Abstract
Plant-specific NAC transcription factors (TFs) play important roles in regulating diverse biological processes, including development, senescence, growth, cell division and responses to environmental stress stimuli. Within the soybean genome, we identified 152 full-length GmNAC TFs, including 11 membrane-bound members. In silico analysis of the GmNACs, together with their Arabidopsis and rice counterparts, revealed similar NAC architecture. Next, we explored the soybean Affymetrix array and Illumina transcriptome sequence data to analyse tissue-specific expression profiles of GmNAC genes. Phylogenetic analysis using stress-related NAC TFs from Arabidopsis and rice as seeding sequences identified 58 of the 152 GmNACs as putative stress-responsive genes, including eight previously reported dehydration-responsive GmNACs. We could design gene-specific primers for quantitative real-time PCR verification of 38 out of 50 newly predicted stress-related genes. Twenty-five and six GmNACs were found to be induced and repressed 2-fold or more, respectively, in soybean roots and/or shoots in response to dehydration. GmNAC085, whose amino acid sequence was 39%; identical to that of well-known SNAC1/ONAC2, was the most induced gene upon dehydration, showing 390-fold and 20-fold induction in shoots and roots, respectively. Our systematic analysis has identified excellent tissue-specific and/or dehydration-responsive candidate GmNAC genes for in-depth characterization and future development of improved drought-tolerant transgenic soybeans.
Keywords: soybean, NAC transcription factors, dehydration, sequence analysis, expression analysis
1. Introduction
Cultivated soybean (Glycine max L.), which provides an abundant source of oil and protein-rich food for human consumption and animal feed, is one of the major and most important legume crops native to East Asia. Soybean growth, productivity and seed quality are adversely affected by a wide range of environmental stresses.1,2 Among the adverse environmental factors, drought is considered the most devastating abiotic stress. Drought stress affects all stages of plant growth and development, resulting in significant yield loss by ∼40%; as well as severely impacting seed quality.2 In response to drought stress, plants activate a number of defence mechanisms that function to increase tolerance to water deficit.3,4 The early events of the adaptation of plants to drought stress includes the perception of stress signals and subsequent signal transduction, leading to the activation of various physiological and metabolic responses.3,5,6 Within the signal transduction networks that are involved in the conversion of stress signal perception to stress-responsive gene expression, various transcription factors (TFs) and cis-acting elements contained in stress-responsive promoters function not only as molecular switches for gene expression, but also as terminal points of signal transduction in the signalling processes. The identification and molecular tailoring of novel TFs have the potential to overcome a number of important limitations involved in the generation of transgenic crop plants with superior yield under stress conditions.
Within higher plants, ∼7%; of their genomes encodes for putative TFs.7 Typically, the TFs contain a distinct type of DNA-binding domain and transcriptional regulation region (TRR) and are capable of activating or repressing the transcription of multiple target genes.3,5,8,9 The NAC TFs contain a highly conserved N-terminal DNA-binding NAC domain and a variable C-terminal TRR.10 Research in Arabidopsis has indicated that there are at least five different target DNA-binding sites for the NAC TFs. These include the drought-responsive NAC recognition sequence (NACRS) containing the CACG core motif; the iron deficiency-responsive IDE2 motif containing the core sequence CA(A/C)G(T/C)(T/C/A)(T/C/A); the CBNACBS-binding site of the Arabidopsis calmodulin-binding CBNAC protein having the GCTT as core-binding motif; the secondary wall NAC-binding element (SNBE) with 19-bp consensus sequence (T/A)NN(C/T) (T/C/G)TNNNNNNNA(A/C)GN(A/C/T) (A/T); and the 21-bp segment of the 35S promoter (−83 to −63) containing two core sequences CGTA and CGTC.11–17 In addition to DNA binding, the NAC domain also possesses the capacity for mediating protein:protein interactions.13,18 The highly variable C-terminal TRRs of NAC TFs can act as either a transcriptional activator or a repressor.11,14,19 Interestingly, the C-terminal domains of numerous NAC TFs also exhibit protein-binding activity.14 On the other hand, the C-terminal regions of some NAC TFs also contain transmembrane motifs (TMs) which are responsible for the anchoring to the plasma membrane. These NAC TFs are classified as membrane-associated and are designated as NTL (NTM1-Like or ‘NAC with Transmembrane Motif 1’-Like) TFs.20,21
NAC TFs have been shown to regulate a number of biological processes, including those which protect plants under water stress conditions.10,22,23 There are at least 105 ANAC and 140 ONAC members in Arabidopsis and rice (Oryza sativa), respectively.24–26 The first evidence demonstrating the involvement of NAC TFs in the improvement of drought tolerance in plants was reported in Arabidopsis by the identification and functional analyses of the ANAC019, ANAC055 and ANAC072 genes. Following this work, a number of studies on abiotic stress-related functions of NAC TFs in various plant species have been reported.2,4,11,19,27 The recent completion of the soybean genomic sequence has facilitated the prediction of at least 61 TF families in soybean, among which the plant-specific NAC TF family consists of more than 180 putative members.28–32 Given the importance of NAC TFs in diverse biological and physiological processes and their potential application for the development of improved drought-tolerant transgenic crop plants, we carried out a systematic analysis of the soybean NAC TF family in the present study. Putative GmNAC TFs predicted by genome-wide surveys of the soybean genomic sequence (Glyma 1 model) and those provided by various databases have been carefully analysed and subjected to phylogenetic analyses with their Arabidopsis and rice counterparts. These comparisons have enabled the identification of gene orthologs and clusters of orthologous groups that can be studied for further functional characterization. Taking advantage of the wealth of available expression data, which were generated by either high-throughput microarray expression profiling experiments or Illumina transcriptome sequencing, we also performed a comprehensive analysis of tissue-specific expression of all GmNAC genes, which in turn provided important complementary datasets to assist in the elucidation of their function. Furthermore, we used a time-coursed dehydration stress treatment and subsequent quantitative real-time PCR (qRT-PCR) analysis as a precise mechanism for detailing the root- and shoot-related expression patterns of predicted stress-responsive GmNAC genes. The results of this systematic analysis of the GmNAC family have enabled us to identify appropriate root- or shoot-related and/or dehydration-responsive GmNAC candidate genes and their respective promoters to be used as candidates in further in planta functional analyses. Ultimately, these findings will lead to potential applications for the improvement of drought resistance in soybean via genetic engineering.
2. Materials and methods
2.1. Plant growth, treatments and collection of tissues
Soybean cv. Williams 82 seeds were germinated in 6-l pots containing vermiculite and were well-watered and grown under greenhouse conditions (continuous 30°C temperature, photoperiod of 12 h/12 h, 80 µmol m−2 s−1 photon flux density and 60%; relative humidity). For tissue-specific expression profiling of GmNAC genes, root and shoot tissues were collected from 12-day-old soybean plants in three biological replicates. For expression profiling of GmNAC genes under dehydration stress, the dehydration treatment was carried out in time-course experiments to identify dynamic changes in transcripts in response to dehydration stress as previously described.33 Root and shoot tissues were collected separately in three biological replicates for the expression profiling studies.
