Abstract
Raphanus sativus (2n = 2x = 18) is a widely cultivated member of the family Brassicaceae, for which genomic resources are available only to a limited extent in comparison to many other members of the family. To promote more genetic and genomic studies and to enhance breeding programmes of R. sativus, we have prepared genetic resources such as complementary DNA libraries, expressed sequences tags (ESTs), simple sequence repeat (SSR) markers and a genetic linkage map. A total of 26 606 ESTs have been collected from seedlings, roots, leaves, and flowers, and clustered into 10 381 unigenes. Similarities were observed between the expression patterns of transcripts from R. sativus and those from representative members of the genera Arabidopsis and Brassica, indicating their functional relatedness. The EST sequence data were used to design 3800 SSR markers and consequently 630 polymorphic SSR loci and 213 reported marker loci have been mapped onto nine linkage groups, covering 1129.2 cM with an average distance of 1.3 cM between loci. Comparison of the mapped EST-SSR marker positions in R. sativus with the genome sequence of A. thaliana indicated that the Brassicaceae members have evolved from a common ancestor. It appears that genomic fragments corresponding to those of A. thaliana have been doubled and tripled in R. sativus. The genetic map developed here is expected to provide a standard map for the genetics, genomics, and molecular breeding of R. sativus as well as of related species. The resources are available at http://marker.kazusa.or.jp/Daikon.
Keywords: comparative map, expressed sequence tag (EST), genetic linkage map, Raphanus sativus, simple sequence repeat (SSR)
1. Introduction
DNA markers are essential tools for plant genetics. In early studies, restriction fragment length polymorphism (RFLP), amplified fragment length polymorphism (AFLP) and random amplified polymorphic DNA (RAPD) techniques were widely used since no sequence information was required for their development. Single nucleotide polymorphism (SNP) is the most abundant source of genomic variation, and genetic studies using large-scale genotyping of SNPs have been performed with several plant species.1–5 However, SNP markers only detect two alleles and biases in allelic frequency reflect the targeted population structure,5 making it difficult to develop SNP markers that widely function across species. Although no similar high-throughput genotyping system has been developed for simple sequence repeats (SSR), these markers have several advantages over SNPs, including multi-allelic detection, high-transferability across species and flexibility with various laboratory systems.6 SSR markers can be classified into two categories: genomic SSRs and expressed sequence tag (EST)-SSRs, which are designed from intergenic and intragenic sequences, respectively. Even though the polymorphic ratio of EST-SSR markers is sometimes lower than that of genomic SSRs, EST-SSRs can be expected to have greater transferability between species than genomic SSRs, since genic regions are more likely to be conserved among related species/genera.5
Comparative genomics has had a significant impact on the fields of plant genetics and genomics. It has provided information on genomic evolution through the identification of chromosomal rearrangements and duplication/deletion events, as well as enabling large amounts of knowledge gained by studying model plants to be transferred to crops. The dicot species Arabidopsis thaliana (2n = 10) is a member of the family Brassicaceae. A. thaliana is the most studied model plant and extensive genomic resources have been developed in this species, including genomic sequences, EST information, mutant lines, DNA libraries and molecular markers, all of which are available to the research community (http://www.arabidopsis.org).7 Similarly, the Brassica belong to the Brassicaceae and many Brassica species are cultivated as vegetable and oil crops. Commercially important Brassica species include broccoli, cabbage, cauliflower, Chinese cabbage, turnip, oilseed, and black mustard. Therefore, it is not surprising that the genomes of many members of the Brassica have been well studied. In particular, karyotype analysis has shown that B. rapa (2n = 20), B. nigra (2n = 16), and B. oleracea (2n = 18) are diploids with AA, BB, and CC genomes, respectively. In contrast, B. juncea (AABB: 2n = 36), B. carinata (BBCC: 2n = 34) and B. napus (AACC: 2n = 38) are amphidiploids, which were derived from combinations of two of the diploids.8 Comparative genomics between A. thaliana and different members of the Brassica have revealed syntenies in which regions of the A. thaliana genome have doubled or tripled.9–11 In B. rapa, comparative studies have enabled map-based or candidate-gene cloning strategies to identify several quantitative trait loci (QTLs) and genes for agronomically important traits, such as flowering time, leaf morphology, and disease resistance.12,13
Raphanus sativus (2n = 2x = 18) is also a member of the Brassicaceae and it is used around the world as a vegetable crop, i.e. the radish or Japanese daikon. In particular, R. sativus is a common crop in East Asia and is the second most produced vegetable in Japan (www.e-stat.go.jp). The genus Raphanus is originated from coastal regions along the Mediterranean and Black Seas and it is classified into two sections that comprise six species.14 R. sativus was thought to have derived by hybridization between R. maritimus and R. landra, exhibiting an RR genome of 468–662 Mb.14–16 Raphanus sativus genomic research has not progressed as far as for members of the Brassica, possibly because the genus Raphanus is less speciose and less economically important. Several genetic maps of R. sativus have been constructed using RFLP, AFLP, and RAPD markers, and these have been applied to QTL identification of disease and pest resistance, the shape and pigmentation of roots, and the flowering time.14,17–19 However, few studies have reported comparative genomics between R. sativus and A. thaliana because the R. sativus maps have no sequence-tagged markers. Kamei et al.18 used genomic SSR markers for B. rapa to perform a genetic analysis of R. sativus, but found low transferability of markers between the two species. Therefore, further advances in the genetic and genomic analyses of R. sativus will require the development of genic sequence-based markers.
In the present study, genomic resources were developed for R. sativus. These resources include cDNA libraries, EST sequences, EST-SSR markers, and a genetic map, all of which have great advantages to subsequent genomic studies in comparison with genomic SSR markers. Comparison with the A. thaliana genome indicated that the two genomes differentiated from a common ancestor and that A. thaliana genomic fragments had been doubled and tripled in the R. sativus genome. This comparative map could be used for the identification of candidate genes in QTLs from R. sativus.
