Abstract
We report a recurrent 680-kb deletion within chromosome 15q13.3 in ten individuals, from four unrelated families, with neurodevelopmental phenotypes including developmental delay, mental retardation and seizures. This deletion likely resulted from nonallelic homologous recombination between low-copy repeats on the normal and inverted region of chromosome 15q13.3. Although this deletion also affects OTUD7A, accumulated data suggest that haploinsufficiency of CHRNA7 is causative for the majority of neurodevelopmental phenotypes in the 15q13.3 microdeletion syndrome.
Recurrent deletions of chromosome 15q13.3 were not recognized as pathological until 2008, when they were first reported in individuals with mental retardation, seizures, autism, schizophrenia and bipolar disorder1–6. This deletion is also found in ~1% of individuals with idiopathic generalized epilepsy7,8. In contrast to what occurs in many analogous deletion syndromes, which arise as de novo events, most affected subjects have inherited the mutation from a parent, who may have any of the known phenotypic manifestations or occasionally a normal phenotype. Incomplete penetrance has been described in several of those families. The reciprocal duplication of 15q13.3 was also reported at a lower frequency; however, there is currently no clear evidence of whether the duplication is invariably benign or whether it may cause phenotypic abnormalities.
The critical 1.5-Mb deletion of 15q13.3 likely arises through non-allelic homologous recombination (NAHR) between low-copy repeat (LCR) sequences designated as breakpoints 4 and 5 (BP4 and BP5), telomeric to the greater Prader-Willi and Angelman syndromes domain. This chromosomal region encompasses six RefSeq genes: MTMR15, MTMR10, TRPM1, KLF13, OTUD7A and CHRNA7. CHRNA7 encodes the α7 subunit of the neuronal nicotinic acetylcholine receptor, which is highly expressed in the brain. Attention has focused on CHRNA7 because mutations in ion channel genes, possibly including CHRNA7 (ref. 9), have been associated frequently with epilepsy, and because of a possible implication of CHRNA7 in schizophrenia10,11. A fusion gene, CHRFAM7A, mapping in BP4 comprises exons A–E of FAM7A1 and FAM7A2 and exons 5–10 of CHRNA7. Any attempts at sequencing to detect mutations must distinguish the sequences for exons 5–10 from CHRNA7 and CHRFAM7A.
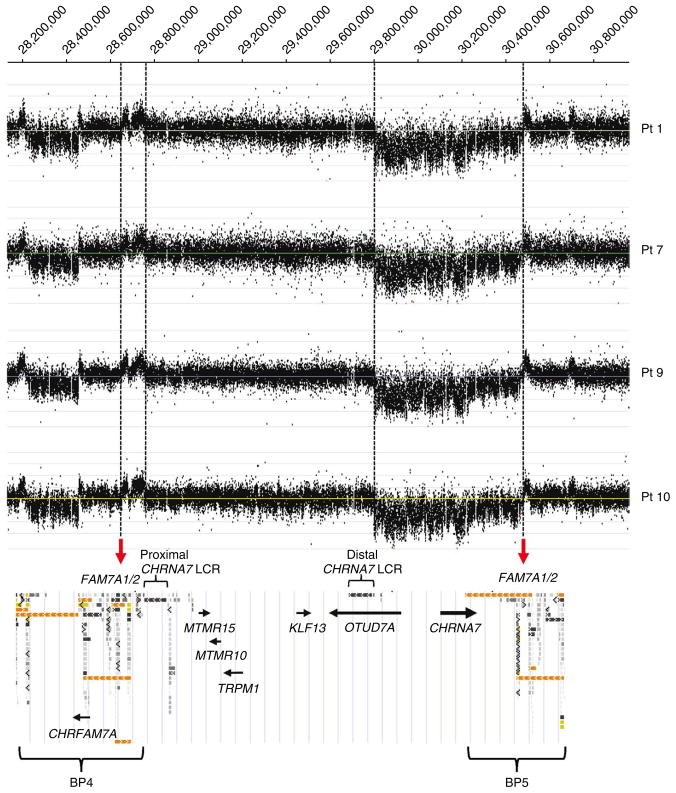
We now report ten individuals from four unrelated families with a smaller, ~680-kb deletion that lies within the 1.5-Mb deletion of 15q13.3 and encompasses the entire CHRNA7 gene and the first exon of one of the isoforms of OTUD7A (Fig. 1, Supplementary Fig. 1a–d and Supplementary Methods).
Figure 1.
