Abstract
Array comparative genomic hybridization (aCGH) is a powerful tool for the molecular elucidation and diagnosis of disorders resulting from genomic copy-number variation (CNV). However, intragenic deletions or duplications—those including genomic intervals of a size smaller than a gene—have remained beyond the detection limit of most clinical aCGH analyses. Increasing array probe number improves genomic resolution, although higher cost may limit implementation, and enhanced detection of benign CNV can confound clinical interpretation. We designed an array with exonic coverage of selected disease and candidate genes and used it clinically to identify losses or gains throughout the genome involving at least one exon and as small as several hundred base pairs in size. In some patients, the detected copy-number change occurs within a gene known to be causative of the observed clinical phenotype, demonstrating the ability of this array to detect clinically relevant CNVs with subkilobase resolution. In summary, we demonstrate the utility of a custom-designed, exon-targeted oligonucleotide array to detect intragenic copy-number changes in patients with various clinical phenotypes.
Keywords: comparative genomic hybridization, CGH, intragenic rearrangements, exon mutations, copy-number variation, CNV
Introduction
High-resolution human genome analysis by array comparative genomic hybridization (aCGH) has revolutionized our ability to identify both benign copy-number variation (CNV) [Conrad et al., 2010b; Iafrate et al., 2004; Redon et al., 2006; Sebat et al., 2004] as well as pathogenic copy-number changes associated with genomic disorders [Lupski, 1998, 2009]. The pathogenic mechanism for these disorders, which involve genomic losses or gains of various sizes, is often dosage sensitivity of one or more of the genes within the rearranged genomic interval, but gene interruption, gene fusions, and position effects are increasingly recognized mechanisms mediating downstream effects of CNVs [Lupski and Stankiewicz, 2005]. Array CGH has enabled the detection of submicroscopic CNV (i.e., microdeletions and microduplications). To date, dozens of disorders have been ascribed to this type of genomic aberration [Mefford and Eichler, 2009; Stankiewicz and Lupski, 2010]. Recurrent microdeletions and microduplications occur via nonallelic homologous recombination (NAHR), with the “fixed” size of the reciprocal rearrangements reflecting the genomic positions of flanking, directly oriented repeat sequences utilized as homologous recombination substrates [Stankiewicz and Lupski, 2002]. In contrast, nonrecurrent rearrangements vary in size from genomic alterations involving megabases of DNA, to single-gene duplication/triplication, to CNV of single exons [Zhang et al., 2009a, 2010b]. Such nonrecurrent CNV occur by nonhomologous end joining (NHEJ) or by the recently described replication-based mechanisms of fork stalling and template switching/microhomology-mediated break induced replication (FoSTeS/MMBIR) [Hastings et al., 2009a,b; Lee et al., 2007].
Deletion or addition of one or more exons in a gene can have varied molecular and phenotypic consequences. A shift in reading frame can result in a premature termination codon, typically followed by nonsense-mediated decay (NMD) to create a loss of function allele [Maquat 1995]. Escape from NMD is possible, which may cause disease by gain of function [Ben-Shachar et al., 2009; Inoue et al., 2004]. Rarely, premature stop codons may also promote exon skipping (nonsense-associated altered splicing; NAS), which has the potential to restore the reading frame [Dietz et al., 1993; J. Wang et al., 2002]. An in-frame loss or gain may result in an altered [Yatsenko et al., 2003] or fused [Lifton et al., 1992; Miyahara et al., 1992] protein product with reduced or novel function. Thus, although haploinsufficiency may result from exonic CNV [Zhang et al., 2009b, 2010a], novel hypomorphic, antimorphic, and even neomorphic mutant alleles may be generated.
Exon-targeted aCGH (i.e., aCGH using an array with probes concentrated disproportionately in exons) can have either genome-wide or focused coverage. Genome-wide exonic arrays have been used to measure mRNA expression [Kapur et al., 2007], which unlike traditional 3′ expression arrays allows alternative splicing to be assessed [Clark et al., 2007; Gardina et al., 2006; Thorsen et al., 2008; Yeo et al., 2007]. This technique has enabled the discovery of tissue- and tumor-specific splice variants. Similar studies have been performed using locus-specific expression exon arrays [Labeit et al., 2006].
In addition to assessing gene expression, exon arrays have been used to assess genomic content. Bailey et al. [2008] studied nine healthy HapMap individuals using an array with exonic coverage for 2,790 genes. This study uncovered substantial CNV, disproportionately localized to regions containing segmental duplications. Although a catalog of benign intragenic CNV has been found by this and other studies [Conrad et al., 2010b], array-based detection of clinically relevant intragenic CNV remains in its infancy.
Hegde et al. [2008], del Gaudio et al. [2008], and Bovolenta et al. [2008] each designed a genomic microarray spanning the length of the dystrophin (DMD) gene. Although these single-locus arrays were not strictly exon targeted, the density and distribution of probes were sufficient to detect exonic (and intronic) CNV within the DMD locus in patients suspected of having mutations in this gene. Wong et al. [2008] also detected exonic CNV, using an array with dense coverage of 130 nuclear genes implicated in mitochondrial and metabolic disorders. This demonstrated the utility of a single nonexon-targeted array to detect intragenic CNV in multiple related genes.
Dhami et al. [2005] constructed an exon-specific array with coverage for 162 exons of five genes implicated in unrelated conditions (COL4A5, DMD, NF2, PLP1, and PMP22). Similarly, Saillour et al. [2008] performed aCGH to assess copy-number variation among 158 exons in eight disease genes (CFTR, DMD, and six sarcoglycan genes), as did Staaf et al. [2008] for the exons of six cancer-related genes (BRCA1, BRCA2, MSH2, MLH1, PTEN, and CDKN2A). Tayeh et al. [2009] constructed a targeted array with exonic (and intronic, with slightly diminished resolution) coverage of 71 disease genes, predominantly implicated in lysosomal storage and metabolic disorders. Significantly, this array was used in a clinical diagnostic setting in cases where gene sequencing failed to detect a mutation or mutations sufficient to explain a patient’s disease. The aforementioned studies provided proof-of-concept that a targeted exon array could be used to diagnose disparate disorders caused by intragenic copy-number changes. Yet, as the patients assessed in these studies were a selected population of previously-diagnosed (either clinically or molecularly) individuals, an array-based methodology to detect clinically relevant exonic copy-number changes genome-wide in unscreened or undiagnosed individuals has not yet been described.
As part of our continuing effort to clinically implement high resolution human genome analysis [Cheung et al., 2005, 2007; Lu et al., 2007, 2008; Ou et al., 2008; Shao et al., 2008], we sought to indentify CNV of smaller sizes (i.e., kilobasepairs in length, containing only one or a few exons) in functionally relevant regions of the human genome. To do this, we designed and developed a whole-genome microarray with coverage of approximately 24,000 exons in over 1,700 clinically relevant and candidate disease genes. This approach enables detection of intragenic copy-number changes in patients with varied clinical presentations that would otherwise be missed by traditional aCGH and would not be detected by gene-specific diagnostic DNA sequencing.
Materials and Methods
Array Design
V8 OLIGO is a custom-designed array with approximately 180,000 interrogating oligonucleotides, manufactured by Agilent Technologies, Inc. (Santa Clara, CA). This array contains the “best-performing” oligonucleotides (oligos) selected from Agilent’s online library (eArray; https://earray.chem.agilent.com/earray/) and has been further empirically optimized. Genomic features of the V8 OLIGO design include interrogation of all known microdeletion and microduplication syndrome regions as well as pericentromeric and subtelomeric regions and computationally predicted NAHR-mediated genomic instability regions flanked by low-copy repeats (LCR) as previously described [El-Hattab et al., 2009]. In addition, ~1,700 selected known or candidate disease genes have exonic coverage (101,644 probes in 24,319 exons; average of 4.2 probes/exon) as well as introns greater than 10 kb. The entire nuclear genome is covered with an average resolution of 30 kb, excluding LCRs and other repetitive sequences. Six hundred seventy probes interrogating the mitochondrial genome (average resolution of 25 bp) are also included. Further details are available at https://www.bcm.edu/geneticlabs/.
All genomic coordinates are based on the March 2006 assembly of the reference genome (NCBI36/hg18).
Human Subjects
Clinical aCGH was performed on 3,743 samples referred to the Medical Genetics Laboratory at Baylor College of Medicine (BCM) from June 2009 to March 2010. Cases 1 and 2, reported herein, were analyzed on our V7.4 OLIGO array prior to this period, and cases 15 and 29 were analyzed subsequent to it. Informed consent, approved by the Institutional Review Board for Human Subject Research at Baylor College of Medicine, was obtained in cases for which an image of the subject is provided.
DNA Isolation
DNA was extracted from whole blood using the Puregene DNA Blood Kit (Gentra, Minneapolis, MN) according to the manufacturer’s instructions.
Chromosomal Microarray Analysis (CMA)
The procedures for DNA digestion, labeling, and hybridization for the oligo arrays were performed according to the manufacturers’ instructions, with minor modifications [Ou et al., 2008]. Slides were scanned into image files using the Agilent G2565 Microarray Scanner. Scanned images were quantified using Agilent Feature Extraction software (v9.0), then analyzed for copy-number change using our in-house analysis package, as described previously [Cheung et al., 2005; Ou et al., 2008; Shaw et al., 2004].
Fluorescence In Situ Hybridization (FISH)
FISH analyses were performed with probes derived from bacterial artificial chromosomes (BACs) or fosmids using standard procedures [Shaffer et al., 1997]. Probe IDs are listed as part of the cytogenetic diagnoses provided as Supp. Data.
Multiplex Ligation-Dependent Probe Amplification (MLPA) Analysis
MLPA analysis was performed using the SALSA MLPA kit (MRC-Holland, Amsterdam, The Netherlands), according to the manufacturer’s instructions. Probe sets are described in the Supp. Methods. Additional information about commercially available probe sets is available at http://www.mrc-holland.com.
Polymerase Chain Reaction (PCR)
Long-range PCR was performed using the TaKaRa LA PCR Kit (TaKaRa Bio, Inc., Shiga, Japan). Reaction volume was 25 μl, containing 100 ng DNA, 0.5 μM of each primer, 400 μM of each dNTP, and 1.5 units TaKaRa LA Taq in 1 × LA PCR Buffer II. Primer sequences are provided in the Supp. Methods. The PCR was performed in a thermal cycler using the following conditions: 94°C × 1 min; 30 cycles of either 94°C × 30 sec followed by 68°C × 7 min, or 98°C × 5 sec followed by 68°C × 15 min; 72°C × 10 min. Agarose gel electrophoresis of amplification products enabled a comparison of amplicon sizes using patient DNA to those using control DNA from normal individuals.
DNA Sequencing
PCR products were cleaned with ExoSAP-IT (USB, Cleveland, OH), according to the manufacturer’s instructions, and nucleotide sequences determined by Sanger di-deoxynucleotide sequencing (Lone Star Labs, Houston, TX).
X-Chromosome Inactivation (XCI) Analysis
XCI analysis was performed as in Erez et al. [2009], based upon the method described by Allen et al. [1992].