2. Materials and methods
2.1. Plant materials
Previously, an F2 mapping population derived from a cross between two inbred lines, ‘GSK3-1’ and ‘HA2’, was used for construction of a genetic map of R. sativus.14 ‘GSK3-1’ is a selfed progeny from a leading Japanese variety of R. sativus known as Utsugi-Gensuke and ‘HA2’ is a Tokinashi type that exhibits late bolting. In the present study, F8–10 progeny (n = 155) generated by single seed descent were used as recombinant inbred lines for linkage analysis. Genomic DNA from each line was extracted using the DNeasy Plant Mini kit (Qiagen, Germany).
2.2. Development of EST-derived SSR markers
Plant RNA Purification Reagent (Invitrogen, USA) was used to extract total RNA from 5 g samples of seedlings, roots, leaves, and flowers of R. sativus ‘GSK3-1’. cDNA libraries were constructed for each organ and then sequencing analysis and data processing were performed as described previously.20 High-quality reads that comprised >50 bp of contiguous sequence were submitted to the DDBJ/EMBL/GenBank databases with the accession numbers FY428055 to FY454660. BLASTN was used to perform similarity searches between non-redundant R. sativus ESTs that clustered using the PHRAD program and the UniGene data sets for R. sativus, A. thaliana, B. napus, B. oleracea, B. rapa, Lotus japonicas, and Oryza sativa (http://www.ncbi.nlm.nih.gov/unigene). BLASTX searches against amino acid sequences in the KOG set (http://www.ncbi.nlm.nih.gov/COG) enabled EST contigs to be classified into KOG categories.21 These sequence similarities were judged to be significant when the E-value was less than 1e−10.
Microsatellites or SSRs of ≥15 nucleotides in length were identified. All possible combinations of di- (NN), tri- (NNN), and tetra-nucleotide (NNNN) repeats were represented. Primer pairs were designed against the flanking sequences of each SSR, as described previously.22
PCR reactions were performed using 0.5 ng genomic DNA in each 5 µl reaction. In addition to template, PCR reaction mixes contained 1 × PCR buffer (BIOLINE, UK), 3 mM MgCl2, 0.04U BIOTAQ™ DNA polymerase (BIOLINE, UK), 0.2 mM dNTPs, and 0.8 µM of each primer. The thermal cycling conditions were as follows: 1 min denaturation at 94°C; 35 cycles of 30 s denaturation at 94°C, 30 s annealing at 60°C and 1 min extension at 72°C; and a final 3 min extension at 72°C. The PCR products were separated by 10% polyacrylamide gel electrophoresis in TBE buffer, according to the standard protocol, or with a Type 3730 DNA fragment analyzer (Applied Biosystems, USA). In the latter case, the data were analysed using GeneMapper software (Applied Biosystems, USA).
2.3. Genotyping and map construction
Linkage analysis was performed with genotypic data derived using the EST-SSR markers developed in this study, as well as segregation data generated from 223 published RAPD, RFLP, and trait marker loci,14,23,24 using JoinMap® software version 4.25 The marker loci were classified roughly into nine linkage groups using the JoinMap® grouping module and LOD scores of 4.0–10.0. The mapped positions of the RAPD, RFLP, and trait marker loci were used as a frame for linkage analysis. Marker order and genetic distance were calculated using a regression mapping algorithm and the following parameters: Haldane's mapping function, recombination frequency ≤0.35 and LOD score ≥2.0.
2.4. Synteny analysis
R. sativus amino acid sequences were deduced from mapped ESTs. BLASTX was used to compare predicted R. sativus sequences against the TAIR9-predicted protein database (http://www.arabidopsis.org) with a threshold E-value of 1e−50. Because Kim et al.11 reported a B. rapa genetic map constructed with EST-based markers, a comparative map between A. thaliana and B. rapa was also constructed by locating nucleotide sequences of marker loci from B. rapa on the A. thaliana genome.
3. Results
3.1. Features of R. sativus ESTs
A total of 32 256 cDNA clones, including 9216 clones from a seedling library, 7680 clones from a root library, 7680 clones from a leaf library, and 7680 clones from a flower library, were sequenced from the 5′end. A total of 26 606 sequences (8458, 6320, 6721, and 5107 from seedling, root, leaf, and flower libraries, respectively) consisting of 19 391 246 qualified bases were obtained, and the average EST length was 729 bp.
The PHRAD program was used to cluster EST sequences in order to identify the number of independent EST species. These analyses indicated 10 381 potential non-redundant EST sequences, which included 6625 contigs and 3756 singletons with an average GC content of 45.6%. Non-redundant EST sequences were compared against the Unigene data of R. sativus, A. thaliana, B. napus, B. oleracea, B. rapa, L. japonicas, and O. sativa (http://www.ncbi.nlm.nih.gov/unigene). These comparisons indicated 9758 non-redundant ESTs with significant similarity (E-value <1e−10) to registered sequences of the seven species and the remaining 623 non-redundant ESTs were unique to the data from this study.
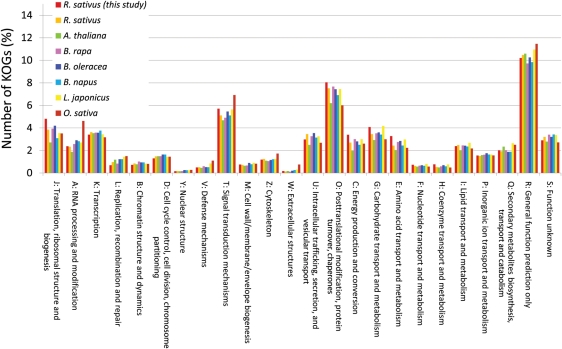
To investigate the functional classification of R. sativus ESTs, BLASTX was used to compare non-redundant EST sequences with eukaryotic clusters of orthologous groups (KOGs) and these ESTs were then classified into KOG categories.21 Among the 10 381 non-redundant R. sativus EST sequences, 6265 showed similarities to KOG sequences that have functional classification, and their distribution is similar to those of the seven species (Fig. 1).
Figure 1.