2.2. Identification of the GmNAC members in soybean
All GmNAC members predicted in soybean were collected for manual analysis,28,30,31 and only those GmNAC proteins containing full open reading frames (ORFs), as predicted by Glyma1 model, were used for further analyses. We used the TMHHM server ver. 2.0 (http://www.cbs.dtu.dk/services/TMHMM/) to enable the prediction of the membrane-bound GmNAC members.
2.3. Phylogenetic analysis
Sequence alignments of related proteins belonging to each class from Arabidopsis, rice and soybean proteins were performed with a gap open penalty of 10 and a gap extension penalty of 0.2 using ClustalW implemented on MEGA 4 software.34,35 The alignments were subsequently visualized using GeneDoc (http://www.nrbsc.org/gfx/genedoc/) as presented in Supplementary Fig. S1.36 The sequence alignments were also used to construct the unrooted phylogenetic trees by the neighbour-joining method using MEGA4 software. The confidence level of monophyletic groups was estimated using a bootstrap analysis of 10 000 replicates. Only bootstrap values higher than 40%; are displayed next to the branch nodes.
2.4. Soybean affymetrix microarray data analysis
Gene expression data available for each putative soybean GmNAC gene were retrieved from the soybean gene expression data housed within the Genevestigator database by correspondences between soybean genes from the Glyma1 model and probe identifiers from Affymetrix GeneChip probes.37 The respective model IDs used in the Glyma1 model for the respective model IDs that were used in the Affymetrix Genechip were identified by Soybase (http://soybase.org/AffyChip/index.php).38 A total of 61 probes were found and corresponded to 48 GmNAC genes in this study. These probes were subsequently used for data retrieval and analysis.
2.5. Soybean Illumina expression data analysis
We utilized Illumina transcriptome sequencing data that was previously generated and analysed by Libault et al.39,40 to evaluate the expression of GmNAC genes. Sequencing data included transcriptome analyses from 13 different conditions, including nodules, roots, root tips, leaves, flowers, green pods, apical meristem, mock-inoculated and Bradyrhizobium japonicum-infected root hair cells harvested at 12, 24 and 48 h after inoculation.
2.6. RNA isolation, DNaseI treatment, cDNA synthesis
Plant tissue samples were ground into a fine powder using a mortar and pestle. Total RNA was isolated using the TRIZOL reagent according to the manufacturer's supplied protocol (Invitrogen). RNA concentration, DNaseI treatment and cDNA synthesis were performed as previously described.33
2.7. qRT-PCR and statistical analyses
Gene-specific primers for soybean GmNAC genes were designed using the Primer3 software.41 Primer specificity was first confirmed by blasting each primer sequence against the soybean genome (Glyma1 model).32 We performed subsequent analysis of melting curves and visualization of amplicon fragments. Primers were found gene-specific if corresponding melting curves yielded a single sharp peak and if the primers exhibited an electrophoresis pattern of a single amplicon with the correct predicted length. Under these strict criteria, we were able to design primers for 42 out of 58 putative stress-related GmNAC genes (Supplementary Table S1). As previously described, the CYP2 gene was selected as a reference gene for the expression profiling of soybean genes.42 qRT-PCR reactions and data analyses were performed as previously described.33 When appropriate, a Student's t-test (one tail, unpaired, equal variance) was used to determine the statistical significance of the differential expression patterns between tissues and/or between treatments. Differential expression data were regarded as statistically significant only when passing the t-test with a P-value of <0.05. Considering the biological significance of the differential expression in this study, we adopted a cut-off value of 3-fold for tissue-specific expression and 2-fold when analysing stress induction or repression. The expression levels were designated as ‘different’, ‘induced’ or ‘repressed’ only if such differences met the above criteria and passed the Student's t-test.
3. Results and discussion
3.1. Identification and chromosomal distribution of the GmNAC members in soybean
The GmNAC members in soybean have been predicted by three independent groups using genome-wide screening of soybean genomic sequences (Glyma1 model). However, each group provided different numbers of putative GmNAC TFs within their public databases. The highest number of GmNAC TFs (226) was reported by SoyDB, while SoybeanTFDB and PlantTFDB predicted 205 and 183 putative GmNACs, respectively.28,30,31,43,44 As an initial step, we collected the sequences for all of the putative GmNAC TFs from three databases for sequence comparison. Among all of the predicted putative GmNAC TFs, 152 members were found to contain full ORFs as predicted by the Glyma1 model. Only these predicted full-length GmNAC TFs were used for further analyses. If Glyma1 predicted several splice variants for a given gene, all of the alternative splice variants were carefully checked. Splice variants encoding the longest reading frames were selected as representatives for subsequent sequence alignments and phylogenetic analyses. For these studies, we also utilized the soybean FL-cDNA information that is also publicly available (http://rsoy.psc.riken.jp/).
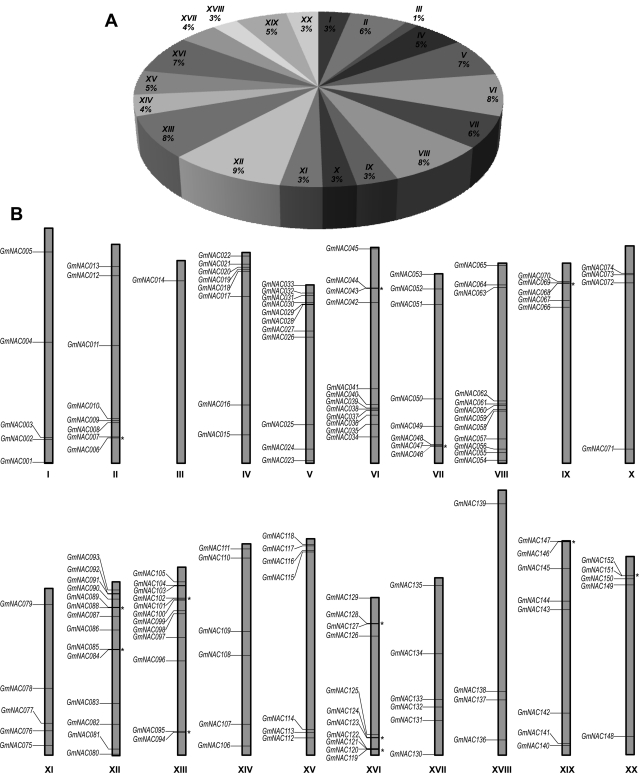
The GmNAC genes are distributed on every chromosome in soybean (Fig. 1A). Chromosome 12 contains the highest number of GmNACs, with14 out of 152 members (∼9%), while chromosomes 3 only contains one member (<1%; Fig. 1A). The relative locations of the GmNAC genes were indicated on their respective chromosome and genes located within 20 loci from each other were marked with a star to indicate possible tandem duplications (Fig. 1B). Supplementary Table S2 provided significant information including gene IDs as defined by the Glyma1 model for each predicted GmNAC TF, lengths of amino acid sequences and corresponding available full-length cDNA (FL-cDNA) accession numbers (RIKEN) for the identified 152 full-length GmNAC TFs. Additionally, the cDNAs and protein sequences annotated by the Glyma1 model of these 152 GmNAC TFs are provided as additional data that can be easily downloaded for convenient use (Supplementary Dataset 1). A uniform nomenclature for all the GmNAC genes identified in this work and those, which were previously characterized,42,45,46 has been adopted taking into account the order of the chromosomes to facilitate scientific communication (Supplementary Table S2).