Results of array CGH analysis with a 15q13.3 region-specific high-resolution 135K oligonucleotide microarray (NimbleGen). We identified the same apparent patterns of CNV changes in all four individuals with the 680-kb deletion, indicating that these deletions arose as a result of NAHR on chromosomes 15 with the same or nearly identical BP4-BP5 inversion.
Our subjects with the 680-kb deletion presented with phenotypes falling within the range of those seen for the 15q13.3 deletion syndrome (Table 1). The 680-kb deletion was inherited in both of two families for which both parents were available for study. Patient 1, the proband for his family, was an 8-year-old male with obesity (BMI = 30.6) and severe mental retardation (Kaufman Assessment Battery for Children score of 21). Mild facial dysmorphisms in this individual included bilateral epicanthal folds, anteverted nares and a thin upper lip. This individual had no history of seizures but had an abnormal electroencephalogram (EEG). The deletion was present in his mother, two siblings, maternal aunt and maternal grandmother (patients 2–6; Table 1 and Supplementary Fig. 1e). The individual’s mother and her sister have a history of mental retardation and epilepsy, and his siblings have global developmental delay. The family is of European ancestry and there were no family members in whom the deletion was nonpenetrant, but this is not substantially different from the published data of a 65–70% penetrance rate for 15q13.3 deletions (Supplementary Table 1). Patient 7 is a 21-month-old female proband with impaired growth and severe global developmental delay. Her deletion was maternally inherited, and her mother (patient 8) was reported to have normal intelligence but also a history of epilepsy since the age of 5. Patient 9 is a 16-year-old African-American male with mild mental retardation, attention deficit hyperactivity disorder and aggressive behavior. He has no history of seizures, and his EEG was reported as normal. His growth parameters are between the 50th and 75th percentiles. This individual’s father does not carry the deletion, but his biological mother was unavailable for additional studies. Patient 10 is an 8-month-old Hispanic male with global developmental delay, hypotonia and failure to thrive. The inheritance of the 680-kb deletion is unknown for this individual. In summary, four out of ten subjects with the 680-kb deletion manifested seizures or EEG abnormalities, and nine out of ten showed developmental delay and/or mental retardation. The variable expressivity in these subjects may relate to common polymorphisms on the remaining hemizygous allele, different genotypes elsewhere in the genome, or sex- and age-dependent penetrance for specific traits.
Table 1.
Phenotypic features of ten individuals from four unrelated families with a 680-kb deletion within chromosome 15q13.3
| Pt 1 | Pt 2 (brother of 1) | Pt 3 (sister of 1) | Pt 4 (mother of 1) | Pt 5 (maternal aunt of 1) | Pt 6 (MGM of 1) | Pt 7 | Pt 8 (mother of 7) | Pt 9 | Pt 10 | |
|---|---|---|---|---|---|---|---|---|---|---|
| Age at diagnosis | 8 yr | 4 yr | 3 yr | 30 yr | 21 yr | 52 yr | 21 mo | 23 yr | 16 yr | 8 mo |
| Sex | Male | Male | Female | Female | Female | Female | Female | Female | Male | Male |
| Ancestrya | E | E | E | E | E | E | E | E | AA | H |
| Cognitive function | Severe MR, IQ = 21 | Global DD | Global DD | Mild MR | Mild MR | Mild MR | Global DDb | Normal | Mild MR, IQ = 57 | Global DD |
| Seizures/EEG | None/abnormal EEGc | None | None | Absence epilepsy | Generalized epilepsy since childhood | None | None | Epilepsy since5 yr | None | None |
| Inheritance of CHRNA7 del.d | Mat | Mat | Mat | Mat | Mat | Unkn | Mat | Unkn | Unkn (not pat) | Unkn |
| Other CMA abnormal. | None | Unkn | Unkn | None | Unkn | Unkn | Xq26.2 dupe | Unkn | None | None |
CMA abnormal., abnormalities on chromosomal microarray analysis; DD, developmental delay; del., deletion; dup, duplication; EEG, electroencephalography; IQ, intelligence quotient; MGM, maternal grandmother; mo, months old; MR, mental retardation; pt, patient; yr, years old.
AA, African-American; E, European; H, Hispanic.
No words and not walking at 21 mo; all growth parameters below the 3rd percentile.
Right temporo-occipital delta-wave deceleration and rhythmic 3–4-s delta activity.
Mat, maternal; pat, paternal; unkn, unknown.
The ~0.3-Mb duplication in Xq26.2 (chr. X: 131,336,391–131,600,358) is also maternally inherited.