Functional classification of non-redundant R. sativus EST sequences and unigenes of the seven plant species into KOG categories. BLASTX was used to compare non-redundant EST sequences with the KOG sequence set. EST sequences were then classified into the KOG categories with the most similar sequences.
3.2. SSR motifs in R. sativus ESTs
In silico data mining of the 26 606 sequences yielded 3800 SSR markers, which were designated as RSS (Raphanus sativus EST-derived SSR) markers (Supplementary Table S1). Out of the 3800 SSR markers, 3733 markers (98.2%) had motif lengths ranging from 20 to 35 bases (Supplementary Table S1). The RSS marker motifs contained 3094 (81.4%), 381 (10.0%), and 325 (8.6%) tri-, di-, and tetra-nucleotide repeats, respectively (Table 1). The most abundant trinucleotide repeats were poly (AAG)n (21.4%), poly (GGA)n (14.2%), and poly (ATC)n (10.1%). Four types of dinucleotide repeats were observed and the poly (AG)n motifs were the most abundant (6.8%). The tetranucleotide repeats poly (AAAG)n (2.7%), poly (AAAC)n (2.1%) and poly (AAAT)n (1.4%) were more frequently observed than the other motifs. Among the 3800 SSR motifs, 1604 and 242 were located on coding and untranscribed regions, respectively (Table 1, Supplementary Table S1). SSRs on coding regions consisted of 30 di-, 1533 tri-, and 41 tetra-nucleotide repeat motifs, while those on untranscribed regions included 96 di-, 104 tri-, and 42 terta-nucleotide repeat motifs. The other 1954 SSRs were not assigned to the regions because the length of each EST was not enough to predict open reading frames.
Table 1.
Number of SSR motifs in the RSS markers
| Motif | Designed SSR markers |
Polymorphic SSR markers |
||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Total | %a | Coding region | %a | UTR | %a | Unassigned regionb | %a | Total | Polymorphic ratio (%)c | Coding region | Polymorphic ratio (%)c | UTR | Polymorphic ratio (%)c | Unassigned regionb | Polymorphic ratio (%)c | |
| Dinucleotide | ||||||||||||||||
| AC | 60 | 1.6 | 3 | 0.1 | 7 | 0.2 | 50 | 1.3 | 14 | 23.3 | 2 | 66.7 | 1 | 14.3 | 11 | 22.0 |
| AG | 257 | 6.8 | 25 | 0.7 | 83 | 2.2 | 149 | 3.9 | 48 | 18.7 | 4 | 16.0 | 22 | 26.5 | 22 | 14.8 |
| AT | 63 | 1.7 | 2 | 0.1 | 6 | 0.2 | 55 | 1.4 | 14 | 22.2 | 1 | 50.0 | 1 | 16.7 | 12 | 21.8 |
| GC | 1 | 0.0 | 0 | 0.0 | 0 | 0.0 | 1 | 0.0 | 0 | 0.0 | 0 | — | 0 | — | 0 | 0.0 |
| Total | 381 | 10.0 | 30 | 0.8 | 96 | 2.5 | 255 | 6.7 | 76 | 19.9 | 7 | 23.3 | 24 | 25.0 | 45 | 17.6 |
| Trinucleotide | ||||||||||||||||
| AAC | 305 | 8.0 | 140 | 3.7 | 9 | 0.2 | 156 | 4.1 | 53 | 17.4 | 27 | 19.3 | 1 | 11.1 | 25 | 16.0 |
| AAG | 815 | 21.4 | 374 | 9.8 | 49 | 1.3 | 392 | 10.3 | 119 | 14.6 | 55 | 14.7 | 7 | 14.3 | 57 | 14.5 |
| AAT | 103 | 2.7 | 13 | 0.3 | 10 | 0.3 | 80 | 2.1 | 20 | 19.4 | 2 | 15.4 | 2 | 20.0 | 16 | 20.0 |
| ACG | 158 | 4.2 | 74 | 1.9 | 6 | 0.2 | 78 | 2.1 | 23 | 14.6 | 9 | 12.2 | 1 | 16.7 | 13 | 16.7 |
| ACT | 65 | 1.7 | 22 | 0.6 | 0 | 0.0 | 43 | 1.1 | 11 | 16.9 | 3 | 13.6 | 0 | — | 8 | 18.6 |
| AGC | 293 | 7.7 | 180 | 4.7 | 1 | 0.0 | 112 | 2.9 | 58 | 19.8 | 34 | 18.9 | 0 | 0.0 | 24 | 21.4 |
| ATC | 385 | 10.1 | 192 | 5.1 | 14 | 0.4 | 179 | 4.7 | 69 | 17.9 | 37 | 19.3 | 4 | 28.6 | 28 | 15.6 |
| GGA | 541 | 14.2 | 310 | 8.2 | 8 | 0.2 | 223 | 5.9 | 89 | 16.5 | 53 | 17.1 | 1 | 12.5 | 35 | 15.7 |
| GGC | 154 | 4.1 | 95 | 2.5 | 1 | 0.0 | 58 | 1.5 | 22 | 14.3 | 17 | 17.9 | 0 | 0.0 | 5 | 8.6 |
| GGT | 275 | 7.2 | 133 | 3.5 | 6 | 0.2 | 136 | 3.6 | 40 | 14.5 | 18 | 13.5 | 1 | 16.7 | 21 | 15.4 |
| Total | 3094 | 81.4 | 1533 | 40.3 | 104 | 2.7 | 1457 | 38.3 | 504 | 16.3 | 255 | 16.6 | 17 | 16.3 | 232 | 15.9 |
| Tetranucleotide | ||||||||||||||||
