Abstract
Numerous microbes inhabit the mammalian intestinal track and strongly impact host physiology; however, our understanding of this ecosystem remains limited owing to the high complexity of the microbial community and the presence of numerous non-culturable microbes. Segmented filamentous bacteria (SFBs), which are clostridia-related Gram-positive bacteria, are among such non-culturable populations and are well known for their unique morphology and tight attachment to intestinal epithelial cells. Recent studies have revealed that SFBs play crucial roles in the post-natal maturation of gut immune function, especially the induction of Th17 lymphocytes. Here, we report the complete genome sequence of mouse SFBs. The genome, which comprises a single circular chromosome of 1 620 005 bp, lacks genes for the biosynthesis of almost all amino acids, vitamins/cofactors and nucleotides, but contains a full set of genes for sporulation/germination and, unexpectedly, for chemotaxis/flagella-based motility. These findings suggest a triphasic lifestyle of the SFB, which comprises two types of vegetative (swimming and epicellular parasitic) phases and a dormant (spore) phase. Furthermore, SFBs encode four types of flagellin, three of which are recognized by Toll-like receptor 5 and could elicit the innate immune response. Our results reveal the non-culturability, lifestyle and immunostimulation mechanisms of SFBs and provide a genetic basis for the future development of the SFB cultivation and gene-manipulation techniques.
Keywords: segmented filamentous bacterium, non-culturable bacteria, genome, flagella
1. Introduction
The mammalian digestive tract harbours diverse microbes, with populations as high as 100 trillion organisms. Their metabolic activities, such as energy harvesting from otherwise indigestible dietary polysaccharides and the synthesis of essential vitamins, and colonization resistance against enteric pathogens are considered beneficial for the host.1,2 The gut microbiota play pivotal roles in the maturation and differentiation of the gut immune system. However, alteration of the gut microbial composition is often linked to various pathological conditions, such as inflammatory bowel diseases, colon cancer and obesity.3–7
Our understanding of gut microbiota has been severely hampered by its extreme complexity; however, the recent advent of culture independent techniques that employ 16S rRNA gene sequencing has revealed that mammalian gut microbiota comprise several hundred or more species, and the majority of the constituents are yet to be cultured or characterized.8–11 Moreover, 20–80% of gut microbes are predicted to be non-culturable by current cultivation techniques. Therefore, genomic information of these non-culturable populations is essential to a more thorough understanding of the biology of gut microbiota; however, genome sequencing of non-culturable microbes is still a significant scientific challenge even with metagenome analysis or single cell sequencing using new sequencing technologies.8–12
Segmented filamentous bacteria (SFBs), which are clostridia-related, spore-forming, Gram-positive bacteria, are well-known members of these non-culturable populations. SFBs inhabit the terminal ilea of a wide range of mammals.13,14 Their characteristic prevalence at weaning periods and unique morphology of being tightly attached to the epithelial cells around Peyer's patches strongly suggest their involvement in the maturation of the gut immune system.15,16 In fact, it was found that SFB colonization enhances the luminal IgA production.17 More recently, several groups have shown that SFBs play a crucial role in the post-natal maturation of gut immune function, especially in the induction of a specific subset of proinflammatory T helper lymphocytes, Th17 cells.18,19 Thus, more attention is being paid to SFB-derived molecules and the signal transduction pathways responsible for such immunomodulatory effects; however, no technique is available to cultivate and genetically manipulate the SFB in vitro.
Here, we report the genome sequence determination of mouse SFBs, the first complete genome of a non-culturable mammalian gut microbe, using SFB cells grown in germ-free mice. An analysis of the genome sequence revealed the genomic features underlying the non-culturability of SFBs and their unique triphasic lifestyle, which would help us to establish an SFB cultivation protocol. In addition, our results are the first to provide insight into the mechanisms of the immunostimulating activities of SFBs and some of the genomic information relevant for human-associated SFBs.
2. Materials and methods
2.1. Preparation of the genomic DNA
SFB-gnotobiotic mice were prepared according to the method described by Umesaki et al.15 Briefly, the terminal ileum of an 8-week-old male BALB/c mouse (Clea Japan) was cut into 1-cm pieces. Mucosal epithelial cells were separated from the lamina propria (LP) as described previously.20 The epithelial cells were transferred to a fresh tube containing 1 ml of phosphate-buffered saline (PBS) and homogenized using a Teflon grinder. The homogenate was treated with 3% chloroform to lyse the host epithelial cells and non-spore-forming bacteria. After removing the chloroform by bubbling with carbon dioxide, 5-week-old male germ-free BALB/c mice (Clea Japan) that had been kept in vinyl isolators were orally inoculated with the sample. At 2 weeks after inoculation, caecal contents were collected and bacterial DNA was extracted as described previously.21
2.2. Preparation of mouse LP lymphocytes
The small intestines were opened longitudinally, washed with PBS and incubated for 20 min at 37°C in a FACS buffer [PBS containing 10% fetal bovine serum (FBS), 10 mM EDTA, 20 mM HEPES, 1 mM sodium pyruvate, 10 µg/ml polymyxin B sulphate, 100 units/ml penicillin and 100 µg/ml streptomycin]. The intestines were then cut into small pieces and incubated with RPMI1640 containing 10% FBS, 400 units/ml collagenase D and 100 µg/ml DNase I (all from Roche Diagnostics) for 1 h at 37°C. The digested tissues were washed with the FACS buffer, resuspended in 10 ml of 40% Percoll (GE Healthcare) and overlaid on 5 ml of 80% Percoll to perform Percoll gradient separation. Interface cells [LP lymphocytes (LPLs)] were collected and suspended in PBS containing 2% FBS and 0.05% NaN3. For IFN-γ and interleukin (IL)-17A detection, the LPLs were stimulated for 4 h with 50 ng/ml of phorbol 12-myristate 13-acetate, 1 µg/ml of ionomycin and 2 µM monensin (all from Sigma). Cells were first stained for surface CD4 and then for intracellular IFN-γ and IL-17A after fixation with a 4% paraformaldehyde phosphate buffer solution and washing with a 0.1% saponin-containing buffer. Antibodies used in this part of the study included biotinylated anti-CD4 monoclonal antibody (mAb), FITC-labelled anti-IFN-γ mAb, PE-labelled anti-IL-17A mAb and streptavidin-PE-Cy7 (all from eBioscience). Flow cytometry analysis was performed using a FACSCanto II with the FACSDiva software (BD Biosciences).