Results
Exon-Targeted aCGH Detects Intragenic CNV
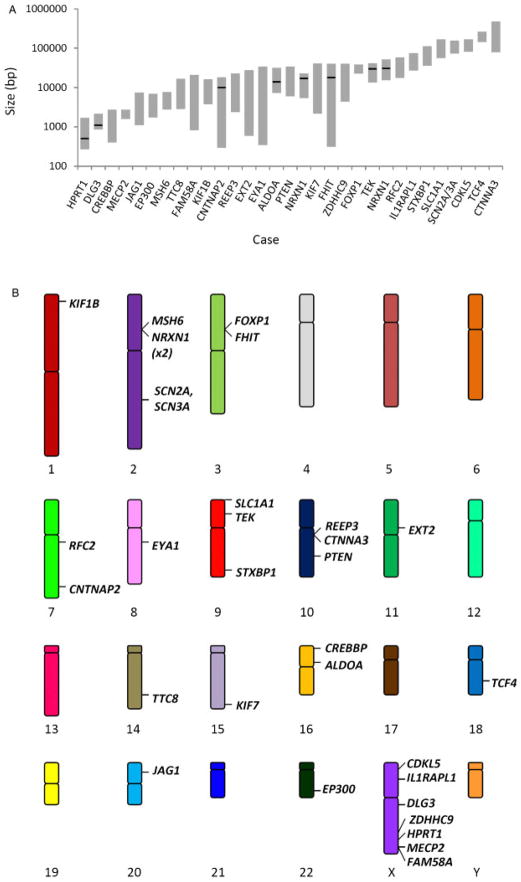
Of 3,743 aCGH analyses performed, the most common finding was a normal result, consistent with our previous experience with unfiltered clinical samples referred to a genetic diagnostic laboratory [Lu et al., 2007]. In addition to detecting many large genomic deletions and duplications, more than 40 cases of intragenic copy-number changes—deletions and duplications spanning a portion of a gene—were identified, a subset of which are presented in this report (Table 1). These 31 CNVs (30 copy-number losses and one gain) range in size from less than 1 kb to more than 105 bp (Fig. 1A); in fact, the smallest CNV analyzed by DNA sequencing was 502 bp (see below). The CNVs were found throughout the genome—on 14 of 22 autosomes and on the X chromosome (Fig. 1B). Observed CNVs appear to be overrepresented on the X chromosome (7/31 CNVs). This finding is consistent with: (1) the large number of X chromosome genes having enhanced exon coverage on our array (163; more than for any other chromosome), (2) the large number of confirmed, disease-associated loci on the X chromosome listed in OMIM (135; more than for any other chromosome except chromosome 1; ftp://ftp.ncbi.nih.gov/repository/OMIM/genemap), and (3) hemizygous expression of most X chromosome genes. Three of these X chromosome CNVs were found in females (cases 1, 4, and 7), each of which occurred in a gene implicated in X-linked dominant disease, while the rest occurred in males. Twenty-nine CNVs spanned one or several exons; 5′, 3′, and central exons were all represented among them. Two CNVs encompass a single intron of a gene. The CNVs of interest in three patients (cases 4, 12, and 13) were found by FISH to be mosaic (Table 1 and Supp. Data). All array findings summarized in Table 1 have been independently identified by an alternative molecular technique, including FISH (see Supp. Data), PCR (Supp. Figs. S1–S5), or MLPA (Supp. Figs. S6–S7). None of these findings represent known benign CNVs listed in the Database of Genomic Variants (DGV; http://projects.tcag.ca/variation/). Although multiple instances of copy-number change may occur in a single patient, only the deletions or duplications considered most likely to be clinically relevant are detailed in Table 1. Additional CNVs are listed in Supp. Table S1. In eight instances, PCR was followed by DNA sequencing of the breakpoints of the rearrangement, providing further confirmation and inference by conceptual translation as to how gene structure and genetic information might have been disrupted, thus aiding the elucidation of the molecular mechanism of disease.
Table 1.
Selected Cases of Intragenic Copy-Number Variation (CNV)
| Case No. | Gene(s) | Exon(s)a | Copy No. change | Chromosomal location | Inheritance | Size of loss/gain (kb)b | No. probes | Mean log2 | CNV describedc |
|---|---|---|---|---|---|---|---|---|---|
| 1 | MECP2 | 3–4 | Loss | Xq28 | De novo | 2 | 25 | −0.50 | Schollen et al., 2003 |
| 2 | PTEN | 3–5 | Loss | 10q23.31 | Not Matd | 8–26 | 14 | −0.75 | – |
| 3 | EP300 | 24–27 | Loss | 22q13.2 | De novo | 2–5 | 13 | −0.83 | Tsai et al., in presse |
| 4 | CDKL5 | 1–3 | Lossf | Xp22.13 | De novo | 105–128 | 28 | −0.43 | Erez et al., 2009,e |
| 5 | CREBBP | 27–28 | Loss | 16p13.3 | De novo | 0.5–2 | 10 | −0.43 | Tsai et al., in presse |
| 6 | ZDHHC9 | 10–11 | Loss | Xq25 | Mat | 6–31 | 10 | −1.27 | – |
| 7 | FAM58A | 5 | Loss | Xq28 | De novo | 1–16 | 5 | −0.64 | Unger et al., 2008 |
| 8 | HPRT1 | 9 | Loss | Xq26.2 | Mat | 502 bp | 3 | −2.48 | Yang et al., 1984; Gibbs et al., 1990 |
| 9 | SCN2A/SCN3A | 1–3/1 | Loss | 2q24.3 | De novo | 94–121 | 26 | −0.75 | Bartnik et al., in presse |
| 10 | STXBP1 | 1–4 | Loss | 9q34.11 | De novo | 47–86 | 16 | −0.73 | e |
| 11 | NRXN1 | 17–18 | Loss | 2p16.3 | Pat | 17,121 bp | 7 | −1 | e |
| 12 | TCF4 | 3–8 | Lossg | 18q21.2 | Unknown | 185–205 | 35 | −0.54 | e |
| 13 | IL1RAPL1 | 7–9 | Lossh | Xp21.2 | Unknown | 35–58 | 13 | −0.98 | – |
| 14 | EYA1 | 12 | Loss | 8q13.3 | Mati | 0.4–27 | 4 | −1.03 | Sanchez-Valle et al., in presse |
| 15 | JAG1 | 6–8 | Loss | 20p12.2 | De novo | 1–6 | 9 | −0.62 | – |
| 16 | RFC2 | 1–5 | Loss | 7q11.23 | Pat | 23–46 | 17 | −0.79 | – |
| 17 | SLC1A1 | 1 | Loss | 9p24.2 | Pat | 73–129 | 9 | −0.76 | – |
| 18 | CNTNAP2 | 8 | Loss | 7q35 | Unknown | 9,958 bp | 4 | −0.79 | – |
| 19 | ALDOA | 1–4j | Loss | 16p11.2 | Unknown | 13,885 bp | 18 | −0.63 | – |
| 20 | CTNNA3 | 10–11 | Loss | 10q21.3 | Pat | 101–367 | 6 | −1.01 | – |
| 21 | FHIT | 4 | Gain | 3p14.2 | Pat | 17,936 bp | 6 | 0.83 | – |
| 22 | TEK | 3–9 | Loss | 9p21.2 | Unknown | 29,964 bp | 27 | −0.53 | – |
| 23 | EXT2 | 12 | Loss | 11p11.2 | Unknown | 0.8–22 | 5 | −0.82 | – |
| 24 | TTC8 | 3–6 | Loss | 14q31.3 | Unknown | 4–13 | 9 | −0.87 | – |
| 25 | REEP3 | 7–8 | Loss | 10q21.3 | Pat | 3–18 | 13 | −0.9 | – |
| 26 | MSH6 | 4–10 | Loss | 2p16.3 | Unknown | 4–6 | 15 | −0.8 | – |
| 27 | FOXP1 | 14–21 | Loss | 3p14.1 | Unknown | 29–30 | 27 | −0.49 | – |
| 28 | KIF7 | 1–2 | Loss | 15q26.1 | Pat | 3–32 | 4 | −0.96 | – |
| 29 | KIF1B | 37–41 | Loss | 1p36.22 | Pat | 5–12 | 16 | −0.64 | – |
| 30 | DLG3 | Intron 18 | Loss | Xq13.1 | Mat | 1,099 bp | 3 | −0.79 | – |
| 31 | NRXN1 | Intron 3 | Loss | 2p16.3 | Not Mat | 30,792 bp | 3 | −0.83 | e |
Exon and intron numbers are based on the longest RefSeq transcript listed in the UCSC Genome Browser (http://genome.ucsc.edu), with the exception of CDKL5, for which exon numbering is as in Erez et al. [2009]. Exons listed are those encompassed by the maximum range predicted by aCGH, unless MLPA or DNA sequencing has provided a more accurate count. Partial overlap of exons is not specifically noted.
Size ranges represent the minimum and maximum deletions predicted by aCGH. DNA sequencing of some of the cases allows the size of the copy change to be known to the base pair.
References indicate previous reports of identical exon losses. A hyphen in this column indicates that our report is the first of this particular exonic deletion.
Father declined testing. See text.
Detailed molecular features reported in the listed reference or in a manuscript in preparation.
Mosaic. 24% deleted (12/50 cells) by FISH.
Mosaic. 16% deleted (32/200 cells) by FISH.
Mosaic. 56% deleted (112/200 cells) by FISH.
Mother has hearing loss, simple cup-shaped ears, and mild facial palsy.
The first four (noncoding) exons of transcript variant 1 (RefSeq: NM_000034.2) are deleted. Transcript variants 2–4 are not affected.
Pat = paternally inherited; Mat = maternally inherited.
Figure 1.
Intragenic CNVs from <103 bp to >105 bp in size have been detected throughout the genome. A: Sizes of intragenic losses and gains. Each position along the abscissa represents a unique case, labeled by the gene containing an intragenic CNV. Gray bars span the size range predicted by aCGH. Cases for which DNA sequencing has been performed are marked by a black line at the exact size of the rearrangement. B: A schematic karyogram demonstrating the genomic location of each described CNV.
Many Intragenic CNVs Are Concordant with Clinical Presentation
In 15 cases, a robust genotype–phenotype correlation could be established (Table 2); mutations in the gene disrupted by a CNV in these subjects are known to cause a disease that matches their clinical phenotypes. Of 12 cases for which parents were available for testing, the CNV was found to be de novo in eight, a maternally inherited, X-linked CNV in a male in two, and an autosomal CNV inherited from an affected parent in one. In the remaining case (case 11; NRXN1 deletion), a CNV was found to be inherited from a parent who did not share the patient’s clinical presentation, suggesting either reduced penetrance or undetected potential mosaicism in the tissue of clinical interest. Many of the CNVs found in these patients are novel, adding to the spectrum of mutations associated with their respective disease phenotypes.
Table 2.
Cases Exhibiting Genotype–Phenotype Concordance
| Case No. | Gene(s) | Patient age | Patient gender | Clinical indication | Molecular diagnosis |
|---|---|---|---|---|---|
| 1 | MECP2 | 14 y | F | Epilepsy, scoliosis, developmental regression, absent verbal skills, inability to walk | Rett syndrome (MIM# 312750) |
| 2 | PTEN | 8 y | F | Macrocephaly, enlarged thyroid, abnormal teeth, bifid uvula, nevi | Bannayan-Riley-Ruvalcaba syndrome (MIM# 153480) |
| 3 | EP300 | 2 y | F | Moderate DD, short stature, DF, microcephaly, behavioral problemsa | Rubinstein-Taybi syndrome (MIM# 180849) |
| 4 | CDKL5 | 15 mo | F | Severe DD, possible developmental regression, seizures | Infantile spasm syndrome, X-linked (MIM# 308350) |
| 5 | CREBBP | 1 d | M | DF, broad and angulated thumbs, persistent fetal pads, broad halluces, undescended testis, micropenis, microcephalya | Rubinstein-Taybi syndrome (MIM# 180849) |
| 6 | ZDHHC9 | 4 y | M | Mild DD/mental retardation | X-linked mental retardation, syndromic, ZDHHC9-related (MIM# 300799) |
| 7 | FAM58A | 1 d | F | Syndactyly, vertebral fusions, telecanthus, low set ears, imperforate anus, ambiguous genitalia, common cloaca, vesicoureteral reflux, ASD, VSD | STAR syndrome (MIM# 300707) |
| 8 | HPRT1 | 13 mo | M | Moderate DD, FTT | Lesch-Nyhan syndrome (MIM# 300322) |
| 9 | SCN2A/SCN3A | 24 y | F | MR, behavioral abnormalities, infantile seizuresa | Convulsions, benign familial infantile, 3 (MIM# 606052); seizures, afebrile (MIM# 604233); seizures, benign familial neonatal-infantile (MIM# 607745); [Chen et al., 2010] |
| 10 | STXBP1 | 12 y | F | Seizures, hypotonia, developmental delay and regression, ataxiaa,b | Epilepsy, MR, tremors, hypotonia [Hamdan et al., 2009]; Epileptic encephalopathy, early infantile, 4 (MIM# 612164) |
| 11 | NRXN1 | 5 y | F | Global DD, ADHD, seizuresa | Autism, susceptibility to, 5 (MIM# 606053; 209850) |
| 12 | TCF4 | 15 y | M | DF, ADHD, anxiety, self-mutilation, stiff jointsa | Pitt-Hopkins syndrome (MIM# 610954) |
| 13 | IL1RAPL1 | 6 y | M | DD, pervasive developmental disorder | X-linked mental retardation, 21/34 (MIM# 300143) |
| 14 | EYA1 | 4 y | M | Tetralogy of Fallot, hearing loss, facial nerve palsy, cleft palate, cup-shaped earsa | Branchiootic syndrome (MIM# 601653)/Branchiootorenal syndrome (MIM# 113650) |
| 15 | JAG1 | 8 mo | F | MCA, DF, peripheral pulmonic stenosis, multiple butterfly vertebrae | Alagille syndrome (MIM# 118450) |
Detailed clinical features reported in the reference listed in Table 1 for this case or in a manuscript in preparation.