| AAAC | 81 | 2.1 | 3 | 0.1 | 11 | 0.3 | 67 | 1.8 | 17 | 21.0 | 1 | 33.3 | 2 | 18.2 | 14 | 20.9 |
| AAAG | 101 | 2.7 | 13 | 0.3 | 15 | 0.4 | 73 | 1.9 | 14 | 13.9 | 2 | 15.4 | 2 | 13.3 | 10 | 13.7 |
| AAAT | 53 | 1.4 | 3 | 0.1 | 4 | 0.1 | 46 | 1.2 | 14 | 26.4 | 1 | 33.3 | 1 | 25.0 | 12 | 26.1 |
| AACG | 1 | 0.0 | 0 | 0.0 | 0 | 0.0 | 1 | 0.0 | 0 | 0.0 | 0 | — | 0 | — | 0 | 0.0 |
| AAGC | 11 | 0.3 | 3 | 0.1 | 1 | 0.0 | 7 | 0.2 | 2 | 18.2 | 1 | 33.3 | 0 | 0.0 | 1 | 14.3 |
| AATC | 24 | 0.6 | 4 | 0.1 | 4 | 0.1 | 16 | 0.4 | 6 | 25.0 | 0 | 0.0 | 3 | 75.0 | 3 | 18.8 |
| AATG | 9 | 0.2 | 1 | 0.0 | 1 | 0.0 | 7 | 0.2 | 1 | 11.1 | 0 | 0.0 | 0 | 0.0 | 1 | 14.3 |
| AATT | 7 | 0.2 | 0 | 0.0 | 1 | 0.0 | 6 | 0.2 | 1 | 14.3 | 0 | — | 0 | 0.0 | 1 | 16.7 |
| AGGC | 1 | 0.0 | 1 | 0.0 | 0 | 0.0 | 0 | 0.0 | 0 | 0.0 | 0 | 0.0 | 0 | — | 0 | — |
| GACG | 2 | 0.1 | 1 | 0.0 | 1 | 0.0 | 0 | 0.0 | 0 | 0.0 | 0 | 0.0 | 0 | 0.0 | 0 | — |
| GAGC | 3 | 0.1 | 1 | 0.0 | 1 | 0.0 | 1 | 0.0 | 1 | 33.3 | 0 | 0.0 | 0 | 0.0 | 1 | 100.0 |
| GATC | 2 | 0.1 | 0 | 0.0 | 1 | 0.0 | 1 | 0.0 | 0 | 0.0 | 0 | — | 0 | 0.0 | 0 | 0.0 |
| GGAC | 1 | 0.0 | 0 | 0.0 | 0 | 0.0 | 1 | 0.0 | 0 | 0.0 | 0 | — | 0 | — | 0 | 0.0 |
| GGAT | 8 | 0.2 | 2 | 0.1 | 0 | 0.0 | 6 | 0.2 | 3 | 37.5 | 1 | 50.0 | 0 | — | 2 | 33.3 |
| GGCT | 1 | 0.0 | 0 | 0.0 | 0 | 0.0 | 1 | 0.0 | 0 | 0.0 | 0 | — | 0 | — | 0 | 0.0 |
| GGGA | 18 | 0.5 | 8 | 0.2 | 2 | 0.1 | 8 | 0.2 | 3 | 16.7 | 2 | 25.0 | 0 | 0.0 | 1 | 12.5 |
| GGGT | 2 | 0.1 | 1 | 0.0 | 0 | 0.0 | 1 | 0.0 | 0 | 0.0 | 0 | 0.0 | 0 | — | 0 | 0.0 |
| Total | 325 | 8.6 | 41 | 1.1 | 42 | 1.1 | 242 | 6.4 | 62 | 19.1 | 8 | 19.5 | 8 | 19.0 | 46 | 19.0 |
| Total | 3800 | 100.0 | 1604 | 42.2 | 242 | 6.4 | 1954 | 51.4 | 642 | 16.9 | 270 | 16.8 | 49 | 20.2 | 323 | 16.5 |
aPercentage to a total of 3800 SSR markers.
bThe position of SSRs was not assigned because of the length of each EST was not enough to predict open reading frames.
cPercentage to the designed SSR markers.
3.3. Linkage analysis and map construction
All 3800 SSR markers were used for polymorphic analysis of parental lines from the mapping population and 642 (16.9%) SSR markers were selected (Table 1, Supplementary Table S1). These markers identified 18 double polymorphic loci and one triple polymorphic locus and, consequently, 662 polymorphic loci were generated from the 642 SSR markers. Among the 662 SSR loci, 545 and 117 identified co-dominant and dominant loci, respectively. Null alleles were observed in 98 and 19 of the 117 dominant markers from ‘HA2’ and ‘GSK3-1’, respectively. The polymorphic ratios of the di-, tri-, and tetra-nucleotide repeats were 19.9, 16.3, and 19.1%, respectively, and the polymorphic ratio of SSRs on UTRs (20.2%) were higher than that on coding regions (16.8%) (Table 1).
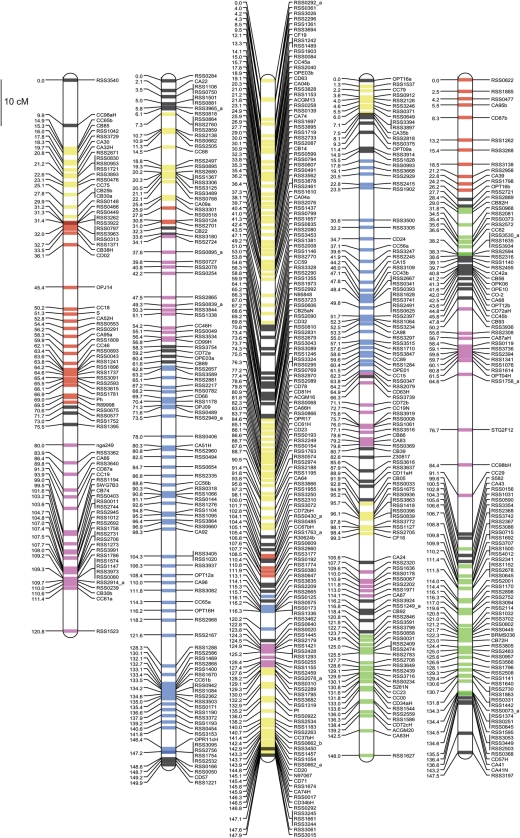
Segregation data were generated from the 662 SSR and 223 RAPD, RFLP, and trait marker loci in the mapping population. Following linkage analysis, 843 loci (630 SSR and 213 RAPD, RFLP and trait loci) were mapped into nine linkage groups, while the remaining 42 loci (32 SSR and 10 RAPD, RFLP, and trait loci) were excluded from the analysis. Linkage groups were named according to the previous report.14 The linkage groups covered 1129.2 cM (Table 2, Fig. 2, and Supplementary Table S2) and the average distance between neighbouring loci was 1.3 cM, with distances ranging from 1.0 cM in linkage group 3 (LG3) to 2.0 cM in LG6. Segregation distortion was observed for 8.9% of the mapped marker loci (Table 2 and Supplementary Table S2). The segregation distortion ratios varied among linkage groups; LG5 showed distortions with 3.7% of the mapped loci, while LG2 showed distortions for 20.2% of the loci.
