Abstract
Arabidopsis belongs to the Brassicaceae family and plays an important role as a model plant for which researchers have developed fine-tuned genome resources. Genome sequencing projects have been initiated for other members of the Brassicaceae family. Among these projects, research on Chinese cabbage (Brassica rapa subsp. pekinensis) started early because of strong interest in this species. Here, we report the development of a library of Chinese cabbage full-length cDNA clones, the RIKEN BRC B. rapa full-length cDNA (BBRAF) resource, to accelerate research on Brassica species. We sequenced 10 000 BBRAF clones and confirmed 5476 independent clones. Most of these cDNAs showed high homology to Arabidopsis genes, but we also obtained more than 200 cDNA clones that lacked any sequence homology to Arabidopsis genes. We also successfully identified several possible candidate marker genes for plant defence responses from our analysis of the expression of the Brassica counterparts of Arabidopsis marker genes in response to salicylic acid and jasmonic acid. We compared gene expression of these markers in several Chinese cabbage cultivars. Our BBRAF cDNA resource will be publicly available from the RIKEN Bioresource Center and will help researchers to transfer Arabidopsis-related knowledge to Brassica crops.
Keywords: Arabidopsis, Brassica rapa, full-length cDNA, jasmonic acid, salicylic acid
1. Introduction
Brassica species include many important crop vegetables and oil seeds, such as cabbage, Brussels sprouts, broccoli, cauliflower, radish, mustard, oilseed rape, kale, and turnip. These species belong to the Brassicaceae (Cruciferae) family, which includes more than 3300 species.1 Arabidopsis thaliana is one of the most important species in the Brassicaceae family as it has been widely studied as an experimental model plant. Many useful genomic resources and other information are now available for A. thaliana (MASC report 2010: http://www.Arabidopsis.org/portals/masc/masc_docs/masc_reports.jsp). Sequencing of the Arabidopsis genome was completed in December 2000 by the Arabidopsis Genome Initiative (AGI 2000).2
Chinese cabbage (Brassica rapa subsp. pekinensis) originated in China and is one of the most important Brassica vegetables found in Asian countries. Presently, genome sequencing of B. rapa is being carried out as a multinational collaboration between China, the UK, Korea, Canada, Australia, and Japan (http://brassica.bbsrc.ac.uk/), and has recently entered the final phase.3 It has been estimated that the Chinese cabbage genome contains ∼46000 genes,4 whereas 27379 genes are currently believed to exist in the Arabidopsis genome (TAIR9 information: http://www.Arabidopsis.org/).
Arabidopsis and Brassica were originally estimated to have diverged from a common ancestor between 14.5 and 20.4 million years ago.5 Recently, Mun et al.6 reported that B. rapa diverged from the Brassica progenitor around 8 million years ago as a result of a whole-genome triplication event. They also performed genetic mapping for genome-wide comparative analyses, which revealed co-linear chromosome segments shared between Arabidopsis and B. rapa. Linkage arrangements between Arabidopsis and Brassica oleracea were also reported.7,8 In the light of these findings, knowledge gained from Arabidopsis becomes very useful for the study of Brassica crops. The development of a catalogue of gene transcripts from Brassica plants will serve as a critical resource for future molecular studies. Currently, 24963 Brassica unigene sequences have been obtained (http://www.brassica-rapa.org/BrEMD/microarray_overview.jsp) from different tissues, organs, or seeds and from different developmental stages of Brassica plants. However, this information is insufficient for the effective transfer of knowledge about Arabidopsis to Brassica crops.
Development of full-length cDNA collections is one of the effective strategies for increasing the catalogue of gene transcripts. These data serve as a valuable resource to describe gene expression profiles and ultimately classify genes into families based on their functions. Full-length cDNA collections can also serve as a powerful tool to facilitate genomic or other -omics research.9 Several techniques have been established to prepare enriched full-length cDNA libraries.10 The usefulness of full-length cDNAs has been confirmed in humans,11 mice,12 and in various plants such as Arabidopsis,13,14 rice,15 poplar,16 maize,17 tomato,18 and soybean.19 A major advantage of this approach is that most of the clones contain the complete coding sequences, in addition to the 5′- and 3′-untranslated regions. Ichikawa et al.20 reported the development of the full-length cDNA overexpressing gene hunting (FOX hunting) system. They reported that 10000 independent Arabidopsis full-length cDNAs were expressed in Arabidopsis plants under the control of the cauliflower mosaic virus (CaMV) 35S promoter. The FOX hunting system has also been applied to full-length cDNA clones from rice.21 This system has aided in the isolation of several important genes that confer tolerance to environmental stresses or resistance to biotic stresses.22–24
In the present study, we prepared a RIKEN BRC B. rapa full-length (BBRAF) cDNA library and cDNA clones, and we isolated 5476 independent sequences from 10 000 clones. In addition, we identified marker genes for the plant defence response in B. rapa based on homology to Arabidopsis genes and compared their expression in several B. rapa cultivars.
2. Materials and methods
2.1. Plant materials and cultivation
We grew Chinese cabbage (B. rapa subsp. pekinensis) plants in soil (Professional soil: Dio Chemicals, Tokyo, Japan). We used KmP02, a clubroot-resistant (CR) double-haploid line derived from a CR F1 cultivar of Chinese cabbage (Hatakeyama et al., unpublished result), for our expression analyses and for constructing the full-length cDNA library. We also used Kyoto No. 3, Kigokoro 85, Okiniiri, Muso (Takii Seed Co. Ltd, Kyoto, Japan), and Chifu hakusai (Tohoku Seed Co. Ltd, Utsunomiya, Japan) for expression analyses. Seeds were sown in sterile soil in pots, moistened, and held at 4°C for 7 days in the dark to synchronize germination. The pots were then transferred to 22°C with a long-day photoperiod (16 h light/8 h dark). Except as noted in subsequent sections, all plants were grown under these conditions for the various stress treatments that we applied. Plants at the four-leaf stage were transferred individually into new pots and grown to the rosette stage.
2.2. Foliar treatment with chemicals
To determine the effects of chemical stress on gene expression, we sprayed the leaves with solutions of 5 mM salicylic acid, 0.5 mM benzothiadiazole, 1 mM ethephon, 0.1 mM methyl jasmonate, 0.025 µM paraquat, 0.1 mM abscisic acid, or 10 mM CuSO4. The plants were then placed in a growth chamber at 22°C under a 16 h light/8 h dark cycle. Leaves were harvested at 5, 10, and 24 h after treatment.
2.3. Drought, salt, and cold stresses
To create drought stress, the seedlings were transferred from the soil pots onto dry filter paper and allowed to air-dry. For salt stress, the seedlings were transferred into 250 mM NaCl solution. For cold stress, seedlings were transferred into a refrigerated chamber, in which the temperature was controlled at 4°C. Leaves were harvested at 5, 10, and 24 h after treatment.
2.4. Wounding
To simulate wounding stress, the leaves were wounded with scissors. The plants were then placed in a growth chamber at 22°C under a 16 h light/8 h dark cycle. Leaves were harvested at 5, 10, and 24 h after treatment.