Figure 1.
Chromosomal distribution of 152 soybean GmNAC genes identified in this study. (A) Abundant distribution of GmNAC genes on each soybean chromosome with indication of percentages of GmNACs located on each chromosome. (B) Graphical representation of locations for putative GmNAC genes on each soybean chromosome. The stars on the right of each chromosome indicate tandem duplicated genes. Greek numbers indicate chromosome numbers.
3.2. Structural and phylogenetic analyses of GmNAC TFs
In order to examine the structure of GmNAC TFs identified in our study, we performed a phylogenetic analysis of their deduced protein sequences together with three representative ANACs of Arabidopsis11 (Supplementary Fig. S1). As expected, most of the GmNAC TFs shared a highly conserved N-terminal DNA-binding domain of the typical NAC domain containing five consensus subdomains and a highly variable C-terminal transcriptional regulation domain. Several GmNACs do not contain the typical NAC domain. For example, Gm04g08320/GmNAC015, Gm13g18620/GmNAC096 and Gm19g08510/GmNAC142 lack the conserved A and B subdomains and Gm08g18050/GmNAC060 lacks the conserved C and D subdomains. In these cases, these proteins are described as NAC-like proteins, according to the classification of ONACs of rice.25 Additionally, all of the examined GmNAC TFs, with the exception of Gm05g32470/GmNAC028 and Gm08g18050/GmNAC060, contain a conserved bipartite nuclear localization signal. This signal sequence has been identified in the D subdomain from many NAC domain proteins among different plant species, suggesting that these GmNACs are nuclear localized (Supplementary Fig. S1).47
To study the evolutionary relationship between the GmNAC TFs and among the NAC TFs from different plant species, all GmNACs and NAC TFs from the dicot (Arabidopsis) and monocot (rice) models systems were subjected to a multiple sequence alignment. The multiple sequence alignment file was then used for the construction of an unrooted phylogenetic tree. As illustrated in Supplementary Fig. S2, the phylogenetic analysis classified the GmNACs into a number of different subgroups together with their ANAC and ONAC orthologs. Similar to Arabidopsis and rice, these data identified the existence of a diversified GmNAC family in soybean with diverse functions.25,26 Interestingly, among the subgroups identified by phylogenetic analysis, one subgroup only contains rice ONACs. This finding suggests that NACs from monocots and dicots are evolutionarily distinct. Specifically, ONACs originate from rice and the ANACs and GmNACs are exclusive to Arabidopsis and soybean, respectively (Supplementary Fig. S2).
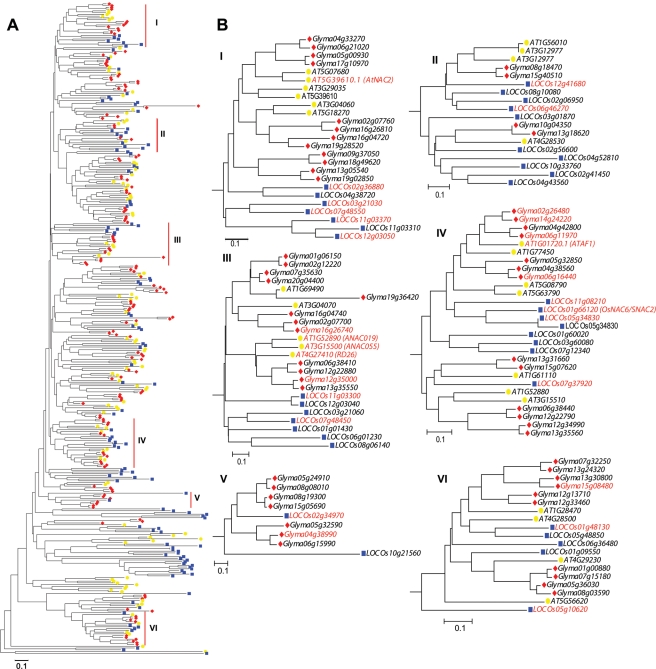
One of our main interests for performing phylogenetic analysis of GmNAC TFs was to enable the prediction of abiotic stress-responsive GmNAC genes that could be subsequently prioritized for further in planta functional studies. Previous reports have provided strong evidence for phylogenetic analysis-based prediction of the stress-related function of several gene families, including TF families. Phylogenetic analysis of the soybean AP2_EREBP and rice ONAC families with their orthologs from other plant species, whose stress-responsive expression patterns and/or functions are known, resulted in a nearly perfect match between sequence conservation and functions or expression patterns.25,48 Among the ANACs and ONACs, many proteins which are involved in the regulation of physiological and biochemical responses are associated with resistance to various abiotic stresses, including drought.10 For instance, Arabidopsis ANAC019, ANAC055 and ANAC072 and rice SNAC1/ONAC002 and OsNAC6/SNAC2/ONAC048 act as positive regulators in drought stress response.11,18,19,27 On the basis of sequence alignments and phylogenetic analyses, which included a number of known stress-related NAC TFs from Arabidopsis and rice, we identified 50 new putative stress-related GmNAC genes. These predicted GmNACs clustered into six monophyletic groups (Fig. 2A and B). Eight of nine GmNAC genes (names are highlighted by red colour), which were previously reported as dehydration-responsive genes,42 were also clustered in the stress-responsive clades III, IV, V and VI. These data suggest that the phylogenetic analysis-based gene identification approach is reliable for rationalizing systematic functional predictions of different TF families (Fig. 2A).42,46,49
Figure 2.
Phylogenetic analysis-based prediction of abiotic stress-related GmNAC genes. (A) Phylogenetic relationship of NAC proteins from Arabidopsis (yellow), rice (blue) and soybean (red). The unrooted phylogenetic tree was constructed using the full ORFs of NAC proteins. Numbers in Greek letters indicate clades with known stress-responsive members. (B) Details of stress-responsive clades. A total of 50 GmNAC new stress-responsive genes were predicted based upon phylogenetic analysis. Known stress-responsive NAC genes, including the eight previously reported dehydration-responsive GmNAC genes, are coloured in red.
3.3. The membrane-bound GmNAC subfamily
It has been established that the activities of TFs are co-ordinately regulated at multiple steps through transcriptional, posttranscriptional, posttranslational and translocational mechanisms.50,51 A number of these proteins are expressed as membrane-bound TFs (MTFs) and are stored in their dormant forms, and the degradation of their cytoplasmic anchors is required for their activation.50 The activated TFs enter the nucleus where they are then capable of functioning to regulate the expression of their respective target genes. There are at least 85 and 45 MTFs in Arabidopsis and rice genomes, respectively, and virtually all of the MTFs exist in all major TF families. Within the NAC family, a genome-wide analysis predicted at least 18 and 5 MTFs in Arabidopsis and rice, respectively.21 Each of the NAC MTFs, which are also called NTLs in other publications, contains an α-helical TM in their C terminal regions. This TM is responsible for anchoring to either plasma membranes or endoplasmic reticulum membranes and is involved in the regulation of activities of NTLs primarily at the processing step.20
Among the 152 soybean GmNACs, 11 GmNAC MTFs were predicted based upon the existence of the TMs identified using the TMHMM server 2.0 (Table 1; Supplementary Table S2). All Arabidopsis and rice NAC MTFs members have been predicted to contain a single TM.21 In contrast, 9 of the 11 identified GmNAC MTFs were found to contain a single TM, whereas the remaining two (GmNAC013 and GmNAC136) contain two TMs (Table 1). A phylogenetic tree of the NAC MTFs from soybean, Arabidopsis and rice was constructed and is visualized in Fig. 3. Strong lines of evidence have indicated that a number of the functionally characterized NAC MTFs are closely related with plant responses to environmental stresses.20,50 In Arabidopsis, studies have shown that at least four NTLs (NTL6/At3g49530, NTL8/At2g27300, NTL9/At4g35580 and NTL12/NTM1/At4g01540) function in stress responses. These NTLs are activated by membrane-associated proteases in the endoplasmic reticulum by liberating the TFs from their TM domain when plants experience environmental stresses.20,52,53 Thus, it is feasible that a number of the GmNAC MTFs identified in this study play an important role in gene regulatory networks that serve as an adaptive strategy for soy plants to survive under adverse growth conditions.