Table 2.
Length, number of mapped loci, and segregation distortion of the genetic map
| Linkage group | Distance (cM) | The number of marker loci |
cM/marker loci | Segregation distortion ratio (%) | ||
|---|---|---|---|---|---|---|
| Total | EST-SSR | RAPD | ||||
| 1 | 120.8 | 82 | 56 | 26 | 1.5 | 7.3 |
| 2 | 149.9 | 109 | 88 | 21 | 1.4 | 20.2 |
| 3 | 147.9 | 155 | 124 | 31 | 1.0 | 7.7 |
| 4 | 148.0 | 114 | 82 | 32 | 1.3 | 4.4 |
| 5 | 147.5 | 107 | 79 | 28 | 1.4 | 3.7 |
| 6 | 153.2 | 77 | 50 | 27 | 2.0 | 5.2 |
| 7 | 100.3 | 84 | 61 | 23 | 1.2 | 10.7 |
| 8 | 85.4 | 65 | 49 | 16 | 1.3 | 7.7 |
| 9 | 76.2 | 50 | 41 | 9 | 1.6 | 16.0 |
| Total | 1129.2 | 843 | 630 | 213 | 1.3 | 8.9 |
Figure 2.
Genetic linkage map of R. sativus. The red, blue, yellow, green and pink lines on each linkage group correspond to chromosomes 1, 2, 3, 4 and 5 of A. thaliana. Detailed information about the map and markers is shown in Supplementary Table S2 and is also available at http://marker.kazusa.or.jp/Daikon.
3.4. Genomic comparison of R. sativus, A. thaliana, and B. rapa
BLASTX was used to compare the nucleotide sequences of 630 ESTs mapped into linkage groups with the predicted-protein database of A. thaliana. Of the 630 ESTs, 358 showed significant homology (≤1e−50 E-value) to 500 A. thaliana genes, and 230, 87, and 22 ESTs mapped onto one, two, and three regions of the A. thaliana genome, respectively, while 19 other ESTs mapped onto four or five regions and 273 ESTs showed no significant homology to the A. thaliana genome (Supplementary Table S2). To compare the genome structures of R. sativus and A. thaliana, syntenic regions were deduced when more than three continuous EST loci on R. sativus linkage groups aligned to a single portion of the genome of A. thaliana. Chromosomal segments of A. thaliana were assigned onto the R. sativus genetic map (Fig. 2). Approximately 65% (739.1 cM) of the genetic map was covered by 35 A. thaliana chromosome segments, which ranged from one segment on LG9 to six segments on LG3 and LG4. The longest segment (84.4 cM) was found on LG2 and it had derived from A. thaliana Chr2. The shortest segment (2.1 cM) was detected on LG7 and it arose from Chr1. The average length of a single segment was 21.1 cM.
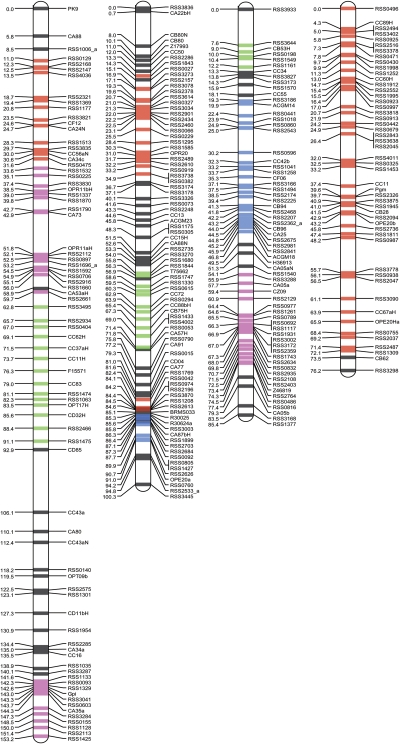
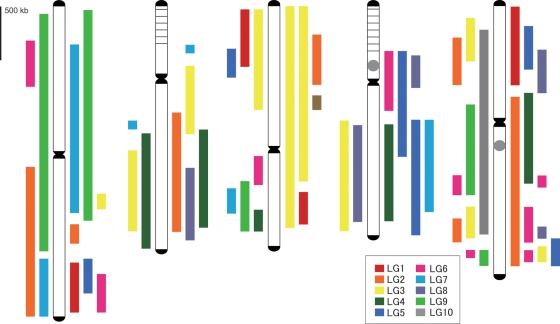
Conversely, the R. sativus genetic map could be assigned to almost the whole A. thaliana genome, with the exception of the ribosomal DNA repeat regions on Chr2 and Chr4 (Fig. 3). R. sativus linkage groups showed average mapped depths of 2.0 on the A. thaliana genome and depths ranged from 1 to 4. Two R. sativus linkage groups were redundantly assigned to 48% of the A. thaliana genome and the remaining 18, 24, and 4% of the A. thaliana genome were covered by single, triple, and quadruple R. sativus linkage groups, respectively. A. thaliana Chr3 was covered with two R. sativus LG3s, each of which was derived from both distal parts of LG3, respectively (Supplementary Table S2). In addition to the R. sativus map, a published genetic map of B. rapa was also assigned onto the A. thaliana genome. This analysis confirmed that the B. rapa genome contains two or three copies of at least 24 genomic segments from A. thaliana, as suggested by a previous report.11 The average mapped depth of B. rapa linkage groups on the A. thaliana genome was 1.4 and ranged from 1 to 3. Forty-eight per cent of the A. thaliana genome was covered by two B. rapa linkage groups, while 24 and 7% of the genome was covered by single and triple B. rapa linkage groups. Large syntenic segments were identified between the three species on the following A. thaliana (At) chromosomes: At Chr1, B. rapa (Br) LG9, R. sativus (Rs) LG7, and Rs LG9; At Chr2, Br LG3, Br LG4, Rs LG2, and LG4; At Chr3, Br LG3 and Rs LG3; At Chr4, Br LG 3, Br LG8, Rs LG4, Rs LG5, and Rs LG7; and At Ch5, Br LG10, and Rs LG2. Scattered syntenic segments were also observed on several genomic regions of A. thaliana. The most overlapping and disrupted syntenic region was identified on a distal part of the long arm of At Chr5.