2.3. Genome sequencing, annotation and comparative genomics
The genome sequence of SFBs was determined using a combined strategy of 454 pyrosequencing (GS FLX; Roche) and Sanger sequencing. A total of 234 915 GS FLX reads were assembled with the GS Assembler software into 242 large contigs (≥500 bp). These contigs were then reassembled using the Phred/Phrap/Consed software (http://www.phrap.com) with 25 257 Sanger sequencing reads from plasmid libraries (3 and 10 kb inserts). Gap closing and the re-sequencing of low-quality regions were performed by sequencing the PCR products and the appropriate plasmid clones. The genome sequence was automatically annotated with the Microbial Genome Annotation Pipeline22 and manually curated using the IMC-GE software (In Silico Biology, Inc.). The genome sequence of SFBs has been deposited in the DDBJ/EMBL/GenBank databases under accession number AP012202.
The SFB gene contents were compared with those of Clostridium difficile strain 630 (CD) and C. novyi strain NT (CN). The orthologous protein coding sequences (CDSs or CDS groups) for CD and CN were taken from the MBGD (http://mbgd.genome.ad.jp/). On the basis of the results of a reciprocal BLASTP search between SFB and CD and between SFB and CN (threshold; ≥25% sequence identity and ≥60% aligned length coverage of a query sequence), we categorized the CDSs (or CDS groups) from the three genomes into groups of ‘conserved in all’, ‘conserved in SFB and CD but not in CN’, ‘conserved in SFB and CN but not in CD’, ‘conserved in CD and CN but not in SFB’ and those unique to each species.
2.4. Analysis of Toll-like receptor 5-linked NF-κB signaling pathway activation
Four flagellin genes were PCR-amplified and cloned into pGEX-6P-1 (GE Healthcare) to generate glutathione-S-transferase (GST)-flagellin fusion genes. GST-fusion proteins were expressed in Escherichia coli BL21 via incubation with 1 mM IPTG at 20°C overnight and purified with Sepharose 4B GST resin (GE Healthcare). The Toll-like receptor 5 (TLR5)-stimulating activities of SFB flagellins were determined by a luciferase reporter assay system as described previously.23 Briefly, the 293T cells were transfected with pNF-κB-Luc (Clontech) and pRL-TK (Promega) in addition to the mouse TLR5-encoding pGA vector or empty vector. pNF-κB-Luc contains the firefly luciferase gene, expression of which is controlled by the upstream NF-κB response elements, and pRL-TK contains the Renilla luciferase gene, which is control by the promoter of the thymidine kinase. At 20 h after transfection, the cells were stimulated with 1 µg/ml of recombinant Salmonella flagellin (IMGENEX Corp.), GST or GST-fusion SFB flagellins for 24 h. After stimulation, the cells were harvested and the luciferase activities in the cell lysates were measured by the Dual Luciferase Reporter Assay System (Promega) using a Lumat LB9507 (Berthold). Luciferase activity was expressed as a ratio of the NF-κB-dependent firefly luciferase activity divided by the control Renilla luciferase activity (relative luciferase units).
2.5. The search for human SFB-derived sequences in human gut metagenome sequences
From the Illumina sequence reads produced in the MetaHIT project,11 we removed reads containing one or more Ns. The remaining 7 345 361 234 reads were trimmed for low-quality sequences and used as queries in a BLAST search against the SFB genome sequence, 1334 RefSeq complete microbial genome sequences (http://www.ncbi.nlm.nih.gov/RefSeq/) and 16 276 contig sequences produced by the Human Microbiome Project (http://www.hmpdacc.org/). If a query sequence returned a top hit to the mouse SFB genome with a ≥95% sequence identity for ≥95% of the query sequence and the difference in the BLAST bit score between the top (to SFB) and second hits was larger than zero, we defined it as a human SFB-related sequence. In the 277 reads we identified, BLASTN bit score differences between the top and second hits ranged from 2.0 to 121.0 (hits for only SFB) and the median value was 11.9, indicating that the 277 sequences showed a significantly higher sequence identity to SFBs than to other known genomes.
2.6. Ethics
All animal experiments were performed at the University of Tokushima according to the university's guidelines for animal experiments. The experimental designs were approved by the university animal committee.
3. Results and discussion
3.1. Genome sequencing
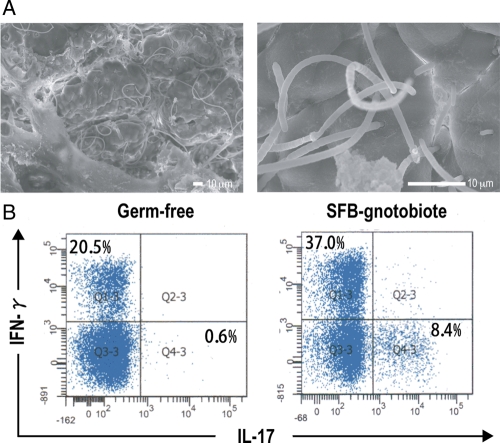
To obtain genomic DNA for sequencing, we generated SFB-associated gnotobiotic mice by orally inoculating SFBs into germ-free BALB/c mice. The SFB cells used for inoculation (mostly in the endospore form) were obtained from the ileal mucosa of an 8-week-old male BALB/c mouse. Monoassociation was confirmed by Gram-staining and sequencing of the 16S rRNA gene. For the latter, a chromatography comprising single peaks was detected. The SFB cells in the gnotobiotic mice had a segmented filamentous morphology and were tightly attached to the gut epithelia with a deep indentation of the epithelial cell surface where the SFB was attached (Fig. 1A). We further confirmed that the colonization of SFBs in the germ-free mouse intestine induced Th17 and Th1 subsets in the gut LP CD4+ lymphocyte preparation as reported previously18,19; the IL-17-producing Th17 population increased from 0.6% (germ-free mouse) to 8.4% (SFB-gnotobiotic mouse) and the IFN-γ-producing Th1 population from 20.5 to 37.0% (Fig. 1B).
Figure 1.
Morphology and immunostimulatory activity of SFBs grown in SFB-monoassociated gnotobiotic mice. (A) Electron microscopic examination of SFBs. BALB/c germ-free mice were colonized with the SFB, and the terminal ilea collected 14 days after inoculation were examined by scanning electron microscopy. SFBs show a characteristic long filamentous morphology and preferentially colonized around Peyer's patches. Epithelial cell surfaces to which SFB attach are deeply indented. (B) Induction of Th1 and Th17 cells by SFB colonization. Intestinal LPLs were isolated from BALB/c germ-free mice or SFB-monoassociated gnotobiotic mice and the percentages of IFN-γ+ or IL-17+ cells within the CD4+ lymphocyte population in each LPL preparation were determined by FACS analysis.
The genomic DNA of SFBs was extracted from the caecal contents of SFB-gnotobiotic mice and used for genome sequencing via a combination of 454 pyrosequencing and Sanger sequencing. Because it is currently impossible to generate SFB-gnotobiotic mice from a single SFB cell, the DNA sample contained DNA molecules derived from multiple SFB clones. In fact, we observed many single-nucleotide polymorphisms and small indels. Therefore, when sequence polymorphisms were observed, we regarded the most frequently appearing sequence as representative of the SFB genome sequence.