A detailed clinical description is reported in [Moretti et al., 2005] (Subject 1) and [Moretti et al., 2008].
DD, developmental delay; DF, dysmorphic features; ASD, atrial septal defect; VSD, ventricular septal defect; FTT, failure to thrive; ADHD, attention deficit/hyperactivity disorder; MCA, multiple congenital anomalies.
Cases involving exonic losses in MECP2, PTEN, ZDHHC9, FAM58A, and HPRT1 are featured in Figures 2–6, respectively, and described in more detail below. These cases are representative of subjects in whom we detected an intragenic CNV causative of the patient’s disease phenotype.
Figure 2.
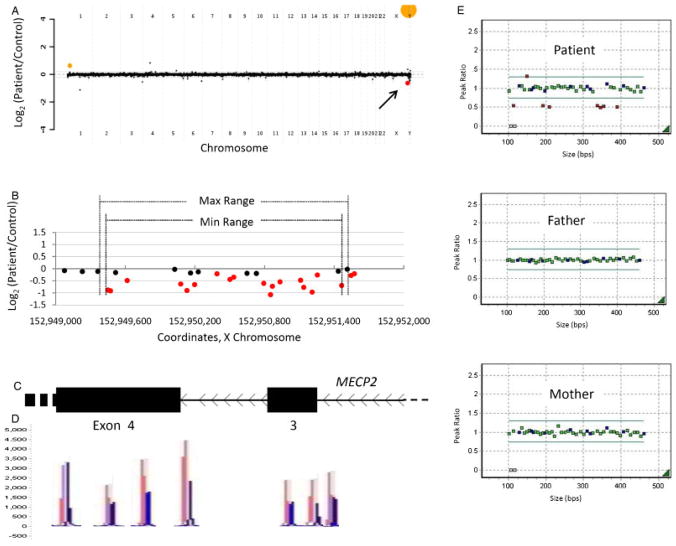
Patient 1 with an intragenic deletion in MECP2. A: Genome-wide view of the aCGH data. The red point (arrow) indicates the copy-number loss of interest (MECP2). Orange points indicate known benign copy-number variation (CNV) that serve as a positive control of hybridization. B–C: Local view of the MECP2 intragenic deletion. These graphics are aligned with one another. B: Plot of individual array probes, from which a “Min Range” and “Max Range”—defining the minimum and maximum expected boundaries of the deletion, respectively—can be established. C: Genomic map of this region of MECP2. D: MLPA trace confirming a deletion of exon 3 and indicating that a portion of exon 4 is also deleted. Control traces are in red, patient traces in blue. Each has been aligned to the genomic location it interrogates. E: Dot plots displaying normalized MLPA results for the patient and parents, demonstrating that this is a de novo copy-number loss. Green probes interrogate exons of MECP2, blue probes are controls, and red probes indicate copy change.
Figure 6.
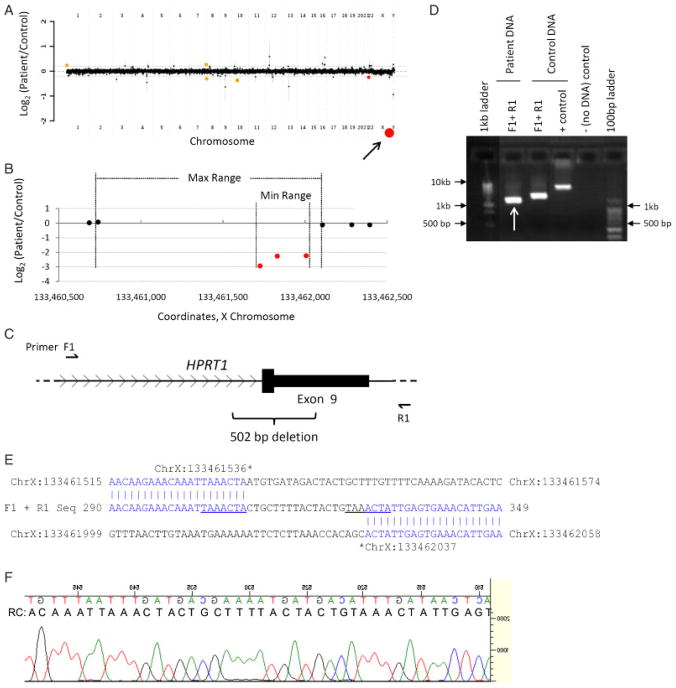
Patient 8 with an intragenic deletion in HPRT1. A: Genome-wide view of aCGH data. Red points indicate copy-number loss. Orange points indicate known benign copy-number variation (CNV). Arrow indicates the copy change of interest (HPRT1). The other loss contains no genes. B–C: Local view of the HPRT1 intragenic deletion. These graphics are aligned with one another. B: Plot of individual array probes, from which a “Min Range” and “Max Range”—defining the minimum and maximum expected boundaries of the deletion, respectively—can be established. C: Genomic map of this region of HPRT1 showing the locations of primers used to confirm the deletion and the exact boundaries of and size of the deletion as determined by DNA sequencing. D: Results of PCR confirming the deletion. Arrow indicates the PCR product that was sequenced. E: Sequencing confirms a partial deletion of exon 9 and identifies an 18 base pair insertion. Sequences of the “upstream” and “downstream” genomic regions, as well as the sequenced PCR product, are displayed. Regions of perfect homology are in blue. A seven base-pair sequence that appears twice at the breakpoint region is underlined. F: Sequencing trace of the breakpoint region. As sequencing was performed in reverse orientation to the reference sequence, a reverse complement (RC) sequence is displayed which matches that in E.
Intragenic Deletion in MECP2
This patient is a 14-year-old female with epilepsy, scoliosis, and absent verbal skills, who lacks the ability to walk. She was born at 40 weeks gestation and developed normally until 6 months of age, after which she lost the ability to sit independently and ceased babbling. An abnormal electroencephalogram was noted at 2 years of age, and frank seizures began at age 5. Scoliosis was noted, and corrective surgery was performed at age 10. DNA sequencing of methyl CpG binding protein 2 (MECP2) was performed at age 10, with no detectable mutations at that time. A karyotype was normal (46,XX), as were the results of multiple biochemical tests. A recent MRI was unremarkable. Currently, the patient is wheelchair bound, but can stand and move her legs with significant support. She is intolerant of heat, and is treated with supplementary vitamin D for osteopenia. Physical exam revealed a nondysmorphic girl with a height of 153.5 cm (~10th centile), weight of 40 kg (~10th centile), and head circumference 53 cm (~25th centile). Some residual scoliosis was noted. Fingers were tapered, and the left third finger was in a swan neck position. The first toes were long bilaterally, and a cutaneous 2–3 toe syndactyly was noted. Muscle bulk was appropriate, although tone was increased in both the upper and lower extremities.
Array CGH revealed a heterozygous genomic loss of about 1 kb, spanning exon 3 and part of exon 4 of MECP2 (Fig. 2A–C). The deletion was confirmed with MLPA (Fig. 2C–E). MLPA of parental samples demonstrated that this is a de novo loss. Both the molecular evidence and the patient’s clinical history and physical are consistent with Rett syndrome (MIM#312750). Heterozygous deletion of exon 3 and part of exon 4 has been previously described as an etiology for this X-linked dominant condition [Schollen et al., 2003].
Intragenic Deletion in PTEN
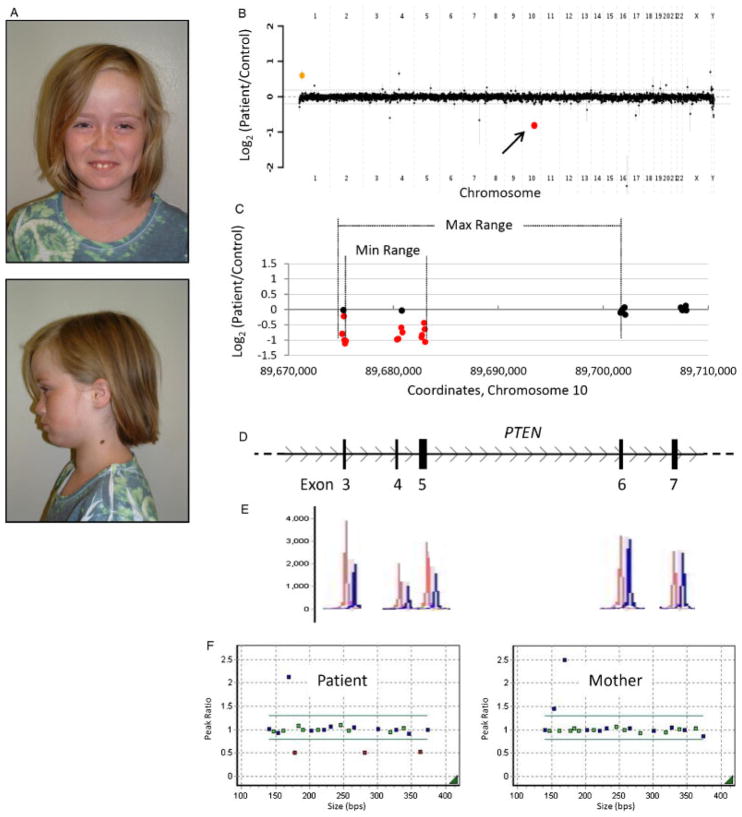
This patient is an 8-year-old female who presented with new-onset joint pain. She was born at 36 weeks gestation, following maternal preeclampsia beginning at 28 weeks. She required resuscitation at birth and phototherapy for neonatal jaundice. Amblyopia was noted at 3 years of age, now treated with corrective lenses. At 4 years of age, she began to lose deciduous teeth, which were described as having “no roots.” At age 8 years, she developed persistent knee and ankle pain, which improved somewhat with mechanical support and crutches. A previous skeletal survey found no abnormalities. She also has been diagnosed with anemia and eczema. The patient has a paternal half-sister, reportedly diagnosed with Proteus syndrome (MIM# 176920). Her paternal half-brother has macrocephaly and autism, by report. Physical exam revealed a well-developed girl with age-appropriate behavior. She had a high forehead and frontal bossing (Fig. 3A), with head circumference 59 cm (>98th centile). Height and weight were at the 95th and 90th centiles, respectively. Additionally, she had a bifid uvula, high arched palate, prominent tongue papillae, mild micrognathia, an enlarged, tender thyroid, and two cervical nevi (Fig. 2A). Examination of the extremities revealed a tender right knee and ankle, tapered fingers, and decreased range of motion at the dorsal interphalangeal joints. A thyroid ultrasound showed gland enlargement but no nodules. Further evaluations are underway.
Figure 3.
Patient 2 with an intragenic deletion in PTEN. A: Photos of patient 2, a 9-year-old female with features of Bannayan-Riley-Ruvalcaba syndrome. B: Genome-wide view of aCGH data. The red point (arrow) indicates the copy-number loss of interest (PTEN). The orange point indicates known benign copy-number variation (CNV). C–D: Local view of the PTEN intragenic deletion. These graphics are aligned with one another. C: Plot of individual array probes, from which a “Min Range” and “Max Range”—defining the minimum and maximum expected boundaries of the deletion, respectively—can be established. D: Genomic map of this region of PTEN. E: MLPA trace confirming a deletion of exons 3–5. Control traces are in red, patient traces in blue. Each has been aligned to the exon it interrogates. F: Dot plots displaying normalized MLPA results for the patient and mother, demonstrating that this copy-number loss was not maternally inherited. The father declined to be tested. Green probes interrogate exons of PTEN, blue probes are controls, and red probes indicate copy change.