The 680-kb deletion was found in the Medical Genetics Laboratories at Baylor College of Medicine in 3 of 8,882 affected individuals, for a frequency of 1 in 2,960. This is far lower than the incidence of the larger 15q13.3 deletions, which were found at frequencies of 1 in 350 among mentally retarded individuals in the first published report1 and 1 in ~550 samples referred to the Medical Genetics Laboratories at Baylor College of Medicine. Our collection of subjects referred for chromosome microarray analysis includes mostly children with various combinations of developmental delay, mental retardation, autism and birth defects; a small percentage has seizures, but rarely as an isolated finding12. In a series of 3,699 controls (1,202 Germans and 2,497 North Americans of predominantly European ancestry) screened for copy number changes at CHRNA7 using SNP-based arrays and MLPA, no such deletions were reported7. Notably, a previously reported individual with paranoid schizophrenia and poor learning performance (case 15 in ref. 6) may have the 680-kb deletion, based on data reported in that study. Also of note, we identified a similar-sized duplication involving CHRNA7 in 59 unrelated cases among 8,882 subjects screened, for a frequency of 1 in ~170, which is close to the frequency of 1 in 185 among controls7. In all 21 instances of duplication in our series for which both parents were studied, the duplication was inherited by the child. Notably, in the same cohort, we observed only three 1.5-Mb BP4-BP5 duplications. The high frequency of duplications compared to deletions (59 compared to 3) and lack of de novo cases suggest that the duplication is either benign, associated with relatively high reproductive fitness or both. The proportion of inherited cases for both the larger 15q13.3 deletion (73% of all previously reported deletions) and the smaller 680-kb deletion is in marked contrast to those for other deletions with a similar spectrum of phenotypes. For example, the majority of 16p11.2 deletion cases are of de novo origin (Supplementary Note and Supplementary Table 1).
In-depth analysis of this genomic region revealed very complex arrangements of LCRs12,13. The distal breakpoints of the 680-kb deletions map within BP5, and the proximal breakpoints map between exons 1 and 2 of OTUD7A at the distal CHRNA7-LCR (Supplementary Fig. 1a–c). Notably, the 680-kb deletion is always accompanied by an ~90-kb duplication within BP4 (Fig. 1 and Supplementary Fig. 1b,c). We propose that the 680-kb deletions are due to NAHR between the CHRNA7-LCR copies on the normal and inverted chromosome 15q13.3 in a cell heterozygous for the inversion (Supplementary Fig. 1f); the inversion was shown to be present in 44% of individuals of varied ethnicities in the study population1. To verify this hypothesis, we designed CHRNA7-LCR–specific long-range PCR primers that would not amplify the original CHRNA7-LCR copies but would amplify the predicted junction between them (Supplementary Table 2). In patients 7 and 9, we amplified and sequenced the patient-specific PCR products, allowing us to narrow the NAHR site regions between two informative cis-morphic nucleotides within 49 bp (chr. 15: 28,815,284–28,815,332; chr. 15: 29,749,424–29,749,472) and the adjacent 67 bp (chr. 15: 28,815,334–28,815,400; chr. 15: 29,749,356–29,749,422) in the proximal and distal CHRNA7-LCRs, respectively. In patient 1, we narrowed the NAHR site to within 217 bp (chr15: 28,776,996-28,777,212; chr15: 29,787,470-29,787,686) in the proximal and distal CHRNA7-LCRs, respectively (Supplementary Fig. 1f and Supplementary Fig. 2). We propose that the BP4-BP5 inversion favors meiotic NAHR between the CHRNA7-LCRs, perhaps explaining the high frequency of CHRNA7 duplications. Until now, the BP4-BP5 inversion breakpoints have not been mapped1. The analyses of the distal breakpoints of all four 680-kb deletions and the proximal breakpoints of the 90-kb duplication fragments within BP4 allowed us to narrow the recombination site regions of the BP4-BP5 inversions to within the FAM7A1 and FAM7A2 genes, suggesting that they are potential recombinogenic sites (Fig. 1).