Figure 3.
Synteny map of A. thaliana and R. sativus or B. rapa. A. thaliana chromosomes are shown with white bars. Telomeric and centromeric regions are indicated in black. Heterochromatic knobs and rDNA repeat regions are identified by grey circles and horizontal lines, respectively. The colored bars on the right and left sides of the A. thaliana chromosomes indicate linkage groups from R. sativus and B. rapa, respectively.
4. Discussion
A genetic map of R. sativus was constructed using 630 EST-SSR and 213 RAPD, RFLP, and trait marker loci. This map comprised nine linkage groups, which corresponded to the number of chromosomes in haploid R. sativus (n = 9). The genetic map was calculated to cover 1129.2 cM and the average interval between marker loci was 1.3 cM. To our knowledge, this map comprises the highest number and density of marker loci of all the published R. sativus genetic maps.14,17–19,26,27 However, the genetic map generated in this study was shorter than the map reported by Budahn et al.,17 which was 1517 cM. That map was constructed using 245 F2 plants and most of mapped markers were dominant, i.e. AFLPs and RAPDs. In contrast, the map from this study was developed using 155 RILs, with a large number of co-dominant SSR markers. These differences in length can be accounted for by the differences in size of mapping population, as well as in the number and types of DNA markers.28 Another contributing factor could be differences in the frequency of chromosome recombination caused by environmental factors and genetic diversity of the mapping parents.29,30
EST-SSR markers are often preferable to RFLP, AFLP, and RAPD markers, since EST nucleotide sequences generally show higher levels of similarity across different species, genera, and families than sequences from intergenic regions. Therefore, the primers of EST-SSR markers are often directly applicable to PCR analyses of related species, although the efficiency of transferability depends upon the genetic distance between the species.6 Moreover, the multi-allelic nature of SSRs makes them more efficient than SNP markers for polymorphic analysis of diverse accessions. A preliminary experiment showed that R. sativus EST-SSR markers could be transferred to a mapping population derived from a cross between a Japanese daikon and a rat-tailed radish. PCR amplified specific DNA fragments in 665 (98%) of 676 tested EST-SSR markers and 291 (43%) markers showed polymorphism between the two lines. The order of mapped markers on this linkage map matched the order on the high-density linkage map from this study (data not shown).
EST-based markers are also valuable for comparisons of genome structure between different plant species. To demonstrate their utility, R. sativus and A. thaliana were compared using homoeologies between amino acid sequences predicted from the EST sequences and their locations on the present linkage map. The genetic map of R. sativus was covered by at least 35 disrupted A. thaliana genomic fragments and 90% of the genetic map corresponded to regions of the A. thaliana genome that had been doubled, tripled, or quadrupled. All ESTs that mapped onto LG9 in the R. sativus map corresponded to ca. 70% of A. thaliana Chr5. However, no perfectly homologous linkage groups were observed within the genetic map of R. sativus. These results suggest that the tripling of the R. sativus genome occurred at some point after divergence from its common ancestor with A. thaliana, and that this amplification was followed by structural rearrangements, including deletion of parts of the chromosomes. Alternatively, one part of the genome may have undergone a duplication event, while another part tripled. These events may have occurred at or after the time of divergence between R. sativus and A. thaliana, ca. 20 million years ago.31,32
In addition to comparative analysis between R. sativus and A. thaliana, the genome structure of B. rapa was compared with R. sativus via the A. thaliana genome. The two species shared large syntenic regions across the A. thaliana genome, which suggests that large regions of the R. sativus and B. rapa genomes are conserved. Several scattered syntenic regions were also observed on the A. thaliana genome, e.g. the distal part of the long arm of Chr5. However, there was no conservation in the order or composition of the corresponding genomic segments between R. sativus and B. rapa (Fig. 3),11 which suggests no chromosomal synteny in this region among the three species. Absence of chromosomal synteny was also reported between A. thaliana and B. oleracea or B. nigra.9,10 It has been suggested that the three Brassica diploids, i.e. B. rapa, B. oleracea, and B. nigra, evolved via chromosomal structural rearrangements and/or polyploidy.9–11 The results from this study confirm the hypothesis that R. sativus and the three Brassica diploids differentiated independently from a common ancestor with A. thaliana and that this divergence occurred via genome rearrangement and/or polyploidy events at and/or after differentiation.
Since the first report for artificial amphidiploids between R. sativus and B. oleracea,33 amphidiploids derived from crosses between several combinations of Brassicaceae have been synthesized.34 Because the amphidiploids were important materials for plant breeding due to their vigorous growths and adaptability for alien chromosomes, the mechanisms for the hybridization and the amphidiploid formation have been investigated by cytogenetic, epigenetic, and genomic approaches.33 According to our results of the comparative genomics of the Brassicaceae, chromosomal synteny is unnecessary for hybridization and the amphidiploid formation, suggesting that genetic factors like Ph1 reported in wheat,35 micro-syntenies, and/or nuclear-organella interactions might be required.