Array CGH revealed a heterozygous genomic loss of 8–26 kb, spanning exons 3–5 of phosphatase and tensin homolog (PTEN) (Fig. 3B–D). Deletion of these exons was confirmed with MLPA (Fig. 3D–F). No deletion was present in the patient’s mother, who is also macrocephalic (head circumference 59 cm, >98th centile). The patient’s father declined to be tested. The clinical features of this patient are consistent with Bannayan-Riley-Ruvalcaba syndrome (BRRS; MIM# 153480), owing to the early clinical presentation and family history, however Cowden syndrome (MIM# 158350) remains a possible diagnosis. Both BRRS and Cowden disease are dominant genetic conditions resulting from mutations in PTEN [Liaw et al., 1997; Marsh et al., 1997]. Deletion of exons 3–5 has not been described previously in patients with BRRS. Complete deletion of exons 3–5 is predicted to remove 328 nucleotides from PTEN mRNA, resulting in a frame shift and premature termination codon. It is of note that the patient’s paternal half-sister has been reportedly diagnosed with Proteus syndrome (MIM# 176920) and that the paternal half-brother is reported to be affected by macrocephaly and autism. It is possible that, despite differing clinical signs, they share the mutant PTEN allele. Point mutations in PTEN have been described in patients with Proteus syndrome and/or Proteus-like syndrome [Smith et al., 2002; Zhou et al., 2001]. Some authors, however, failed to find intraexonic point mutations in Proteus syndrome patients [Barker et al. 2001; Biesecker et al., 2001; Thiffault et al., 2004], although they did not test the possibility of other types of mutations, including CNV.
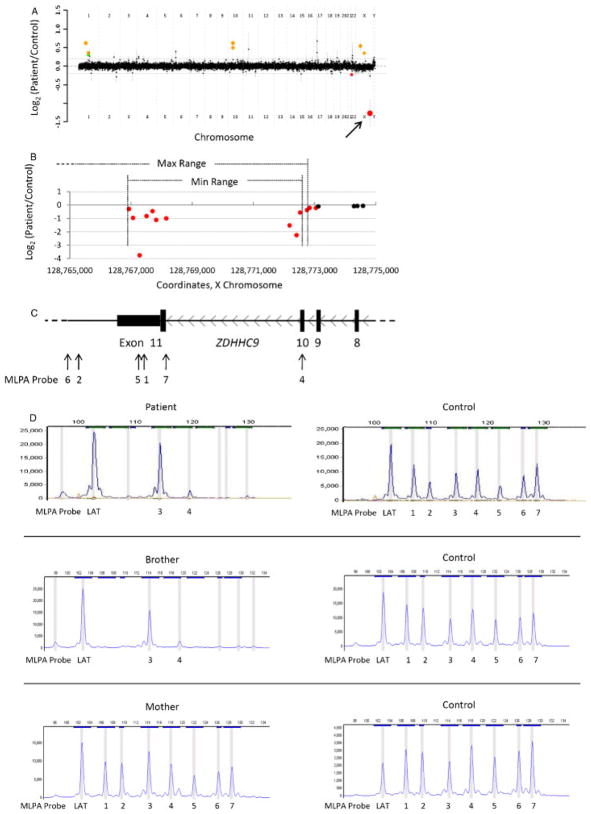
Intragenic deletion in ZDHHC9
This patient is a 4-year-old male, who presented for evaluation secondary to developmental delay. He had significant behavioral problems, including aggressiveness to others and himself and head banging. He also had significant speech delay. A developmental evaluation, however, revealed no autistic features. He had sleep difficulties with disturbances initiating sleep and frequent awakening. On physical exam, the boy had normal growth parameters and was free of dysmorphic features. He was noted to have esotropia. A brain MRI performed at 4 years of age showed a paucity of white matter, as well as patchy white matter hyperintensities on T2 weighted images.
Array CGH revealed a complete genomic loss of 6–31 kb, spanning exons 10–11 of the zinc finger, DHHC-type containing 9 (ZDHHC9) gene (Fig. 4A–C), which encodes a palmitoyltransferase [Swarthout et al., 2005]. Deletion of these exons in the patient was confirmed using MLPA (Fig. 4C–D), which did not exclude a deletion of other ZDHHC9 exons. The array, molecular, and clinical findings for this patient are consistent with ZDHHC9-related X-linked syndromic mental retardation (MIM# 300799). This syndrome was first reported by Raymond et al. [2007], associated with hemizygous point mutations (one frameshift, two missense, and one splice site) in ZDHHC9. Four families with X-linked mental retardation were described, three of which had the additional clinical feature of Marfanoid habitus. Behavioral problems and schizophrenia were also described in one patient. Tarpey et al. [2009] found point mutations in ZDHHC9 in two families (one frameshift and one splice site mutation) and two other individuals (two missense mutations) with X-linked mental retardation, some of which had Marfanoid habitus.
Figure 4.
Patient 6 with an intragenic deletion in ZDHHC9. A: Genome-wide view of aCGH data. Red points indicate copy-number loss. The arrow indicates the copy change of interest (ZDHHC9), while the other indicated loss contains no genes. The green point indicates a copy-number gain that contains no genes. Orange points indicate known benign copy-number variation (CNV). B–C: Local view of the ZDHHC9 intragenic deletion. These graphics are aligned with one another. B: Plot of individual array probes, from which a “Min Range” and “Max Range”—defining the minimum and maximum expected boundaries of the deletion, respectively—can be established. C: Local genomic map of ZDHHC9 showing the locations of MLPA probes used to confirm the deletion. D: MLPA traces confirming a deletion of exons 10 and 11 in the patient, his brother, and their mother. Patient traces are on the left; control traces for each are on the right. “LAT” and “3” are control probes; see Supp. Methods for sequences and genomic coordinates of these probes.
To date, no genomic deletion has been reported in ZDHHC9. MLPA identified the same genomic deletion in the patient’s brother, who has a milder developmental delay, and in their mother, who is a carrier for this X-linked recessive disorder (Fig. 4C–D). Although the patient we describe lacks a Marfanoid habitus, at his young age (younger than any patient described by Raymond et al. [2007]), this feature may not have yet manifested.
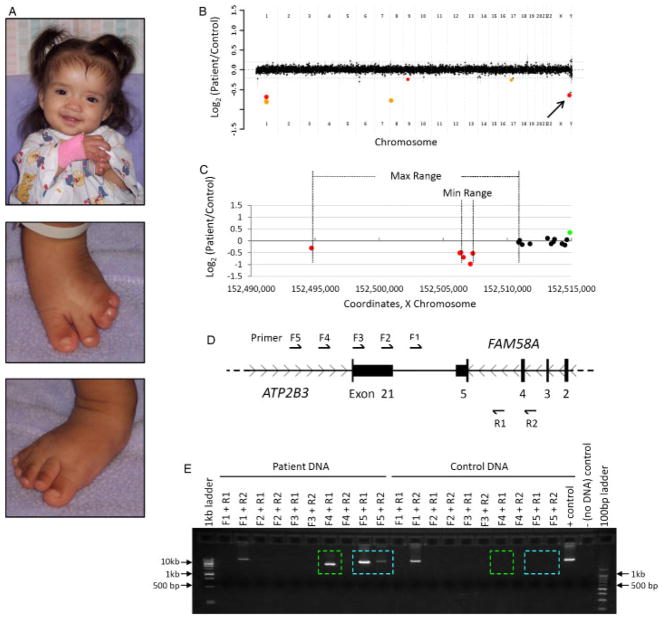
Intragenic Deletion in FAM58A
This newborn female was evaluated secondary to multiple congenital anomalies. She was born to a 25-year-old female by spontaneous vaginal delivery. Ventriculomegaly and the presence of an abdominal mass were noted in utero. At the time of birth she was noted to have dysmorphic features (Fig. 5A), including telecanthus, a wide nasal bridge, abnormally shaped and low set ears, and a right ear pit. Cardiac exam revealed a 2/6 systolic murmur heard at the left sternal border. She had an imperforate anus and developed abdominal distension after the first feed, for which a colostomy was subsequently placed. Additionally, she had an enlarged clitoris that raised concerns for ambiguous genitalia, and limb abnormalities. Syndactyly was present in both hands and feet and clinodactyly of the left fifth digit was also noted. A skeletal survey showed multiple abnormalities including fusion of vertebral spine from C3 to C4 and S3 through S5, 11 pair of ribs, an absent middle phalanx of the left fifth finger, absent ossification on the middle phalanges of the feet, and soft tissue fusion extending from the third to fifth toes bilaterally. An echocardiogram showed both atrial and ventricular septal defects. A voiding uretero-cystogram demonstrated grade V vesicoureteral reflux on the right and grade II reflux on the left, for which she was placed on antibiotic treatment prophylaxis for urinary tract infections.
Figure 5.
Patient 7 with an intragenic deletion in FAM58A. A: Photos of patient 7, a female with features of STAR syndrome, at nine months of age. B: Genome-wide view of aCGH data. Red points indicate copy-number losses. The point indicated by an arrow is the copy change of interest. The other two losses contain no genes. Orange points indicate known benign copy-number variation (CNV). C–D: Local view of the FAM58A deletion. These graphics are aligned with one another. C: Plot of individual array probes, from which a “Min Range” and “Max Range”—defining the minimum and maximum expected boundaries of the deletion, respectively—can be established. The maximum range also encompasses the final exon of ATP2B3. D: Local genomic map of this region of the X chromosome showing the locations of primers used to confirm the deletion. E: Results of PCR confirming the deletion. Colored boxes indicate PCR products amplified from patient DNA but not from control DNA. In addition to exon 5 of FAM58A, part or all of exon 21 of ATP2B3 is also deleted.
Array CGH revealed a heterozygous genomic loss of 1–16 kb, spanning exon 5 of the family with sequence similarity 58, member A (FAM58A) gene (Fig. 5B–D). The copy-number loss was confirmed by PCR (Fig. 5D–E), which enabled its size to be estimated at about 8–10 kb. The final exon of ATPase, Ca(2+)-transporting, plasma membrane, 3 (ATP2B3), which is not a known or suspected disease gene, is also involved in the deletion. Array CGH of both parents did not detect the deletion, indicating that this is a de novo mutation. The clinical and molecular features of this patient are consistent with STAR syndrome (toe Syndactyly, Telecanthus, and Anogenital and Renal malformations; MIM# 300707). Deletion of exon 5 of FAM58A has been described previously as an etiology for this X-linked dominant condition [Unger et al., 2008]. All six of the patients studied by these authors, including two patients originally described by Green et al. [1996], were female. Our patient, the seventh described with this genetic syndrome, is also female, suggesting further that similar mutations in FAM58A in males may be lethal. Unger et al. [2008] described complete or near-complete skewing of X-chromosome inactivation (XCI) in all patients studied. This finding, coupled with in vitro experiments, suggests that inactivating mutations of FAM58A may result in a cell-autonomous proliferation defect during fetal development. X-inactivation studies performed on our patient also revealed a complete skewing of XCI (data not shown).
Intragenic Deletion in HPRT1
This patient is a 13-month-old male with moderate developmental delay and failure to thrive. No additional clinical details could be obtained. Array CGH revealed a genomic loss of 0.3–1 kb, spanning part of exon 9 of hypoxanthine phosphoribosyltransferase 1 (HPRT1) (Fig. 6A–C). The deletion was identified also with PCR (Fig. 6C–D), and biochemical analysis confirmed an absence of detectable HPRT activity (0 nmol/min/g Hb; normal range = 400–2,200 nmol/min/g Hb). DNA sequencing of the breakpoint region demonstrated a 502 bp deletion with an 18-bp insertion (Fig. 6E–F). This 18-bp sequence is not found in the reference human genome. Interestingly, 7 bp which roughly flank each side of the deletion breakpoint are homologous to one another (Fig. 6E). These features suggest NHEJ and/or replication slippage may be responsible for the formation of this genomic rearrangement.
The genomic deletion was found by aCGH to be maternally inherited (data not shown), indicating that the patient’s mother is a carrier for this X-linked recessive disorder. The array, molecular, and biochemical findings for this patient are consistent with Lesch-Nyhan syndrome (MIM# 300322). Deletion of exon 9, the final exon of HPRT1, has been reported previously in the context of this disorder [Gibbs et al., 1990; Yang et al., 1984].
Cases of Uncertain Clinical Significance
Table 3 lists cases for which a genotype–phenotype correlation is less certain. This uncertainty may be on account of limited clinical information (e.g., in case 19 it is not known whether the patient has specific features of aldolase A deficiency [MIM# 103850]/glycogen storage disease XII [MIM# 611881]), a paucity of published data linking the gene to dominant disease (e.g., case 18), or age-dependent penetrance of the associated condition (e.g., case 23; this patient is likely too young for exostoses to be present). Additionally, two patients with intronic deletions are described in Table 3 (cases 30 and 31). Proposing a genotype–phenotype correlation in these cases is somewhat speculative, although intronic rearrangements—including those near splice sites [Higgins et al., 2001; Zhuang et al., 1993], those affecting splicing by constraining intron size [L. Wang et al., 2002], and deep intronic deletions and duplications [Bovolenta et al., 2008, 2010]—have been associated with disease phenotypes. Although mutations in the genes listed in Table 3 have been described, none of the specific intragenic CNVs listed therein has been reported previously. Thus, they may represent novel mutations in known genetic disorders, or they may define novel genetic syndromes. The latter possibility is particularly intriguing and suggests that our approach may enable elucidation of gene function for some of large number of genes in the human genome that do not have a confirmed role in human phenotypic variability or disease (ftp://ftp.ncbi.nih.gov/repository/OMIM/genemap).
