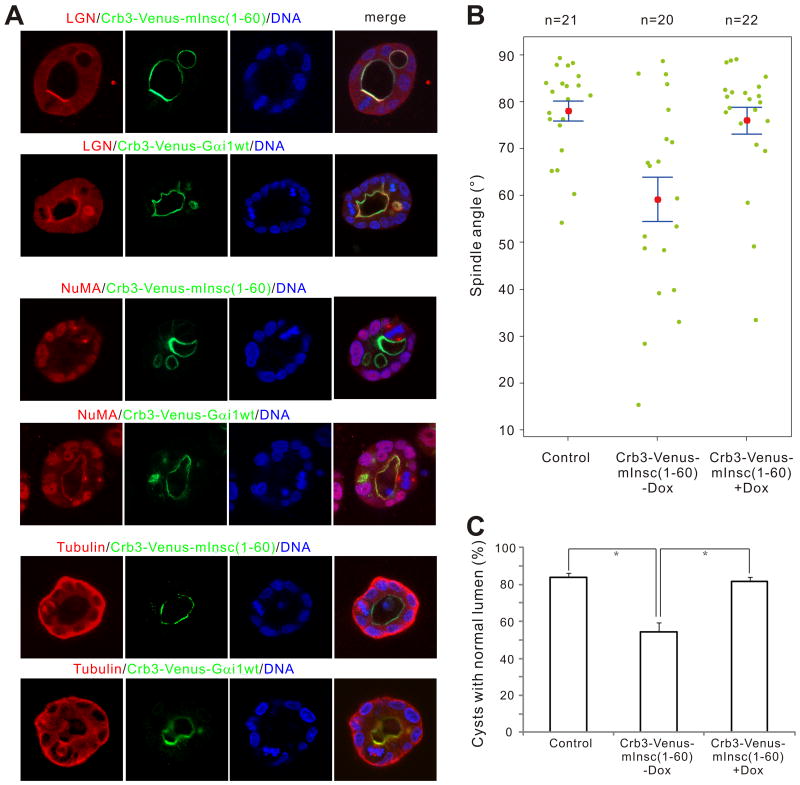
We recently showed that forced recruitment of endogenous LGN to the apical cell cortex via Crumbs3 (Crb3)-mediated apical targeting of the wild-type Gαi1 led to a nearly 90 degree rotation of the mitotic spindle and profound defects in cystogenesis (Zheng et al., 2010). We applied a similar approach to test whether artificial targeting of mInsc to apical cell cortices could change the localization of endogenous LGN, and whether this would have any effects on spindle orientation and cystogenesis. We fused the N-terminal 60-residue fragment of mInsc to the C-terminal tail of Venus-Crb3, an apical maker in polarized MDCK cells (Roh et al., 2003), and stable Tet-Off cell lines expressing Crb3-Venus-mInsc(1-60) were established. As predicted, the resulting Crb3-Venus-mInsc(1-60) was targeted predominantly to the apical sides of cell cortices when expressed in 3D cultured MDCK cysts (Fig. 6A). The apical targeting of mInsc(1-60) led to the concurrent apical targeting of endogenous LGN, similar to Crb3-Venus-Gαi1 expressing cells (Fig. 6A, top two panels). However, the localization patterns of endogenous NuMA in Crb3-Venus-mInsc(1-60) and Crb3-Venus-Gαi1 expressing cells are very different. The apical expression of Crb3-Venus-Gαi1 also leads to the apical localization of NuMA (Fig. 6A, middle panel), and this can be easily explained by the binding of NuMA to the TPR repeats of LGN. In contrast, no apical localization of NuMA could be detected in Crb3-Venus-mInsc(1-60)-expressing cells (Fig. 6A), suggesting that when LGN is bound to mInsc, it fails to recruit NuMA, further supporting our hypothesis that mInsc and NuMA bind to LGN in a mutually exclusive manner.
Figure 6. Apical targeting of mInsc(1-60) leads to apical recruitment of endogenous LGN and subsequent spindle misorientation and defective cystogenesis in MDCK cysts.