As has been observed with the genetic and genomic studies of Brassica species,12,13 a comparative map between R. sativus and the A. thaliana genome will accelerate gene identification in Raphanus species. In particular, it will aid candidate gene isolation since a large number of genes/traits have already been characterized and mapped onto the A. thaliana genome. Kaneko et al.14 identified a QTL associated with resistance to yellow disease caused by Fusarium oxysporum f. sp. raphani, and this QTL was linked to the OPJ14 locus on LG1. According to our comparative map between R. sativus and A. thaliana, this locus is predicted to correspond to the long arm of A. thaliana Chr1, 27 Mb from the top of the chromosome (Supplementary Table S2). TIR-NBS genes have been implicated in disease resistance in many plant species.36 A cluster of these genes was identified in the chromosomal region of A. thaliana that corresponds to resistance to yellow disease in the comparative map, a finding which suggests that a member of the TIR-NBS gene family is responsible for F. oxysporum resistance in R. sativus.
The R. sativus EST data, SSR markers and genetic map developed in this study can be used for various genetic analyses including gene mapping, QTL analysis, population genetics, marker-assisted breeding, and whole-genome sequencing studies. This study has provided a standard map for genomics, genetics, and breeding of R. sativus and related species.
Supplementary data
Supplementary data are available at www.dnaresearch.oxfordjournals.org.
Funding
This work was supported by the Kazusa DNA Research Institute and the Mikado Kyowa Seed Co., Ltd.
Supplementary Material
Acknowledgements
We are grateful to Prof. Kazunari Nomura (Nihon University, Japan) for providing the rat-tailed radish.
References
- 1.Deulvot C., Charrel H., Marty A., et al. Highly-multiplexed SNP genotyping for genetic mapping and germplasm diversity studies in pea. BMC Genomics. 2010;11:468. doi: 10.1186/1471-2164-11-468. doi:10.1186/1471-2164-11-468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Hyten D.L., Song Q., Choi I.Y., et al. High-throughput genotyping with the GoldenGate assay in the complex genome of soybean. Theor. Appl. Genet. 2008;116:945–52. doi: 10.1007/s00122-008-0726-2. doi:10.1007/s00122-008-0726-2. [DOI] [PubMed] [Google Scholar]
- 3.Hyten D.L., Song Q., Fickus E.W., et al. High-throughput SNP discovery and assay development in common bean. BMC Genomics. 2010;11:475. doi: 10.1186/1471-2164-11-475. doi:10.1186/1471-2164-11-475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Muchero W., Diop N.N., Bhat P.R., et al. A consensus genetic map of cowpea [Vigna unguiculata (L) Walp.] and synteny based on EST-derived SNPs. Proc. Natl. Acad. Sci. USA. 2009;106:18159–64. doi: 10.1073/pnas.0905886106. doi:10.1073/pnas.0905886106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Shirasawa K., Isobe S., Hirakawa H., et al. SNP discovery and linkage map construction in cultivated tomato. DNA Res. 2010;17:381–91. doi: 10.1093/dnares/dsq024. doi:10.1093/dnares/dsq024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kalia R.K., Rai M.K., Kalia S., Singh R., Dhawan A.K. Microsatellite markers: an overview of the recent progress in plants. Euphytica. 2011;117:309–34. doi:10.1007/s10681-010-0286-9. [Google Scholar]
- 7.The Arabidopsis Genome Initiative. Analysis of the genome sequence of the flowering plant Arabidopsis thaliana. Nature. 2000;408:796–815. doi: 10.1038/35048692. doi:10.1038/35048692. [DOI] [PubMed] [Google Scholar]
- 8.U N. Genome-analysis in Brassica with special reference to the experimental formation of B. napus and peculiar mode of fertilization. Jpn. J. Bot. 1935;7:389–452. [Google Scholar]
- 9.Lagercrantz U. Comparative mapping between Arabidopsis thaliana and Brassica nigra indicates that Brassica genomes have evolved through extensive genome replication accompanied by chromosome fusions and frequent rearrangements. Genetics. 1998;150:1217–28. doi: 10.1093/genetics/150.3.1217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lan T.H., DelMonte T.A., Reischmann K.P., et al. An EST-enriched comparative map of Brassica oleracea and Arabidopsis thaliana. Genome Res. 2000;10:776–88. doi: 10.1101/gr.10.6.776. doi:10.1101/gr.10.6.776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kim J.S., Chung T.Y., King G.J., et al. A sequence-tagged linkage map of Brassica rapa. Genetics. 2006;174:29–39. doi: 10.1534/genetics.106.060152. doi:10.1534/genetics.106.060152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Li F., Kitashiba H., Inaba K., Nishio T. A Brassica rapa linkage map of EST-based SNP markers for identification of candidate genes controlling flowering time and leaf morphological traits. DNA Res. 2009;16:311–23. doi: 10.1093/dnares/dsp020. doi:10.1093/dnares/dsp020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Suwabe K., Tsukazaki H., Iketani H., et al. Simple sequence repeat-based comparative genomics between Brassica rapa and Arabidopsis thaliana: the genetic origin of clubroot resistance. Genetics. 2006;173:309–19. doi: 10.1534/genetics.104.038968. doi:10.1534/genetics.104.038968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kaneko Y., Kimizuka-Takagi C., Bang S.W., Matsuzawa Y. Radish. In: Kole C., editor. Genome Mapping and Molecular Breeding in Plants. vol. 5. New York: Springer; 2007. pp. 141–60. [Google Scholar]