Table 3.
Cases Involving Intragenic CNVs of Uncertain Clinical Significance
| Case No. | Gene(s) | Patient age | Patient gender | Clinical indication | Suspected/related diagnosis |
|---|---|---|---|---|---|
| 16 | RFC2 | 8 y | M | Static encephalopathy | – |
| 17 | SLC1A1 | 18 mo | M | Moderate DD | – |
| 18 | CNTNAP2 | 22 mo | M | DD, cyanotic episodes | – |
| 19 | ALDOA | 11 mo | Ma | FTT | Aldolase A deficiency (MIM# 103850), Glycogen storage disease XII (MIM# 611881) |
| 20 | CTNNA3 | 13 mo | F | Congenital heart disease | – |
| 21 | FHIT | 3 mo | M | Macrocephaly, microretrognathia, minor dysmorphism, hypertonia, bilateral neonatal strokes | – |
| 22 | TEK | 9 y | M | Fetal alcohol syndrome | – |
| 23 | EXT2 | 11 mo | F | Mild DD, nystagmus, optic nerve hypoplasia | – |
| 24 | TTC8 | 3 y | M | Mild DD, mental retardation | – |
| 25 | REEP3 | 9 mo | F | Reactive airway disease exacerbation | – |
| 26 | MSH6 | 7 y | M | Mild DD | – |
| 27 | FOXP1 | 7 y | F | Severe DD | – |
| 28 | KIF7 | 2 y | F | MCA, global DD | – |
| 29 | KIF1B | 2 y | M | Short stature | – |
| 30 | DLG3 | 3 y | M | Moderate DD, DF, FTT | Mental retardation, X-linked-90 (MIM# 300189) |
| 31 | NRXN1 | 6 y | F | Global DD, microcephaly, autismb | Autism, susceptibility to, 5 (MIM#s 606053, 209850) |
Patient has a 47,XXY karyotype.
Manuscript in preparation. Detailed clinical features of this case will be reported therein.
DD, developmental delay; FTT, Failure to thrive; MCA, multiple congenital anomalies; DF, dysmorphic features.
Breakpoint Sequencing
Breakpoint regions were sequenced for seven deletion CNVs (cases 8, 11, 18, 19, 22, 30, and 31; Supp. Table S2), revealing microhomology of 2–4 bp in four cases, extended microhomology (62 bp) with breakpoints in Alu elements in one case, and an insertion of 7 and 18 bases in one case each. Each instance of copy number loss was a “simple” (i.e., not complex) deletion. Breakpoints were also mapped and sequenced in the case of a copy-number gain (case 21; Supp. Table S2), which revealed no microhomology. This is a tandem gain of at least one additional copy, and perhaps more (PCR and DNA sequencing do not distinguish between duplications, triplications, etc.). Breakpoint coordinates are listed in Supp. Table S1. Seven of 16 breakpoints in the aforementioned cases localize to repetitive sequences (Supp. Table S2).
Discussion
Exon-Targeted aCGH Detects Clinically Relevant Intragenic CNV
The development of aCGH with exon coverage has enabled the detection of intragenic deletions and duplications throughout the entire human genome. We describe multiple cases involving the deletion or duplication of one or more exons, 15 of which exhibit an obvious concordance with the patient’s phenotype. These rearrangements are consistent with autosomal dominant (cases 2, 3, 5, 9–12, 14, and 15), X-linked recessive (cases 6, 8, and 13), and X-linked dominant (cases 1, 4, 7) disorders and predisposition to disease. Further, a variety of disease processes are represented, including neurodevelopmental disorders (cases 1, 3–6, 9–13), an enzyme deficiency syndrome (case 8), and other recognizable patterns of human malformation (case 2, Bannayan-Riley-Ruvalcaba syndrome; case 7, STAR syndrome; case 14, branchiootic syndrome; and case 15, Alagille syndrome). A subset of these phenotypically concordant genomic alterations has not been reported previously, including a loss of exons 3–5 of PTEN associated with Bannayan-Riley-Ruvalcaba syndrome; losses of exons 10–11 of ZDHHC9, and of exons 7–9 of IL1RAPL1, each independently associated with mental retardation in males; loss of exons 1–4 of STXBP1 associated with childhood epilepsy and other features; and a loss of exons 6–8 of JAG1 associated with Alagille syndrome (MIM numbers associated with these conditions are listed in Table 1). Thus, our approach allows new mutations to be described for known genetic conditions.
Three of the clinically correlated CNVs (cases 4, 12, and 13) were mosaic, demonstrating the ability of our methodology to detect mosaic copy-number changes with exonic resolution. The limited availability of clinical information precludes objective assessment of clinical severity in comparison to patients with nonmosaic CNVs.
For 16 cases, no firm genotype–phenotype relationship exists. Such ambiguity can result from either an incomplete clinical history or an absence of published literature describing a clear phenotypic consequence of mutations in the gene of interest. Overwhelmingly, the genomic aberrations in these cases are previously undescribed, and as such may define novel genetic syndromes. Most of the copy-number changes we describe are heterozygous. Thus, correlation with a known disease state is only possible when a dominant condition has been described in the literature. However, it is possible that alleles that were previously described as causing recessive disease act as dominant alleles in a milder or alternative disease state, for example, splenic syndrome at altitude or sudden death in sickle cell trait [Kark et al., 1987; Lane and Githens, 1985] and intermediate defects in cholesterol regulation in individuals heterozygous for mutations in low-density lipoprotein receptor (LDLR) [Brown and Goldstein, 1974]. Single-copy deletions or duplications may also cause disease as compound heterozygotes, with a CNV on one allele “unmasking” a single nucleotide variant (SNV), for example, on the other allele [Borg et al., 2009; Kurotaki et al., 2005; Tayeh et al., 2009]. In this case, sequencing of the other allele is necessary to find the full genetic cause of recessive disease. Such a mechanism is suspected in cases of inherited copy-number changes where disease is not seen in the parent transmitting the CNV-containing allele. Further, more complex inheritance schemes, for example, a two-hit model [Girirajan et al., 2010; Lupski 2007] or the co-occurrence of two or more conditions in a single patient, each attributable to an independent genomic rearrangement [Potocki et al., 1999], are also possible.
Molecular Mechanisms and Consequences of Intragenic CNV
Although exon deletions and duplications can disrupt a gene, causing loss of function, they may also constitute gain of function [Bochukova et al., 2009] or dominant negative [Inoue et al., 2004] mutations with unexpected phenotypic consequences. In addition, copy-number changes in other types of conserved sequences, for example, introns, promoters, and enhancers, can have pathogenic consequences [Bovolenta et al., 2008, 2010; Higgins et al., 2001; Lee et al., 2006; Smyk et al., 2007; L. Wang et al., 2002; Weterman et al., 2010; Zhang et al., 2010b; Zhuang et al. 1993]. We have sequenced the breakpoint regions of eight intragenic CNVs (Supp. Table S2). Each was a “simple” copy-number loss or gain, as no evidence of complex genomic rearrangement [Zhang et al., 2009a] was found. The molecular consequences of each CNV, predicted by conceptual translation, are listed in Supp. Table S2. These CNVs are anticipated to have variable, and in several cases uncertain, effects on gene expression. In four of seven sequenced intragenic deletions, microhomology ranging from two to four base pairs exists between the “upstream” and “downstream” deletion breakpoints (Supp. Table S2). This is characteristic of either NHEJ or the replication-based FoSTeS/MMBIR mechanisms [Gu et al., 2008], which have been implicated in genomic, genic, and exonic copy-number changes [Zhang et al., 2009b]. The substantial representation of breakpoint microhomology among this small group of samples and other exonic deletions [Zhang et al., 2010a] hints at the importance of replication-based mechanisms in causing clinically relevant exonic deletion syndromes throughout the genome. Microhomology at deletion breakpoints is also a common feature of benign CNV, found in 219/315 (70%) of breakpoints described by Conrad et al. [2010a]. The finding of mosaicism in three samples, for which a sequence was not obtained, is consistent with a mitotic, postzygotic mechanism of CNV generation (e.g., the replication-based MMBIR/FoSTeS). In one case (case 19), 62 bp of perfect homology is seen at deletion breakpoints that localize to two Alu elements in the same genomic orientation (Supp. Table S2). As these elements belong to differing Alu subfamilies (AluSq and AluSx) and share only 80% identity over 245 bp, NAHR (Alu– Alu recombination) [Lehrman et al., 1987] is improbable. Rather, Alu-specific microhomology-mediated deletion is likely to have generated this and other recently described CNV with breakpoints in Alu elements [Erez et al., 2009; Stankiewicz et al., 2009; Vissers et al., 2009; Zhang et al., 2009a]. The remaining two deletions contain insertions at their breakpoints of 18 (case 8) and 7 (case 30) bp in length. The 18-bp inserted sequence is not found in the human genome, whereas the 7-bp insertion is found throughout the genome, including within the 1,099-bp region deleted in patient 30. Interestingly, in both of these cases involving insertions, there exist 7-bp sequences that are repeated at each side of the deletion breakpoint. The above features of these two cases are consistent with NHEJ, with a possible contribution of replication slippage. In the final case, a tandem intragenic copy-number gain (case 21) displayed no microhomology (Supp. Table S2), suggestive of NHEJ.
In the case that an intragenic deletion spans the final exon of a gene, the possibility exists that a fusion transcript is made that incorporates part of a downstream gene. If the stop codon of the upstream gene is involved and the reading frame is preserved upon splicing to or via direct fusion with an exon of the downstream gene (exon accretion), a fusion protein may be made, although this is likely only if the two genes share the same genomic orientation [Walsh et al., 2008]. In six of the reported deletion cases (cases 6–8 and 25–27), the final exon of the gene of interest is included in the region of copy-number loss, although in each case the most proximal downstream gene is in an opposing orientation (Supp. Table S1). Thus, fusion proteins, which can impart novel functions, may be less likely in these cases. However, a theoretical new gene could be created using the complementary exon strands as was shown for the evolution of human HREP [Inoue et al., 2001; Inoue and Lupski, 2002]. Such novel genes may encode new proteins with either neomorphic or antimorphic activities. Furthermore, in one case (case 28) it is possible that PEX11A or PLIN1, genes that map upstream of the gene of interest (KIF7) in the same orientation and the 3′ ends of which may be deleted, splice to KIF7. DNA sequencing of the deletion breakpoint and further molecular experiments would be necessary to determine if in fact a fusion transcript is produced.
Exon-Targeted aCGH Detects CNVs That Would Be Missed by Nonexon-Targeted Arrays
We hypothesized that by concentrating array probes in exons of known or putative disease genes, we would be able to detect small genomic rearrangements that would escape detection with standard genomic arrays of similar probe number. Our exon-targeted array contains more probes in each CNV of known clinical significance (cases 1–15) than standard 105,000- (105 k), 180,000- (180 k), or 244,000-probe (244 k) arrays (Agilent Technologies) (Table 4). In eight of 15 cases, the 180 k standard array, which contains approximately the same number of total probes as our V8 OLIGO array, includes either one or no probes in the CNV of interest. Thus, the detected “exonic CNVs” would be missed using standard aCGH analysis performed with the nonexon-targeted 180 k array.
Table 4.