2.5. Ultraviolet radiation
Ultraviolet (UV-C) irradiation (0.1 kJ/m2) was supplied by a UV-C light. The plants were then placed in a growth chamber at 22°C under a 16 h light/8 h dark cycle. Leaves were harvested at 5, 10, and 24 h after treatment.
2.6. Root treatment with chemicals
The seedlings were transferred from soil pots into 0.5 mM benzothiadiazole solution or 1 mM ethephon solution. The roots were harvested 24 h after treatment.
2.7. RNA preparation
Total RNA was isolated using a modification of the method of Chirgwin et al.25 The tissues were ground to a fine in the presence of liquid nitrogen. The powder was then mixed with 10 volumes of GTC solution containing 4 M guanidinium thiocyanate, 25 mM sodium citrate (pH 7.0), 0.5% sodium N-lauroylsarcosine, and 0.1 M 2-mercaptoethanol. The cellular debris was pelleted out (14000g for 10 min at 4°C). About 2.4 ml of the supernatant was layered on top of 1.1 ml of 5.7 M CsCl cushion solution (5.7 M CsCl, 0.1 M EDTA) to create a step gradient and centrifuged at 240000g for 2 h at 20°C. The RNA pellet was dissolved in a mixture of 10 mM Tris–HCl (pH 7.5) and 5 mM EDTA (pH 8.0). The supernatant was extracted with successive, equal volumes of phenol–chloroform and chloroform. The upper phase was collected and mixed with one-third of its volume of 8 M LiCl. The RNA was precipitated at 4°C overnight and centrifuged at 14 000g for 30 min. The pellet was washed with 70% ethanol and dissolved in DEPC-treated water. Poly(A) + RNA was isolated by using Oligo(dT)-Latex (OligotexTM-dT30-Super; Roche, Tokyo, Japan) by following the manufacturer's instructions.
2.8. Construction of the cDNA library
Aliquots of total RNA extracted from the plant materials after each treatment were mixed equally to obtain one composite sample per treatment. The RNA mixture was used for the construction of an enriched full-length cDNA library based on the method of Kato et al.26 by using a vector-capping method. The resultant double-stranded cDNAs were ligated into a pGCAP10 vector. The primary library size was estimated at 2.4 × 106 colony-forming units (cfu) in the Chinese cabbage library.
2.9. Sequencing of both ends of the BBRAF cDNA clones
The plasmid DNA of each clone was prepared from bacterial cultures by using a NucleoSpin Multi-96 Plus Plasmid kit (Nihon Genetix Co.). End sequencing of 10 000 clones was carried out with an ABI 3700 capillary sequencer (Applied Biosystems, Foster City, CA, USA). The BBRAFpGCAP10F primer (5′-ACTGCTCCTCAGTGGATGTT-3′) and the BBRAF-polyT primer [mixture of equal amounts of 5′-T(30)AA-3′, 5′-T(30)AT-3′, 5′-T(30)AC-3′, 5′-T(30)AG-3′, 5′-T(30) CA-3′, 5′-T(30)CT-3′, 5′-T(30)CC-3′, 5′-T(30)CG-3′, 5′-T(30)GA-3′, 5′-T(30)GT-3′, 5′-T(30)GC-3′, and 5′-T(30)GG-3′] were used for forward and reverse sequencing, respectively.
2.10. Trimming and assembly of sequence data
Raw sequence data (chromatograms) were base-called using version X of the Phred software and vector sequences were then detected by using cross_ match. We trimmed off low-quality regions (for which the average quality score of a five-base window was <20) and the vector sequences of both ends of each read. Poly-A sequences of 15 or more bases were also trimmed off if found at the end of 5′ expressed sequence tags. Sequences that were shorter than 30 bases and left behind after the trimming process were also omitted from further analysis. The expressed sequence tags were assembled using version X of the PCAP.REP software without mate-pair constraints27 as the insert size of each clone could not be assigned a priori. All sequences were submitted to the DNA Databank of Japan (DDBJ).
2.11. Annotation of the sequences
We aligned the above sequences with known Arabidopsis sequences in the TAIR9 transcript database (http://www.Arabidopsis.org/) using a BLASTN search (e-value < 0.1) to estimate the degree of homology to Arabidopsis genes. To detect genes that did not exist in the Arabidopsis database, we used a BLASTN search with e-values >0.1.
2.12. Quantitative real-time reverse-transcription PCR
We treated 2-week-old Chinese cabbage plants at the rosette stage with 100 μM jasmonic acid or 1 mM salicylic acid for 2, 5, 10, or 24 h (n = 5 per duration, with two replicates). The plants were then frozen in liquid nitrogen and total RNA (2 µg) was isolated with Trizol reagent (Invitrogen, Carlsbad, CA, USA) and an RNeasy MinElute Cleanup Kit (Qiagen, Valencia, CA, USA). Quantitative real-time reverse-transcription PCR was carried out with the Power SYBR Green PCR Master Mix (Applied Biosystems) by using the first-strand cDNA as a template and an ABI Prism 7900HT sequence detector (Applied Biosystems). Expression of BrACT2 was used for normalization (i.e. values for all other genes were divided by the activity of this gene). Nucleotide sequences for the B. rapa gene-specific primers were as follows: BrVSP2 (forward, 5′-GACTC CAAAACGGTGTGCAAA-3′; reverse, 5′-AGGGTCTCG TCAAGGTCAAAGA-3′); BrLOX2 (forward, 5′-TCCCCA CTTCCGCTACACC-3′; reverse, 5′-AATACTTTCCGGGC CAGAAAC-3′); BrPR1 (forward, 5′-TACGCTCAAAACTA CGCCGA-3′; reverse, 5′-GAAAGGTCCCCGCTACTTCC-3′); BrBGL2 (forward, 5′-GCAGAACATCGATAGAGC GGT-3′; reverse, 5′-TGAATGTCCCACTCGAAGGC-3′); and BrACT2 (forward, 5′-ACCCAAAGGCCAACAGA GAG-3′; reverse, 5′-CTGGCGTAAAGGGAGAGAACA-3′).
2.13. Accession numbers
The GenBank accession numbers for the genes mentioned in Section 2.12 are as follows: BrPR1 (BBRAF03K11), BrBGL2 (BBRAF10P08), BrVSP2 (EX103556), BrLOX2 (EX100417), and BrACT2 (BBRAF03F20).
3. Results and discussion
3.1. Construction and sequencing of the BBRAF clones
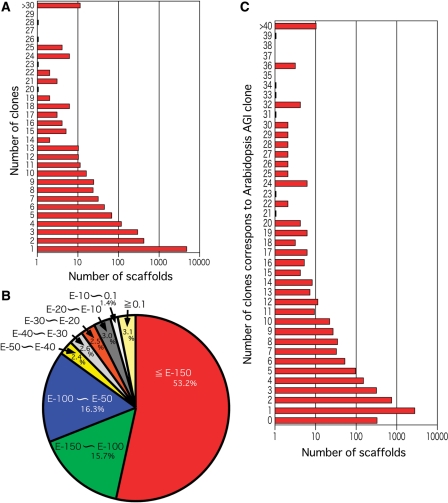
We used the CR breeding line, KmP02, for constructing the RIKEN BRC B. rapa full-length cDNA (BBRAF cDNA) library, because the resistance to clubroot disease is one of the most important breeding aim for Chinese cabbage. The library was constructed by using plants subjected to the biotic and abiotic stresses summarized in Table 1. We then determined the 5′ and 3′ sequences from 10 000 independent BBRAF clones, resulting in 4251 full-length sequences that corresponded to 2621 independent clones. The average length of these BBRAF clones was 900 bp. Further analyses will be required to determine the full-length sequences of the remaining 5749 BBRAF clones. The number of independent B. rapa cDNAs was 5476 from 10000 BBRAF clones. We built a cluster profile representing the number of each independent clones (Fig. 1A). The most frequently encountered gene was the ribulose-1,5-bisphosphate carboxylase oxygenase (RuBisCO) small subunit 2B (rbcs-2B) gene with 87 BBRAF clones.