(A) MDCK cells stably expressing Crb3-Venus-mInsc(1-60) or Crb3-Venus-Gαi1 were cultured in matrigel. After culturing for 4 days in the absence of doxycycline, cysts were fixed and stained with anti-LGN, anti-NuMA or anti-Tubulin antibodies, respectively. DNA was stained with Hoechst 33342. Single confocal images from the middle of the cysts are shown. (B) Scatter diagram of the metaphase spindle angles in control or Crb3-Venus-mInsc(1-60) cysts in the absence (-Dox) or presence (+Dox) of 20 ng/ml of doxycycline. Results were from three independent experiments. Pink dots indicate mean values, green dots indicate individual data points, and error bars represent the SEM of the total number of spindles analyzed. (C) Quantification of cysts with single normal lumen from MDCK cells expressing Venus (control) or Crb3-Venus-mInsc(1-60) in the absence (-Dox) or presence (+Dox) of 20 ng/ml of doxycycline. Values are mean ± SD from three independent experiments, n > 100 cysts/experiment. *P < 0.01. Also see Fig. S6.
As one would expect, the expression of Crb3-Venus-mInsc(1-60) leads to defective cystogenesis, with a significant proportion of cysts (45.5± 4.8 %) containing abnormal lumens (Figs. 6A&C). The cystogenesis defects in Crb3-Venus-mInsc(1-60) cysts are similar to those in LGN knockdown cysts but are not as severe as the defects in Crb3-Venus-Gαi1 cysts (Zheng et al., 2010). Forced apical targeting of endogenous LGN by Crb3-Venus-Gαi1 rotates the spindle by nearly 90 degrees relative to the apical-basal axis, suggesting that LGN functions with Gαi to direct spindle orientation, probably via TRP repeats-mediated binding to NuMA (Zheng et al., 2010). However, in cells expressing Crb3-Venus-mInsc(1-60), although LGN was also efficiently recruited to the apical cortex, spindle orientations are largely randomized (Fig. 6B, control cysts, mean angle =78.0±2.2°; Crb3-Venus-mInsc(1-60) cysts, mean angle =59.2±4.2°), likely as a result of the mislocalization of endogenous LGN as well as the inhibitory effect of mInsc on the binding of NuMA to LGN. Importantly, spindle mis-orientation and defective cystogenesis essentially disappeared when Crb3-Venus-mInsc(1-60) cells were cultured in the presence of doxycycline, indicating that both spindle mis-orientation and defective cytogenesis were caused by the specific ectopic expression of Crb3-Venus-mInsc(1-60) (Figs. 6B&C).
Discussion
The crystal structure of LGN/mInsc and LGN/NuMA complexes solved in this work provide the first structural information regarding the molecular mechanism of LGN-mediated polarity establishment and spindle positioning. The N-terminal fragment of mInsc binds to the inner convex surface of LGN TPR4-7 through concentrated hydrophobic and charge-charge interactions (Fig. 4), whereas an extended C-terminal fragment of NuMA occupies all of the LGN TPR0-7 repeats, interacting with LGN via dispersed hydrogen bonds and salt bridges (Figs. 1&2). Given the extremely high amino acid sequence conservation of mInsc, LGN, and NuMA (the corresponding regions involved in the formation of the mInsc/LGN and NuMA/LGN complexes in particular) throughout evolution (Figs. 2&4 and Fig. S1), it is reasonably safe to assume that the structural and biochemical features of the mInsc/LGN and NuMA/LGN complexes described here are shared by the complexes formed by the orthologs of these three mammalian proteins in other species.
