Abstract
How cells coordinate the immune system activities is important for potentially life-saving organ or stem cell transplantations. Polymorphic immunoregulatory genes, many of them located in the human major histocompatibility complex, impact the process and assure the proper execution of tolerance-versus-activity mechanisms. In haematopoietic stem cell transplantation, on the basis of fully human leukocyte antigen (HLA)-matched donor–recipient pairs, adverse effects like graft versus leukaemia and graft versus host are observed and difficult to handle. So far, high-resolution HLA typing was performed with Sanger sequencing, but for methodological reasons information on additional immunocompetent major histocompatibility complex loci has not been revealed. Now, we have used microarray sequence capture and targeted enrichment combined with next generation pyrosequencing for 3.5 million base pair human major histocompatibility complex resequencing in a clinical transplant setting and describe 3025 variant single nucleotide polymorphisms, insertions and deletions among recipient and donor in a single sequencing experiment. Taken together, the presented data show that sequence capture and massively parallel pyrosequencing can be used as a new tool for risk assessment in the setting of allogeneic stem cell transplantation.
Keywords: transplantation, MHC, human resequencing
1. Introduction
Haematopoietic stem cell transplantation (HSCT) is a well-established treatment in various haematologic malignancies. Advances in clinical practice and improvements in donor–patient matching have produced significant better outcomes in related and unrelated allogeneic HSCT. Especially, refinements in high-resolution DNA-based tissue typing and knowledge which transplantation antigens are most significant when selecting donors for recipients helped to improve and prolong survival rates.1 The classical transplantation antigens defined as human leukocyte antigens (HLA) are physically located within a ∼4 Mb highly polymorphic multigene region, known as the major histocompatibility complex (MHC) on chromosome 6.2–4 Many of the genes annotated inside the MHC have immune-related functions, for instance genes involved in HLA expression and transport or antigen degradation and presentation. Hence, MHC-located genes with distinct extensive polymorphisms play a fundamental role in transplantation medicine and moreover in autoimmune, inflammatory and infection diseases in humans.5,6
From the clinical perspective, exact HLA typing for class I and class II genes is a necessity for haematologic stem cell transplantation.7,8 In addition to HLA, MHC-resident unidentified transplantation determinants recently were proposed to be responsible for graft-versus-host disease (GVHD).9 Petersdorf et al. identified significantly increased risks of severe acute GVHD and lower incidence of disease recurrence in haplotype-mismatched HLA-matched unrelated HSCT. Extended haplotypes of unrelated donors and recipients were allusively defined by applying a mapping approach using the physical linkage of HLA-A, -B and -DRB1. Another methodological approach, measuring risks associated with donor–recipient MHC microsatellite marker matches or mismatches, showed increased or decreased risk for death and GVHD development.10
In case of yet unknown transplantation determinants located inside the MHC region, new technologies are essential for providing direct insights. Current approaches to define MHC-resident disease association are generally based on analytical methods for indirect haplotype determination. Definition of haplotype blocks11 with linked dedicated genetic variants is an effective tool, however phasing of long-distance segments is required and comes with several challenging issues and systemic limitations.11 Due to the long genetic distances between MHC loci and the necessity for phasing of uneven distributed single nucleotide polymorphisms (SNPs), long-range haplotyping is hard to achieve.
Now, we hypothesize that applying targeted sequence capture to the human MHC, ideally combined and enhanced by HLA typing12–14 in one single diagnostic sequencing experiment, provides additional information on transplant outcome.
Here we describe for the first time NimbleGen sequence capture on solid surface for targeted MHC resequencing by 454 sequencing in a clinical transplant setting. Moreover, to address the accuracy of the sequencing approach, we analysed approximately 1500 SNP markers mapping along the sequenced MHC region using genome-wide SNP arrays. For this study, we selected a patient who underwent unrelated HLA-matched allogeneic stem cell transplantation, the fully matched unrelated donor and also the parents of the patient. The patient developed acute GVHD after transplantation and died 1.5 months later.
Thus, we show that MHC resequencing, although applied to two HLA-identical genomes, reveals a large number of genetic differences and variants potentially linked to GVHD development. Various differences causing non-synonymous amino acid exchanges are discussed for their influence on GVHD development or drug efficacy and donor selection. In contrast to microarray-based SNP analysis or haplotype association studies, direct resequencing offers the near-complete information on genotypic differences for important transplantation outcome determinants at the single nucleotide resolution level.
2. Material and methods
2.1. Clinical setting and experimental design
We performed a comprehensive MHC resequencing approach and analysed a patient with acute myeloid leukaemia undergoing HLA-matched allergenic HSCT, the unmatched donor and the patient's parents. Initial donor selection was performed using molecular high-resolution HLA typing by Sanger sequencing. Written consent was obtained from all patients and donors as required by the institutional review board in accordance with the Declaration of Helsinki. The transplant was matched for 10 out of 10 alleles. The patient underwent a myeloablative busulfan/cyclophosphamide-based conditioning regime before transplantation. Short-course methotrexate combined with mycophenolate mofetil was used as a standard GVHD prophylaxis, due to intolerance to calcineurin inhibitors. On Day +21 after transplantation the patient developed acute GVHD Grade IV (skin IV, liver IV, gut II; Seattle criteria) and died 3 weeks later.
After microarray sequence capture and enrichment (SCE), MHC-targeted resequencing of 3.5 million base pairs for all four DNA samples was performed on a single Genome Sequencer instrument run. Additionally, microarray based genome-wide SNP analysis (Affymetrix Genome-wide Human SNP Nsp/Sty 6.0) was performed for donor and recipient for further MHC and genome-wide SNP analysis.