Table 1.
Putative membrane-bound soybean GmNAC TFs
| Names | Old namesa | Gene model | Length (a.a) | Transmembrane sequencesb |
|
|---|---|---|---|---|---|
| GmNAC012 | GmNAC027 | Glyma02g38710.1 | 589 | 565–587 | |
| GmNAC013 | Glyma02g40750.1 | 584 | 508–527 | 561–578 | |
| GmNAC021 | GmNAC025 | Glyma04g40450.1 | 603 | 579–601 | |
| GmNAC036 | GmNAC026 | Glyma06g14290.1 | 598 | 574–596 | |
| GmNAC074 | Glyma10g36360.1 | 560 | 529–551 | ||
| GmNAC103 | Glyma13g39090.1 | 422 | 330–352 | ||
| GmNAC110 | Glyma14g36840.1 | 590 | 566–588 | ||
| GmNAC111 | Glyma14g39080.1 | 600 | 524–543 | ||
| GmNAC136 | Glyma18g05020.1 | 631 | 533–552 | 609–628 | |
| GmNAC149 | GmNAC024 | Glyma20g31210.1 | 549 | 518–540 | |
| GmNAC151 | Glyma20g33390.1 | 609 | 584–606 | ||
aOld names were given as in Tran et al.42
bTransmembrane segments were predicted using the TMHMM server 2.0 (http://www.cbs.dtu.dk/services/TMHMM/).
Figure 3.
Phylogenetic relationship of NAC MTFs from Arabidopsis, rice and soybean. The unrooted phylogenetic tree was constructed using the full ORFs of NAC proteins. The bar indicates the relative divergence of the sequences examined and bootstrap values are displayed next to the branch.
3.4. Analysis of expression patterns of GmNAC genes using Affymetrix arrays
Tissue-specific expression profiles are useful data because they identify the genes which are involved in defining the precise nature of individual tissues. It is well established that the mechanisms controlling drought resistance are either associated with root- and/or shoot-related traits.1 For instance, an extensive fibrous root system can be useful for foraging subsoil surface moisture and nutrients such as phosphorus. In addition, plants can adapt to drought stress by developing a longer taproot which enables the plant to reach lower soil layers where water is more readily available. On the other hand, a restraint of shoot growth has been shown to be advantageous in adverse environments by minimizing evaporative leaf surface area. Hence, the appropriate control of shoot- and root-related morphological traits is a promising approach for developing drought resistance in a number of crops, including soybean.1,54,55 In Arabidopsis, the CUC1, CUC2 and CUC3 (CUP-SHAPED COTYLEDON) NAC TFs have been shown to be involved in shoot apical meristem formation and development,22,56 while NAC1 and AtNAC2 are involved in the regulation of lateral root development.13,57 Overexpression of the NAC1 and AtNAC2 genes, which are preferentially expressed in roots, promoted lateral root development in Arabidopsis.13,57 These data suggest that tissue-specifically expressed NAC genes have the potential to be used for the genetic engineering of specific traits.
As a result, we first utilized the Affymetrix array data housed within Genevestigator to examine the specific expression patterns of GmNAC genes.37 This was our first approach towards the identification of candidate genes that could be potentially used for enhancing drought resistance by altering shoot and/or root growth when overexpressed or repressed in transgenic plant systems. The Affymetrix Soybean Array GeneChip was specifically designed to analyse ∼37 500 soybean, 15 800 Phytophthora sojae as well as 7500 Heterodera glycines transcripts. The current Affymetrix array data available at Genevestigator contain measurements of transcript levels for 35 different organs and tissues. The respective model IDs of GmNAC genes used in the Glyma1 model for the respective model IDs that were used in the Affymetrix Genechip were identified by Soybase (http://soybase.org/AffyChip/index.php).38
We confirmed that probes exist for a total of 48 GmNAC genes on the soybean GeneChip. The heat map shown in Supplementary Fig. S3 displays the patterns of expression of these GmNAC genes within 35 major organs and tissues. Clustering analysis of the expression data indicates high variability in transcript abundance of the GmNAC genes. Among the 48 GmNAC genes, many are highly and specifically expressed in roots and/or leaves, suggesting that these could be candidate genes for the potential engineering of plant responses within those specific tissue types. Only a small portion of GmNAC genes were found to be ubiquitously expressed in all of the examined tissues. The data supplied here are useful to assess the extent of GmNAC gene expression, as they provide the first line of temporal and spatial evidence which links them to putative in planta functions. With respect to the response to P. sojae infections, our clustering analysis detected a group of 10 GmNAC genes which showed strong induction in all of the studies examined (Supplementary Fig. S4). These data suggest that these GmNACs potentially function in response to P. sojae infections.