One of the key findings of this study is that mInsc and NuMA bind to LGN in a mutually exclusive manner, and that mInsc can displace NuMA from LGN even though their macroscopic binding constants are comparable. The structures of the mInsc/LGN and NuMA/LGN complexes provide clear mechanistic explanations for this mutual exclusivity. Importantly, our biochemical and structural data argues against a commonly accepted model of ACD in Drosophila NB: that Insc functions as the linker to connect the Baz/Par-6/aPKC cortical polarity complex with the Pins/Gαi/Mud spindle orientation regulatory complex. Although Insc can constitutively associate with Baz via its C-terminal PDZ domain binding motif (Fig. S7), the mutually exclusive binding of Insc and Mud to Pins implies that the Baz/Par-6/aPKC/Insc complex cannot simultaneously interacts with both Pins and Mud. Supporting the above notion, it is known that the Par/Insc interaction pathway and the astral microtubule/Mud/Gαi pathway function independently, albeit with some overlapping/redundant functions, in regulating the polarized cortical localization of Pins (Izumi et al., 2006; Izumi et al., 2004; Siegrist and Doe, 2005; Siller et al., 2006). The dominance of mInsc over NuMA in binding to LGN also correlates well with the timing of localization of these proteins in the apical cortices of asymmetrically dividing cells. It is known that the Baz/Par-6/aPKC/Insc complex appears earlier (starting at the delamination) than the Pins/Mud/Gαi complex (formed in the metaphase) during mitosis (Kraut et al., 1996; Parmentier et al., 2000; Schaefer et al., 2000; Siegrist and Doe, 2006; Siller et al., 2006). We also demonstrate in this study that over-expression of mInsc or apical targeting of a LGN binding fragment of mInsc(1-60) in MDCK cells leads to the randomization of spindle orientation (Fig. 6; Fig. S6), and this is likely due to the disruption of the interaction between NuMA and LGN. The dominant binding of mInsc over NuMA to LGN also suggests that both the level and timing of mInsc expression during development can serve as important regulatory factors for asymmetric division in stem/progenitor cells. In agreement with our prediction, a very recent study showed that low-level over-expression of mInsc is sufficient to drive ACD, but prolonged over-expression of mInsc uncouples NuMA and spindle axes from asymmetrically localized mInsc/LGN (Poulson and Lechler, 2010), likely as a consequence of the disruption of the endogenous LGN/NuMA interaction by over-expressed mInsc. Our results also suggest that caution should be taken in interpreting the over-expression phenotypes of mInsc (Konno et al., 2008; Poulson and Lechler, 2010).
Taken together, we suggest a revised sequential binding model for Drosophila neuroblast asymmetric cell division. The establishment of the Baz/Par-6/aPKC/Insc cortical polarity complex at the delamination/late interphase recruits Pins to the apical cortex. This is consistent with the finding that mInsc and Par3 bind to distinct regions of LGN (Fig. S7). The targeting of Pins at the apical cortices provides anchoring points for the astral microtubules at the prometaphase via the Mud/Pins interaction. In order for this interaction to occur, Pins has to be freed from Insc; there are a number of possible ways in which this could occur, including Insc degradation, accumulation of higher than stoichiometric amounts of Pins, and regulated dissociation of the Insc/Pins complex. The stable apical localization of Insc-free Pins, prior to its binding to Mud, is likely the result of its binding to the membrane-bound Gαi via the C-terminal GoLoco motifs of Pins. In return, Gαi promotes the Pins/Mud interaction, as the binding of Gαi to the GoLoco motifs of Pins relieves its auto-inhibited conformation (Du and Macara, 2004; Nipper et al., 2007). Interestingly, the binding of Insc to Pins does not require the Gαi-mediated opening of the auto-inhibited conformation of Pins (i.e., Insc binds to Pins equally well with and without the presence of Gαi, our unpublished data), further suggesting that Insc is capable of targeting Pins to apical cortices at early stages in asymmetric cell division regardless of the presenceof Gαi. The above binding model also provides a mechanistic explanation for the observed co-localization of Insc, Pins, Gαi, and Mud (and their vertebrate counterparts) in the apical cortices of asymmetrically dividing cells.
Experimental Procedures
Protein Expression and Purification
The mouse LGN TPR repeats (Fig. 1A), the mouse Insc N-terminal fragments (Fig. 4B) and the human NuMA C-terminal fragments (Fig. 1B) were individually cloned into a modified version of pET32a vector. All the mutations were created using the standard PCR-based method and confirmed by DNA sequencing. Recombinant proteins were expressed in Escherichia coli BL21 (DE3) host cells at 16°C and were purified using a Ni2+-NTA agarose affinity chromatography followed by a size-exclusion chromatography. For in vitro biochemical analysis, LGN15-350 was expressed as the GST-fused protein and purified by GSH-sepharose affinity chromatography.
Crystallography
Crystals of the LGN/mInsc and LGN/NuMA complexes were obtained by the hanging drop vapor diffusion method at 18 °C. Freshly purified LGN fragments were concentrated to 0.5 mM before a saturating amount of mInsc or NuMA peptide were added respectively. The LGN/mInsc complex crystals were grown in 1.5 M NaCl, 10% Ethanol, and the LGN/NuMA crystals were from 0.02 M MgCl2, 0.1 M HEPES pH 7.5, 22% w/v polyacrylic acid 5100 buffer. Crystals were soaked in crystallization solution containing 20% glycerol for cryoprotection. Molecular replacement was used to solve the structure of the LGN/mInsc complex. The initial model was rebuilt manually and then refined using REFMAC (Murshudov et al., 1997) against the 1.1-Å resolution data set. Then, anisotropic refinement was applied using SHELXL (Sheldrick, 2008). Further manual model building and adjustment were completed using COOT (Emsley and Cowtan, 2004). For the LGN/NuMA complex, the LGN/mInsc complex structure was used as the search model for molecular replacement against the 2.3-Å resolution data set. Further refinement procedure was the same as that of the LGN/mInsc complex.