2.2. DNA samples
Genomic DNA was isolated and purified from venous blood samples using the magnetic bead extracting MagNaPure Compact instrument (Roche Diagnostics, Mannheim, Germany) following the manufactu-rer's instructions. DNA concentration and quality were photometrically determined (BioPhotometer, Eppendorf, Hamburg, Germany).
2.3. High-resolution sequence-based HLA typing (SBT)
The samples were typed initially for loci A, C, DQB1 and DRB1 using a commercial typing kit (S4 HLA-DRB1 and S3 HLA-A/C/DQB1; Protrans, Hockenheim, Germany) and HLA-B (AlleleSEQR HLA-B; Atria Genetics, South San Francisco, CA). PCR products were treated with ExoSAP-iT (Amersham Biosciences, Freiburg, Germany) to remove primers and dNTPs and purified by size exclusion chromatography with Sephadex® G-50 (Sigma-Aldrich, Steinheim, Germany). Direct cycle sequencing was performed in a 9800 Fast Thermal Cycler (Applied Biosystems, Foster City, CA) using the BigDyeTerminator™ v1.1 chemistry (Applied Biosystems) with the respective sequencing primers. The sequencing platform was an ABI Prism 3100 Genetic Analyzer (Applied Biosystems). The sequences representing frequent and well-known alleles were analysed by the Assign 3.5 software (Conexio Genomics) and presented in Table 1.
Table 1.
HLA typing results
| Parent 1 | Parent 2 | ||||
| A* | 29 | 01 | A* | 02 | 32 |
| B* | 44 | 57 | B* | 44 | 14 |
| C* | 06 | 06 | C* | 05 | 08 |
| DRB1* | 03 | 07 | DRB1* | 04 | 07 |
| DQB1* | 03 | 03 | DQB1* | 03 | 02 |
| DPB1* | 04 | 04 | DPB1* | 04 | 04 |
| Recipient | Donor | ||||
| A* | 01:01 | 02:01 | A* | 01:01 | 02:01 |
| B* | 57:01 | 44:02 | B* | 57:01 | 44:02 |
| C* | 06:02 | 05:01 | C* | 06:02 | 05:01 |
| DRB1* | 07:01 | 04:01 | DRB1* | 07:01 | 04:01 |
| DQB1* | 03:03 | 03:02 | DQB1* | 03:03 | 03:02 |
| DPB1* | 04:01 | 04:01 | DPB1* | 04:01 | 04:01 |
*DNA based typing.
2.4. Affymetrix genome-wide SNP analysis
For genome-wide SNP determination, an Affymetrix Genome-wide Human SNP Array (Nsp/Sty Assay Kit 5.0/6.0; carrying 909,622 SNP markers) processing with sample preparation, digestion, ligation, amplification, labelling, hybridization, staining and scanning were conducted according to the manufacturer's instructions. For donor and recipient, 250 ng of double-stranded genomic DNA starting material were digested with Nsp I and Sty I (New England BioLabs GmbH, Frankfurt, Germany). Hybridizations were conducted with one replicate of all times and treatments concurrently. Each array image was visually screened to count for general signal quality and then submitted to Genotyping Console 4.0 (Affymetrix). Genotype calling was determined by the Birdseed version 2.0 using 90 HapMap reference samples for cluster analysis.
2.5. Sequence capture and enrichment
Design and production of a GS FLX Titanium optimized sequence capture array (385k) was done at NimbleGen (Roche NimbleGen, Madison, WI) targeting 5 Mb reference sequence (human hg18) spanning the MHC complex on chromosome 6 (NCBI: chr6 NGS primary_target_region 29,594,756-33,046,546). Targeted sequence capture, with pre-capture ligation-mediated PCR (LM-PCR), hybridization, washing, elution and quantitative PCR (qPCR) to assess capture success, was performed according to the manufacturer's instructions. According to ENSEMBL v49 release (HG 18 in UCSC nomenclature) reference genome, the final target bases covering the complete MHC class I, II and III regions were defined to be 3 451 791 bp; of those 2 922 962, target bases (84.7%) were covered by capture oligonucleotides as defined by NimbleGens default settings for probe selection. 528 829 bp (15.3%) of the initial target region were omitted due to reasons of specificity and uniqueness.
2.6. MHC genotyping by Roche/454 Genome Sequencer FLX sequencing
Using the titanium-optimized sequence, capture protocol provides the captured DNA ready for use in 454 emPCR because required adaptor sequences are integrated during the LM-PCR step. The four captured DNA-sequencing libraries were prepared separately and each one loaded on one of the four-lane gasket PicoTiterPlate device (PTP; 70×75 mm; Roche/454), respectively. Each library was quantitated by Pico-Green® (Quant-iT™, Molecular Probes, Invitrogen), diluted to 1 × 105 molecules/µl and used for emPCR at a ratio of 0.1 copies of library fragments per DNA capture bead. After the DNA capture bead recovery, DNA capture bead enrichment, to separate DNA-carrying beads from non-DNA carrying beads, was performed. Finally, we loaded 490 000–690 000 then single-strand DNA carrying beads on the PTP. The sequencing itself was performed on a Genome Sequencer FLX system using titanium chemistry and standard Roche 454 protocols (Lib-L SV; Rev. Jan 2010). Sequence information is available on NCBI (release on 2011-11-24; Accession nb: SRP004546).