3.5. Analysis of expression patterns of GmNAC genes using Illumina transcriptome data
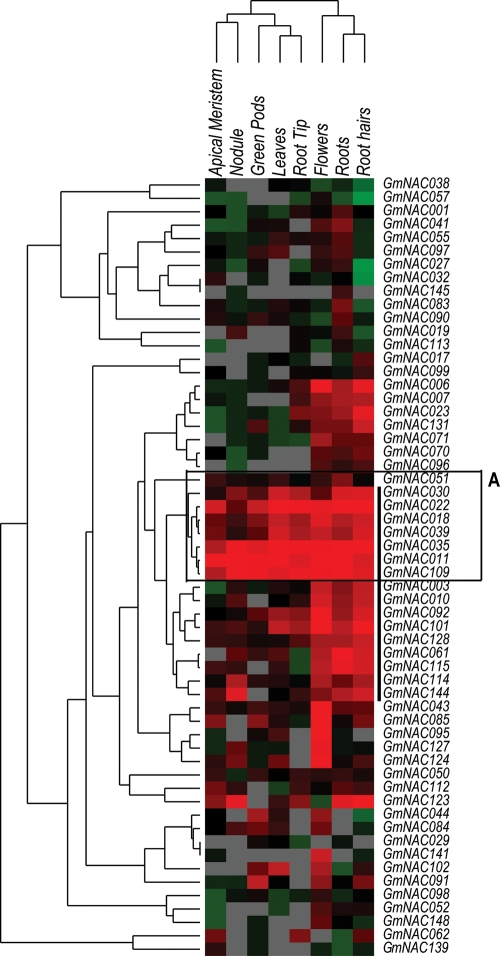
Since the current soybean Affymetrix Genechip platform was limited and did not enable analysis for all 152 GmNAC genes, we also utilized transcriptome data derived from Illumina sequencing of soybean short transcripts to assess the expression patterns of all GmNAC genes. This transcriptome atlas provided expression data for 55 616 putative soybean genes in eight types of tissues and organs, including root tips, roots, root hairs, nodules, leaves, shoot apical meristems, flowers and green pods.40 Although there were fewer tissues and organs examined in comparison to Affymetrix Genechip data, expression profiles for all 152 GmNAC genes could be investigated. Consistent with observations from Affymetrix Genechip data, the results shown in Supplementary Fig. S5A indicate high variability in the transcript abundance of GmNAC genes. As shown in the heat maps, the spatial expression patterns of numerous GmNAC genes are tissue-specific, while others are ubiquitous. These observations indicate that the functions of the GmNACs are diversified in a similar manner as that of their Arabidopsis counterparts.10,22,58 Figure 4 highlighted the expression patterns of 58 GmNAC genes, which were predicted as stress-related genes by phylogenetic analysis. Eight genes were expressed strongly and ubiquitously in all eight tissues (Fig. 4, box A), while 16 genes were preferentially expressed in roots, root hairs and flowers (Fig. 4, black bar at right). Interestingly, among these 16 genes, only seven genes were also found to be preferentially transcribed in root tips (Fig. 4). The tissue-specific and stress-related genes could serve as good candidates for the engineering of stress-related traits under stress conditions. Additionally, for those who have an interest in studying the functions of GmNACs in response to B. japonicum innoculations, expression data for all 152 GmNAC genes in mock-inoculated and B. japonicum-infected root hair cells harvested at 12, 24 and 48 h after inoculation are summarized and provided on Supplementary Fig. S4. The expression of a number of GmNAC genes was remarkably altered after the infection with the bacterium.
Figure 4.
Heat map representation for tissue-specific expression of 50 predicted stress-responsive and 8 previously reported dehydration-responsive GmNAC genes. Expression patterns of 58 GmNAC genes were analysed using Illumina transcriptome data. The colours indicate expression intensity (red, high expression; green, low expression; grey, no expression). Box A indicates a group of ubiquitously expressed GmNAC genes in the eight types of tissues examined. The black bar indicates a group of 16 GmNAC genes highly expressed in roots, root hairs and flowers.
The tissue-specific expression data analysed using Affymetrix Genechip data and Illumina sequencing of soybean short transcripts can be used to address the combinatorial usage of GmNAC TFs, enabling great precision and flexibility in dictating the transcriptional program of different tissues. On the other hand, ubiquitously expressed GmNACs alone, in isolation or in combination with each other or with other type(s) of TFs, might control general cellular machinery. Combinations of specific and/or stress-related GmNACs with other type(s) of TFs might regulate tissue-specific and/or stress-responsive downstream genes. Alternatively, ubiquitous GmNACs might serve as a platform to regulate a broad set of genes which are subsequently fine-tuned by specific regulators. Molecular dynamics involving extensive protein–protein interactions, such as specific homodimerizations and heterodimerizations, as well as modular flexibility and posttranslational modifications, have been shown to determine the functional specificity of TFs. Analysis of such interactions will help elucidate patterns of combinatorial regulation and will ultimately help define the regulatory functions of the GmNAC TFs themselves.3,18,59,60
3.6. Expression patterns of predicted GmNAC genes in roots and shoots during dehydration stress
In a previous section, we used sequence similarity comparisons and phylogenetic analyses to predict 58 stress-related members, of which 50 were new, among all 152 GmNAC genes (Fig. 2). With the goal of identifying candidate dehydration-responsive GmNAC genes for the engineering of soybean plants with improved drought resistance, we aimed to first perform systematic expression profiling of GmNACs prior to launching laborious in planta functional studies for multiple GmNAC genes. Specifically, we employed qRT-PCR analysis of 38 predicted stress-related GmNAC genes with gene-specific primers in root and shoot tissues of 12-day-old soybean plants subjected to 2 and 10 h dehydration stress. Five (GmNAC011, GmNAC019, GmNAC 043, GmNAC092 and GmNAC109) and three (GmNAC041, GmNAC061 and GmNAC102) GmNAC genes previously reported as dehydration-responsive and dehydration-unresponsive, respectively, were also included in the qRT-PCR experiment as decoys to verify the positive and negative discovery rates (Supplementary Table S2).42 Additionally, the evaluation of expression patterns in individual stressed tissues, rather than whole plants, might provide information on the mode of action of stress-responsive genes in specific tissues.
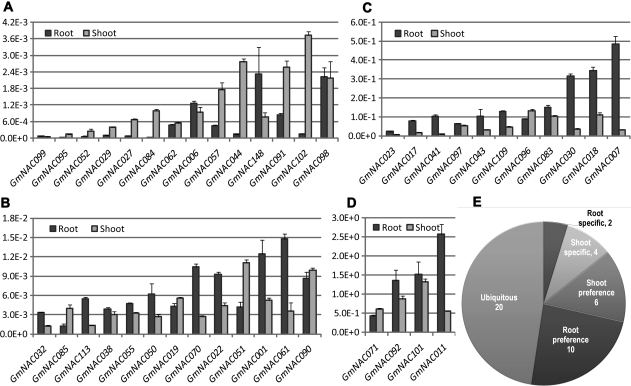
On the other hand, in comparison with the in silico expression analyses based on either the Affymetrix Genechip platform or Illumina sequencing, this experimental design also provided us with more reliable information on the expression patterns of these 42 GmNAC genes in root and shoot tissues of soybean seedlings grown under normal conditions. The expression of all 42 genes was detectable under our experimental conditions and the qRT-PCR results allowed us to classify the 42 GmNAC genes into four groups based on their transcript abundance detected in roots and shoots (Fig. 5A–D). Twenty-two of the 42 GmNAC genes were preferentially or specifically expressed in either roots or shoots of seedlings according to the criteria defined for the analysis of tissue-specific expression (Fig. 5E).40 Specifically, 12 and 10 GmNAC genes were preferably or specifically expressed in root and shoot tissues, respectively, whereas 20 out of 42 GmNAC genes tested were ubiquitously expressed in roots and shoots (Fig. 5).
Figure 5.
Expression patterns of 38 predicted stress-responsive and 4 decoy dehydration-responsive GmNAC genes in root and shoot tissues of soybean seedlings under normal conditions. (A–D) The GmNAC genes were classified into four groups based upon their expression levels. (E) Number of GmNAC genes expressed ubiquitously or tissue-specifically in roots or shoots.