Fluorescence Assay
Fluorescence assay were performed on a PerkinElmer LS-55 fluorimeter equipped with an automated polarizer at 25 °C. In a typical assay, a FITC-labeled peptide (∼1 μM) was titrated with a binding partner in 50 mM Tris pH 8.0 buffer containing 100 mM NaCl, 1 mM DTT, and 1 mM EDTA.
GST Pull-Down Assay
For GST pull-down assay, GST or GST-tagged proteins (50 μl from 1 mg/ml stock solutions) were first loaded to 30 μl GSH-sepharose 4B slurry beads in an assay buffer (50 mM Tris pH 8.0, 100 mM NaCl, 1 mM DTT, and 1 mM EDTA). The GST fusion protein loaded beads were then mixed with potential binding partners, and the mixtures were incubated for 2 hrs at 4°C. After three times washing, proteins captured by affinity beads were eluted by boiling, resolved by 15% SDS-PAGE, and detected by Coomassie blue staining.
Analytical gel filtration chromatography
Analytical gel filtration chromatography was carried out on an AKTA FPLC system (GE Healthcare). Proteins were loaded on to a Superose 12 10/300 GL column 20 (GE Healthcare) equilibrated with a buffer containing 50 mM Tris-HCl pH 8.0, 100 mM NaCl, 1 mM DTT and 1 mM EDTA.
Cell culture, cystogenesis, measurement of spindle angles and imaging
MDCK cells were cultured in Dulbecco's modified Eagle's medium supplemented with 10% fetal calf serum and penicillin–streptomycin (100 IU/ml and 100 mg/ml, respectively) at 37 °C in a humidified 5% CO2 atmosphere.
Stable Tet-off inducible MDCK cell lines expressing Crb3-Venus-Gαi1 was described previously (Zheng et al., 2010). Stable Tet-off inducible MDCK cell lines expressing Venus-mInsc-FL and Crb3-Venus-mInsc(1-60) were generated as described (Du et al., 2001). Briefly, cDNA encoding mInsc-FL or mInsc(1-60) was inserted into pTRE2-Venus or pTRE2Crb3-Venus (Zheng et al., 2010) to generate pTRE2-Venus-mInsc-FL or pTRE2Crb3-Venus-mInsc(1-60), respectively. The plasmids were transfected into MDCK T23 cells and stable clones were isolated as described (Du et al., 2001). The 3D culture of MDCK cells in matrigel and the measurement of spindle angles were performed as described (Zheng et al., 2010), and details can be found in the legends of Fig. 6, and Fig. 6S.
Coordinates
The atomic coordinates of the LGN/mInsc and LGN/NuMA complexes have been deposited at the Protein Data Bank under the accession code of 3RO3 and 3RO2, respectively.