2.7. Data analysis
2.7.1. Image processing to gene annotation
Image processing and base calling was performed with GS FLX software (Roche/454 Life Sciences). For 100% identity contiguous sequence production, the GS Reference Mapper (Roche/454 Life Sciences) was used. The MHC Haplotype Project with the analysis of eight MHC haplotypes provided a comprehensive reference sequence database, which is consequently used for variation assessments in the present study4. Haplotype (A3-B7-DR15) of the cell line PGF was designated as the new MHC reference sequence, representing the reference sequence for the used capture oligonucleotide design (NCBI human HG 18). Genes were annotated according to the Vertebrate Genome Annotation database (http://www.VEGA.sanger.ac.uk) on the basis of the referenced PGF cell line.
2.7.2. Gene annotation to variant analysis
A series of Perl scripts was developed and applied to determine the type (substitute, insertion, deletion, synonymous or non-synonymous) and quantity (number of reads per variant) of variation between the four samples and the reference cell line and in between donor and recipient. A simple, but clear-cut ratio of the number of matched or mismatched variants in relation to the total number of sequenced base pairs defines the similarity of actual or potential donor and recipient MHC, reliably enabling fast qualification of possible donor-recipient matches with a single quality factor over and above the obligatory HLA match.
2.8. Variant analysis to graft-versus-host or disease susceptibility
Additional series of Perl scripts allow to search and compare for known disease associations or other valued medical information. Parameters such as intergenic, intronic, coding/non-coding, synonymous/non-synonymous and coverage as well as prevalence are used to define a list of SNPs to start batch analysis in NCBI's OMIM or related databases. Once the sequence data were generated by the sequencing instrument, it took just 2 h for the data analysis programs to analyse the raw data and generate report variation and disease association information. A summary table with all prominent types of SNPs in resequenced regions with at least 20-fold coverage is presented in Table 2.
Table 2.
MHC resequencing: summary of insertions, deletions and mismatches for Parents 1 and 2, donor and recipient compared with the reference cell line PGF
| 3.5 Mb human MHC resequencing |
||||||
|---|---|---|---|---|---|---|
| Parent 1 | Parent 2 | Donor | Recipient | Sum | (%) | |
| Exon | ||||||
| Insertion | 18 | 28 | 28 | 23 | 97 | (5.0) |
| Deletion | 4 | 4 | 3 | 3 | 14 | (0.7) |
| Mismatch | 548 | 518 | 459 | 316 | 1841 | (94.3) |
| Sum (%) | 570 (10.2) | 550 (13.3) | 490 (10.2) | 342 (7.3) | 1952 | (100) |
| Intron | ||||||
| Insertion | 122 | 95 | 110 | 107 | 434 | (7.3) |
| Deletion | 16 | 12 | 11 | 14 | 53 | (0.9) |
| Mismatch | 1664 | 1109 | 1406 | 1278 | 5457 | (91.8) |
| Sum (%) | 1802 (32.3) | 1216 (29.4) | 1527 (31.9) | 1399 (30.1) | 5944 | (100) |
| Intergenic | ||||||
| Insertion | 138 | 134 | 137 | 152 | 561 | (5.0) |
| Deletion | 17 | 14 | 22 | 26 | 79 | (0.7) |
| Mismatch | 3050 | 2225 | 2605 | 2740 | 10 620 | (94.3) |
| Sum (%) | 3205 (57.5) | 2373 (57.3) | 2764 (57.8) | 2918 (62.6) | 11 260 | (100) |
| Total | 5577 | 4139 | 4781 | 4659 | ||
3. Results and Discussion
3.1. Efficiency of the enrichment process
When estimating the efficiency of the enrichment process, the four different probes (NSC0237, NSC0247, NSC0268 and NSC0272) included in the sequence capture assay were quantified by qPCR before and after the enrichment process revealing that ∼337-fold enrichment was achieved (ranging from 164- to 851-fold). This is clearly above the quality standard of 100-fold enrichment, which should be realized when using microarray-based sequence capture systems.
3.2. Specificity of the enrichment process
According to PGF reference alignment observations, a total of 706 431 reads representing 78% of the 908 152 high-quality reads produced were located inside the specified MHC target region. 61 455 reads (8.7%) were rejected due to length (<50 b), repetitive or chimeric characteristics.
3.3. 454 pyrosequencing
Libraries 1–4 (MHC Parent 1, MHC Parent 2, MHC donor, MHC recipient) showed a post-emPCR and recovery enrichment efficiency of 2.5, 4.5, 5.7 and 5.8%. As a consequence 490 450, 544 500, 682 950 and 690 900 DNA-capture beads per lane were loaded to the GS-PTP (PicoTiterPlate) regions 1–4. The number of individual read sequences which passed internal quality control filters, with a mean read length of 350 bp, were 223 005, 225 385, 226 501 and 234 017, respectively, for each region.