- 15.Kitamura S. Cultivars of radish and their change. In: Nishiyama I., editor. Japanese Radish. Tokyo: Japan Society for the Promotion of Science; 1958. pp. 1–19. (in Japanese) [Google Scholar]
- 16.Arumuganathan K., Earle E.D. Nuclear DNA content of some important plant. Plant Mol. Biol. Rep. 1991;9:208–18. doi:10.1007/BF02672069. [Google Scholar]
- 17.Budahn H., Peterka H., Mousa M.A., Ding Y., Zhang S., Li J. Molecular mapping in oil radish (Raphanus sativus L.) and QTL analysis of resistance against beet cyst nematode (Heterodera schachtii) Theor. Appl. Genet. 2009;118:775–82. doi: 10.1007/s00122-008-0937-6. doi:10.1007/s00122-008-0937-6. [DOI] [PubMed] [Google Scholar]
- 18.Kamei A., Tsuro M., Kubo N., et al. QTL mapping of clubroot resistance in radish (Raphanus sativus L.) Theor. Appl. Genet. 2010;120:1021–7. doi: 10.1007/s00122-009-1230-z. doi:10.1007/s00122-009-1230-z. [DOI] [PubMed] [Google Scholar]
- 19.Tsuro M., Suwabe K., Kubo N., Matsumoto S., Hirai M. Mapping of QTLs controlling root shape and red pigmentation in radish, Raphanus sativus L. Breed. Sci. 2008;58:55–61. doi:10.1270/jsbbs.58.55. [Google Scholar]
- 20.Sato S., Isobe S., Asamizu E., et al. Comprehensive structural analysis of the genome of red clover (Trifolium pratense L.) DNA Res. 2005;12:301–64. doi: 10.1093/dnares/dsi018. doi:10.1093/dnares/dsi018. [DOI] [PubMed] [Google Scholar]
- 21.Tatusov R.L., Fedorova N.D., Jackson J.D., et al. The COG database: an updated version includes eukaryotes. BMC Bioinformatics. 2003;4:41. doi: 10.1186/1471-2105-4-41. doi:10.1186/1471-2105-4-41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Shirasawa K., Asamizu E., Fukuoka H., et al. An interspecific linkage map of SSR and intronic polymorphism markers in tomato. Theor. Appl. Genet. 2010;121:731–9. doi: 10.1007/s00122-010-1344-3. doi:10.1007/s00122-010-1344-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Botella M.A., Coleman M.J., Hughes D.E., Nishimura M.T., Jones J.D., Somerville S.C. Map positions of 47 Arabidopsis sequences with sequence similarity to disease resistance genes. Plant J. 1997;12:1197–211. doi: 10.1046/j.1365-313x.1997.12051197.x. doi:10.1046/j.1365-313X.1997.12051197.x. [DOI] [PubMed] [Google Scholar]
- 24.Fourmann M., Barret P., Froger N., et al. From Arabidopsis thaliana to Brassica napus: development of amplified consensus genetic markers (ACGM) for construction of a gene map. Theor. Appl. Genet. 2002;105:1196–206. doi: 10.1007/s00122-002-1040-z. doi:10.1007/s00122-002-1040-z. [DOI] [PubMed] [Google Scholar]
- 25.Van Ooijen J.W. Software for the Calculation of Genetic Linkage Maps in Experimental Populations. Wageningen, The Netherlands: Kyazma, B.V; 2006. JoinMap®4. [Google Scholar]
- 26.Bett K.E., Lydiate D.J. Genetic analysis and genome mapping in Raphanus. Genome. 2003;46:423–30. doi: 10.1139/g03-026. doi:10.1139/g03-026. [DOI] [PubMed] [Google Scholar]
- 27.Tsuro M., Suwabe K., Kubo N., Matsumoto S., Hirai M. Construction of a molecular linkage map of radish (Raphanus sativus L.), based on AFLP and Brassica-SSR markers. Breed. Sci. 2005;55:107–11. doi:10.1270/jsbbs.55.107. [Google Scholar]
- 28.Gosselin I., Zhou Y., Bousquet J., Isabel N. Megagametophyte-derived linkage maps of white spruce (Picea glauca) based on RAPD, SCAR and ESTP markers. Theor. Appl. Genet. 2002;104:987–97. doi: 10.1007/s00122-001-0823-y. doi:10.1007/s00122-001-0823-y. [DOI] [PubMed] [Google Scholar]
- 29.Allard R.W. Evidence for genetic restriction of recombination in the lima bean. Genetics. 1963;48:1389–95. doi: 10.1093/genetics/48.10.1389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Axelsson T., Bowman C.M., Sharpe A.G., Lydiate D.J., Lagercrantz U. Amphidiploid Brassica juncea contains conserved progenitor genomes. Genome. 2000;43:679–88. [PubMed] [Google Scholar]
- 31.Yang Y.W., Lai K.N., Tai P.Y., Li W.H. Rates of nucleotide substitution in angiosperm mitochondrial DNA sequences and dates of divergence between Brassica and other angiosperm lineages. J. Mol. Evol. 1999;48:597–604. doi: 10.1007/pl00006502. doi:10.1007/PL00006502. [DOI] [PubMed] [Google Scholar]
- 32.Koch M.A., Haubold B., Mitchell-Olds T. Comparative evolutionary analysis of chalcone synthase and alcohol dehydrogenase loci in Arabidopsis, Arabis, and related genera (Brassicaceae) Mol. Biol. Evol. 2000;17:1483–98. doi: 10.1093/oxfordjournals.molbev.a026248. [DOI] [PubMed] [Google Scholar]
- 33.Karpechenko G.D. Poliploid hybrids Raphanus sativus L. times Brassica oleracea L. Bull. Appl. Bot. 1927;17:305–410. [Google Scholar]
- 34.Snowdon R.J. Cytogenetics and genome analysis in Brassica crops. Chromosome Res. 2007;15:85–95. doi: 10.1007/s10577-006-1105-y. doi:10.1007/s10577-006-1105-y. [DOI] [PubMed] [Google Scholar]
- 35.Riley R., Chapman V. Genetic control of the cytologically diploid behaviour of hexaploid wheat. Nature. 1959;182:713–5. doi:10.1038/182713a0. [Google Scholar]
- 36.Meyers B.C., Kozik A., Griego A., Kuang H., Michelmore R.W. Genome-wide analysis of NBS-LRR-encoding genes in Arabidopsis. Plant Cell. 2003;15:809–34. doi: 10.1105/tpc.009308. doi:10.1105/tpc.009308. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.