Table 1.
The list of plant treatments used for preparation of total RNA
| Treatment | Condition | Tissue |
|---|---|---|
| 5 mM salicylic acid | Sprayed to aerial part | Aerial parts |
| 1 mM ethephone | Dipped whole pot into solution | Aerial parts, roots |
| 100 μM jasmonic acid | Sprayed to aerial part | Aerial parts |
| 100 mM copper sulphate | Sprayed to aerial part | Aerial parts |
| 500 μM benzothiadiazole | Dipped whole pot into solution | Aerial parts, roots |
| 25 μM paraquat | Sprayed to aerial part | Aerial parts |
| 100 μM abscisic acid | Sprayed to aerial part | Aerial parts |
| 250 mM sodium chloride | Sprayed to aerial part | Aerial parts |
| 0.1 kJ/m2 UV-C | Set under the UV light | Aerial parts |
| Wounding | Cut by scissors | Aerial parts |
| Dehydration | Transfer to a filter paper | Aerial parts |
| Cold | 4°C | Aerial parts |
Figure 1.
Clustering of the BBRAF Chinese cabbage (B. rapa subsp. pekinensis) full-length cDNA clones and homology to Arabidopsis transcripts. (A) Distribution of the number of BBRAF clones in each cluster of the sequence assembly. We clustered a total of 10 000 BBRAF clones into 5476 independent clones. The sequence assembly was performed using the PCAP.REP software. (B) Pie chart of the distribution of sequence homology between the BBRAF clones and Arabidopsis TAIR9 transcripts. The e-value of the BLASTN analyses is shown. (C) Distribution of the number of BBRAF clones homologous to Arabidopsis genes in the TAIR9 transcript database.
3.2. Comparative analyses between the BBRAF clones and Arabidopsis genes
Chinese cabbage belongs to the same family (Brassicaceae) as Arabidopsis, an experimental plant with a large body of functional information and expression data for each gene. It is therefore useful to take advantage of the Arabidopsis-related information to serve as a reference for Brassica genes. To do so, we searched for Arabidopsis homologues of each BBRAF clone using the BLASTN software and the TAIR9 Arabidopsis DNA database (ftp://ftp.Arabidopsis.org/home/tair/Genes/TAIR9_genome_release/), which includes coding sequences.28
The BLASTN search revealed that 85.2 and 53.2% of the BBRAF sequences corresponded to Arabidopsis sequences with e-values <1e − 50 and <1e − 150, respectively (Fig. 1B). This analysis revealed that B. rapa cDNA showed high homology to Arabidopsis cDNA sequences and suggested the facile formation of counterparts between Chinese cabbage and Arabidopsis. On the other hand, the BBRAF library also contained clones with no homology to Arabidopsis genes. When we used an e-value of >0.1 to identify such clones, we found that 3.1% of the BBRAF clones lacked any Arabidopsis homologues (Fig. 1B). We then carried out BLASTX analyses to search for homologues in the DDBJ plant protein sequence database (http://blast.ddbj.nig.ac.jp/top-j.html).
We found the 30 clones with homology to some protein in BLASTX search (e-value <0.001) and have summarized the information on these clones in Table 2. The 16 clones of which were homologous to genes encoding proteins from species other than Arabidopsis. The remaining 14 clones were homologous to genes that encoded Arabidopsis proteins in BLASTX search (Table 2). In Fig. 1C, we have summarized the distribution of gene annotations for the BBRAF clones by using Arabidopsis TAIR9 annotations. Table 3 summarizes the 20 Arabidopsis genes with the largest number of BBRAF counterparts. The two most frequently observed Arabidopsis annotations were for ribulose bisphosphate carboxylase small chain 2B (rbcs-2B, At5g38420) and ribulose bisphosphate carboxylase small subunit 3B (rbcs-3B, At5g38410) in that order. In addition, At1g79040, At2g34430, At2g34420, and At2g39730 were also frequently observed photosynthesis-related annotations.
Table 2.
The list of the BBRAF clones that showed no homology to Arabidopsis genes (e-value > 0.1) in BLASTN searches
| BBRAF ID | Accession no. | Description | E-value of BLASTX | Organism species |
|---|---|---|---|---|
| BBRAF43N02 | Q4P4F2 | Hypothetical protein | 1.00E−70 | Ustilago maydis |
| BBRAF40E12 | Q8LDM9 | Unknown proytein, At4g02370 | 7.00E−62 | Arabidopsis thaliana |
| BBRAF42P02 | Q9FMZ7 | Agenet domain-containing protein, At5g42670 | 9.00E−55 | Arabidopsis thaliana |
| BBRAF09O19 | 3015358B | Unknown protein, At1g14870 | 1.00E−36 | Arabidopsis thaliana |
| BBRAF07K19 | D7MM84 | Predicted protein | 2.00E−36 | Arabidopsis lyrata |
| BBRAF33N04 | Q9T0D8 | Hypothetical protein AT4g11710 | 4.00E−34 | Arabidopsis thaliana |
| BBRAF42F03 | D7KJA6 | Predicted protein | 1.00E−33 | Arabidopsis lyrata |
| BBRAF34O11 | Q3E7N8 | Protein, At1g77030 | 1.00E−30 | Arabidopsis thaliana |
| BBRAF05C14 | D7MWJ1 | Putative uncharacterized protein | 4.00E−20 | Arabidopsis lyrata |
| BBRAF33F04 | O80793 | Putative Ta11-like non-LTR retroelement protein, At2g07760 | 1.00E-18 | Arabidopsis thaliana |
| BBRAF40I14 | Q9SKI0 | Expressed protein, At2g10940 | 2.00E−18 | Arabidopsis thaliana |
| BBRAF10M12 | Q9C6L3 | Hypothetical protein F2J7.11 | 5.00E−18 | Arabidopsis thaliana |
| BBRAF01D10 | D7LWS2 | Putative uncharacterized protein | 8.00E−13 | Arabidopsis lyrata |
| BBRAF03H10 | D7MJE7 | Expressed protein | 4.00E−12 | Arabidopsis lyrata |
| BBRAF02C18 | D7LZP8 | Putative uncharacterized protein | 9.00E−11 | Arabidopsis lyrata |
| BBRAF38D06 | Q8GYP2 | Hypothetical protein, At1g24145 | 1.00E−10 | Arabidopsis thaliana |
| BBRAF04D10 | Q0WR19 | Hypothetical protein | 2.00E−10 | Arabidopsis thaliana |
| BBRAF08F12 | Q8GXY9 | Hypothetical protein, At5g38980 | 6.00E−09 | Arabidopsis thaliana |
| BBRAF01L23 | Q9LZD9 | Hypothetical protein, At5g03480 | 2.00E−08 | Arabidopsis thaliana |
| BBRAF08M11 | D7MQG1 | Putative uncharacterized protein | 7.00E−08 | Arabidopsis lyrata |
| BBRAF09O16 | 1209325C | ORF 3 | 2.00E−07 | Nicotiana sp. |
| BBRAF36N16 | Q2A9K2 | Hypothetical protein | 2.00E−06 | Brassica oleracea |
| BBRAF11H13 | D7L4Q6 | Putative uncharacterized protein | 2.00E−06 | Arabidopsis lyrata |
| BBRAF01A14 | D7MKH7 | Protease inhibitor/seed storage/lipid transfer protein family protein | 2.00E−05 | Arabidopsis lyrata |
| BBRAF02F22 | A7T6Z1 | Predicted protein | 2.00E−04 | Nematostella vectensis |
| BBRAF42I09 | Q8LAL6 | Uncharacterized protein, At1g24575 | 2.00E−04 | Arabidopsis thaliana |
| BBRAF02M20 | D7KPQ5 | ATP binding protein | 3.00E−04 | Arabidopsis lyrata |
| BBRAF12G14 | Q2A9U9 | Putative uncharacterized protein | 9.00E−04 | Brassica oleracea |
| BBRAF32K11 | Q8LEP7 | Putative uncharacterized protein, At4g39675 | 0.001 | Arabidopsis thaliana |
| BBRAF04I02 | D7KQS0 | Putative uncharacterized protein | 0.001 | Arabidopsis lyrata |
Table 3.