Supplementary Material
Acknowledgments
We thank Mr. Anthony Zhang for editing the manuscript, Dr. Duane Compton for the anti-NuMA antibody, the Shanghai Synchrotron Radiation Facility BL17U for X-ray beam time. This work was supported by the National High Technology Research Program (2006AA02A320), the National Major Basic Research Program (2009CB918600, 2011CB808505), National Science Foundation of China (30970574, 20973040, 31070642), the Shanghai Rising-Star Program (10QA1400700), Science & Technology Commission of Shanghai Municipality (08DZ2270500), Shanghai Leading Academic Discipline Project (B108) to W Wen and W Wang, National Institute of Health grant (GM079506) and American Cancer Society grant (RSG0717601CSM) to QD, and RGC grants (663808, 664009, 660709, 663610, HKUST6/CRF/10, SEG_HKUST06, AoE/B-15/01-II, and AoE/M-04/04) to MZ.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Bowman SK, Neumuller RA, Novatchkova M, Du Q, Knoblich JA. The Drosophila NuMA Homolog Mud regulates spindle orientation in asymmetric cell division. Dev Cell. 2006;10:731–742. doi: 10.1016/j.devcel.2006.05.005. [DOI] [PubMed] [Google Scholar]
- Bryant DM, Datta A, Rodriguez-Fraticelli AE, Peranen J, Martin-Belmonte F, Mostov KE. A molecular network for de novo generation of the apical surface and lumen. Nat Cell Biol. 2010;12:1035–1045. doi: 10.1038/ncb2106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cowan CR, Hyman AA. Asymmetric cell division in C. elegans: cortical polarity and spindle positioning. Annu Rev Cell Dev Biol. 2004;20:427–453. doi: 10.1146/annurev.cellbio.19.111301.113823. [DOI] [PubMed] [Google Scholar]
- Das AK, Cohen PW, Barford D. The structure of the tetratricopeptide repeats of protein phosphatase 5: implications for TPR-mediated protein-protein interactions. Embo J. 1998;17:1192–1199. doi: 10.1093/emboj/17.5.1192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Du Q, Macara IG. Mammalian Pins is a conformational switch that links NuMA to heterotrimeric G proteins. Cell. 2004;119:503–516. doi: 10.1016/j.cell.2004.10.028. [DOI] [PubMed] [Google Scholar]
- Du Q, Stukenberg PT, Macara IG. A mammalian Partner of inscuteable binds NuMA and regulates mitotic spindle organization. Nat Cell Biol. 2001;3:1069–1075. doi: 10.1038/ncb1201-1069. [DOI] [PubMed] [Google Scholar]
- Du Q, Taylor L, Compton DA, Macara IG. LGN blocks the ability of NuMA to bind and stabilize microtubules. A mechanism for mitotic spindle assembly regulation. Curr Biol. 2002;12:1928–1933. doi: 10.1016/s0960-9822(02)01298-8. [DOI] [PubMed] [Google Scholar]
- Emsley P, Cowtan K. Coot: model-building tools for molecular graphics. Acta Crystallographica Section D-Biological Crystallography. 2004;60:2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- Gonczy P. Mechanisms of asymmetric cell division: flies and worms pave the way. Nat Rev Mol Cell Biol. 2008;9:355–366. doi: 10.1038/nrm2388. [DOI] [PubMed] [Google Scholar]
- Hao Y, Du QS, Chen XY, Zheng Z, Balsbaugh JL, Maitra S, Shabanowitz J, Hunt DF, Macara IG. Par3 Controls Epithelial Spindle Orientation by aPKC-Mediated Phosphorylation of Apical Pins. Curr Biol. 2010;20:1809–1818. doi: 10.1016/j.cub.2010.09.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horvitz HR, Herskowitz I. Mechanisms of asymmetric cell division: two Bs or not two Bs, that is the question. Cell. 1992;68:237–255. doi: 10.1016/0092-8674(92)90468-r. [DOI] [PubMed] [Google Scholar]
- Izumi Y, Ohta N, Hisata K, Raabe T, Matsuzaki F. Drosophila Pins-binding protein Mud regulates spindle-polarity coupling and centrosome organization. Nat Cell Biol. 2006;8:586–593. doi: 10.1038/ncb1409. [DOI] [PubMed] [Google Scholar]
- Izumi Y, Ohta N, Itoh-Furuya A, Fuse N, Matsuzaki F. Differential functions of G protein and Baz-aPKC signaling pathways in Drosophila neuroblast asymmetric division. J Cell Biol. 2004;164:729–738. doi: 10.1083/jcb.200309162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jinek M, Rehwinkel J, Lazarus BD, Izaurralde E, Hanover JA, Conti E. The superhelical TPR-repeat domain of O-linked GlcNAc transferase exhibits structural similarities to importin alpha. Nat Struct Mol Biol. 2004;11:1001–1007. doi: 10.1038/nsmb833. [DOI] [PubMed] [Google Scholar]