Given (i) the known function of CHRNA7, (ii) the phenotypic overlap among individuals with the larger 15q13.3 deletion and the 680-kb deletion, (iii) the cosegregation of the 680-kb deletion with the abnormal neurobehavioral phenotype in a family with six affected individuals and (iv) the fact that this deletion was not seen in 3,699 control samples7, we propose that the phenotypes associated with the larger 15q13.3 deletions and the 680-kb deletion are equivalent or nearly so. Four lines of argument suggest that haploinsufficiency for CHRNA7 and not for OTUD7A is causing the phenotype, although both are expressed in brain. First, OTUD7A is a putative deubiquitinase14, and haploinsufficiency for enzymes is usually recessive and not deleterious. Second, CHRNA7 encodes an ion channel, and mutations in ion channel genes have been associated frequently with epilepsy. Third, there is some evidence linking CHRNA7 to epilepsy and schizophrenia in humans9–11. Fourth, a heterozygous missense mutation in Chrna7 causes increased susceptibility to nicotine-induced seizures in mice15. Identifying disease-associated point mutations in CHRNA7 may eventually completely resolve the question of the genetic basis of neurodevelopmental phenotypes.
If haploinsufficiency for CHRN7A conveys most or all of the phenotypic abnormalities associated with 15q13.3 deletions, we hypothesize that treatment with an agonist to CHRNA7 (for example, chantix (varenicline)) might be beneficial in such individuals. Further discussion is provided in a Supplementary Note.
Supplementary Material
Acknowledgments
We thank all families who participated in this study; C. Carvalho, W. Gu, J.R. Lupski, C. Shaw and M. Strivens for helpful discussion; and P. Eng, Z. Ou and M. Suzuki for technical assistance. A.L.B. was supported by US National Institutes of Health grants HD-037283, M01-RR00188 (General Clinical Research Center), HD-024064 (Mental Retardation and Developmental Disabilities Research Center) and RR-019478 (Rare Disease Clinical Research Consortia) and by generous support from the William Stamps Farish Fund. P.S. was supported in part by grant R13-0005-04/2008 from the Polish Ministry of Science and Higher Education.
Footnotes
Note: Supplementary information is available on the Nature Genetics website.
AUTHOR CONTRIBUTIONS
B.L., H.S.H. and L.L.I. provided clinical information. M.S., C.P.S., S.N., G.S.P., C.N.-S. and A.L.B. coordinated clinical data collection. M.S., S.S.B., S.N., C.N.-S., Z.X., J.R.G. and P.S. performed array CGH, DNA sequencing and bioinformatic analyses. M.S., C.P.S., A.P., S.W.C., A.L.B. and P.S. interpreted the data, performed critical revisions and wrote the manuscript. M.S., C.P.S., A.L.B. and P.S. oversaw manuscript preparation and revision.
COMPETING INTERESTS STATEMENT
The authors declare competing financial interests: details accompany the full-text HTML version of the paper at http://www.nature.com/naturegenetics/.
Reprints and permissions information is available online at http://npg.nature.com/reprintsandpermissions/.
References
- 1.Sharp AJ, et al. Nat Genet. 2008;40:322–328. doi: 10.1038/ng.93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Miller DT, et al. J Med Genet. 2009;46:242–248. doi: 10.1136/jmg.2008.059907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ben-Shachar S, et al. J Med Genet. 2009;46:382–388. doi: 10.1136/jmg.2008.064378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.van Bon BW, et al. J Med Genet. 2009;46:511–523. doi: 10.1136/jmg.2008.063412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Stefansson H, et al. Nature. 2008;455:232–236. doi: 10.1038/nature07229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.International Schizophrenia Consortium. Nature. 2008;455:237–241. doi: 10.1038/nature07239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Helbig I, et al. Nat Genet. 2009;41:160–162. doi: 10.1038/ng.292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Dibbens LM, et al. Hum Mol Genet. 2009;18:3626–3631. doi: 10.1093/hmg/ddp311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Taske NL, et al. Epilepsy Res. 2002;49:157–172. doi: 10.1016/s0920-1211(02)00027-x. [DOI] [PubMed] [Google Scholar]
- 10.Leonard S, Freedman R. Biol Psychiatry. 2006;60:115–122. doi: 10.1016/j.biopsych.2006.03.054. [DOI] [PubMed] [Google Scholar]
- 11.Stephens SH, et al. Schizophr Res. 2009;109:102–112. doi: 10.1016/j.schres.2008.12.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lu X, et al. PLoS One. 2007;2:e327. doi: 10.1371/journal.pone.0000327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Makoff AJ, Flomen RH. Genome Biol. 2007;8:R114. doi: 10.1186/gb-2007-8-6-r114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kayagaki N, et al. Science. 2007;318:1628–1632. doi: 10.1126/science.1145918. [DOI] [PubMed] [Google Scholar]
- 15.Broide RS, et al. Mol Pharmacol. 2002;61:695–705. doi: 10.1124/mol.61.3.695. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.