3.2. General genome features
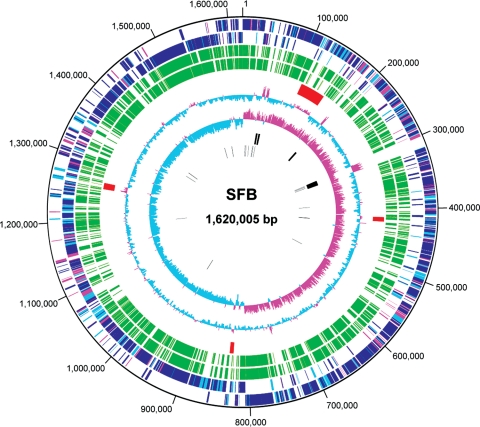
The genome of SFBs comprises a single circular chromosome of 1 620 005 bp with an average 28.8% GC content (Table 1). The chromosome encodes 1491 CDSs, 6 rRNA (rrn) operons, 37 tRNA genes and only 24 pseudogenes. Of the 1491 CDSs, 994 (66.7%) were functionally assigned. Like other low GC content Gram-positive bacteria, the SFB chromosome exhibits clear GC skew transitions at the replication origin and terminus and a strong coding bias with 82.4% of the CDSs being encoded on the leading strand (Fig. 2). Chromosome regions showing GC bias anomalies correspond to rrn or prophage regions.
Table 1.
General features of the SFB chromosome
| Size (bp) | 1 620 005 |
| G + C content (%) | 28.3 |
| CDSs | 1491a |
| Coding density | 89.8 |
| Average CDS length (bp) | 960 |
| rRNA operons | 6 |
| tRNA genes | 37 |
| Prophages | 4 |
aTwenty-four pseudogenes are not included.
Figure 2.
Circular map of the SFB chromosome. From the outside in: the first circle depicts the nucleotide sequence positions (in bp). The second and third depict CDSs that are transcribed clockwise and counterclockwise, respectively, and CDSs were coloured according to the results of the BLASTP homology search of each CDS (E-value <1e−5) against the RefSeq database at the NCBI (blue, top hit to order Clostridiales; light blue, top hit to other species; red, no hit). The fourth and fifth depict CDSs conserved in C. difficile and C. novyi, respectively (≥25% sequence identity and ≥60% aligned length coverage of a query sequence). The sixth depicts prophages, the seventh GC content, the eighth GC skew, the ninth rRNA operons and the tenth tRNA genes.
The SFB contains no IS elements but four prophages (Fig. 2). Among the four prophages, one (SFBMP01) is apparently intact, whereas the others (SFBMP02∼SFBMP04) are highly degraded phage remnants and SFBMP02 is similar to a part of SFBMP01 (Supplementary Fig. S1). The presence of multiple prophages suggests that phages are the main driving force for horizontal gene transfer between SFBs and other microbes. Frequent exposure to invading foreign DNAs is also suggested by the presence of three loci of clustered, regularly interspaced and short palindromic repeats (CRISPRs) in the SFB genome (Supplementary Fig. S1). CRISPRs are involved in a recently discovered microbial adaptive immunity system that protects microbial cells from invading foreign genetic elements, such as bacteriophages and conjugative plasmids.24,25 CRISPR loci typically consist of several non-contiguous direct repeats separated by stretches of variable sequences called spacers, which correspond to segments of captured phage and plasmid sequences. The CRISPR sequences thus provide an adaptive, heritable record of past infections and express small RNAs that target invading nucleic acids. One of the CRISPR loci in the SFB genome (CRISPR3) contains 13 spacer-repeat units, which are preceded by a gene cluster encoding six CRISPR-associated (Cas) proteins. Interestingly, the other two loci (each contains three and four spacer-repeat units, respectively) are located on prophage SFBMP01. Neither is associated with cas genes, but they share the same 160-bp leader sequence and 32-bp repeat sequences with CRISPR3, suggesting that the precursor RNAs derived from these prophage-encoded CRISPRs may also be matured to short CRISPR RNAs by the CRISPR3-associated Cas proteins. Prophage-encoded CRISPRs were previously described in C. difficille.26 Any spacer sequences found in the three CRISPR loci of SFBs showed no significant homology to the sequences in databases.
3.3. The phylogenetic position of SFBs
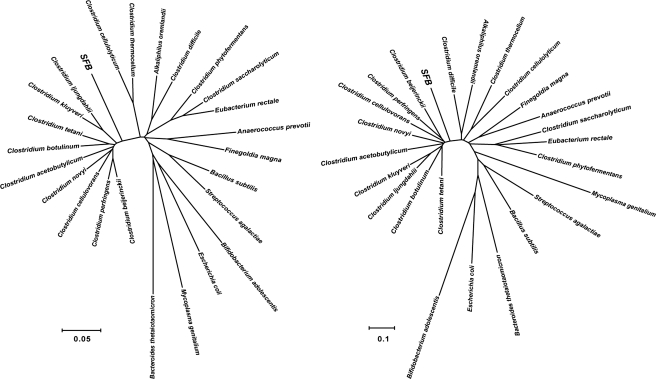
In our initial BLAST analysis against the RefSeq database, 66.2% of the SFB CDSs had best-hit homologues of the order Clostridiales but with relatively low levels of sequence identity (Fig. 2 and Supplementary Table S1). These results are consistent with the phylogenetic position of SFBs previously inferred by 16S rRNA sequence analysis; SFBs belong to a distinct lineage within the Clostridium subphylum at the rank of genus, whose proposed genus name was Candidatus Arthromitus (Fig. 3A).27 We further confirmed this phylogenetic position by constructing a genomic phylogenetic tree based on the sequences of 31 universal genes28 from fully sequenced species belonging to the order Clostridiales (Fig. 3B).
Figure 3.
The phylogenetic position of SFBs among the fully sequenced Clostridium species. Phylogenetic trees constructed based on the 16S rDNA sequences (A) and the concatenated sequences of 31 universal genes28 (B) are shown.