The 20 Arabidopsis genes with the largest number of BBRAF homologues in the BLASTN analyses
| Ranking | Accession no. | Description | Number of BBRAF clones |
|---|---|---|---|
| 1 | AT5G38420a | RBCS-2B (RuBisCO small subunit 2B) | 87 |
| 2 | AT5G38410a | RBCS-3B (RuBisCO small subunit 3B) | 73 |
| 3 | AT5G42530 | Unknown protein | 65 |
| 4 | AT5G44430b | PDF1.2C (PLANT DEFENSIN 1.2C) | 62 |
| 5 | AT2G33830 | Dormancy/auxin-associated family protein | 57 |
| 6 | AT1G79040a | PSBR (photosystem II subunit R) | 50 |
| 7 | AT5G59310b | LTP4 (LIPID TRANSFER PROTEIN 4) | 48 |
| 8 | AT2G34430a | LHCB1 (LIGHT-HARVESTING CHLOROPHYLL-PROTEIN COMPLEX II SUBUNIT B1) | 44 |
| 9 | AT1G62380b | ACO2 (ACC OXIDASE 2) | 42 |
| 9 | AT2G34420a | LHB1B2 (Photosystem II light harvesting complex gene 1.5) | 42 |
| 11 | AT5G19140b | Auxin/aluminium-responsive protein | 39 |
| 12 | AT4G02520b | ATGSTF2 [Arabidopsis thaliana Glutathione S-transferase (class phi) 2] | 36 |
| 12 | AT1G72290b | Trypsin and protease inhibitor family protein | 36 |
| 12 | AT2G39730a | RCA (RUBISCO ACTIVASE) | 36 |
| 15 | AT2G05380b | GRP3S (GLYCINE-RICH PROTEIN 3 SHORT ISOFORM) | 34 |
| 16 | AT2G21660b | ATGRP7 (COLD, CIRCADIAN RHYTHM, AND RNA BINDING 2) | 33 |
| 17 | AT3G15353b | MT3 (METALLOTHIONEIN 3) | 32 |
| 17 | AT3G09390b | MT2A (METALLOTHIONEIN 2A) | 32 |
| 17 | AT1G20620b | CAT3 (CATALASE 3) | 32 |
| 17 | AT5G66400b | RAB18 (RESPONSIVE TO ABA 18) | 32 |
aPhotosynthesis-related genes.
bStress-related genes.
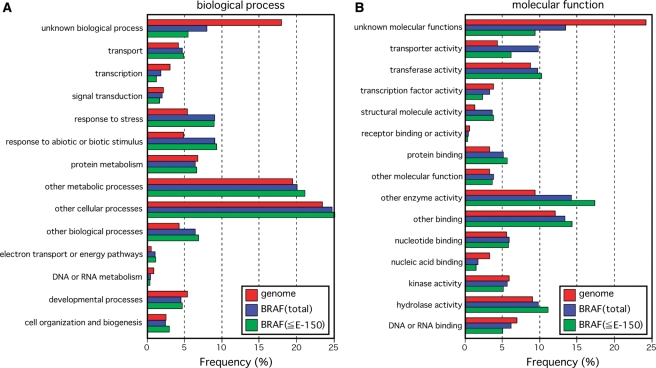
We used many stress conditions (Table 1) to prepare the full-length cDNA library. Importantly, 12 of the 20 most frequently observed annotations summarized in Table 3 were stress-related. To further understand the characteristics of the stress-related clones, we compared the results of searches for the biological functions and molecular processes involving the BBRAF clones in the Arabidopsis gene ontology database (http://www.Arabidopsis.org/tools/bulk/go/index.jsp) with those for the Arabidopsis whole-genome categorization (Fig. 2). Gene ontology searches showed that the BBRAF clones tended to be those with selective responses to abiotic or biotic stimuli (Fig. 2A). As seen in Fig. 2B, the frequency of clones of unknown molecular function was also lower in the BBRAF library than in the Arabidopsis whole-genome library (Fig. 2B). In addition, unknown molecular function was observed at a lower frequency for BBRAF clones with higher homology to Arabidopsis genes (≤1e − 150) than for BBRAF clones with lower homology to Arabidopsis genes (Fig. 2B).
Figure 2.
Functional annotation of the BBRAF Chinese cabbage (B. rapa subsp. pekinensis) full-length cDNA clones using the Arabidopsis gene ontology database. Functional annotations are presented in relation to (A) the biological process and (B) the molecular function. The distribution is shown for the total BBRAF clones (total), BBRAF clones with high homology to Arabidopsis genes (≤1e − 150), and the whole-genome Arabidopsis (TAIR9 transcripts genome).
3.3. Evaluation of the quality of the BBRAF clones and comparison of the BBRAF and Arabidopsis RuBisCO small subunit gene families
The quality of the BBRAF library was evaluated based on the presence of a start ATG codon in the 5′ sequence of the 20 most frequently observed BBRAF clones in the Arabidopsis annotations (Table 3). The start ATG codon was estimated using the position of the ATG codon in the corresponding Arabidopsis gene. We found that 93.9% of the corresponding BBRAF clones had ATG codons, indicating that the quality of our BBRAF clones was sufficient.