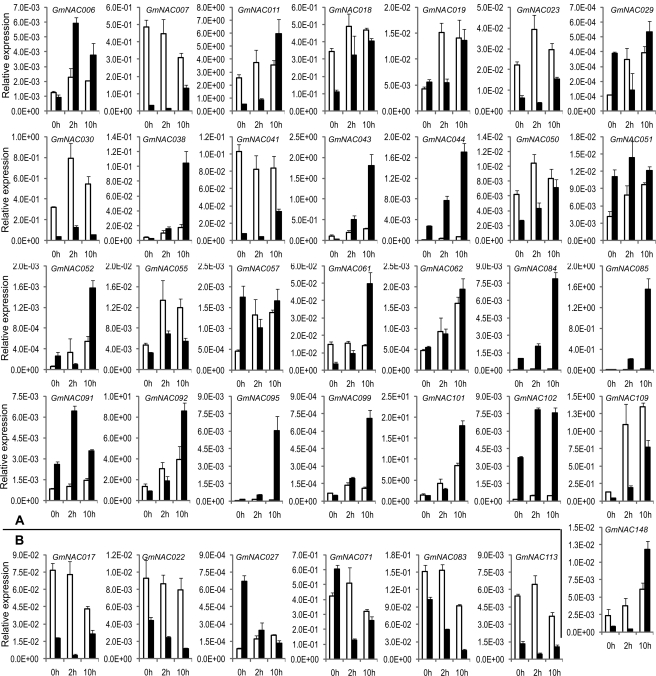
As for dehydration-responsive expression, a significant number of soybean GmNAC genes were found to be dehydration-responsive in either roots or shoots or both tissues (Fig. 6). A total of 29 induced and six repressed GmNAC genes were identified with a fold change of two or more, representing 83%; of the 42 genes examined being stress-related. Among 29 GmNAC genes, including four decoy dehydration-responsive genes, 4, 13 and 12 genes were induced in root, shoot and both tissues, respectively (Fig. 6A). As for the repressed GmNAC genes, expression of all six genes was down-regulated by dehydration in the shoots (Fig. 6B). Expression levels of GmNAC genes that did not respond to dehydration were not shown. Overall, the qRT-PCR verification demonstrated that the sequence similarity-based method has an accuracy rate of 83%; for the stress-related GmNAC genes. This rate suggests that this sequence similarity-based targeted gene identification approach has great potential for genome-wide prediction for stress-related TFs in plants or other species. Additionally, among the induced GmNAC genes, GmNAC085 was the most induced by dehydration with 390-fold and 20-fold induction in shoots and roots, respectively. The protein encoded by GmNAC085 exhibited 39%; and 50%; identity and similarity, respectively, to the most extensively characterized rice NAC TF (SNAC1/ONAC02) which conferred drought tolerance to transgenic rice plants under field conditions.19 GmNAC085, therefore, appears to be an excellent candidate for further in planta studies in soybean.
Figure 6.
Identification of dehydration-responsive GmNAC genes. Expression patterns of 38 predicted stress-responsive and 4 decoy dehydration-responsive GmNAC genes were analysed in dehydration-treated root (white bars) and shoot (black bars) tissues of soybean seedlings. Illustrated genes were either up-regulated by at least 2-fold (A) or down-regulated at least 2-fold (B) by dehydration stress in root and/or shoot tissues.
3.7. Conclusions
The focusing of research efforts on uncharacterized TFs using high-throughput genomic surveys and the analysis of huge resources of available expression data to describe key features of novel TFs, in combination with a detailed examination using traditional molecular approaches, will undoubtedly accelerate our functional understanding of these important regulatory genes. This report has provided the comprehensive identification and characterization of the soybean NAC TF family, with a special emphasis on the relation to dehydration stress responsiveness. Additionally, our results have provided useful information by identifying candidate tissue-specific and/or dehydration-responsive GmNAC genes. By combining these genes with their associated dehydration-responsive promoters, scientists will be able utilize these resources to engineer soybean plants for enhanced stress resistance. The foundation of knowledge presented in this work has revealed the diverse functions of the GmNAC TFs in different biological aspects. Future follow-up studies will rapidly improve our understanding of the regulatory function of NAC members. A greater understanding of how TFs operate will be subsequently translated into their potential applications to enhance plant productivity.
Supplementary data
Supplementary Data are available at www.dnaresearch.oxfordjournals.org.
Funding
D.T.L. is supported by the RIKEN Foreign Postdoctoral Fellowship. This work was funded by the Grants-in-Aid (Start-up) for Scientific Research (21870046) from the Ministry of Education, Culture, Sports, Science and Technology of Japan to L.-S.T.
Supplementary Material
References
- 1.Manavalan L.P., Guttikonda S.K., Tran L.S., Nguyen H.T. Physiological and molecular approaches to improve drought resistance in soybean. Plant Cell Physiol. 2009;50:1260–76. doi: 10.1093/pcp/pcp082. [DOI] [PubMed] [Google Scholar]
- 2.Tran L.S., Mochida K. Functional genomics of soybean for improvement of productivity in adverse conditions. Funct. Integr. Genomics. 2010;10:447–62. doi: 10.1007/s10142-010-0178-z. [DOI] [PubMed] [Google Scholar]
- 3.Yamaguchi-Shinozaki K., Shinozaki K. Transcriptional regulatory networks in cellular responses and tolerance to dehydration and cold stresses. Annu. Rev. Plant. Biol. 2006;57:781–803. doi: 10.1146/annurev.arplant.57.032905.105444. [DOI] [PubMed] [Google Scholar]
- 4.Hadiarto T., Tran L.S. Progress studies of drought-responsive genes in rice. Plant Cell Rep. 2011;30:297–310. doi: 10.1007/s00299-010-0956-z. [DOI] [PubMed] [Google Scholar]
- 5.Tran L.S., Nakashima K., Shinozaki K., Yamaguchi-Shinozaki K. Plant gene networks in osmotic stress response: from genes to regulatory networks. Methods Enzymol. 2007a;428:109–28. doi: 10.1016/S0076-6879(07)28006-1. [DOI] [PubMed] [Google Scholar]
- 6.Valliyodan B., Nguyen H.T. Understanding regulatory networks and engineering for enhanced drought tolerance in plants. Curr. Opin. Plant Biol. 2006;9:189–95. doi: 10.1016/j.pbi.2006.01.019. [DOI] [PubMed] [Google Scholar]
- 7.Udvardi M.K., Kakar K., Wandrey M., et al. Legume transcription factors: global regulators of plant development and response to the environment. Plant Physiol. 2007;144:538–49. doi: 10.1104/pp.107.098061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Yamaguchi-Shinozaki K., Shinozaki K. Organization of cis-acting regulatory elements in osmotic- and cold-stress-responsive promoters. Trends Plant Sci. 2005;10:88–94. doi: 10.1016/j.tplants.2004.12.012. [DOI] [PubMed] [Google Scholar]
- 9.Nakashima K., Ito Y., Yamaguchi-Shinozaki K. Transcriptional regulatory networks in response to abiotic stresses in Arabidopsis and grasses. Plant Physiol. 2009;149:88–95. doi: 10.1104/pp.108.129791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Tran L.S., Nishiyama R., Yamaguchi-Shinozaki K., Shinozaki K. Potential utilization of NAC transcription factors to enhance abiotic stress tolerance in plants by biotechnological approach. GM Crops. 2010;1:32–9. doi: 10.4161/gmcr.1.1.10569. [DOI] [PubMed] [Google Scholar]