3.4. Comparative genomics
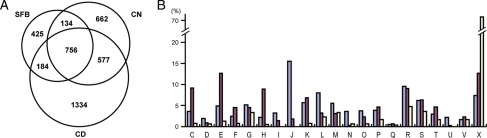
Because no closely related species has yet been sequenced, we chose CD and CN for genomic comparison because both are gut-associated clostridia and they have the largest or smallest genome among the fully sequenced Clostridiales (4.3 and 2.5 Mb, respectively).26,29 Based on the orthologous gene group classification of CD and CN genes in the MBGD database (http://mbgd.genome.ad.jp/), in which 3741 CDSs of CD were grouped into 2851 CDSs (or CDS groups) and 2315 CDSs of CN were grouped into 2129 CDSs (or CDS groups), we compared the gene content of SFBs with these two gut-associated clostridia via an all-to-all BLAST analysis. The results revealed that 940 and 890 of the SFB CDSs (or CDSs groups) are conserved in CD and CN, respectively; 756 are conserved in both strains, whereas 425 are unique to SFB (Fig. 4A). Most of the unique genes are of unknown function. More importantly, we identified 577 CDSs that are shared by CD and CN but not SFB. The distribution of the 577 CDSs into the functional categories of the clusters of orthologous groups (COGs) of proteins indicates that, in comparison to CD and CN, the SFB lacks remarkable numbers of genes involved in the following processes: ‘energy production and conversion (C in Fig. 4B)’, ‘amino acid transport and metabolism (E)’, ‘nucleotide transport and metabolism (F)’ and ‘coenzyme metabolism (H)’. Significant portions of the CD/CN shared genes that are related to ‘carbohydrate transport and metabolism (G)’, ‘transcription (K)’ and ‘inorganic ion transport and metabolism (P)’ are also absent in SFBs. These results and the smaller genome size of SFBs suggest that during its strict niche specialization, genome reduction has taken place and many metabolic functions have been lost (see below).
Figure 4.
A comparison of SFB gene content to C. difficile and C. novyi. (A) A Venn diagram showing the conservation of orthologous CDSs (or CDS groups) between SFBs, CD and CN. The numbers of orthologous CDSs in each category are shown. (B) The functional distributions of orthologous CDSs belonging to three conservation categories; ‘conserved in the three bacteria’ (left bars), ‘conserved in CD and CN but not in SFB’ (middle) and ‘unique to SFB’ (right). The functional classification follows the COG categories (http://www.ncbi.nlm.nih.gov/COG/) (C: energy production and conversion; D: Cell cycle control, mitosis and meiosis; E: amino acid transport and metabolism; F: nucleotide transport and metabolism; G: carbohydrate transport and metabolism; H: coenzyme transport and metabolism; I: lipid transport and metabolism; J: translation; K: transcription; L: replication, recombination and repair; M: cell wall/membrane biogenesis; N: cell motility; O: posttranslational modification, protein turnover, chaperones; P: inorganic ion transport and metabolism; Q: secondary metabolites biosynthesis, transport and catabolism; R: general function prediction only; S: function unknown; T: signal transduction mechanisms; U: intracellular trafficking and secretion; V: defense mechanisms) and ‘X’ indicates CDSs that are not assigned to any category. Each bar represents the percentage of CDSs belonging to each functional category in each of three conservation categories.
3.5. Metabolic capabilities
Consistent with the prediction that SFBs are strict anaerobes, no CDS for tricarboxylic acid cycle- or respiratory chain-related proteins were identified, whereas the SFB contains two catalases and one peroxidase. They may confer some level of oxygen tolerance, which is required for the SFB to pursue an epicellular life on intestinal epithelia where a microaerobic environment is formed. However, SFBs contain a complete enzyme set for the glycolytic pathway from glucose to pyruvate (Supplementary Fig. S2A). SFBs can further metabolize pyruvate using the pyruvate:ferredoxin oxidoreductase–hydrogenase system to produce acetate or ethanol but not butyrate and butanol. Pyruvate can also be metabolized to lactate. These are the primary energy production pathways for the SFB, like many other clostridia.30 Furthermore, SFBs possess a D-ribose ATP-binding cassette (ABC) transporter, ribokinase and transketolase and can, therefore, uptake extracellular ribose and metabolize it to glyceraldehyde-3-phosphate, an intermediate of the glycolytic pathway. Although the pentose phosphate pathway (PPP) is conserved, components for the de novo synthesis of purine and pyrimidine nucleotides from phosphoribosyl pyrophosphate (PRPP) are missing. Thus, it is not likely that the PPP of SFBs is involved in nucleotide biosynthesis.
A more striking metabolic feature of SFBs is the almost complete lack of de novo biosynthesis pathways for amino acids, vitamins/cofactors and nucleotides (Table 2). As for amino acid biosynthesis, we identified only 20 genes related to this function. Genes for lysine biosynthesis are relatively well conserved, but they probably just function to synthesize diaminopimeric acid as a component of peptidoglycan. Similarly, SFBs possess only 14 genes for vitamin and cofactor biosynthesis, and almost all the genes required for the biosynthesis of thiamin, riboflavin, pyridoxine, nicotinamide, pantothenate, coenzyme A, biotin, folate and vitamin B12 are absent. As mentioned above, SFBs are unable to synthesize purines and pyrimidines from PRPP. In addition, key enzymes for nucleotide interconversion, such as thymidylate synthase and guanine monophosphate reductase, are also absent in the SFB.
Table 2.
Gene repertoires for amino acid and vitamin/cofactor biosynthesis in C. difficile (CD), C. novyi (CN) and SFB
| Organisms | CD | CN | SFB |
|---|---|---|---|
| Amino acids | |||
| Glycine, serine and threonine | serA | serA | serA |
| serB | serB | ||
| glyA | glyA | glyA | |
| thrBC | thrBC | ||
| hom | hom | ||
| Cysteine and methionine | cysEK | cysEK | |
| metAY | metAY | ||
| metC | metC | ||
| metH | metH | ||
| metK | metK | metK | |
| Valine, leucine and isoleucine | ilvA | ilvA | |
| ilvBCD | ilvBCD | ||
| ilvE | ilvE | ||
| leuABCD | |||
| Lysine | dapABCDEFG | dapABCDEFG | dapABDFG |
| lysA | lysA | lysA | |
| glnA | glnA | glnA | |
| Alanine, aspartate and glutamate | asnA | asnA | |
| asnB | asnB | ||
| gltD | gltD | ||
| Arginine and proline | aspC | aspC | |
| aspD | aspD | ||
| proC | proABC | ||
| argBCDEFJ | argBCDFJ | ||
| argGH | argGH | ||
| Histidine | hisABCDFGHIZ | hisABCDFGIZ | |
| hisK | hisK | ||
| Phenylalanine, tyrosine and tryptophan | aroABCEGK | aroABCEGK | aroABCEGK |
| aroD | aroD | ||
| trpB | trpB | ||
| tyrA | tyrA | ||
| pheA | pheA | ||
| Vitamins and cofactors | |||
| Thiamin | thiCFGH | thiCFGH | thiH |
| thiD | thiD | ||
| thiI | thiI | ||
| thiN | thiN | thiN | |
| isc | isc | isc | |
| Riboflavin | ribABDH | ribABDH | |
| ribC | ribC | ribC | |
| Vitamin B6 | pdxA | ||
| pdxK | |||
| pdxS | pdxS | ||
| Nicotinate and nicotinamide | nadABC | nadABC | |
| nadD | nadD | nadD | |
| nadE | |||
| Pantothenate | panBC | panBCD | |
| panE | panE | ||
| Coenzyme A | coaX | coaX | |
| coaBC | coaBC | ||
| coaD | coaD | coaD | |
| coaE | coaE | coaE | |
| Biotin | bioA | bioAD | |
| bioB | bioB | ||
| birA | birA | birA | |
| Folate | pabABC | pabABC | pabC |
| folA | folA | ||
| folBEKP | folBEKP | ||
| folC | folC | ||
| Vitamin B12 | hemBCD | hemABCDL | |
| hemN | hemN | ||
| cbiABCDEFGHJKLPT | cbiABCDEFGHJKLPT | cbiA | |
| cobCDSTU | cobDSTU | cobC | |
| cobO | cobO | ||
| eutT | |||
Severe deficiency in the biosynthesis capability of amino acids, vitamins/cofactors and nucleotides suggests that SFBs uptake these compounds from extracellular environments. Consistent with this, at least 103 genes are devoted to transport functions (Supplementary Table S2). They include ABC transporter systems for oligopeptides, amino acids, sugars, phosphate, phosphonate, iron compounds and cobalt. SFBM_1365, SFBM_1366 and SFBM_1367 may constitute an energy-coupling factor transporter to uptake vitamins and cofactors,31 although the substrate-binding components therein have yet to be identified. In addition, SFBs possess nine phosphotransferase systems to uptake various simple sugars and several other types of transport systems for nucleotides, amino acids and other compounds.