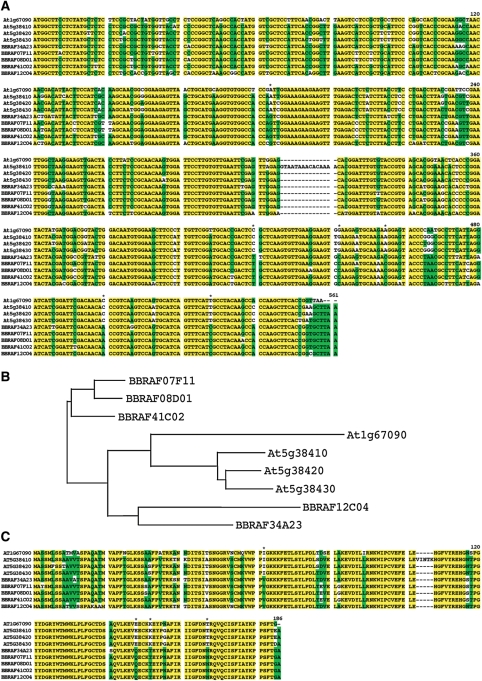
To understand more about the BBRAF clones and the degree of similarity between BBRAF clones and Arabidopsis genes, we compared the CDS nucleic acid sequence and the amino acid sequence of rbcs2b and rbcs3b, the two most frequently observed Arabidopsis annotations in the BBRAF clones. RBCS encodes the small subunit of RuBisCo, which is one of the most strongly conserved and well-analysed plant proteins because it catalyses carbon fixation, the first major step in the Calvin cycle.29 In addition, the expression of rbcs is well known to be differentially regulated by light, sugar, and abscisic acid.30,31 In the present study, rbcs-2B was the most frequently observed BBRAF clone (87 clones, Table 3). Comparison of the sequences of the open reading frames (ORFs) of the rbcs gene families in Chinese cabbage and Arabidopsis revealed four rbcs genes in the Arabidopsis genome versus five independent rbcs genes from Chinese cabbage in the BBRAF clones. Sequence alignment (Fig. 3) revealed that six DNA residues at positions located at 184 (A/G), 420 (C/T), 442 (G/C), 455 (A/C), 500 (C/A), and 528 (T/C), and four amino acid residues, located at positions 62 (I/V), 223 (E/Q), 227 (K/T), and 242 (T/N), distinguished Arabidopsis rbcs from Chinese cabbage rbcs (Fig. 3A and C).
Figure 3.
Comparison of the Chinese cabbage (B. rapa) and Arabidopsis RuBisCo small subunit 2B genes. (A) Sequence comparison of five Chinese cabbage (B. rapa subsp. pekinensis) and four Arabidopsis rbcs genes. Fully conserved bases are shown in yellow. The conserved positions for more than five genes are shown in green. (B) The phylogenetic tree for the five Chinese cabbage and four Arabidopsis rbcs genes. (C) Comparison of the amino acid sequences of the five Chinese cabbage and four Arabidopsis RBCS proteins. Fully conserved bases are shown in yellow. The conserved positions for more than five genes are shown in green.
Our phylogenetic analyses (Fig. 3B) indicated that the four Arabidopsis rbcs genes belonged to the same branch, although At5g38410, At5g38420, and At5g38430 were more closely related to each other than to At1g67090. Chinese cabbage rbcs could be grouped into two classes: one is the Arabidopsis-related class (BBRAF12C04, BBRAF34A23) and the other class is weakly related (BBRAF07F11, BBRAF08D01, BBRAF41C02). Genkov and Spreitzer32 reported that the primary residues (Leu-73, Tyr-87, Glu-98, and Trp-127) that are responsible for RBCS protein stability and function were also conserved in all of the Chinese cabbage and Arabidopsis RBCS proteins. Most of the BBRAF clones contained whole-ORF sequences, as described above. We believe that it will be important to determine the full-length sequence of every BBRAF clone. In the future, comparative analyses of Brassica and Arabidopsis proteins at the amino acid sequence level would help us to understand the key residues, important domains, and important functions of these proteins.
3.4. Identification of Brassica defence response genes
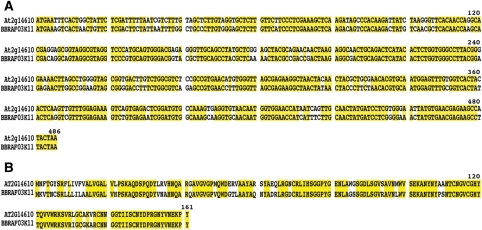
It is important to compare the changes in gene expression of Arabidopsis and Brassica crops to understand their common and species-specific defence mechanisms. To identify the relevant marker genes in Chinese cabbage, we searched for homologues to Arabidopsis marker genes for the salicylic acid and jasmonic acid pathways. We found putative B. rapa counterpart genes for AtPR1 and AtBGL2 (i.e. BrPR1 and BrBGL2) for the salicylic acid pathway. We compared the DNA sequences of AtPR1 and BrPR1 (Fig. 4A) and the amino acid sequences of their respective proteins, ATPR1 and BRPR1 (Fig. 4B). The DNA sequence identity between AtPR1 and BrPR1 was lower (83.5%) than that between Atrbcs and Brrbcs, which had more than 90% homology (data not shown). We also identified putative B. rapa counterpart genes for AtLOX2 and AtVSP2 (i.e. BrLOX2 and BrVSP2), which are marker genes for the jasmonic acid pathway.
Figure 4.
Comparison of Chinese cabbage (B. rapa subsp. pekinensis) and Arabidopsis PR1 marker genes. (A) DNA sequence comparison. Conserved base positions are shown in yellow. (B) Amino acid sequence comparison. Conserved base positions are shown in yellow.
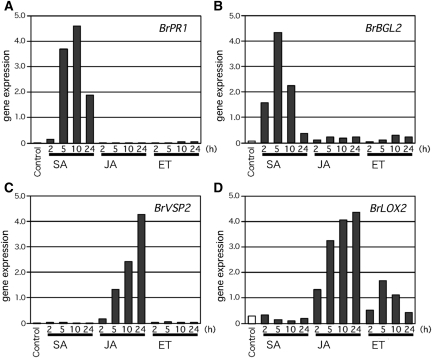
Next, we analysed the effects of salicylic and jasmonic acids on the expression of these Brassica putative marker genes by quantitative real-time RT–PCR. Gene expression of BrPR1 and BrBGL2 was induced by salicylic acid treatment but not by treatment with jasmonic acid or the ethylene precursor, 1-aminocyclopropane-carboxylic acid treatment (Fig. 5A and B), as is the case for Arabidopsis AtPR1 and AtBGL2. BrLOX2 and BrVSP2 gene expression was induced by jasmonic acid but not by salicylic acid, as is the case for Arabidopsis AtLOX2 and AtVSP2. Interestingly, BrLOX2 was slightly induced by 1-aminocyclopropane-carboxylic acid treatment, but this response was not detected for BrVSP2.
Figure 5.