3.4. MHC-resequencing coverage
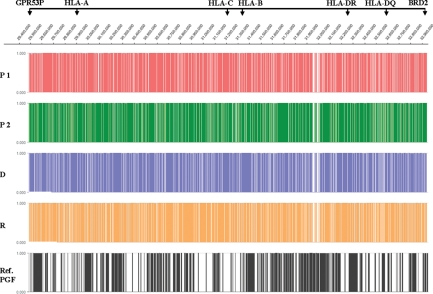
In total, we were able to annotate 273 genes to our MHC sequence data. For samples 1–4 (Parent 1, Parent 2, donor and recipient) 165 956, 167 376, 178 588 and 194 511 reads were MHC matched (Table 3). For contig analysis (parameters: 90% homology; three reads minimum for base pair position), a sum of 1246 (3 009 032 bp, 87% of target), 1287 (2 931 090 bp, 85% of target), 1073 (3 036 931 bp (88% of target) and 1129 (3 048 429 bp, 88% of target) contigs with on average 140, 131, 174 and 179 reads per contig were generated. In total, 4735 contigs with 12 025 482 bp and 706 431 reads were submitted for further analysis. On average, 13% of the targeted 3.5 Mb MHC were missed by this resequencing approach. For an annotated gene list of 273 genes (pseudogenes included) from the telomeric gene DAQB-12N14.5 (MHC-extended class I) to the centromeric gene BRD2 (MHC-class II) for which we have full sequence, the coverage is on average 17-, 18-, 19- and 20-folds for each position for Parent 1, Parent 2, donor and recipient, respectively (Table 3 and Fig. 1).
Table 3.
Summary of 454 MHC resequencing data after sequence capture microarray enrichment for parents, donor and recipient
| 3.5 Mb human MHC resequencing |
|||||||
|---|---|---|---|---|---|---|---|
| 454 genome sequencer 1× instrument run (titanium chemistry) | Contigs (n)a | Reads (n) passed filter (%) | Reads (n) p. Contig (n) | Coverage read length (bp) | MHC covered (%)b | MHC not covered (%)b | |
| Region 1 | Parent 1 | 1246 | 174 382 (66%) | 140 | 17× (342) | 3 009 032 (87%) | 438 685 (13%) |
| Region 2 | Parent 2 | 1287 | 175 749 (66%) | 137 | 18× (347) | 2 931 090 (85%) | 518 853 (15%) |
| Region 3 | Donor | 1073 | 186 819 (58%) | 174 | 19× (352) | 3 036 931 (88%) | 410 878 (12%) |
| Region 4 | Recipient | 1129 | 202 358 (62%) | 179 | 20× (340) | 3 048 429 (88%) | 400 206 (12%) |
| Sum | 4735 | 739 311 | 12 025 485 | 1 768 (623) | |||
| Mean | 1184 | 184 827 (63%) | 157 | 18.5× (345) | 3 006 371 (87%) | 442 156 (13%) | |
aContigs match parameter: 90% homology, minimum three reads for a given position.
bPer cent of complete target length 3 451 791 bp.
Figure 1.
MHC resequencing results for parents, donor and recipient compared with annotated gene loci. Graphs are representing at least 3-fold coverage (present = 1) or gaps (absent = 0) for the complete 3.5 Mb spanning MHC region of the four sequenced DNA samples (P1, parent 1), (P2, parent 2), (D, donor) and (R, recipient). Gaps present in P1 to R are a consequence of repeat rich regions and poor sequence capture or failed amplification. Graph for reference cell line PGF show known annotated gene loci in black (gene annotation = 1).
3.5. MHC-resequencing variation analysis
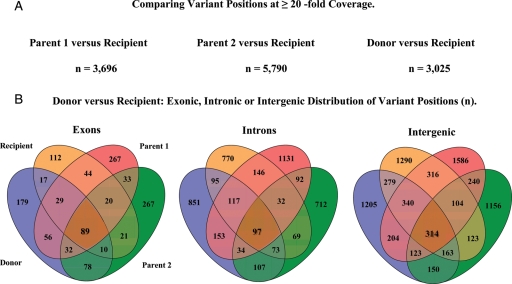
When comparing the sequence data of the four individual MHC regions to PGF, a significant number of known (dbSNP included) and new (non-dbSNP included) variant positions were detected. For parent 1 5 577, for parent 2 4139, for donor 4781 and for the bone marrow transplanted patient 4659, variant positions were detected. Among these 6% are insertions, 1% are deletions and 93% represent substitutions. An estimated 10% of all detected variants affiliate to exonic, 31% to intronic and 59% to intergenic regions. The complete numbers are shown in Table 1.
Comparing the MHC sequences of donor and recipient with the reference PGF cell line, in sum 32 586 variant positions were detected with a 3-fold coverage. Analysing donor and recipient sequences directly to each other, with the restriction to 10- or 20-fold coverage these numbers drop to 15 699 and 4596, respectively. Taking the mean of 3-, 10- and 20-fold coverage numbers, these variant positions are equally distributed and located 59% to intergenic, 7% to exonic and 34% to intronic regions.
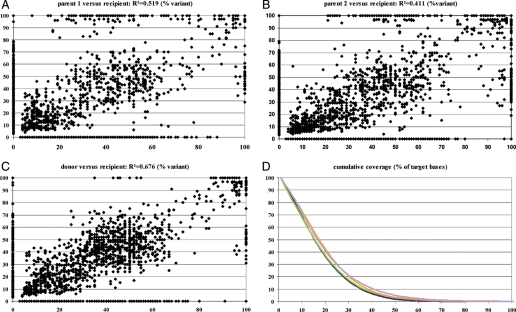
Now, in the case of comparing the MHC sequences of donor and recipient directly to each other, 3025 different positions were observed with coverage greater or equal to 20-fold. Additional 1517 positions are shared among donor and recipient, but differ with PGF sequences. Comparing Parent 1 to recipient, 3639 variant positions differ. Comparing parent 2 and recipient showed 5790 differences. Analogous, 1574 and 2276 positions shared differ in comparison to the referenced cell line. Figure 2 shows the graphical interpretation of similarities and dissimilarities among possible donor and recipient genotypes. Although, being far from perfect, donor versus recipient correlation compared with recipient versus parent correlation show the influence of the HLA match. Ultra-deep sequencing would be necessary to bring the data closer to the homogenous or heterogenous distribution values (50, 100%).