Although the Tat protein secretion system is not present in the SFB, the Sec system is conserved. It may be noteworthy that, among the eight glycosyl hydrolases of SFBs (Supplementary Table S3), only SFBM_0066 was predicted as an extracellular enzyme by the PSORT program (http://psort.hgc.jp/). This suggests that SFBs have a very limited ability to digest food- or host-derived polysaccharides by themselves and, instead, primarily uptake simple sugars present in ready-for-use states in food or provided by the host or other gut bacteria. In contrast, 41 protease/peptidases are encoded by SFB. Of these, 4 are extracellular enzymes and 20 are membrane-associated (Supplementary Table S3). In SFBs, it is very likely that these enzymes, together with transporters for oligopeptides and amino acids, contribute to the acquisition of amino acids derived from external polypeptides. Similarly, two extracellular nucleases (SFBM_0242 and SFBM_0633) probably contribute to nucleotide acquisition from external sources.
In summary, SFB growth heavily relies on the uptake of essential compounds from extracellular environments, some of which may be supplied by host cells that SFB tightly attaches to. These metabolic features form the basis of the non-culturability of SFBs in vitro and would be valuable information for developing culture media and protocols for the SFB.
3.6. Sporulation and germination
During the epicellular parasitic phase, SFB probably obtain various nutritional components via intimate interactions with host cells; however, once they are detached from the host cells or SFB-attached host cells slough off in the process of epithelial cell turnover, such nutrition supply from the host cells would be shutdown, which probably induces the sporulation process in the SFB. We identified 66 genes related to sporulation or germination in the SFB, which are dispersed throughout the chromosome (Supplementary Table S4). They include key proteins for the sporulation signalling cascade, such as sporulation sigma factors, and other stage-specific sporulation proteins (Supplementary Fig. S2B); however, in common with other sequenced clostridia, SFBs lack several proteins that have been shown in B. subtilis to be involved in the phosphorelays that trigger sporulation, such as Spo0F, Spo0B and the Kin proteins.32,33 In B. subtilis, all the five sensor histidine kinases (KinA to KinE) are orphan kinases. Among the 10 sensor kinases identified in the SFB (Supplementary Table S5), SFBM_0504 is the sole orphan kinase and has a PAS domain-like domain, like the B. subtilis KinA protein32 and, thus, could be a candidate sensor kinase for sporulation initiation.
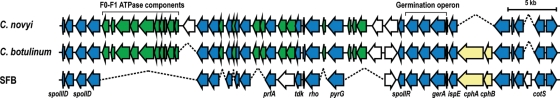
Key elements in the germination process involve germinant receptors that sense appropriate germinants, such as specific amino acids, to trigger germination. The GerA family of germinant receptors are encoded by tricistronic ger operons,34 and bacilli and clostridia often contain multiple ger operons to sense different germinants. For example, CN and C. botulinum (CB) possess two and three operons, respectively (Fig. 5), although the ger operon was not identified in the CD genome.26 In SFBs, we identified only one ger operon (Fig. 5), indicating that SFBs can sense limited types of germinants. A comparison of the analogous ger operon loci between SFB, CN and CB have revealed that, although many genes downstream of the ger operon are missing in the SFB, the overall genetic organization of this locus is conserved.
Figure 5.
Comparison of the SFB ger operon locus with C. novyi (CN) and C. botulinum (CB). SFB contains a single ger operon encoding a germinant receptor, whereas CN and CB possess two and three ger operons, respectively. Among these, only the loci analogous to that of SFBs are shown. The genes conserved in the three species are depicted by blue arrows, those conserved in the CN and CB but not in SFB by green and those unique to each species by white. Regions missing in each genome are indicated by a dotted line.