Gene expression analyses of gene homologues for Arabidopsis salicylic acid (SA) and jasmonic acid (JA) marker genes. ET, 1-aminocyclopropane-carboxylic acid treatment. (A and B) Chinese cabbage (B. rapa subsp. pekinensis) homologues to Arabidopsis PR1 and BGL2 marker genes for the salicylic acid pathway. (C and D) VSP2 and LOX2 as marker genes for the jasmonic acid pathway. We used 2-week-old plants grown in a single pot in this experiment (n = 5, with two replicates). The plants were treated with 1 mM salicylic acid, 100 μM jasmonic acid, and 50 μM 1-aminocyclopropane-carboxylic acid for 2, 5, 10, or 24 h. The total RNA was extracted, and first-strand cDNA was synthesized for expression analysis. The expression level of each gene was normalized to the expression of BrACT2, which is constitutively expressed and shown as the reference value.
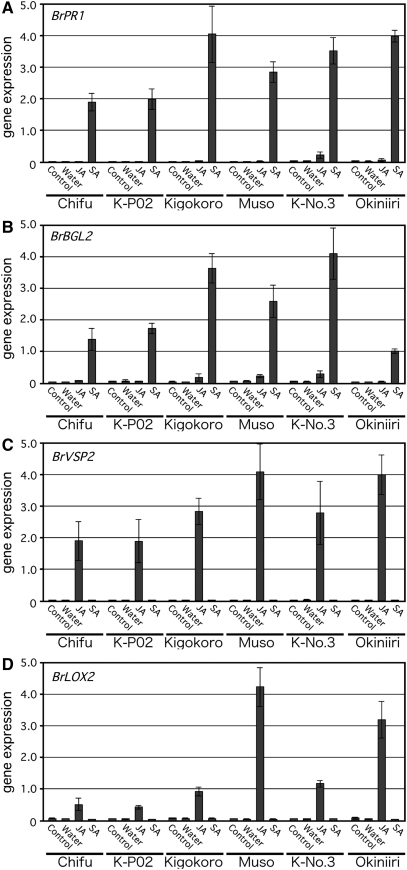
Next, we compared the effects of salicylic and jasmonic acids on gene expression in several Chinese cabbage cultivars (Kyoto No. 3, Muso, Kigokoro 85, Okiniiri, Chifu hakusai, and KmP02) (Fig. 6). We observed expression of all four marker genes in each of the varieties, which suggests that these genes will be useful in future research on other B. rapa varieties. Interestingly, we detected a stronger response in Muso to jasmonic acid, which regulates resistance to insects such as thrips.33 This suggested that it would be useful to analyse the relationship between this higher sensitivity to jasmonic acid and herbivore resistance in Muso. However, when we compared the resistance of Kyoto No. 3 and Muso with thrips, we found no significant difference (data not shown). Insect resistance is a primary breeding objective but is difficult to improve because it is controlled by a complex signalling cascade. Further trials are needed to identify the Brassica genes involved in jasmonic acid-regulated plant defence responses so as to determine whether the cultivars differ in their resistance to caterpillar and leafminer feeding as well as thrips feeding. Similarly, the Brassica genes involved in salicylic acid-regulated plant defence responses would also be important breeding targets as this pathway is involved in disease resistance.
Figure 6.
Gene expression analyses of Chinese cabbage (B. rapa subsp. pekinensis) marker genes for salicylic acid and jasmonic acid response in major cultivars. Cultivars used in this experiment: Chifu, Chifu hakusai; K-P02, KmP02; Kigokoro, Kigokoro 85; Muso; K-No.3, Kyoto No. 3; Okiniiri. (A and B) BrPR1 and BrBGL2 as marker genes for the salicylic acid pathway; (C and D) BrVSP2 and BrLOX2 as marker genes for the jasmonic acid pathway. We used 2-week-old plants (n = 3, with three replicates), grown in a single pot, and treated them with water, 100 μM jasmonic acid, and 1 mM salicylic acid for 24 h. The total RNA was extracted and first-strand cDNA was synthesized for the PCR analysis. The expression level of each gene was normalized with respect to the expression of BrACT2 (control).
3.5. BBRAF as the Brassica genome resource
Many uses have been reported for full-length cDNA clones in -omics analyses9 as full-length cDNA is a powerful tool to create overexpressors that can be used in research. It is particularly worth noting that several important genes involved in plant stress tolerance have been isolated using the FOX hunting system.34 As in other plant species, several transformation systems have been reported in B. rapa.35–37 Min et al.38 developed the mannose selection system, which is based on tolerance of phosphomannose isomerase. Using a B. rapa transformation system, several researchers have attempted to create stress- and disease-tolerant plants through the overexpression of transgenes.39,40 The BBRAF resource described in the present study would be a good tool for the identification of genes that will be useful for creating Brassica crops with special characteristics, such as insect and disease resistance.
Molecular markers have gained considerable importance in plant science and breeding. Among the different classes of existing markers, single sequence repeat (SSR) markers are known to be optimal for plant breeding41 and several studies to develop the SSR markers for B. rapa have been reported.42–45 However, most of these SSRs were genomic SSRs and were frequently located in heterochromatin regions of the chromosomes. Therefore, EST-derived SSRs are preferable because they tend to be randomly distributed along the chromosomes.46 Full-length cDNA contains 5′- and 3′-untranslated regions in addition to complete coding sequences. Parida et al.47 reported that interspecific polymorphism between B. napus and B. rapa detected in silico for the unigne-derived SSR markers was 4.16 times higher in untranslated regions than in coding sequences. The BBRAF clones could therefore be good materials to develop the new SSR markers for B. rapa. Development and mapping of SSRs derived from the BBRAF clones are currently in progress, and some of these SSR markers have already been used for mapping of the late-bolting characteristic of B. rapa.48
Developing and enhancing such resources will support and accelerate many aspects of research on B. rapa, for which the genome sequencing of which is now entering its final phase. The importance of such genome resources will increase in the future. For example, Yu et al.49 recently reported the development of 3400 T-DNA insertion mutagenesis lines in Chinese cabbage. Stephenson et al.50 also reported the development of the TILLING platform (Targeting Induced Local Lesions in Genomes), which consists of 9216 M2 plants. Interestingly, the self-fertile line R-o-18 was used to develop the TILLING platform. Self-infertility of Brassica crops is one of the major obstacles in Brassica research. The availability of a self-fertile line could therefore be a breakthrough for Brassica research.
We report here the development of new B. rapa genomic resources, the RIKEN BBRAF. The BBRAF resource will be publicly available from the RIKEN Bioresource Center (http://www.brc.riken.go.jp/lab/epd/Eng/). The BBRAF library was constructed using many stress-treated plants. Gene ontology searches showed that BBRAF clones were successfully collected counterpart genes of stress-related Arabidopsis genes. Further transcriptomic data, in addition to the sequence data described in this paper, are required to improve our understanding of the relationships between B. rapa cDNAs and Arabidopsis genes. We are now developing an informatics resource based on the BBRAF clones that will function as a bridge between information for the model plant Arabidopsis and Brassica research. We hope that the BBRAF collection will be a useful tool that can be used to accelerate Brassica research.
Funding
This work was supported in part by the Program for Promotion of Basic and Applied Researches for Innovations in Bio-oriented Industry (BRAIN), provided by the Industrial Technology Research Grant Program from the New Energy and Industrial Technology Development Organization (NEDO) of Japan.