The V8 OLIGO Exon-Targeted Array Contains More Probes in Each Clinically Significant CNV Than Standard 105,000-(105 k), 180,000- (180 k), or 244,000-Probe (244 k) Arrays
| Case No. | Gene(s) | Number of probes
|
|||
|---|---|---|---|---|---|
| 105 k | 180 k | 244 k | V8 OLIGO | ||
| 1 | MECP2 | 0 | 0 | 0 | 25 |
| 2 | PTEN | 1 | 3 | 3 | 14 |
| 3 | EP300 | 0 | 0 | 0 | 13 |
| 4 | CDKL5 | 5 | 11 | 13 | 28 |
| 5 | CREBBP | 0 | 0 | 0 | 10 |
| 6 | ZDHHC9 | 3 | 3 | 6 | 10 |
| 7 | FAM58A | 0 | 0 | 3 | 5 |
| 8 | HPRT1 | 1 | 1 | 1 | 3 |
| 9 | SCN2A/3A | 5 | 9 | 11 | 26 |
| 10 | STXBP1 | 5 | 7 | 9 | 16 |
| 11 | NRXN1 | 1 | 1 | 3 | 7 |
| 12 | TCF4 | 8 | 16 | 24 | 35 |
| 13 | IL1RAPL1 | 2 | 4 | 7 | 13 |
| 14 | EYA1 | 1 | 1 | 3 | 4 |
| 15 | JAG1 | 0 | 0 | 0 | 9 |
105 k, Agilent Human Genome CGH Microarray 105A; 180 k, Agilent SurePrint G3 Human CGH Microarray 180 K; 244 k, Agilent Human Genome CGH Microarray 244A.
Conclusions
One strategy for improving the resolution of traditional aCGH is to increase the number of array probes. However, this is not without increased cost and results in detection of a greater number of clinically irrelevant copy-number changes. Indeed, high-resolution whole-genome arrays have demonstrated an immense CNV load among normal individuals [Conrad et al., 2010b]. Uncertainty as to the pathogenicity of newly discovered copy-number variants may linger and will likely be of even greater importance as new methods—for example, higher density arrays, whole-exome arrays, “conservome” arrays (those with increased coverage of conserved noncoding regions of the genome), and next generation sequencing—are increasingly used to link copy-number variation to disease. By supplementing our whole genome array with dense coverage of the exons of known and suspected disease genes, we have focused on what we hypothesized to be the most clinically relevant and interpretable genomic copy-number changes. Our approach improves the resolution of aCGH to the level of the exon while excluding much of the noise inherent in other strategies.
The ability to detect single-exon copy-number changes by aCGH provides new opportunities for genetic research and diagnosis. Nevertheless, interpretation of such rearrangements may still present a challenge, as the functional impact of these genomic alterations is not always well understood. Determining their significance, especially in the case of previously unreported variations, involves investigative teamwork between the laboratory and the clinician. Despite these challenges, this method provides a screening method to detect, with subkilobase resolution, genomic rearrangements of clinical import and research significance. Furthermore, such an approach may enable the elucidation of the function of some of the majority of the predicted genes in the human genome for which a function remains enigmatic.
Supplementary Material
Acknowledgments
We are indebted to the patients and families who participated in this study. We thank Jonathan Berg and Nicola Brunetti for assistance in array design. F.J.P. holds a Career Award for Medical Scientists from the Burroughs Wellcome Fund. The Department of Molecular and Human Genetics at Baylor College of Medicine derives revenue from the chromosomal microarray analysis offered in the Medical Genetics Laboratory.
Grant sponsor: The Baylor College of Medicine Medical Scientist Training Program; Grant number: T32GM007330-34 (to P.M.B.); Grant sponsor: The National Institute of Neurological Disorders and Stroke (NINDS, NIH); Grant number: R01NS058529 (to J.R.L.); Grant sponsor: The Polish Ministry of Science and Higher Education; Grant number: R13-0005-04/2008 (to P.S.).
Footnotes
Communicated by Michael Dean
Additional Supporting Information may be found in the online version of this article.
References
- Allen RC, Zoghbi HY, Moseley AB, Rosenblatt HM, Belmont JW. Methylation of HpaII and HhaI sites near the polymorphic CAG repeat in the human androgen–receptor gene correlates with X chromosome inactivation. Am J Hum Genet. 1992;51:1229–1239. [PMC free article] [PubMed] [Google Scholar]
- Bailey JA, Kidd JM, Eichler EE. Human copy number polymorphic genes. Cytogenet Genome Res. 2008;123:234–243. doi: 10.1159/000184713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barker K, Martinez A, Wang R, Bevan S, Murday V, Shipley J, Houlston R, Harper J. PTEN mutations are uncommon in Proteus syndrome. J Med Genet. 2001;38:480–481. doi: 10.1136/jmg.38.7.480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartnik M, Tsai AC-H, Xia Z, Cheung SW, Stankiewicz P. Disruption of the SCN2A and SCN3A genes in a patient with mental retardation, neurobehavioral and psychiatric abnormalities, and a history of infantile seizures. Clin Genet. doi: 10.1111/j.1399-0004.2010.01526.x. (in press) [DOI] [PubMed] [Google Scholar]
- Ben-Shachar S, Khajavi M, Withers MA, Shaw CA, van Bokhoven H, Brunner HG, Lupski JR. Dominant versus recessive traits conveyed by allelic mutations—to what extent is nonsense-mediated decay involved? Clin Genet. 2009;75:394–400. doi: 10.1111/j.1399-0004.2008.01114.x. [DOI] [PubMed] [Google Scholar]
- Biesecker LG, Rosenberg MJ, Vacha S, Turner JT, Cohen MM. PTEN mutations and proteus syndrome. Lancet. 2001;358:2079–2080. doi: 10.1016/S0140-6736(01)07109-4. [DOI] [PubMed] [Google Scholar]
- Bochukova EG, Roscioli T, Hedges DJ, Taylor IB, Johnson D, David DJ, Deininger PL, Wilkie AO. Rare mutations of FGFR2 causing Apert syndrome: identification of the first partial gene deletion, and an Alu element insertion from a new subfamily. Hum Mutat. 2009;30:204–211. doi: 10.1002/humu.20825. [DOI] [PubMed] [Google Scholar]
- Borg K, Stucka R, Locke M, Melin E, Åhlberg G, Klutzny U, von der Hagen M, Huebner A, Lochmüller H, Wrogemann K, Thornell L-E, Blake DJ, Schoser B. Intragenic deletion of TRIM32 in compound heterozygotes with sarcotubular myopathy/LGMD2H. Hum Mutat. 2009;30:E831–E844. doi: 10.1002/humu.21063. [DOI] [PubMed] [Google Scholar]
- Bovolenta M, Neri M, Fini S, Fabris M, Trabanelli C, Venturoli A, Martoni E, Bassi E, Spitali P, Brioschi S, Falzarano MS, Rimessi P, Ciccone R, Ashton E, McCauley J, Yau S, Abbs S, Muntoni F, Merlini L, Gualandi F, Ferlini A. A novel custom high density-comparative genomic hybridization array detects common rearrangements as well as deep intronic mutations in dystrophinopathies. BMC Genomics. 2008;9:572. doi: 10.1186/1471-2164-9-572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bovolenta M, Neri M, Martoni E, Urciuolo A, Sabatelli P, Fabris M, Grumati P, Mercuri E, Bertini E, Merlini L, Bonaldo P, Ferlini A, Gualandi F. Identification of a deep intronic mutation in the COL6A2 gene by a novel custom oligonucleotide CGH array designed to explore allelic and genetic heterogeneity in collagen VI-related myopathies. BMC Med Genet. 2010;11:44. doi: 10.1186/1471-2350-11-44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown MS, Goldstein JL. Expression of the familial hypercholesterolemia gene in heterozygotes: mechanism for a dominant disorder in man. Science. 1974;185:61–63. doi: 10.1126/science.185.4145.61. [DOI] [PubMed] [Google Scholar]
- Chen C-P, Lin S-P, Chern S-R, Chen Y-J, Tsai F-J, Wu P-C, Wang W. Array-CGH detection of a de novo 2.8 Mb deletion in 2q24.2→q24.3 in a girl with autistic features and developmental delay. Eur J Med Genet. 2010 doi: 10.1016/j.ejmg.2010.03.006. [DOI] [PubMed] [Google Scholar]
- Cheung SW, Shaw CA, Scott DA, Patel A, Sahoo T, Bacino CA, Pursley A, Li J, Erickson R, Gropman AL, Miller DT, Seashore MR, Summers AM, Stankiewicz P, Chinault AC, Lupski JR, Beaudet AL, Sutton VR. Microarray-based CGH detects chromosomal mosaicism not revealed by conventional cytogenetics. Am J Med Genet A. 2007;143A:1679–1686. doi: 10.1002/ajmg.a.31740. [DOI] [PubMed] [Google Scholar]
- Cheung SW, Shaw CA, Yu W, Li J, Ou Z, Patel A, Yatsenko SA, Cooper ML, Furman P, Stankiewicz P, Lupski JR, Chinault AC, Beaudet AL. Development and validation of a CGH microarray for clinical cytogenetic diagnosis. Genet Med. 2005;7:422–432. doi: 10.1097/01.gim.0000170992.63691.32. [DOI] [PubMed] [Google Scholar]
- Clark TA, Schweitzer AC, Chen TX, Staples MK, Lu G, Wang H, Williams A, Blume JE. Discovery of tissue-specific exons using comprehensive human exon microarrays. Genome Biol. 2007;8:R64. doi: 10.1186/gb-2007-8-4-r64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conrad DF, Bird C, Blackburne B, Lindsay S, Mamanova L, Lee C, Turner DJ, Hurles ME. Mutation spectrum revealed by breakpoint sequencing of human germline CNVs. Nat Genet. 2010a;42:385–391. doi: 10.1038/ng.564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conrad DF, Pinto D, Redon R, Feuk L, Gokcumen O, Zhang Y, Aerts J, Andrews TD, Barnes C, Campbell P, Fitzgerald T, Hu M, Ihm CH, Kristiansson K, MacArthur DG, MacDonald JR, Onyiah I, Pang AWC, Robson S, Stirrups K, Valsesia A, Walter K, Wei J, Tyler-Smith C, Carter NP, Lee C, Scherer SW, Hurles ME Wellcome Trust Case Control Consortium. Origins and functional impact of copy number variation in the human genome. Nature. 2010b;464:704–712. doi: 10.1038/nature08516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- del Gaudio D, Yang Y, Boggs BA, Schmitt ES, Lee JA, Sahoo T, Pham HT, Wiszniewska J, Chinault AC, Beaudet AL, Eng CM. Molecular diagnosis of Duchenne/Becker muscular dystrophy: enhanced detection of dystrophin gene rearrangements by oligonucleotide array-comparative genomic hybridization. Hum Mutat. 2008;29:1100–1107. doi: 10.1002/humu.20841. [DOI] [PubMed] [Google Scholar]
- Dhami P, Coffey AJ, Abbs S, Vermeesch JR, Dumanski JP, Woodward KJ, Andrews RM, Langford C, Vetrie D. Exon array CGH: detection of copy-number changes at the resolution of individual exons in the human genome. Am J Hum Genet. 2005;76:750–762. doi: 10.1086/429588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dietz HC, Valle D, Francomano CA, Kendzior RJ, Jr, Pyeritz RE, Cutting GR. The skipping of constitutive exons in vivo induced by nonsense mutations. Science. 1993;259:680–683. doi: 10.1126/science.8430317. [DOI] [PubMed] [Google Scholar]
- El-Hattab AW, Smolarek TA, Walker ME, Schorry EK, Immken LL, Patel G, Abbott M-A, Lanpher BC, Ou Z, Kang S-HL, Patel A, Scaglia F, Lupski JR, Cheung SW, Stankiewicz P. Redefined genomic architecture in 15q24 directed by patient deletion/duplication breakpoint mapping. Hum Genet. 2009;126:589–602. doi: 10.1007/s00439-009-0706-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Erez A, Patel AJ, Wang X, Xia Z, Bhatt SS, Craigen W, Cheung SW, Lewis RA, Fang P, Davenport SLH, Stankiewicz P, Lalani SR. Alu-specific microhomology-mediated deletions in CDKL5 in females with early-onset seizure disorder. Neurogenetics. 2009;10:363–369. doi: 10.1007/s10048-009-0195-z. [DOI] [PubMed] [Google Scholar]