As shown in Fig. 5, the ispE and cphAB genes are located just upstream of the ger operon in the SFB. The presence of these genes may be noteworthy in terms of the metabolism of SFBs. The ispE gene encodes 4-(cytidine-5′-diphospho)-2-C-methyl-d-erythritol kinase, a component of the methyl-erythritol phosphate (MEP) pathway to synthesize terpenoids that are essential for membrane and peptidoglycan biogenesis. Because other components for the pathway were also identified (Supplementary Fig. S2C), the MEP pathway is completely conserved in the SFB. The cphAB genes encode cyanophycin synthase and cyanophycinase (Fig. 5). Cyanophycin is a prokaryote-specific copolymer of aspartate and arginine, which has been shown in many cyanobacteria to serve as a storage compound for nitrogen, carbon and energy.35 The presence of the cphAB and isoaspartyl dipeptidase (SFBM_1358) genes suggests that SFBs can synthesize cyanophycin and may utilize it as an amino acid pool in the germination process. In a previous analysis of 570 microbial genomes, it was predicted that only 24 strains, which include several clostridia (C. beijerinckii, C. botulinum, C. perfringens and C. thermocellum), can synthesize cyanopycin.36
3.7. Chemotaxis and flagella-mediated motility
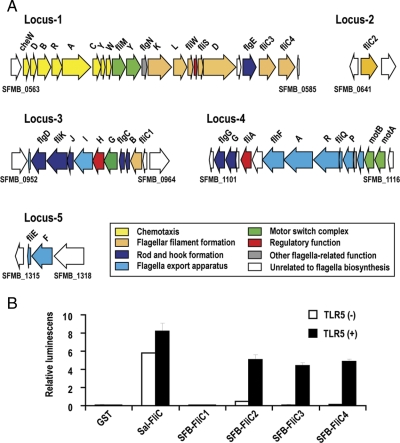
SFBs possess a full set of genes for the chemotaxis and flagella-mediated motility system. These genes are separately located in five genomic loci (Locus-1 to Locus-5; Fig. 6A and Supplementary Table S4). In addition, two methyl-accepting chemotaxis proteins (receptors for attractants) are encoded in other loci (SFBM_0442 and SFBM_1032). Flagellated SFB cells were not observed in our electron microscopic examination of SFB-monoassociated gnotobiotic mice (from 3 days to 2 weeks after oral inoculation) as well as in previous examinations.13,37,38 Thus, it is possible that in SFBs, the system has been inactivated or is used for some other purposes, such as a type III secretion system (flagella type); however, the presence of a complete set for chemotaxis genes and four flagellin genes and the absence of recognizable pseudogenes in the gene set (except for one flagellin gene; see below) strongly suggest that SFBs possess an active chemotaxis and flagella-mediated motility system. This information provides us a previously unknown aspect of SFB biology and life cycle. It is unknown at which life cycle stage SFBs produce flagella, but the most plausible scenario may be that after germination, SFBs swim in the intestine, by sensing some chemoattractant(s), to the distal parts of the small intestine where they colonize. In this regard, it is worth mentioning that SFBs contain only two methyl-accepting chemotaxis proteins (SFBM_0442 is probably the intracellular type). This may suggest that SFBs can sense very limited types of attractants.
Figure 6.
Chemotaxis/flagella biosynthesis gene loci and the activation of the TLR5-linked NF-κB signalling pathway by SFB flagellins. (A) Genetic structures of five SFB genomic loci containing genes for chemotaxis and flagella-mediated motility are shown. Two loci containing genes for methyl-accepting chemotaxis proteins are not included in this figure. See Supplementary Table S4 for detail on each gene. (B) Luciferase activities in the 293T cells that express (black bar) or do not express mouse TLR5 (white bar) after stimulation with the recombinant Salmonella flagellin, recombinant GST fusion proteins of SFB flagellins (FliC1, FliC2, FliC3 or FliC4) or GST are shown. Results are presented as fold-increase relative to the control value.
3.8. Multiple flagellins and immunostimulation activity
A unique feature of the SFB flagella system is the presence of four types of flagellins (FliC1 to FliC4). Of these, FliC3 and FliC4 are encoded in tandem in Locus-1 (Fig. 6A) and show a high sequence similarity (82.3% identity). All contain conserved N- and C-terminal flagellin domains, but FliC1 lacks a variable region between the N- and the C-terminal domains.
Flagellins are recognized by TLR5 as pathogen-associated molecular patterns (PAMPs) and induce cytokine production to activate the host innate immune system.39 TLR5 is highly expressed in LP in the small intestine, wherein dendritic cells are the dominant antigen-presenting cells and only CD11chiCD11bhi LPDCs express TLR5.40–42 These flagellin-exposed LPDCs produce IL-6 and IL-12, which results in the generation of IgA+ plasma cells and polarization towards the Th17 and Th1 functions.42 Therefore, the identified flagellins could be the key molecules responsible for the unique immunostimulation activity of SFBs. To test this presumption, we prepared recombinant proteins for each type of the SFB flagellins and examined whether they could activate the TLR5-linked NF-κB signalling pathway. As shown in Fig. 6B, although FliC1 was unable to activate the pathway, the other three activated it like that achieved by Salmonella flagellin. These results indicate that the immunostimulation activity of SFBs is attributable to the FliC2, FliC3 and FliC4 flagellins, although other molecules, such as cyanophycin and sporulation-related byproducts, may also be involved.
The tight association of SFBs to Peyer's patches, which could make LPDCs more easily accessible to SFB-derived PAMPs, may also directly or indirectly contribute to innate immunity activation. In this regard, the presence of a fibronectin-binding protein (SFBM_0986) and a phosphatidylinositol-specific phospholipase C (PI-PLC; SFBM_0755) is noteworthy. The elevation of intracellular free calcium levels induced by PI-PLC from various pathogens plays key roles in various processes of microbe–host cell interaction, such as bacterial internalization and host cell actin rearrangement.43,44 Therefore, SFB PI-PLC may also be involved in the formation of dense aggregates consisting of actin-like microfibrils beneath the host cell membrane where the SFB attached.13 Other unidentified factors are also probably required for the intimate binding of SFBs to host epithelial cells, and many of such factors may be encoded by SFB-specific genes, most of which are of unknown function (Fig. 4).
3.9. Human SFB-related sequences in the human gut metagenome sequence data
SFBs have also been found in humans, but SFBs are known to have strict host specificities and the SFB from a different host species may represent a different species.13,14 The availability of the mouse SFB genome sequence provided us a first opportunity to search for human SFB-related sequences in the large data set of the human gut metagenome sequences.11 By searching 7 345 361 234 high-quality Illumina sequences in the data set, we identified 277 reads that are highly homologous to the mouse SFB sequence (sequence identity; ≥95%, alignment length; ≥95% of the read, excluded reads related to rRNA and tRNA sequences) (Supplementary Table S6). Only a small number of reads corresponding to 0.31% of the mouse SFB genome were identified. This may be because non-mapped regions exhibit significant sequence divergence between mouse and human SFB (our search was conservative) or, more likely, because the currently available metagenome sequence data set does not include samples from children at weaning periods. In mice, SFBs become prevalent at this age. Thus, the metagenome sequence data from such children would be required to obtain more genomic sequence information on human SFB.
4. Conclusion
The whole-genome sequence of mouse SFBs revealed that this unique gut microbe has a very limited range of metabolic capabilities similar to those of obligate intracellular parasites. In particular, SFBs lack nearly all de novo biosynthesis pathways for amino acids, vitamins and cofactors and nucleotides. These features well explain the non-culturability of SFBs and further suggest that they acquire these compounds, probably from the host intestinal epithelia to which SFBs are tightly attached, via well-conserved transport systems. In addition to a full set of genes for sporulation and germination, SFBs possess a chemotaxis and flagella-based motility system. This finding suggests a triphasic lifestyle of SFBs that comprises swimming, epicellular and dormant (spore) phases. Moreover, SFBs encode four different types of flagellins, and three of these are capable of activating the TLR5-linked NF-κB signalling pathway, which could promote the luminal IgA production and the induction of particular T helper cell subclasses (Th1 and Th17). Thus, SFB genome sequence determination discloses the genetic basis for their non-culturability, which could help us to develop SFB-specific cultivation techniques. In addition, it reveals a previously unknown lifestyle including a flagellated swimming phase, and it provides a first glimpse into understanding the molecular basis for SFB-induced immunostimulation. Furthermore, through a systematic search of the human gut metagenome sequence database, the first pieces of the genomic information on human-associated SFB were obtained.