Figure 2.
MHC resequencing results. Variant correlation of parents, donor and recipient. (A–C) Correlating all detected variant positions in percentage of reads per position for two individuals visualize the degree of sequence match. A clear reduction of variant positions is desplayed in (C). (D) Cumulative coverage in percentage of target bases (blue p1, green p2, lila donor, red recipient).
Analysis of the variant list with equal or greater than 20× coverage in donor and recipient showed, that 1492 are located in intergenic regions, 1173 in intronic regions and 360 in exons. These 360 exon variants represent, with 6.6 variant positions per gene, a list of 59 genes (Fig. 3).
Figure 3.
MHC resequencing results. Summary of variant positions between parents, donor and recipient.
3.6. Variation analysis SNP microarray
SNP microarray variants (Affymetrix) with at least 10-fold coverage for recipient and donor with 1345 and 1382 SNPs show that ∼93.38% (n= 1,256) for recipient and 94.07% (n= 1,300) for donor are validated by GS-FLX data, or vice versa. SNPs with no coverage with GS-FLX data represent 1.3%, meaning, that only 1.3% fall in gap regions, a value acceptable to be low. Among those SNPs which differ with PGF, 46% are heterozygous and 54% are homozygous. Data for SNPs outside MHC comprise 885 576 specified SNPs; among these, 551 954 (62.33%) are concordant between donor and recipient. In contrast inside MHC, 1365 (86.6%) SNPs match.
Taken together, here we report the performance of newly developed methods for human MHC locus resequencing, namely targeted microarray-mediated SCE by NimbleGen and massively-parallel clonal pyrosequencing with 454's next generation sequencing technology. The genomes we partly resequenced represent a typical HSCT situation with 10 out of 10 HLA-loci matched. The procedures we used were tested with respect to efficiency and specificity of on-surface sequence capture, and the ability to discover genetic variation in coding and non-coding regions comparing the genotypes with single-base resolution for selected 3.5 Mb MHC sequence. In total, four MHC captures from four different samples were performed using DNA extracted from peripheral blood, prepared for 454 sequencing separately and subjected to one of the four sequencing regions on a single picotitre plate. The intention of the study was not to look at known or unknown GVHD-determining parameters, but to show whether available technologies and analytical software were powerful enough to start considering a study that addressed this problem comprehensively.
3.7. Enrichment, sequencing and recipient-donor pairing
In total, a mean of 337-fold enrichment achieved, among these sequences 83% could be adjusted to the targeted MHC sequences. We qualified both values (enrichment and specificity) highly sufficient for sound comparison of sequence among samples. 454-pyrosequencing gave approximately und genau? 200 000 reads per sample. A mean of 87% of the MHC was resequenced with at least 3-fold coverage. The MHC-resequencing variation analysis we chose resulted in a simple but clear score for comparing genotype variants. When comparing two individuals on the genotype level, we, first, describe and second, simply count the numbers of single nucleotide variant positions, including insertions or deletions neglecting any known functional background and score a simple number to each possible transplant-pair donor versus recipient 3025, Parent 1 versus recipient 3696 and Parent 2 versus recipient 5790. Applying this type of analysis can reveal the best fit donor out from a series of equally well fully HLA-matched candidates. As already mentioned, we cannot yet discriminate between expected transplant outcome, but we can analyse each MHC with single nucleotide resolution what can help with dependence on known disease association or possible immunologic activity generating as well as drug-acceptance potential for each and any SNP to select the best-suited donor candidate.
3.8. Genes and SNPs
On the basis of this individual MHC resequencing data and gene annotation, all database including genetic variant information for a total of 273 genes was gathered. To date numerous SNPs are described for these regions. Among these, all variants located in exons of the ∼45 MHC resident genes with known immunologic function, presented in Supplementary data only, show that a huge amount of possible proteinous structural variation, which could be responsible for inflammation induction when encountered by immunologic competent cells not tolerant against them. In total, we have 30 protein-coding genes with 56 non-synonymous (44%) and 71 synonymous (56%) sequence variants in exonic regions, three RNA genes (HCG17, HCG27 and NCRNA00171) and one transposable element (TIGD1L). Genes with non-synonymous amino acid exchanges are TUBB, GNL1, PRR3, HCG17, MDC1, HCG27, UBD, PSMB9, HSPA1L, TAP2, PPP1R10, MSH5, CLIC, CFB, C6orf10, TRIM40, CSNK2B, C6orf136 and BAT3. The list of protein-coding genes with synonymous amino acid exchanges, includes the genes PPP1R11, TRIM10, MICB, C6orf205, TRIM26, TCF19, ZFP57, NEU1, KIAA1949, GABBR1, DDR1, C6orf26 and AIF1 (Table 4).
Table 4.
List of genes with variant positionsa between donor and recipient
| Non-synonymous amino acid changes: TUBB, GNL1, PRR3, HCG17, MDC1, HCG27, UBD, PSMB9, HSPA1L, TAP2, PPP1R10, MSH5, CLIC, CFB, C6orf10, TRIM40, CSNK2B, C6orf136 and BAT3 |
| Synonymous amino acid changes: PPP1R11, TRIM10, MICB, C6orf205, TRIM26, TCF19, ZFP57, NEU1, KIAA1949, GABBR1, DDR1 C6orf26 and AIF1 |
aVariants located in exonic regions only.