- Gardina PJ, Clark TA, Shimada B, Staples MK, Yang Q, Veitch J, Schweitzer A, Awad T, Sugnet C, Dee S, Davies C, Williams A, Turpaz Y. Alternative splicing and differential gene expression in colon cancer detected by a whole genome exon array. BMC Genomics. 2006;7:325. doi: 10.1186/1471-2164-7-325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gibbs RA, Nguyen P-N, Edwards A, Civitello AB, Caskey CT. Multiplex DNA deletion detection and exon sequencing of the hypoxanthine phosphoribosyl-transferase gene in Lesch-Nyhan families. Genomics. 1990;7:235–244. doi: 10.1016/0888-7543(90)90545-6. [DOI] [PubMed] [Google Scholar]
- Girirajan S, Rosenfeld JA, Cooper GM, Antonacci F, Siswara P, Itsara A, Vives L, Walsh T, McCarthy SE, Baker C, Mefford HC, Kidd JM, Browning SR, Browning BL, Dickel DE, Levy DL, Ballif BC, Platky K, Farber DM, Gowans GC, Wetherbee JJ, Asamoah A, Weaver DD, Mark PR, Dickerson J, Garg BP, Ellingwood SA, Smith R, Banks VC, Smith W, McDonald MT, Hoo JJ, French BN, Hudson C, Johnson JP, Ozmore JR, Moeschler JB, Surti U, Escobar LF, El-Khechen D, Gorski JL, Kussmann J, Salbert B, Lacassie Y, Biser A, McDonald-McGinn DM, Zackai EH, Deardorff MA, Shaikh TH, Haan E, Friend KL, Fichera M, Romano C, Gécz J, DeLisi LE, Sebat J, King M-C, Shaffer LG, Eichler EE. A recurrent 16p12.1 microdeletion supports a two-hit model for severe developmental delay. Nat Genet. 2010;42:203–209. doi: 10.1038/ng.534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Green AJ, Sandford RN, Davison BCC. An autosomal dominant syndrome of renal and anogenital malformations with syndactyly. J Med Genet. 1996;33:594–596. doi: 10.1136/jmg.33.7.594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gu W, Zhang F, Lupski JR. Mechanisms for human genomic rearrangements. PathoGenetics. 2008;1:4. doi: 10.1186/1755-8417-1-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamdan FF, Piton A, Gauthier J, Lortie A, Dubeau F, Dobrzeniecka S, Spiegelman D, Noreau A, Pellerin S, Côté M, Henrion E, Fombonne É, Mottron L, Marineau C, Drapeau P, Lafrenière RG, Lacaille JC, Rouleau GA, Michaud JL. de novo STXBP1 mutations in mental retardation and nonsyndromic epilepsy. Ann Neurol. 2009;65:748–753. doi: 10.1002/ana.21625. [DOI] [PubMed] [Google Scholar]
- Hastings PJ, Ira G, Lupski JR. A microhomology-mediated break-induced replication model for the origin of human copy number variation. PLoS Genet. 2009a;5:e1000327. doi: 10.1371/journal.pgen.1000327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hastings PJ, Lupski JR, Rosenberg SM, Ira G. Mechanisms of change in gene copy number. Nat Rev Genet. 2009b;10:551–564. doi: 10.1038/nrg2593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hegde MR, Chin ELH, Mulle JG, Okou DT, Warren ST, Zwick ME. Microarray-based mutation detection in the dystrophin gene. Hum Mutat. 2008;29:1091–1099. doi: 10.1002/humu.20831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Higgins JJ, Loveless JM, Goswami S, Nee LE, Cozzo C, De Biase A, Rosen DR. An atypical intronic deletion widens the spectrum of mutations in hereditary spastic paraplegia. Neurology. 2001;56:1482–1485. doi: 10.1212/wnl.56.11.1482. [DOI] [PubMed] [Google Scholar]
- Iafrate AJ, Feuk L, Rivera MN, Listewnik ML, Donahoe PK, Qi Y, Scherer SW, Lee C. Detection of large-scale variation in the human genome. Nat Genet. 2004;36:949–951. doi: 10.1038/ng1416. [DOI] [PubMed] [Google Scholar]
- Inoue K, Dewar K, Katsanis N, Reiter LT, Lander ES, Devon KL, Wyman DW, Lupski JR, Birren B. The 1.4-Mb CMT1A duplication/HNPP deletion genomic region reveals unique genome architectural features and provides insights into the recent evolution of new genes. Genome Res. 2001;11:1018–1033. doi: 10.1101/gr.180401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inoue K, Khajavi M, Ohyama T, Hirabayashi S, Wilson J, Reggin JD, Mancias P, Butler IJ, Wilkinson MF, Wegner M, Lupski JR. Molecular mechanism for distinct neurological phenotypes conveyed by allelic truncating mutations. Nat Genet. 2004;36:361–369. doi: 10.1038/ng1322. [DOI] [PubMed] [Google Scholar]
- Inoue K, Lupski JR. Molecular mechanisms for genomic disorders. Annu Rev Genomics Hum Genet. 2002;3:199–242. doi: 10.1146/annurev.genom.3.032802.120023. [DOI] [PubMed] [Google Scholar]
- Kapur K, Xing Y, Ouyang Z, Wong WH. Exon arrays provide accurate assessments of gene expression. Genome Biol. 2007;8:R82. doi: 10.1186/gb-2007-8-5-r82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kark JA, Posey DM, Schumacher HR, Ruehle CJ. Sickle-cell trait as a risk factor for sudden death in physical training. N Engl J Med. 1987;317:781–787. doi: 10.1056/NEJM198709243171301. [DOI] [PubMed] [Google Scholar]
- Kurotaki N, Shen JJ, Touyama M, Kondoh T, Visser R, Ozaki T, Nishimoto J, Shiihara T, Uetake K, Makita Y, Harada N, Raskin S, Brown CW, Höglund P, Okamoto N, Lupski JR. Phenotypic consequences of genetic variation at hemizygous alleles: Sotos syndrome is a contiguous gene syndrome incorporating coagulation factor twelve (FXII) deficiency. Genet Med. 2005;7:479–783. doi: 10.1097/01.gim.0000177419.43309.37. [DOI] [PubMed] [Google Scholar]
- Labeit S, Lahmers S, Burkart C, Fong C, McNabb M, Witt S, Witt C, Labeit D, Granzier H. Expression of distinct classes of titin isoforms in striated and smooth muscles by alternative splicing, and their conserved interaction with filamins. J Mol Biol. 2006;362:664–681. doi: 10.1016/j.jmb.2006.07.077. [DOI] [PubMed] [Google Scholar]
- Lane PA, Githens JH. Splenic syndrome at mountain altitudes in sickle cell trait: its occurrence in nonblack persons. J Am Med Assoc. 1985;253:2251–2254. [PubMed] [Google Scholar]
- Lee JA, Carvalho CMB, Lupski JR. A DNA replication mechanism for generating nonrecurrent rearrangements associated with genomic disorders. Cell. 2007;131:1235–1247. doi: 10.1016/j.cell.2007.11.037. [DOI] [PubMed] [Google Scholar]
- Lee JA, Madrid RE, Sperle K, Ritterson CM, Hobson GM, Garbern J, Lupski JR, Inoue K. Spastic paraplegia type 2 associated with axonal neuropathy and apparent PLP1 position effect. Ann Neurol. 2006;59:398–403. doi: 10.1002/ana.20732. [DOI] [PubMed] [Google Scholar]
- Lehrman MA, Russell DW, Goldstein JL, Brown MS. Alu–Alu recombination deletes splice acceptor sites and produces secreted low density lipoprotein receptor in a subject with familial hypercholesterolemia. J Biol Chem. 1987;262:3354–3361. [PubMed] [Google Scholar]
- Liaw D, Marsh DJ, Li J, Dahia PLM, Wang SI, Zheng Z, Bose S, Call KM, Tsou HC, Peacocke M, Eng C, Parsons R. Germline mutations of the PTEN gene in Cowden disease, an inherited breast and thyroid cancer syndrome. Nat Genet. 1997;16:64–67. doi: 10.1038/ng0597-64. [DOI] [PubMed] [Google Scholar]
- Lifton RP, Dluhy RG, Powers M, Rich GM, Cook S, Ulick S, Lalouel J-M. A chimaeric 11β-hydroxylase/aldosterone synthase gene causes glucocorticoid-remediable aldosteronism and human hypertension. Nature. 1992;355:262–265. doi: 10.1038/355262a0. [DOI] [PubMed] [Google Scholar]
- Lu X, Shaw CA, Patel A, Li J, Cooper ML, Wells WR, Sullivan CM, Sahoo T, Yatsenko SA, Bacino CA, Stankiewicz P, Ou Z, Chinault AC, Beaudet AL, Lupski JR, Cheung SW, Ward PA. Clinical implementation of chromosomal microarray analysis: summary of 2513 postnatal cases. PLoS One. 2007;2:e327. doi: 10.1371/journal.pone.0000327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu X-Y, Phung MT, Shaw CA, Pham K, Neil SE, Patel A, Sahoo T, Bacino CA, Stankiewicz P, Kang S-HL, Lalani S, Chinault AC, Lupski JR, Cheung SW, Beaudet AL. Genomic imbalances in neonates with birth defects: high detection rates by using chromosomal microarray analysis. Pediatrics. 2008;122:1310–1318. doi: 10.1542/peds.2008-0297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lupski JR. Genomic disorders: structural features of the genome can lead to DNA rearrangements and human disease traits. Trends Genet. 1998;14:417–422. doi: 10.1016/s0168-9525(98)01555-8. [DOI] [PubMed] [Google Scholar]
- Lupski JR. Structural variation in the human genome. N Engl J Med. 2007;356:1169–1171. doi: 10.1056/NEJMcibr067658. [DOI] [PubMed] [Google Scholar]
- Lupski JR. Genomic disorders ten years on. Genome Med. 2009;1:42. doi: 10.1186/gm42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lupski JR, Stankiewicz P. Genomic disorders: molecular mechanisms for rearrangements and conveyed phenotypes. PLoS Genet. 2005;1:e49. doi: 10.1371/journal.pgen.0010049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maquat LE. When cells stop making sense: effects of nonsense codons on RNA metabolism in vertebrate cells. RNA. 1995;1:453–465. [PMC free article] [PubMed] [Google Scholar]
- Marsh DJ, Dahia PLM, Zheng Z, Liaw D, Parsons R, Gorlin RJ, Eng C. Germline mutations in PTEN are present in Bannayan-Zonana syndrome. Nat Genet. 1997;16:333–334. doi: 10.1038/ng0897-333. [DOI] [PubMed] [Google Scholar]
- Mefford HC, Eichler EE. Duplication hotspots, rare genomic disorders, and common disease. Curr Opin Genet Dev. 2009;19:196–204. doi: 10.1016/j.gde.2009.04.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyahara K, Kawamoto T, Mitsuuchi Y, Toda K, Imura H, Gordon RD, Shizuta Y. The chimeric gene linked to glucocorticoid-suppressible hyperaldosteronism encodes a fused P-450 protein possessing aldosterone synthase activity. Biochem Biophys Res Commun. 1992;189:885–891. doi: 10.1016/0006-291x(92)92286-7. [DOI] [PubMed] [Google Scholar]
- Moretti P, Peters SU, del Gaudio D, Sahoo T, Hyland K, Bottiglieri T, Hopkin RJ, Peach E, Min SH, Goldman D, Roa B, Bacino CA, Scaglia F. Brief report: autistic symptoms, developmental regression, mental retardation, epilepsy, and dyskinesias in CNS folate deficiency. J Autism Dev Disord. 2008;38:1170–1177. doi: 10.1007/s10803-007-0492-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moretti P, Sahoo T, Hyland K, Bottiglieri T, Peters S, del Gaudio D, Roa B, Curry S, Zhu H, Finnell RH, Neul JL, Ramaekers VT, Blau N, Bacino CA, Miller G, Scaglia F. Cerebral folate deficiency with developmental delay, autism, and response to folinic acid. Neurology. 2005;64:1088–1090. doi: 10.1212/01.WNL.0000154641.08211.B7. [DOI] [PubMed] [Google Scholar]