Supplemental data
Supplemental data are available at www.dnaresearch.oxfordjournals.org.
Funding
This work was supported by grants-in-aid for Scientific Researches (C) and (B) (to T.K. and T.H., respectively), for Young Scientists (B) (to H.N-I.) and for Scientific Research on Priority Areas ‘Applied Genomics’ (to T.H.) and ‘Comprehensive Genomics’ (to M.H.) from the Ministry of Education, Science and Technology of Japan and by that from the Institute for Bioinformatics Research and Development, the Japan Science and Technology Agency (BIRD-JST) (to K.K.).
Supplementary Material
Acknowledgements
We thank Drs K. J. Ishii (Osaka University) and F. Takeshita (Yokohama City University) for providing the TLR5 cDNA and the NF-κB reporter plasmid, respectively. The 16 276-contig sequences used in this study were generated in part with US federal funds from the NIH Human Microbiome Project, the Common Fund, National Institutes of Health and the Department of Health and Human Services.
References
- 1.Backhed F., Ley R.E., Sonnenburg J.L., Peterson D.A., Gordon J.I. Host-bacterial mutualism in the human intestine. Science. 2005;307:1915–20. doi: 10.1126/science.1104816. [DOI] [PubMed] [Google Scholar]
- 2.Hooper L.V., Midtvedt T., Gordon J.I. How host-microbial interactions shape the nutrient environment of the mammalian intestine. Ann. Rev. Nutr. 2002;22:283–307. doi: 10.1146/annurev.nutr.22.011602.092259. [DOI] [PubMed] [Google Scholar]
- 3.Elson C.O., Cong Y., McCracken V.J., Dimmitt R.A., Lorenz R.G., Weaver C.T. Experimental models of inflammatory bowel disease reveal innate, adaptive, and regulatory mechanisms of host dialogue with the microbiota. Immunol. Rev. 2005;206:260–76. doi: 10.1111/j.0105-2896.2005.00291.x. [DOI] [PubMed] [Google Scholar]
- 4.Sartor R.B. Microbial influences in inflammatory bowel diseases. Gastroenterology. 2008;134:577–94. doi: 10.1053/j.gastro.2007.11.059. [DOI] [PubMed] [Google Scholar]
- 5.Chu F.F., Esworthy R.S., Chu P.G., et al. Bacteria-induced intestinal cancer in mice with disrupted gpx1 and gpx2 genes. Cancer Res. 2004;64:962–8. doi: 10.1158/0008-5472.can-03-2272. [DOI] [PubMed] [Google Scholar]
- 6.Ley R.E., Turnbauch P.J., Klein S., Gordon J.I. Microbial ecology: human gut microbes associated with obesity. Nature. 2006;444:1022–3. doi: 10.1038/4441022a. [DOI] [PubMed] [Google Scholar]
- 7.Turnbaugh P.J., Ley R.E., Mahowald M.A., Magrini V., Mardis E.R., Gordon J.I. An obesity-associated gut microbiome with increased capacity for energy harvest. Nature. 2006;444:1027–31. doi: 10.1038/nature05414. [DOI] [PubMed] [Google Scholar]
- 8.Gill S.R., Pop M., Deboy R.T., et al. Metagenomic analysis of the human distal gut microbiome. Science. 2006;312:1355–9. doi: 10.1126/science.1124234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kurokawa K., Itoh T., Kuwahara T., et al. Comparative metagenomics revealed commonly enriched gene sets in human gut microbiomes. DNA Res. 2007;14:169–81. doi: 10.1093/dnares/dsm018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Castello E.K., Lauber C.L., Hamady M., Fierer N., Gordon J.I., Knight R. Bacterial community variation in human body habitats across space and time. Science. 2009;326:1694–7. doi: 10.1126/science.1177486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Qin J., Li R., Raes J., et al. A human microbial gene catalogue established by metagenomic sequencing. Nature. 2010;464:59–67. doi: 10.1038/nature08821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Marcy Y., Ouverney C., Bik E.M., et al. Dissecting biological ‘dark matter’ with single-cell genetic analysis of rare and uncultivated TM7 microbes from the human mouth. Proc. Natl Acad. Sci. USA. 2007;104:11889–94. doi: 10.1073/pnas.0704662104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Klaasen H.L.B.M., Koopman J.P., Poelma F.G.J., Beynen A.C. Intestinal, segmented, filamentous bacteria. FEMS Microbiol. Rev. 1992;88:165–80. doi: 10.1111/j.1574-6968.1992.tb04986.x. [DOI] [PubMed] [Google Scholar]
- 14.Klaasen H.L.B.M., Koopman J.P., Van den Brink M.E., Bakker M.H., Poelma F.G., Beynen A.C. Intestinal, segmented, filamentous bacteria in a wide range of vertebrate species. Lab. Anim. 1993;27:141–50. doi: 10.1258/002367793780810441. [DOI] [PubMed] [Google Scholar]
- 15.Umesaki Y., Okada Y., Matsmoto S., Imaoka A., Setoyama H. Segmented filamentous bacteria are indigenous intestinal bacteria that activate intraepithelial lymphocytes and induce MHC class II molecules and fucosyl asialo GM1 glycolipids on the small intestinal epithelial cells in the ex-germ-free mouse. Microbiol. Immunol. 1995;39:555–62. doi: 10.1111/j.1348-0421.1995.tb02242.x. [DOI] [PubMed] [Google Scholar]
- 16.Klaasen H.L.B.M., Van der Heijden P.J., Stok W., et al. Apathogenic, intestinal, segmented, filamentous bacteria stimulate the mucosal immune system of mice. Infect. Immun. 1993;61:303–6. doi: 10.1128/iai.61.1.303-306.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Suzuki K., Meek B., Doi Y., et al. Aberrant expansion of segmented filamentous bacteria in IgA-deficient gut. Proc. Natl Acad. Sci. USA. 2004;101:1981–6. doi: 10.1073/pnas.0307317101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ivanov I.I., Atarashi K., Manel N., et al. Induction of intestinal Th17 cells by segmented filamentous bacteria. Cell. 2009;139:485–98. doi: 10.1016/j.cell.2009.09.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gaboriau-Routhiau V., Rakotobe S., Lécuyer E., et al. The key role of segmented filamentous bacteria in the coordinated maturation of gut helper T cell response. Immunity. 2009;31:677–89. doi: 10.1016/j.immuni.2009.08.020. [DOI] [PubMed] [Google Scholar]