The gene most affected by sequence variation is Tubulin beta. It is different in 54 positions in all four exonic regions including 5 insertions (GG, T, TAAGA, AA and TG) and 3 deletions (A, TG and T) with all variants >22-fold coverage, among those positions different 33 distinct variants result in 21 synonymous amino acid changes. The data need further verification with additional bioinformatic tools with the purpose to discriminate among tubulin family members and true variants. But, tubulin beta functions in microtubule formation and is therefore important for many aspects of cell activity, including morphogenesis, intracellular vesicle/signal transport and cell cycle regulation. In breast cancer, tubulin-associated SNPs are known to be responsible for taxol/taxane resistance.15 Another aspect of tubulin-drug interaction is that beside taxane, noscapine, its analoga and colchicine have tubulin-binding properties and influence anti-inflammatory activity by inhibition of cytokine and chemokine release from macrophages. Consequently, if tubulin-variant dependent microtubule dynamics influence cytosolic signalling, thus dampening inflammatory responses the described individual differences might possibly help explain why GVHD signals originate and sprout.16 In that context it is interesting to mention that GVHD often starts in skin, gastrointestinal tract or liver and that tubulin is involved in pathogenic Escherichia coli infections, which cause inflammation and sepsis, therefore it should be allowed to think, that tubulin variants can contribute to GVHD in one or the other way.17,18
TIGD1L (tigger transposable element derived 1-like) is a DNA transposon, or class 2 transposable element, has also 17 genomic variants between recipient and donor—meaning full or not? Generally spoken, the influence of this class of elements on mammalian genome evolution is very interesting and it would be helpful to better understand their influence on the immune system and why some species/individuals are more susceptible to invasion by parasites or principal stress situations than others.19
The next gene with high amounts of SNP differences in our specific recipient and donor setting are the genes for the inhibitory units 11 and 10 of protein phosphatase 1 (PPP1R11; protein phosphatase 1, regulatory (inhibitor) subunit 11 with 9 synonymous exchanges; PPP1R10 1 non-synonymous and 1 synonymous amino acid exchange). Especially, in combination with CsA, most immunosuppressive drugs that support successful allograft survival act by inhibiting or depleting T lymphocytes by inhibiting protein phosphatases.20
Another gene with two non-synonymous, five synonymous amino acid exchange differences between recipient and donor is the gene guanine nucleotide-binding protein-like 1 (GNL1). Recently, guanine nucleotide binding protein-like 3-like (GNL3L), another member of the GNL family was described to bind TRF1 (telomeric repeat-binding factor 1). In addition to GNL, TRF1 can be bound and modulated also by two nucleolar GTP-binding proteins, nucleostemin, which exhibit apparently opposite effects on the protein degradation of TRF1. In particular, in a recent paper from Zhu et al., GNL3L is able to stabilize TRF1 protein during mitosis and promote the metaphase-to-anaphase transition.21 Telomeric repeat binding factor 1 (TRF1) is a component of the multiprotein complex ‘shelterin’, which organizes the telomere into a high-order structure.
TRIM10, TRIM 26, TRIM 40 have 4, 2 and 1 SNPs different with 1 non-synonymous amino acid exchange in TRIM 40. A study using a cDNA expression screening procedure in a patient–donor model with full HLA match, the major target antigen of donor lymphocytes was identified to be TRIM22-442 C, a polymorphic allele of the tripartite motif family member TRIM22 (synonym: STAF50). The authors described that an arginine(R)-to-cysteine(C) exchange at position 442 generated an immunogenic T cell epitope equivalent to a minor histocompatibility antigen (mHag). Approximately 1.3% of Caucasians carry TRIM22-442 C in association with HLA-A*02:01.22
MICB, with only four synonymous exchanges encodes a heavily glycosylated protein, which is a ligand for the NKG2D type II receptor. Binding of the ligand activates the cytolytic response of natural killer (NK) cells, CD8 alpha/beta T cells and gamma/delta T cells, which express the receptor. This protein is stress induced and is similar to MHC class I molecules. MICB seems to have no role in antigen presentation, but acts as a stress-induced self-antigen, a ligand for KLRK1/NKG2D receptor, that is recognized by gamma/delta T cells.23,24
Ubiquitin-binding domains of the gene ubiquitin D (UBD) are modular elements that bind non-covalently to the protein modifier ubiquitin. The sequence context of UBDs and the conformational changes that follow their binding to ubiquitin also contribute to ubiquitin signalling and can therefore influence ubiquitins functions in HLA molecule expression and antigen presentation as well as dendritic cell activation.25
PSMB9 is different at three exonic positions. A recent publication with a rat cardiac allograft model has shown that immunoproteasome beta subunit 10 was found to be specifically increased in the graft and blood samples during chronic active antibody-mediated rejection. Using this model, they found that administration of the proteasome inhibitor, bortezomib, delayed acute rejection and attenuated the humoral response in both the acute phase and established state of this syndrome in a dose-dependent manner.26
3.9. Genome-wide SNP analysis
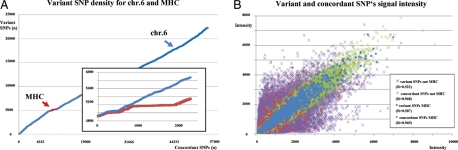
The analysis of the SNPs located inside MHC by next generation sequencing was complemented by whole genome SNP analysis. In total, we compared 885 576 SNPs equally well distributed throughout the genome observing that 62% (551 954) are shared between donor and recipient outside the MHC, whereas 86% (1365) are shared inside the MHC, reflecting the higher linkage equilibrium of MHC localized loci (Fig. 4). Taken into account that great variance exists between persons as well as demographic groups, this analysis can for the time being only be an estimate as a consequence of missing data on an individuals’ variant level. Validation results from Affymetrix-SNP data within the MHC region show 93.38% concordance with the next generation sequence data. On the basis of more individuals' sequence availability for MHC, the data for SNP design will improve accordingly, but whether it will ever meet the power of direct resequencing is an unanswered question.