Acknowledgements
We thank F. Mori and S. Kawamura of RIKEN BRC for their excellent technical assistance.
References
- 1.Hall A.E., Fiebig A., Preuss D. Beyond the Arabidopsis genome: opportunities for comparative genomics. Plant Physiol. 2002;129:1439–47. doi: 10.1104/pp.004051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.AGI. Analysis of the genome sequence of the flowering plant Arabidopsis thaliana. Nature. 2000;408:796–815. doi: 10.1038/35048692. [DOI] [PubMed] [Google Scholar]
- 3.Hong C.P., Kwon S.J., Kim J.S., Yang T.J., Park B.S., Lim Y.P. Progress in understanding and sequencing the genome of Brassica rapa. Int. J. Plant Genomics. 2008;2008:582837. doi: 10.1155/2008/582837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Yang T.J., Kim J.S., Kwon S.J., et al. Sequence-level analysis of the diploidization process in the triplicated FLOWERING LOCUS C region of Brassica rapa. Plant Cell. 2006;18:1339–47. doi: 10.1105/tpc.105.040535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bowers J.E., Chapman B.A., Rong J., Paterson A.H. Unravelling angiosperm genome evolution by phylogenetic analysis of chromosomal duplication events. Nature. 2003;422:433–8. doi: 10.1038/nature01521. [DOI] [PubMed] [Google Scholar]
- 6.Mun J.H., Kwon S.J., Yang T.J., et al. Genome-wide comparative analysis of the Brassica rapa gene space reveals genome shrinkage and differential loss of duplicated genes after whole genome triplication. Genome Biol. 2009;10:R111. doi: 10.1186/gb-2009-10-10-r111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Paterson A.H., Lan T.H., Amasino R., Osborn T.C., Quiros C. Brassica genomics: a complement to, and early beneficiary of, the Arabidopsis sequence. Genome Biol. 2001;2:REVIEWS1011. doi: 10.1186/gb-2001-2-3-reviews1011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Town C.D., Cheung F., Maiti R., et al. Comparative genomics of Brassica oleracea and Arabidopsis thaliana reveal gene loss, fragmentation, and dispersal after polyploidy. Plant Cell. 2006;18:1348–59. doi: 10.1105/tpc.106.041665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Seki M., Shinozaki K. Functional genomics using RIKEN Arabidopsis thaliana full-length cDNAs. J. Plant Res. 2009;122:355–66. doi: 10.1007/s10265-009-0239-3. [DOI] [PubMed] [Google Scholar]
- 10.Seki M., Carninci P., Nishiyama Y., Hayashizaki Y., Shinozaki K. High-efficiency cloning of Arabidopsis full-length cDNA by biotinylated CAP trapper. Plant J. 1998;15:707–20. doi: 10.1046/j.1365-313x.1998.00237.x. [DOI] [PubMed] [Google Scholar]
- 11.Okazaki Y., Furuno M., Kasukawa T., et al. Analysis of the mouse transcriptome based on functional annotation of 60,770 full-length cDNAs. Nature. 2002;420:563–73. doi: 10.1038/nature01266. [DOI] [PubMed] [Google Scholar]
- 12.Imanishi T., Itoh T., Suzuki Y., et al. Integrative annotation of 21,037 human genes validated by full-length cDNA clones. PLoS Biol. 2004;2:e162. doi: 10.1371/journal.pbio.0020162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Seki M., Narusaka M., Kamiya A., et al. Functional annotation of a full-length Arabidopsis cDNA collection. Science. 2002;296:141–5. doi: 10.1126/science.1071006. [DOI] [PubMed] [Google Scholar]
- 14.Alexandrov N.N., Troukhan M.E., Brover V.V., Tatarinova T., Flavell R.B., Feldmann K.A. Features of Arabidopsis genes and genome discovered using full-length cDNAs. Plant Mol. Biol. 2006;60:69–85. doi: 10.1007/s11103-005-2564-9. [DOI] [PubMed] [Google Scholar]
- 15.Ralph S.G., Chun H.J.E., Cooper D., et al. Analysis of 4,664 high-quality sequence-finished poplar full-length cDNA clones and their utility for the discovery of genes responding to insect feeding. BMC Genomics. 2008;9:57. doi: 10.1186/1471-2164-9-57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Jia J.P., Fu J.J., Zheng J., et al. Annotation and expression profile analysis of 2073 full-length cDNAs from stress-induced maize (Zea mays L.) seedlings. Plant J. 2006;48:710–27. doi: 10.1111/j.1365-313X.2006.02905.x. [DOI] [PubMed] [Google Scholar]
- 17.Alexandrov N.N., Brover V.V., Freidin S., et al. Insights into corn genes derived from large-scale cDNA sequencing. Plant Mol. Biol. 2009;69:179–94. doi: 10.1007/s11103-008-9415-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Aoki K., Yano K., Suzuki A., et al. Large-scale analysis of full-length cDNAs from the tomato (Solanum lycopersicum) cultivar Micro-Tom, a reference system for the Solanaceae genomics. BMC Genomics. 2010;11:210. doi: 10.1186/1471-2164-11-210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Umezawa T., Sakurai T., Totoki Y., et al. Sequencing and analysis of approximately 40,000 soybean cDNA clones from a full-length-enriched cDNA library. DNA Res. 2008;15:333–46. doi: 10.1093/dnares/dsn024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ichikawa T., Nakazawa M., Kawashima M., et al. The FOX hunting system: an alternative gain-of-function gene hunting technique. Plant J. 2006;48:974–85. doi: 10.1111/j.1365-313X.2006.02924.x. [DOI] [PubMed] [Google Scholar]
- 21.Kondou Y., Higuchi M., Takahashi S., et al. Systematic approaches to using the FOX hunting system to identify useful rice genes. Plant J. 2009;57:883–94. doi: 10.1111/j.1365-313X.2008.03733.x. [DOI] [PubMed] [Google Scholar]
- 22.Fujita M., Mizukado S., Fujita Y., et al. Identification of stress-tolerance-related transcription-factor genes via mini-scale full-length cDNA Over-eXpressor (FOX) gene hunting system. Biochem. Biophys. Res. Commun. 2007;364:250–57. doi: 10.1016/j.bbrc.2007.09.124. [DOI] [PubMed] [Google Scholar]
- 23.Yokotani N., Ichikawa T., Kondou Y., et al. Tolerance to various environmental stresses conferred by the salt-responsive rice gene ONAC063 in transgenic Arabidopsis. Planta. 2009;229:1065–75. doi: 10.1007/s00425-009-0895-5. [DOI] [PubMed] [Google Scholar]
- 24.Tsutsui T., Kato W., Asada Y., et al. DEAR1, a transcriptional repressor of DREB protein that mediates plant defense and freezing stress responses in Arabidopsis. J. Plant Res. 2009;122:633–43. doi: 10.1007/s10265-009-0252-6. [DOI] [PubMed] [Google Scholar]