- 20.Hase K., Ohshima S., Kawano K., et al. Distinct gene expression profiles characterize cellular phenotypes of follicle-associated epithelium and M cells. DNA Res. 2005;12:127–37. doi: 10.1093/dnares/12.2.127. [DOI] [PubMed] [Google Scholar]
- 21.Morita H., Kuwahara T., Ohshima K., et al. An improved DNA isolation method for metagenomic analysis of the microbial flora of the human intestine. Microb. Environ. 2007;22:214–22. [Google Scholar]
- 22.Sugawara H., Ohyama A., Mori H., Kurokawa K. Microbial Genome Annotation Pipeline (MiGAP) for diverse users. The 20th International Conference on Genome Informatics (GIW2009) Poster and Software Demonstrations (Yokohama), S001-1-2.2009. [Google Scholar]
- 23.Hayashi F., Smith K.D., Ozinsky A., et al. The innate immune response to bacterial flagellin is mediated by Toll-like receptor 5. Nature. 2001;410:1099–103. doi: 10.1038/35074106. [DOI] [PubMed] [Google Scholar]
- 24.Horvath P., Barrangou R. CRISPR/Cas, the immune system of bacteria and archaea. Science. 2010;327:167–70. doi: 10.1126/science.1179555. [DOI] [PubMed] [Google Scholar]
- 25.Marraffini L.A., Sontheimer E.J. CRISPR interference: RNA-directed adaptive immunity in bacteria and archaea. Nat. Rev. Genet. 2010;11:181–90. doi: 10.1038/nrg2749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sebaihia M., Wren B.W., Mullany P., et al. The multidrug-resistant human pathogen Clostridium difficile has a highly mobile, mosaic genome. Nat. Genet. 2006;38:779–86. doi: 10.1038/ng1830. [DOI] [PubMed] [Google Scholar]
- 27.Snel J., Heinen P.P., Blok H.J., et al. Comparison of 16S rRNA sequences of segmented filamentous bacteria isolated from mice, rats, and chickens and proposal of ‘Candidatus arthromitus. Int. J. Syst. Bacteriol. 1995;133:445–50. doi: 10.1099/00207713-45-4-780. [DOI] [PubMed] [Google Scholar]
- 28.Ciccarelli F.D., Doerks T., von Mering C., Creevey C.J., Snel B., Bork P. Toward automatic reconstruction of a highly resolved tree of life. Science. 2006;311:1283–7. doi: 10.1126/science.1123061. [DOI] [PubMed] [Google Scholar]
- 29.Bettegowda C., Huang X., Lin J., et al. The genome and transcriptomes of the anti-tumor agent Clostridium novyi-NT. Nat. Biotechnol. 2006;24:1573–80. doi: 10.1038/nbt1256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Desvaux M. Clostridium cellulolyticum: model organism of mesophilic cellulolytic clostridia. FEMS Microbiol. Rev. 2005;29:741–64. doi: 10.1016/j.femsre.2004.11.003. [DOI] [PubMed] [Google Scholar]
- 31.Rodionov D.A., Hebbeln P., Eudes A., et al. A novel class of modular transporters for vitamins in prokaryotes. J. Bacteriol. 2009;191:42–51. doi: 10.1128/JB.01208-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Piggot P.J., Hilbert D.W. Sporulation of Bacillus subtilis. Curr. Opin. Microbiol. 2004;7:579–86. doi: 10.1016/j.mib.2004.10.001. [DOI] [PubMed] [Google Scholar]
- 33.Paredes C.J., Alsaker K.V., Papoutsakis T.A. Comparative genomic view of clostridial sporulation and physiology. Nat. Rev. Microbiol. 2005;3:969–78. doi: 10.1038/nrmicro1288. [DOI] [PubMed] [Google Scholar]
- 34.Moir A., Corfe B.M., Behravan J. Spore germination. Cell Mol. Life Sci. 2002;59:403–9. doi: 10.1007/s00018-002-8432-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Allen M.M. Cyanobacterial cell inclusions. Annu. Rev. Microbiol. 1984;38:1–25. doi: 10.1146/annurev.mi.38.100184.000245. [DOI] [PubMed] [Google Scholar]
- 36.Fuser G., Steinbuchel A. Analysis of genome sequences for genes of cyanophycin metabolism: identifying putative cyanophycin metabolizing prokaryotes. Macromol. Biosci. 2007;7:278–96. doi: 10.1002/mabi.200600207. [DOI] [PubMed] [Google Scholar]
- 37.Meyerholz D.K., Stabel T.J., Cheville N.F. Segmented filamentous bacteria interact with intraepithelial mononuclear cells. Infect. Immun. 2002;70:3277–80. doi: 10.1128/IAI.70.6.3277-3280.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Yamauchi K., Snel J. Transmission electron microscopic demonstration of phagocytosis and intracellular processing of segmented filamentous bacteria by intestinal epithelial cells of the chick ileum. Infect. Immun. 2000;68:6496–504. doi: 10.1128/iai.68.11.6496-6504.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Smith K.D., Andersen-Nissen E., Hayashi F., et al. Toll-like receptor 5 recognizes a conserved site on flagellin required for protofilament formation and bacterial motility. Nat. Immunol. 2003;4:1247–53. doi: 10.1038/ni1011. [DOI] [PubMed] [Google Scholar]
- 40.Uematsu S., Jang M.H., Chevrier N., et al. Detection of pathogenic intestinal bacteria by Toll-like receptor 5 on intestinal CD11c+ lamina propria cells. Nat. Immunol. 2006;7:868–74. doi: 10.1038/ni1362. [DOI] [PubMed] [Google Scholar]
- 41.Uematsu S., Fujimoto K., Jang M.H., et al. Regulation of humoral and cellular gut immunity by lamina propria dendritic cells expressing Toll-like receptor 5. Nat. Immunol. 2008;9:769–76. doi: 10.1038/ni.1622. [DOI] [PubMed] [Google Scholar]
- 42.Uematsu S., Akira S. Immune responses of TLR5+ lamina propria dendritic cells in enterobacterial infection. J. Gastroenterol. 2009;44:803–11. doi: 10.1007/s00535-009-0094-y. [DOI] [PubMed] [Google Scholar]
- 43.Wadsworth S.J., Goldfine H. Listeria monocytogenes phospholipase C-dependent calcium signaling modulates bacterial entry into J774 macrophage-like cells. Infect. Immun. 1999;67:1770–8. doi: 10.1128/iai.67.4.1770-1778.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ginocchio C., Pace J., Galan J.E. Identification and molecular characterization of Salmonella typhimurium gene involved in triggering the internalization of salmonellae into cultured epithelial cells. Proc. Natl Acad. Sci. USA. 1992;89:5976–80. doi: 10.1073/pnas.89.13.5976. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.