- 11.Tran L.S., Nakashima K., Sakuma Y., et al. Isolation and functional analysis of Arabidopsis stress-inducible NAC transcription factors that bind to a drought-responsive cis-element in the early responsive to dehydration stress 1 promoter. Plant Cell. 2004;16:2481–98. doi: 10.1105/tpc.104.022699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ernst H.A., Olsen A.N., Larsen S., Lo Leggio L. Structure of the conserved domain of ANAC, a member of the NAC family of transcription factors. EMBO Rep. 2004;5:297–303. doi: 10.1038/sj.embor.7400093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Xie Q., Frugis G., Colgan D., Chua N.H. Arabidopsis NAC1 transduces auxin signal downstream of TIR1 to promote lateral root development. Genes Dev. 2000;14:3024–36. doi: 10.1101/gad.852200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kim H.S., Park B.O., Yoo J.H., et al. Identification of a calmodulin-binding NAC protein as a transcriptional repressor in Arabidopsis. J. Biol. Chem. 2007;282:36292–302. doi: 10.1074/jbc.M705217200. [DOI] [PubMed] [Google Scholar]
- 15.Ogo Y., Kobayashi T., Nakanishi Itai R., et al. A novel NAC transcription factor, IDEF2, that recognizes the iron deficiency-responsive element 2 regulates the genes involved in iron homeostasis in plants. J. Biol. Chem. 2008;283:13407–17. doi: 10.1074/jbc.M708732200. [DOI] [PubMed] [Google Scholar]
- 16.Zhong R., Lee C., Ye Z.H. Global Analysis of Direct Targets of Secondary Wall NAC Master Switches in Arabidopsis. Mol. Plant. 2010;3:1087–103. doi: 10.1093/mp/ssq062. [DOI] [PubMed] [Google Scholar]
- 17.Jensen M.K., Kjaersgaard T., Nielsen M.M., et al. The Arabidopsis thaliana NAC transcription factor family: structure-function relationships and determinants of ANAC019 stress signaling. Biochem. J. 2010;426:183–96. doi: 10.1042/BJ20091234. [DOI] [PubMed] [Google Scholar]
- 18.Tran L.S., Nakashima K., Sakuma Y., et al. Co-expression of the stress-inducible zinc finger homeodomain ZFHD1 and NAC transcription factors enhances expression of the ERD1 gene in Arabidopsis. Plant J. 2007;49:46–63. doi: 10.1111/j.1365-313X.2006.02932.x. [DOI] [PubMed] [Google Scholar]
- 19.Hu H., Dai M., Yao J., et al. Overexpressing a NAM, ATAF, and CUC (NAC) transcription factor enhances drought resistance and salt tolerance in rice. Proc. Natl. Acad. Sci. USA. 2006;103:12987–92. doi: 10.1073/pnas.0604882103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Seo P.J., Kim S.G., Park C.M. Membrane-bound transcription factors in plants. Trends Plant Sci. 2008;13:550–6. doi: 10.1016/j.tplants.2008.06.008. [DOI] [PubMed] [Google Scholar]
- 21.Kim S.G., Lee S., Seo P.J., Kim S.K., Kim J.K., Park C.M. Genome-scale screening and molecular characterization of membrane-bound transcription factors in Arabidopsis and rice. Genomics. 2010;95:56–65. doi: 10.1016/j.ygeno.2009.09.003. [DOI] [PubMed] [Google Scholar]
- 22.Olsen A.N., Ernst H.A., Leggio L.L., Skriver K. NAC transcription factors: structurally distinct, functionally diverse. Trends Plant Sci. 2005;10:79–87. doi: 10.1016/j.tplants.2004.12.010. [DOI] [PubMed] [Google Scholar]
- 23.Demura T., Ye Z.H. Regulation of plant biomass production. Curr. Opin. Plant Biol. 2010;13:299–304. doi: 10.1016/j.pbi.2010.03.002. [DOI] [PubMed] [Google Scholar]
- 24.Ooka H., Satoh K., Doi K., et al. Comprehensive analysis of NAC family genes in Oryza sativa and Arabidopsis thaliana. DNA Res. 2003;10:239–47. doi: 10.1093/dnares/10.6.239. [DOI] [PubMed] [Google Scholar]
- 25.Fang Y., You J., Xie K., Xie W., Xiong L. Systematic sequence analysis and identification of tissue-specific or stress-responsive genes of NAC transcription factor family in rice. Mol. Genet. Genomics. 2008;280:547–63. doi: 10.1007/s00438-008-0386-6. [DOI] [PubMed] [Google Scholar]
- 26.Nuruzzaman M., Manimekalai R., Sharoni A.M., et al. Genome-wide analysis of NAC transcription factor family in rice. Gene. 2010;465:30–44. doi: 10.1016/j.gene.2010.06.008. [DOI] [PubMed] [Google Scholar]
- 27.Nakashima K., Tran L.S., Van Nguyen D., et al. Functional analysis of a NAC-type transcription factor OsNAC6 involved in abiotic and biotic stress-responsive gene expression in rice. Plant J. 2007;51:617–30. doi: 10.1111/j.1365-313X.2007.03168.x. [DOI] [PubMed] [Google Scholar]
- 28.Mochida K., Yoshida T., Sakurai T., Yamaguchi-Shinozaki K., Shinozaki K., Tran L.S. In silico analysis of transcription factor repertoire and prediction of stress responsive transcription factors in soybean. DNA Res. 2009;16:353–69. doi: 10.1093/dnares/dsp023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Mochida K., Yoshida T., Sakurai T., Yamaguchi-Shinozaki K., Shinozaki K., Tran L.S. LegumeTFDB: an integrative database of Glycine max, Lotus japonicus and Medicago truncatula transcription factors. Bioinformatics. 2010a;26:290–1. doi: 10.1093/bioinformatics/btp645. [DOI] [PubMed] [Google Scholar]
- 30.Wang Z., Libault M., Joshi T., et al. SoyDB: a knowledge database of soybean transcription factors. BMC Plant Biol. 2010;10:14. doi: 10.1186/1471-2229-10-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.He K., Guo A.Y., Gao G., et al. Computational identification of plant transcription factors and the construction of the PlantTFDB database. Methods Mol. Biol. 2010;674:351–68. doi: 10.1007/978-1-60761-854-6_21. [DOI] [PubMed] [Google Scholar]
- 32.Schmutz J., Cannon S.B., Schlueter J., et al. Genome sequence of the palaeopolyploid soybean. Nature. 2010;463:178–83. doi: 10.1038/nature08670. [DOI] [PubMed] [Google Scholar]
- 33.Le D.T., Nishiyama R., Watanabe Y., et al. Genome-wide expression profiling of soybean two-component system genes in soybean root and shoot tissues under dehydration stress. DNA Res. 2011;18:17–29. doi: 10.1093/dnares/dsq032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Thompson J.D., Gibson T.J., Plewniak F., Jeanmougin F., Higgins D.G. The CLUSTAL_X windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res. 1997;25:4876–82. doi: 10.1093/nar/25.24.4876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Tamura K., Dudley J., Nei M., Kumar S. MEGA4: Molecular Evolutionary Genetics Analysis (MEGA) software version 4.0. Mol. Biol. Evol. 2007;24:1596–9. doi: 10.1093/molbev/msm092. [DOI] [PubMed] [Google Scholar]