- 25.Chirgwin J.M., Przybyla A.E., MacDonald R.J., Rutter W.J. Isolation of biologically active ribonucleic acid from sources enriched in ribonuclease. Biochemistry. 1979;18:5294–9. doi: 10.1021/bi00591a005. [DOI] [PubMed] [Google Scholar]
- 26.Kato S., Ohtoko K., Ohtake H., Kimura T. Vector-capping: a simple method for preparing a high-quality full-length cDNA library. DNA Res. 2005;12:53–62. doi: 10.1093/dnares/12.1.53. [DOI] [PubMed] [Google Scholar]
- 27.Huang X., Yang S.P., Chinwalla A.T., et al. Application of a superword array in genome assembly. Nucleic Acids Res. 2006;34:201–5. doi: 10.1093/nar/gkj419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Rhee S.Y., Beavis W., Berardini T.Z., et al. The Arabidopsis information resource (TAIR): a model organism database providing a centralized, curated gateway to Arabidopsis biology, research materials and community. Nucleic Acids Res. 2003;31:224–8. doi: 10.1093/nar/gkg076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Coen D.M., Bedbrook J.R., Bogorad L., Rich A. Maize chloroplast DNA fragment encoding the large subunit of ribulosebisphosphate carboxylase. Proc. Natl Acad. Sci. USA. 1977;74:5487–91. doi: 10.1073/pnas.74.12.5487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Dedonder A., Rethy R., Fredericq H., Van Montagu M., Krebbers E. Arabidopsis rbcS genes are differentially regulated by light. Plant Physiol. 1993;101:801–8. doi: 10.1104/pp.101.3.801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Acevedo-Hernandez G.J., Leon P., Herrera-Estrella L.R. Sugar and ABA responsiveness of a minimal RBCS light-responsive unit is mediated by direct binding of ABI4. Plant J. 2005;43:506–19. doi: 10.1111/j.1365-313X.2005.02468.x. [DOI] [PubMed] [Google Scholar]
- 32.Genkov T., Spreitzer R.J. Highly conserved small subunit residues influence rubisco large subunit catalysis. J. Biol. Chem. 2009;284:30105–12. doi: 10.1074/jbc.M109.044081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Abe H., Ohnishi J., Narusaka M., et al. Function of jasmonate in response and tolerance of Arabidopsis to thrip feeding. Plant Cell Physiol. 2008;49:68–80. doi: 10.1093/pcp/pcm168. [DOI] [PubMed] [Google Scholar]
- 34.Higuchi M., Kondou Y., Ichikawa T., Matsui M. Full-length cDNA overexpressor gene hunting system (FOX hunting system) Methods Mol. Biol. 2011;678:77–89. doi: 10.1007/978-1-60761-682-5_7. [DOI] [PubMed] [Google Scholar]
- 35.Takasaki T., Hatakeyama K., Ojima K., Watanabe M., Toriyama K., Hinata K. Factors influencing Agrobacterium-mediated transformation of Brassica rapa L. Breed. Sci. 1997;47:127–34. [Google Scholar]
- 36.Zhang F.L., Takahata Y., Watanabe M., Xu J.B. Agrobacterium-mediated transformation of cotyledonary explants of Chinese cabbage (Brassica campestris L. ssp pekinensis) Plant Cell Rep. 2000;19:569–75. doi: 10.1007/s002990050775. [DOI] [PubMed] [Google Scholar]
- 37.Lee M.K., Kim H.S., Kim J.S., Kim S.H., Park Y.D. Agrobacterium-mediated transformation system for large-scale production of transgenic Chinese cabbage (Brassica rapa L. ssp. pekinensis) plants for insertional mutagenesis. J. Plant Biol. 2004;47:300–6. [Google Scholar]
- 38.Min B.W., Cho Y.N., Song M.J., et al. Successful genetic transformation of Chinese cabbage using phosphomannose isomerase as a selection marker. Plant Cell Rep. 2007;26:337–44. doi: 10.1007/s00299-006-0247-x. [DOI] [PubMed] [Google Scholar]
- 39.Park B.J., Liu Z.C., Kanno A., Kameya T. Genetic improvement of Chinese cabbage for salt and drought tolerance by constitutive expression of a B. napus LEA gene. Plant Sci. 2005;169:553–8. [Google Scholar]
- 40.Vanjildorj E., Song S.Y., Yang Z.H., et al. Enhancement of tolerance to soft rot disease in the transgenic Chinese cabbage (Brassica rapa L. ssp. pekinensis) inbred line, Kenshin. Plant Cell Rep. 2009;28:1581–91. doi: 10.1007/s00299-009-0757-4. [DOI] [PubMed] [Google Scholar]
- 41.Gupta P.K., Varshney R.K. The development and use of microsatellite markers for genetic analysis and plant breeding with emphasis on bread wheat. Euphytica. 2000;113:163–85. [Google Scholar]
- 42.Suwabe K., Iketani H., Nunome T., Kage T., Hirai M. Isolation and characterization of micro satellites in Brassica rapa L. Theor. Appl. Genet. 2002;104:1092–8. doi: 10.1007/s00122-002-0875-7. [DOI] [PubMed] [Google Scholar]
- 43.Suwabe K., Iketani H., Nunome T., Ohyama A., Hirai M., Fukuoka H. Characteristics of microsatellites in Brassica rapa genome and their potential utilization for comparative genomics in Cruciferae. Breeding Science. 2004;54:85–90. [Google Scholar]
- 44.Suwabe K., Tsukazaki H., Iketani H., et al. Simple sequence repeat-based comparative genomics between Brassica rapa and Arabidopsis thaliana: the genetic origin of clubroot resistance. Genetics. 2006;173:309–19. doi: 10.1534/genetics.104.038968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Lowe A.J., Moule C., Trick M., Edwards K.J. Efficient large-scale development of microsatellites for marker and mapping applications in Brassica crop species. Theor. Appl. Genet. 2004;108:1103–12. doi: 10.1007/s00122-003-1522-7. [DOI] [PubMed] [Google Scholar]
- 46.Ohyama A., Asamizu E., Negoro S., et al. Characterization of tomato SSR markers developed using BAC-end and cDNA sequences from genome databases. Mol. Breed. 2009;23:685–91. [Google Scholar]
- 47.Parida S.K., Yadava D.K., Mohapatra T. Microsatellites in Brassica unigenes: relative abundance, marker design, and use in comparative physical mapping and genome analysis. Genome. 2010;53:55–67. doi: 10.1139/g09-084. [DOI] [PubMed] [Google Scholar]
- 48.Kakizaki T., Kato T., Fukino N., Ishida M., Hatakeyama K., Matsumoto S. Identification of quantitative trait loci controlling late bolting in Chinese cabbage (Brassica rapa L.) parental line Nou 6 gou. Breed. Sci. 2011;61:151–9. [Google Scholar]
- 49.Yu J.G., Lee G.H., Kim J.S., Shim E.J., Park Y.D. An insertional mutagenesis system for analyzing the Chinese cabbage genome using Agrobacterium T-DNA. Mol. Cells. 2010;29:267–75. doi: 10.1007/s10059-010-0013-3. [DOI] [PubMed] [Google Scholar]
- 50.Stephenson P., Baker D., Girin T., et al. A rich TILLING resource for studying gene function in Brassica rapa. BMC Plant Biol. 2010;10:62. doi: 10.1186/1471-2229-10-62. [DOI] [PMC free article] [PubMed] [Google Scholar]