Figure 4.
Summary of SNP array results for donor and recipient inside and outside MHC. Analysis of Affymetrix microarray HGW SNP 6.0 data for SNP density and signal intensities. (A) The slope of the graph represents the cumulative number of variant SNPs (x = number of SNPs; y = number of variant SNPs). Inset MHC region (Chr.6: 29 000 000–34 999 000 only) and with unrelated not HLA matched control sample (inset blue). (B) Correlation of signal intensities for concordant and variant SNP's inside and outside the MHC region. Taken together, showing that HLA matching in contrast to an unrelated and HLA unmatched control sample significantly reduces the variant SNP density in MHC regions (R = 0.892 versus R = 0.704; z = 6.76) and not in non-MHC regions (R = 0.801 versus R = 0.740; z = −34.53).
5. Conclusions
Rapidly evolving next generation sequencing technologies have reduced the hands on time and the cost per base compared with Sanger sequencing and are beginning to spread into disease prog- and diagnostics. New methodological applications, for intensive characterization of donor and recipient, minor and major histocompatibility antigens or other biomarker candidates affecting transplant outcome can clearly help select the best-matched donor by extending the list of genes screened. The list of candidate genes affecting GVHD induction and maintenance is long and the mechanisms are likely to be diverse. The data presented here demonstrate the utility of targeted sequence capture and massive parallel sequencing in detailed characterization of the genetic diversity of the donor and the recipient MHC repertoire. Such a wealth of information derived from such studies will lead to extended personalized genomic knowledge on individual MHC structures, SNPs disease associations, drug efficacy information, minor histocompatibility antigen polymorphisms or T-cell epitope formation, all crucial for further improvement of donor selection and T cell transfer strategies. Ongoing developments in targeted sequence capture in solution and the scheduled 1 kb read length in 454's pyrosequencing technology together with supplemental analysis for haplotype structures will increase the impact of that type of analysis in the field of transplant medicine.
Acknowledgements: The authors thank Henry A. Erlich for critically reading the manuscript and helpful discussion. For this publication J.P. and M.D. wrote the manuscript and performed research. S.S., K.H., C.H., S.A. and C.G. performed research, M.D., J.P., P.H. and C.G. designed research and critically reviewed the manuscript, N.N. designed and performed all bioinformatic and statistical analyses. H.H., O.K. and C.G. organised sample preparation and critically reviewed the manuscript.
Supplementary data: Supplementary Data are available at www.dnaresearch.oxfordjournals.org.
Funding
This project is supported by the Austrian Research Promotion Agency (FFG grant: Nr. 815468 B1).
References
- 1.Spellman S., Setterholm M., Maiers M., et al. Advances in the selection of HLA-compatible donors: refinements in HLA typing and matching over the first 20 years of the National Marrow Donor Program Registry. Biol. Blood Marrow Transplant. 2008;14:37–44. doi: 10.1016/j.bbmt.2008.05.001. doi:10.1016/j.bbmt.2008.05.001. [DOI] [PubMed] [Google Scholar]
- 2.Complete sequence and gene map of a human major histocompatibility complex. The MHC sequencing consortium. Nature. 1999;401:921–3. doi: 10.1038/44853. doi:10.1038/44853. [DOI] [PubMed] [Google Scholar]
- 3.Stewart C. A., Horton R., Allcock R. J., et al. Complete MHC haplotype sequencing for common disease gene mapping. Genome Res. 2004;14:1176–87. doi: 10.1101/gr.2188104. doi:10.1101/gr.2188104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Horton R., Gibson R., Coggill P., et al. Variation analysis and gene annotation of eight MHC haplotypes: the MHC Haplotype Project. Immunogenetics. 2008;60:1–18. doi: 10.1007/s00251-007-0262-2. doi:10.1007/s00251-007-0262-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Davies J. L., Kawaguchi Y., Bennett S. T., et al. A genome-wide search for human type 1 diabetes susceptibility genes. Nature. 1994;371:130–6. doi: 10.1038/371130a0. doi:10.1038/371130a0. [DOI] [PubMed] [Google Scholar]
- 6.Goulder P. J., Watkins D. I. Impact of MHC class I diversity on immune control of immunodeficiency virus replication. Nat. Rev. Immunol. 2008;8:619–30. doi: 10.1038/nri2357. doi:10.1038/nri2357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lee S. J., Klein J., Haagenson M., et al. High-resolution donor-recipient HLA matching contributes to the success of unrelated donor marrow transplantation. Blood. 2007;110:4576–83. doi: 10.1182/blood-2007-06-097386. doi:10.1182/blood-2007-06-097386. [DOI] [PubMed] [Google Scholar]