- 36.Mochida K., Yoshida T., Sakurai T., Yamaguchi-Shinozaki K., Shinozaki K., Tran L.S. Genome-wide analysis of two-component systems and prediction of stress-responsive two-component system members in soybean. DNA Res. 2010b;17:303–24. doi: 10.1093/dnares/dsq021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hruz T., Laule O., Szabo G., et al. Genevestigator v3: a reference expression database for the meta-analysis of transcriptomes. Adv. Bioinformatics. 2008;2008:420747. doi: 10.1155/2008/420747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Grant D., Nelson R.T., Cannon S.B., Shoemaker R.C. SoyBase, the USDA-ARS soybean genetics and genomics database. Nucleic Acids Res. 2010;38:D843–6. doi: 10.1093/nar/gkp798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Libault M., Farmer A., Brechenmacher L., et al. Complete transcriptome of the soybean root hair cell, a single-cell model, and its alteration in response to Bradyrhizobium japonicum infection. Plant Physiol. 2010;152:541–52. doi: 10.1104/pp.109.148379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Libault M., Farmer A., Joshi T., et al. An integrated transcriptome atlas of the crop model Glycine max, and its use in comparative analyses in plants. Plant J. 2010;63:86–99. doi: 10.1111/j.1365-313X.2010.04222.x. [DOI] [PubMed] [Google Scholar]
- 41.Rozen S., Skaletsky H. Primer3 on the WWW for general users and for biologist programmers. Methods Mol. Biol. 2000;132:365–86. doi: 10.1385/1-59259-192-2:365. [DOI] [PubMed] [Google Scholar]
- 42.Tran L.S., Quach T.N., Guttikonda S.K., et al. Molecular characterization of stress-inducible GmNAC genes in soybean. Mol. Genet Genomics. 2009;281:647–64. doi: 10.1007/s00438-009-0436-8. [DOI] [PubMed] [Google Scholar]
- 43.Tran L.S., Mochida K. Identification and prediction of abiotic stress responsive transcription factors involved in abiotic stress signaling in soybean. Plant Signal Behav. 2010;5:255–7. doi: 10.4161/psb.5.3.10550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Tran L.S., Mochida K. A platform for functional prediction and comparative analyses of transcription factors of legumes and beyond. Plant Signal Behav. 2010;5:550–2. doi: 10.4161/psb.11088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Meng Q., Zhang C., Gai J., Yu D. Molecular cloning, sequence characterization and tissue-specific expression of six NAC-like genes in soybean (Glycine max (L.) Merr.) J. Plant Physiol. 2007;164:1002–12. doi: 10.1016/j.jplph.2006.05.019. [DOI] [PubMed] [Google Scholar]
- 46.Hao Y.J., Song Q.X., Chen H.W., et al. Plant NAC-type transcription factor proteins contain a NARD domain for repression of transcriptional activation. Planta. 2010;232:1033–43. doi: 10.1007/s00425-010-1238-2. [DOI] [PubMed] [Google Scholar]
- 47.Greve K., La Cour T., Jensen M.K., Poulsen F.M., Skriver K. Interactions between plant RING-H2 and plant-specific NAC (NAM/ATAF1/2/CUC2) proteins: RING-H2 molecular specificity and cellular localization. Biochem. J. 2003;371:97–108. doi: 10.1042/BJ20021123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Zhang G., Chen M., Chen X., et al. Phylogeny, gene structures, and expression patterns of the ERF gene family in soybean (Glycine max L.) J. Exp. Bot. 2008;59:4095–107. doi: 10.1093/jxb/ern248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Pinheiro G.L., Marques C.S., Costa M.D., et al. Complete inventory of soybean NAC transcription factors: sequence conservation and expression analysis uncover their distinct roles in stress response. Gene. 2009;444:10–23. doi: 10.1016/j.gene.2009.05.012. [DOI] [PubMed] [Google Scholar]
- 50.Kim S.Y., Kim S.G., Kim Y.S., et al. Exploring membrane-associated NAC transcription factors in Arabidopsis: implications for membrane biology in genome regulation. Nucleic Acids Res. 2007;35:203–13. doi: 10.1093/nar/gkl1068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Qin F., Sakuma Y., Tran L.S., et al. Arabidopsis DREB2A-interacting proteins function as RING E3 ligases and negatively regulate plant drought stress-responsive gene expression. Plant Cell. 2008;20:1693–707. doi: 10.1105/tpc.107.057380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Kim S.G., Lee A.K., Yoon H.K., Park C.M. A membrane-bound NAC transcription factor NTL8 regulates gibberellic acid-mediated salt signaling in Arabidopsis seed germination. Plant J. 2008;55:77–88. doi: 10.1111/j.1365-313X.2008.03493.x. [DOI] [PubMed] [Google Scholar]
- 53.Chen Y.N., Slabaugh E., Brandizzi F. Membrane-tethered transcription factors in Arabidopsis thaliana: novel regulators in stress response and development. Curr. Opin. Plant Biol. 2008;11:695–701. doi: 10.1016/j.pbi.2008.10.005. [DOI] [PubMed] [Google Scholar]
- 54.Achard P., Cheng H., De Grauwe L., et al. Integration of plant responses to environmentally activated phytohormonal signals. Science. 2006;311:91–4. doi: 10.1126/science.1118642. [DOI] [PubMed] [Google Scholar]
- 55.Huang J.G., Yang M., Liu P., Yang G.D., Wu C.A., Zheng C.C. GhDREB1 enhances abiotic stress tolerance, delays GA-mediated development and represses cytokinin signalling in transgenic Arabidopsis. Plant Cell Environ. 2009;32:1132–45. doi: 10.1111/j.1365-3040.2009.01995.x. [DOI] [PubMed] [Google Scholar]
- 56.Souer E., van Houwelingen A., Kloos D., Mol J., Koes R. The no apical meristem gene of Petunia is required for pattern formation in embryos and flowers and is expressed at meristem and primordia boundaries. Cell. 1996;85:159–70. doi: 10.1016/s0092-8674(00)81093-4. [DOI] [PubMed] [Google Scholar]
- 57.He X.J., Mu R.L., Cao W.H., Zhang Z.G., Zhang J.S., Chen S.Y. AtNAC2, a transcription factor downstream of ethylene and auxin signaling pathways, is involved in salt stress response and lateral root development. Plant J. 2005;44:903–16. doi: 10.1111/j.1365-313X.2005.02575.x. [DOI] [PubMed] [Google Scholar]
- 58.Demura T., Fukuda H. Transcriptional regulation in wood formation. Trends Plant Sci. 2007;12:64–70. doi: 10.1016/j.tplants.2006.12.006. [DOI] [PubMed] [Google Scholar]
- 59.Tran L.S., Nakashima K., Shinozaki K., Yamaguchi-Shinozaki K. Plant gene networks in osmotic stress response: from genes to regulatory networks. Methods Enzymol. 2007;428:109–28. doi: 10.1016/S0076-6879(07)28006-1. [DOI] [PubMed] [Google Scholar]
- 60.Vaquerizas J.M., Kummerfeld S.K., Teichmann S.A., Luscombe N.M. A census of human transcription factors: function, expression and evolution. Nat. Rev. Genet. 2009;10:252–63. doi: 10.1038/nrg2538. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.