- Johnston CA, Hirono K, Prehoda KE, Doe CQ. Identification of an Aurora-A/PinsLINKER/Dlg spindle orientation pathway using induced cell polarity in S2 cells. Cell. 2009;138:1150–1163. doi: 10.1016/j.cell.2009.07.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knoblich JA. Mechanisms of asymmetric stem cell division. Cell. 2008;132:583–597. doi: 10.1016/j.cell.2008.02.007. [DOI] [PubMed] [Google Scholar]
- Konno D, Shioi G, Shitamukai A, Mori A, Kiyonari H, Miyata T, Matsuzaki F. Neuroepithelial progenitors undergo LGN-dependent planar divisions to maintain self-renewability during mammalian neurogenesis. Nat Cell Biol. 2008;10:93–101. doi: 10.1038/ncb1673. [DOI] [PubMed] [Google Scholar]
- Kraut R, Campos-Ortega JA. inscuteable, a neural precursor gene of Drosophila, encodes a candidate for a cytoskeleton adaptor protein. Dev Biol. 1996;174:65–81. doi: 10.1006/dbio.1996.0052. [DOI] [PubMed] [Google Scholar]
- Kraut R, Chia W, Jan LY, Jan YN, Knoblich JA. Role of inscuteable in orienting asymmetric cell divisions in Drosophila. Nature. 1996;383:50–55. doi: 10.1038/383050a0. [DOI] [PubMed] [Google Scholar]
- Kuchinke U, Grawe F, Knust E. Control of spindle orientation in Drosophila by the Par-3-related PDZ-domain protein Bazooka. Curr Biol. 1998;8:1357–1365. doi: 10.1016/s0960-9822(98)00016-5. [DOI] [PubMed] [Google Scholar]
- Lin H. The stem-cell niche theory: lessons from flies. Nat Rev Genet. 2002;3:931–940. doi: 10.1038/nrg952. [DOI] [PubMed] [Google Scholar]
- Morrison SJ, Kimble J. Asymmetric and symmetric stem-cell divisions in development and cancer. Nature. 2006;441:1068–1074. doi: 10.1038/nature04956. [DOI] [PubMed] [Google Scholar]
- Morrison SJ, Spradling AC. Stem cells and niches: mechanisms that promote stem cell maintenance throughout life. Cell. 2008;132:598–611. doi: 10.1016/j.cell.2008.01.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murshudov GN, Vagin AA, Dodson EJ. Refinement of macromolecular structures by the maximum-likelihood method. Acta Crystallographica Section D-Biological Crystallography. 1997;53:240–255. doi: 10.1107/S0907444996012255. [DOI] [PubMed] [Google Scholar]
- Neumuller RA, Knoblich JA. Dividing cellular asymmetry: asymmetric cell division and its implications for stem cells and cancer. Genes Dev. 2009;23:2675–2699. doi: 10.1101/gad.1850809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nipper RW, Siller KH, Smith NR, Doe CQ, Prehoda KE. Galphai generates multiple Pins activation states to link cortical polarity and spindle orientation in Drosophila neuroblasts. Proc Natl Acad Sci U S A. 2007;104:14306–14311. doi: 10.1073/pnas.0701812104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parmentier ML, Woods D, Greig S, Phan PG, Radovic A, Bryant P, O'Kane CJ. Rapsynoid/partner of inscuteable controls asymmetric division of larval neuroblasts in Drosophila. J Neurosci. 2000;20:RC84. doi: 10.1523/JNEUROSCI.20-14-j0003.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petronczki M, Knoblich JA. DmPAR-6 directs epithelial polarity and asymmetric cell division of neuroblasts in Drosophila. Nat Cell Biol. 2001;3:43–49. doi: 10.1038/35050550. [DOI] [PubMed] [Google Scholar]
- Poulson ND, Lechler T. Robust control of mitotic spindle orientation in the developing epidermis. The Journal of Cell Biology. 2010 doi: 10.1083/jcb.201008001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roh MH, Fan S, Liu CJ, Margolis B. The Crumbs3-Pals1 complex participates in the establishment of polarity in mammalian epithelial cells. J Cell Sci. 2003;116:2895–2906. doi: 10.1242/jcs.00500. [DOI] [PubMed] [Google Scholar]