- 8.Petersdorf E. W. HLA matching in allogeneic stem cell transplantation. Curr. Opin. Hematol. 2004;11:386–91. doi: 10.1097/01.moh.0000143701.88042.d9. doi:10.1097/01.moh.0000143701.88042.d9. [DOI] [PubMed] [Google Scholar]
- 9.Petersdorf E. W., Malkki M., Gooley T. A., Martin P. J., Guo Z. MHC haplotype matching for unrelated hematopoietic cell transplantation. PLoS Med. 2007;4:e8. doi: 10.1371/journal.pmed.0040008. doi:10.1371/journal.pmed.0040008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gabriel S. B., Schaffner S. F., Nguyen H., et al. The structure of haplotype blocks in the human genome. Science. 2002;296:2225–9. doi: 10.1126/science.1069424. doi:10.1126/science.1069424. [DOI] [PubMed] [Google Scholar]
- 11.Guo Z., Hood L., Malkki M., Petersdorf E. W. Long-range multilocus haplotype phasing of the MHC. Proc. Natl. Acad. Sci. USA. 2006;103:6964–9. doi: 10.1073/pnas.0602286103. doi:10.1073/pnas.0602286103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bentley G., Higuchi R., Hoglund B., et al. High-resolution, high-throughput HLA genotyping by next-generation sequencing. Tissue Antigens. 2009;74:393–403. doi: 10.1111/j.1399-0039.2009.01345.x. doi:10.1111/j.1399-0039.2009.01345.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gabriel C., Danzer M., Hackl C., et al. Rapid high-throughput human leukocyte antigen typing by massively parallel pyrosequencing for high-resolution allele identification. Hum. Immunol. 2009;70:960–4. doi: 10.1016/j.humimm.2009.08.009. doi:10.1016/j.humimm.2009.08.009. [DOI] [PubMed] [Google Scholar]
- 14.Lind C., Ferriola D., Mackiewicz K., et al. Next-generation sequencing: the solution for high-resolution. Unambiguous HLA Typing, Hum. Immunol. 2010;71:1033–42. doi: 10.1016/j.humimm.2010.06.016. [DOI] [PubMed] [Google Scholar]
- 15.Fojo T., Menefee M. Mechanisms of multidrug resistance: the potential role of microtubule-stabilizing agents. Ann. Oncol. 2007;18(Suppl. 5):v3–v8. doi: 10.1093/annonc/mdm172. doi:10.1093/annonc/mdm172. [DOI] [PubMed] [Google Scholar]
- 16.Zughaier S., Karna P., Stephens D., Aneja R. Potent anti-inflammatory activity of novel microtubule-modulating brominated noscapine analogs. PLoS One. 2010;5:e9165. doi: 10.1371/journal.pone.0009165. doi:10.1371/journal.pone.0009165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Penack O., Holler E., van den Brink M. R. Graft-versus-host disease: regulation by microbe-associated molecules and innate immune receptors. Blood. 2010;115:1865–72. doi: 10.1182/blood-2009-09-242784. doi:10.1182/blood-2009-09-242784. [DOI] [PubMed] [Google Scholar]
- 18.Shaw R. K., Smollett K., Cleary J., et al. Enteropathogenic Escherichia coli type III effectors EspG and EspG2 disrupt the microtubule network of intestinal epithelial cells. Infect. Immun. 2005;73:4385–90. doi: 10.1128/IAI.73.7.4385-4390.2005. doi:10.1128/IAI.73.7.4385-4390.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.van Oosterhout C. Transposons in the MHC: the Yin and Yang of the vertebrate immune system. Heredity. 2009;103:190–1. doi: 10.1038/hdy.2009.46. doi:10.1038/hdy.2009.46. [DOI] [PubMed] [Google Scholar]
- 20.Wee Y. M., Choi M. Y., Kang C. H., et al. The synergistic effect of tautomycetin on cyclosporine a-mediated immunosuppression in a rodent islet allograft model. Mol. Med. 2010;16:298–306. doi: 10.2119/molmed.2009.00099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zhu Q., Meng L., Hsu J. K., Lin T., Teishima J., Tsai R. Y. GNL3L stabilizes the TRF1 complex and promotes mitotic transition. J. Cell Biol. 2009;185:827–39. doi: 10.1083/jcb.200812121. doi:10.1083/jcb.200812121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wolfel C., Lennerz V., Lindemann E., et al. Dissection and molecular analysis of alloreactive CD8+ T cell responses in allogeneic haematopoietic stem cell transplantation. Cancer Immunol. Immunother. 2008;57:849–57. doi: 10.1007/s00262-007-0421-1. doi:10.1007/s00262-007-0421-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gonzalez S., Groh V., Spies T. Immunobiology of human NKG2D and its ligands. Curr. Top. Microbiol. Immunol. 2006;298:121–38. doi: 10.1007/3-540-27743-9_6. doi:10.1007/3-540-27743-9_6. [DOI] [PubMed] [Google Scholar]
- 24.Suarez-Alvarez B., Lopez-Vazquez A., Baltar J. M., Ortega F., Lopez-Larrea C. Potential role of NKG2D and its ligands in organ transplantation: new target for immunointervention. Am. J. Transplant. 2009;9:251–7. doi: 10.1111/j.1600-6143.2008.02526.x. doi:10.1111/j.1600-6143.2008.02526.x. [DOI] [PubMed] [Google Scholar]
- 25.Winget J. M., Mayor T. The diversity of ubiquitin recognition: hot spots and varied specificity. Mol. Cell. 2010;38:627–35. doi: 10.1016/j.molcel.2010.05.003. doi:10.1016/j.molcel.2010.05.003. [DOI] [PubMed] [Google Scholar]
- 26.Ashton-Chess J., Mai H. L., Jovanovic V., et al. Immunoproteasome beta subunit 10 is increased in chronic antibody-mediated rejection. Kidney Int. 2010;77:880–90. doi: 10.1038/ki.2010.15. doi:10.1038/ki.2010.15. [DOI] [PubMed] [Google Scholar]