- Schaefer M, Petronczki M, Dorner D, Forte M, Knoblich JA. Heterotrimeric G proteins direct two modes of asymmetric cell division in the Drosophila nervous system. Cell. 2001;107:183–194. doi: 10.1016/s0092-8674(01)00521-9. [DOI] [PubMed] [Google Scholar]
- Schaefer M, Shevchenko A, Shevchenko A, Knoblich JA. A protein complex containing Inscuteable and the Galpha-binding protein Pins orients asymmetric cell divisions in Drosophila. Curr Biol. 2000;10:353–362. doi: 10.1016/s0960-9822(00)00401-2. [DOI] [PubMed] [Google Scholar]
- Scheufler C, Brinker A, Bourenkov G, Pegoraro S, Moroder L, Bartunik H, Hartl FU, Moarefi I. Structure of TPR domain-peptide complexes: critical elements in the assembly of the Hsp70-Hsp90 multichaperone machine. Cell. 2000;101:199–210. doi: 10.1016/S0092-8674(00)80830-2. [DOI] [PubMed] [Google Scholar]
- Schober M, Schaefer M, Knoblich JA. Bazooka recruits Inscuteable to orient asymmetric cell divisions in Drosophila neuroblasts. Nature. 1999;402:548–551. doi: 10.1038/990135. [DOI] [PubMed] [Google Scholar]
- Sheldrick GM. A short history of SHELX. Acta Crystallographica Section A. 2008;64:112–122. doi: 10.1107/S0108767307043930. [DOI] [PubMed] [Google Scholar]
- Siegrist SE, Doe CQ. Microtubule-induced Pins/Galphai cortical polarity in Drosophila neuroblasts. Cell. 2005;123:1323–1335. doi: 10.1016/j.cell.2005.09.043. [DOI] [PubMed] [Google Scholar]
- Siegrist SE, Doe CQ. Extrinsic cues orient the cell division axis in Drosophila embryonic neuroblasts. Development. 2006;133:529–536. doi: 10.1242/dev.02211. [DOI] [PubMed] [Google Scholar]
- Siller KH, Cabernard C, Doe CQ. The NuMA-related Mud protein binds Pins and regulates spindle orientation in Drosophila neuroblasts. Nat Cell Biol. 2006;8:594–600. doi: 10.1038/ncb1412. [DOI] [PubMed] [Google Scholar]
- Siller KH, Doe CQ. Spindle orientation during asymmetric cell division. Nat Cell Biol. 2009;11:365–374. doi: 10.1038/ncb0409-365. [DOI] [PubMed] [Google Scholar]
- Wang J, Dye BT, Rajashankar KR, Kurinov I, Schulman BA. Insights into anaphase promoting complex TPR subdomain assembly from a CDC26-APC6 structure. Nat Struct Mol Biol. 2009;16:987–989. doi: 10.1038/nsmb.1645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams SE, Beronja S, Pasolli HA, Fuchs E. Asymmetric cell divisions promote Notch-dependent epidermal differentiation. Nature. 2011;470:353–358. doi: 10.1038/nature09793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wodarz A, Ramrath A, Grimm A, Knust E. Drosophila atypical protein kinase C associates with Bazooka and controls polarity of epithelia and neuroblasts. J Cell Biol. 2000;150:1361–1374. doi: 10.1083/jcb.150.6.1361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wodarz A, Ramrath A, Kuchinke U, Knust E. Bazooka provides an apical cue for Inscuteable localization in Drosophila neuroblasts. Nature. 1999;402:544–547. doi: 10.1038/990128. [DOI] [PubMed] [Google Scholar]
- Yu F, Morin X, Cai Y, Yang X, Chia W. Analysis of partner of inscuteable, a novel player of Drosophila asymmetric divisions, reveals two distinct steps in inscuteable apical localization. Cell. 2000;100:399–409. doi: 10.1016/s0092-8674(00)80676-5. [DOI] [PubMed] [Google Scholar]
- Zhang Z, Kulkarni K, Hanrahan SJ, Thompson AJ, Barford D. The APC/C subunit Cdc16/Cut9 is a contiguous tetratricopeptide repeat superhelix with a homo-dimer interface similar to Cdc27. Embo J. 2010;29:3733–3744. doi: 10.1038/emboj.2010.247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng Z, Zhu H, Wan Q, Liu J, Xiao Z, Siderovski DP, Du Q. LGN regulates mitotic spindle orientation during epithelial morphogenesis. J Cell Biol. 2010;189:275–288. doi: 10.1083/jcb.200910021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zigman M, Cayouette M, Charalambous C, Schleiffer A, Hoeller O, Dunican D, McCudden CR, Firnberg N, Barres BA, Siderovski DP, Knoblich JA. Mammalian inscuteable regulates spindle orientation and cell fate in the developing retina. Neuron. 2005;48:539–545. doi: 10.1016/j.neuron.2005.09.030. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.