- Ou Z, Kang S-HL, Shaw CA, Carmack CE, White LD, Patel A, Beaudet AL, Cheung SW, Chinault AC. Bacterial artificial chromosome-emulation oligonucleotide arrays for targeted clinical array-comparative genomic hybridization analyses. Genet Med. 2008;10:278–289. doi: 10.1097/GIM.0b013e31816b4420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Potocki L, Chen K-S, Koeuth T, Killian J, Iannaccone ST, Shapira SK, Kashork CD, Spikes AS, Shaffer LG, Lupski JR. DNA rearrangements on both homologues of chromosome 17 in a mildly delayed individual with a family history of autosomal dominant carpal tunnel syndrome. Am J Hum Genet. 1999;64:471–478. doi: 10.1086/302240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raymond FL, Tarpey PS, Edkins S, Tofts C, O’Meara S, Teague J, Butler A, Stevens C, Barthorpe S, Buck G, Cole J, Dicks E, Gray K, Halliday K, Hills K, Hinton J, Jones D, Menzies A, Perry J, Raine K, Shepherd R, Small A, Varian J, Widaa S, Mallya U, Moon J, Luo Y, Shaw M, Boyle J, Kerr B, Turner G, Quarrell O, Cole T, Easton DF, Wooster R, Bobrow M, Schwartz CE, Gecz J, Stratton MR, Futreal PA. Mutations in ZDHHC9, which encodes a palmitoyltransferase of NRAS and HRAS, cause X-linked mental retardation associated with a Marfanoid habitus. Am J Hum Genet. 2007;80:982–987. doi: 10.1086/513609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Redon R, Ishikawa S, Fitch KR, Feuk L, Perry GH, Andrews TD, Fiegler H, Shapero MH, Carson AR, Chen W, Cho EK, Dallaire S, Freeman JL, González JR, Gratacòs M, Huang J, Kalaitzopoulos D, Komura D, MacDonald JR, Marshall CR, Mei R, Montgomery L, Nishimura K, Okamura K, Shen F, Somerville MJ, Tchinda J, Valsesia A, Woodwark C, Yang F, Zhang J, Zerjal T, Zhang J, Armengol L, Conrad DF, Estivill X, Tyler-Smith C, Carter NP, Aburatani H, Lee C, Jones KW, Scherer SW, Hurles ME. Global variation in copy number in the human genome. Nature. 2006;444:444–454. doi: 10.1038/nature05329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saillour Y, Cossée M, Leturcq F, Vasson A, Beugnet C, Poirier K, Commere V, Sublemontier S, Viel M, Letourneur F, Barbot JC, Deburgrave N, Chelly J, Bienvenu T. Detection of exonic copy-number changes using a highly efficient oligonucleotide-based comparative genomic hybridization-array method. Hum Mutat. 2008;29:1083–1090. doi: 10.1002/humu.20829. [DOI] [PubMed] [Google Scholar]
- Sanchez-Valle A, Wang X, Potocki L, Xia Z, Kang S-HL, Carlin ME, Michel D, Williams P, Cabrera-Meza G, Brundage EK, Eifert AL, Stankiewicz P, Cheung SW, Lalani SR. HERV mediated genomic rearrangement of EYA1 in an individual with Branchio-oto-renal syndrome. Am J Med Genet. doi: 10.1002/ajmg.a.33686. (in press) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schollen E, Smeets E, Deflem E, Fryns JP, Matthijs G. Gross rearrangements in the MECP2 gene in three patients with Rett syndrome: implications for routine diagnosis of Rett syndrome. Hum Mutat. 2003;22:116–120. doi: 10.1002/humu.10242. [DOI] [PubMed] [Google Scholar]
- Sebat J, Lakshmi B, Troge J, Alexander J, Young J, Lundin P, Månér S, Massa H, Walker M, Chi M, Navin N, Lucito R, Healy J, Hicks J, Ye K, Reiner A, Gilliam TC, Trask B, Patterson N, Zetterberg A, Wigler M. Large-scale copy number polymorphism in the human genome. Science. 2004;305:525–528. doi: 10.1126/science.1098918. [DOI] [PubMed] [Google Scholar]
- Shaffer LG, Kennedy GM, Spikes AS, Lupski JR. Diagnosis of CMT1A duplications and HNPP deletions by interphase FISH: implications for testing in the cytogenetics laboratory. Am J Med Genet. 1997;69:325–331. [PubMed] [Google Scholar]
- Shao L, Shaw CA, Lu X-Y, Sahoo T, Bacino CA, Lalani SR, Stankiewicz P, Yatsenko SA, Li Y, Neill S, Pursley AN, Chinault AC, Patel A, Beaudet AL, Lupski JR, Cheung SW. Identification of chromosome abnormalities in subtelomeric regions by microarray analysis: a study of 5,380 cases. Am J Med Genet A. 2008;146A:2242–2251. doi: 10.1002/ajmg.a.32399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shaw CJ, Shaw CA, Yu W, Stankiewicz P, White LD, Beaudet AL, Lupski JR. Comparative genomic hybridisation using a proximal 17p BAC/PAC array detects rearrangements responsible for four genomic disorders. J Med Genet. 2004;41:113–119. doi: 10.1136/jmg.2003.012831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith JM, Kirk EPE, Theodosopoulos G, Marshall GM, Walker J, Rogers M, Field M, Brereton JJ, Marsh DJ. Germline mutation of the tumour suppressor PTEN in Proteus syndrome. J Med Genet. 2002;39:937–940. doi: 10.1136/jmg.39.12.937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smyk M, Berg JS, Pursley A, Curtis FK, Fernandez BA, Bien-Willner GA, Lupski JR, Cheung SW, Stankiewicz P. Male-to-female sex reversal associated with an ~250 kb deletion upstream of NR0B1 (DAX1) Hum Genet. 2007;122:63–70. doi: 10.1007/s00439-007-0373-8. [DOI] [PubMed] [Google Scholar]
- Staaf J, Törngren T, Rambech E, Johansson U, Persson C, Sellberg G, Tellhed L, Nilbert M, Borg Å. Detection and precise mapping of germline rearrangements in BRCA1, BRCA2, MSH2, and MLH1 using zoom-in array comparative genomic hybridization (aCGH) Hum Mutat. 2008;29:555–564. doi: 10.1002/humu.20678. [DOI] [PubMed] [Google Scholar]
- Stankiewicz P, Lupski JR. Genome architecture, rearrangements and genomic disorders. Trends Genet. 2002;18:74–82. doi: 10.1016/s0168-9525(02)02592-1. [DOI] [PubMed] [Google Scholar]
- Stankiewicz P, Lupski JR. Structural variation in the human genome and its role in disease. Annu Rev Med. 2010;61:437–455. doi: 10.1146/annurev-med-100708-204735. [DOI] [PubMed] [Google Scholar]
- Stankiewicz P, Sen P, Bhatt SS, Storer M, Xia Z, Bejjani BA, Ou Z, Wiszniewska J, Driscoll DJ, Maisenbacher MK, Bolivar J, Bauer M, Zackai EH, McDonald-McGinn D, Nowaczyk MMJ, Murray M, Hustead V, Mascotti K, Schultz R, Hallam L, McRae D, Nicholson AG, Newbury R, Durham-O’Donnell J, Knight G, Kini U, Shaikh TH, Martin V, Tyreman M, Simonic I, Willatt L, Paterson J, Mehta S, Jones CW, Rajan D, Fitzgerald T, Gribble S, Prigmore E, Patel A, Shaffer LG, Carter NP, Cheung SW, Langston C, Shaw-Smith C. Genomic and genic deletions of the FOX gene cluster on 16q24.1 and inactivating mutations of FOXF1 cause alveolar capillary dysplasia and other malformations. Am J Hum Genet. 2009;84:780–791. doi: 10.1016/j.ajhg.2009.05.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swarthout JT, Lobo S, Farh L, Croke MR, Greentree WK, Deschenes RJ, Linder ME. DHHC9 and GCP16 constitute a human protein fatty acyltransferase with specificity for H- and N-Ras. J Biol Chem. 2005;280:31141–31148. doi: 10.1074/jbc.M504113200. [DOI] [PubMed] [Google Scholar]
- Tarpey PS, Smith R, Pleasance E, Whibley A, Edkins S, Hardy C, O’Meara S, Latimer C, Dicks E, Menzies A, Stephens P, Blow M, Greenman C, Xue Y, Tyler-Smith C, Thompson D, Gray K, Andrews J, Barthorpe S, Buck G, Cole J, Dunmore R, Jones D, Maddison M, Mironenko T, Turner R, Turrell K, Varian J, West S, Widaa S, Wray P, Teague J, Butler A, Jenkinson A, Jia M, Richardson D, Shepherd R, Wooster R, Tejada MI, Martinez F, Carvill G, Goliath R, de Brouwer APM, van Bokhoven H, Van Esch H, Chelly J, Raynaud M, Ropers H-H, Abidi FE, Srivastava AK, Cox J, Luo Y, Mallya U, Moon J, Parnau J, Mohammed S, Tolmie JL, Shoubridge C, Corbett M, Gardner A, Haan E, Rujirabanjerd S, Shaw M, Vandeleur L, Fullston T, Easton DF, Boyle J, Partington M, Hackett A, Field M, Skinner C, Stevenson RE, Bobrow M, Turner G, Schwartz CE, Gecz J, Raymond FL, Futreal PA, Stratton MR. A systematic, large-scale resequencing screen of X-chromosome coding exons in mental retardation. Nat Genet. 2009;41:535–543. doi: 10.1038/ng.367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tayeh MK, Chin ELH, Miller VR, Bean LJH, Coffee B, Hegde M. Targeted comparative genomic hybridization array for the detection of single- and multiexon gene deletions and duplications. Genet Med. 2009;11:232–240. doi: 10.1097/GIM.0b013e318195e191. [DOI] [PubMed] [Google Scholar]
- Thiffault I, Schwartz CE, Der Kaloustian V, Foulkes WD. Mutation analysis of the tumor suppressor PTEN and the glypican 3 (GPC3) gene in patients diagnosed with Proteus syndrome. Am J Med Genet A. 2004;130A:123–127. doi: 10.1002/ajmg.a.30335. [DOI] [PubMed] [Google Scholar]
- Thorsen K, Sørensen KD, Brems-Eskildsen AS, Modin C, Gaustadnes M, Hein A-MK, Kruhøffer M, Laurberg S, Borre M, Wang K, Brunak S, Krainer AR, Tørring N, Dyrskjøt L, Andersen CL, Ørntoft TF. Alternative splicing in colon, bladder, and prostate cancer identified by exon array analysis. Mol Cell Proteomics. 2008;7:1214–1224. doi: 10.1074/mcp.M700590-MCP200. [DOI] [PubMed] [Google Scholar]
- Tsai AC-H, Dossett C, Walton CS, Cramer A, Eng PA, Nowakowska BA, Pursley AN, Stankiewicz P, Wiszniewska J, Cheung SW. Exon deletions of the EP300 and CREBBP genes in two children with Rubinstein-Taybi syndrome detected by aCGH. Eur J Hum Genet. doi: 10.1038/ejhg.2010.121. (in press) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Unger S, Böhm D, Kaiser FJ, Kaulfuss S, Borozdin W, Buiting K, Burfeind P, Böhm J, Barrionuevo F, Craig A, Borowski K, Keppler-Noreuil K, Schmitt-Mechelke T, Steiner B, Bartholdi D, Lemke J, Mortier G, Sandford R, Zabel B, Superti-Furga A, Kohlhase J. Mutations in the cyclin family member FAM58A cause an X-linked dominant disorder characterized by syndactyly, telecanthus and anogenital and renal malformations. Nat Genet. 2008;40:287–289. doi: 10.1038/ng.86. [DOI] [PubMed] [Google Scholar]
- Vissers LELM, Bhatt SS, Janssen IM, Xia Z, Lalani SR, Pfundt R, Derwinska K, de Vries BBA, Gilissen C, Hoischen A, Nesteruk M, Wisniowiecka-Kowalnik B, Smyk M, Brunner HG, Cheung SW, van Kessel AG, Veltman JA, Stankiewicz P. Rare pathogenic microdeletions and tandem duplications are micro-homology-mediated and stimulated by local genomic architecture. Hum Mol Genet. 2009;18:3579–3593. doi: 10.1093/hmg/ddp306. [DOI] [PubMed] [Google Scholar]
- Walsh T, McClellan JM, McCarthy SE, Addington AM, Pierce SB, Cooper GM, Nord AS, Kusenda M, Malhotra D, Bhandari A, Stray SM, Rippey CF, Roccanova P, Makarov V, Lakshmi B, Findling RL, Sikich L, Stromberg T, Merriman B, Gogtay N, Butler P, Eckstrand K, Noory L, Gochman P, Long R, Chen Z, Davis S, Baker C, Eichler EE, Meltzer PS, Nelson SF, Singleton AB, Lee MK, Rapoport JL, King M-C, Sebat J. Rare structural variants disrupt multiple genes in neurodevelopmental pathways in schizophrenia. Science. 2008;320:539–543. doi: 10.1126/science.1155174. [DOI] [PubMed] [Google Scholar]
- Wang J, Chang Y-F, Hamilton JI, Wilkinson MF. Nonsense-associated altered splicing: a frame-dependent response distinct from nonsense-mediated decay. Mol Cell. 2002;10:951–957. doi: 10.1016/s1097-2765(02)00635-4. [DOI] [PubMed] [Google Scholar]
- Wang LL, Worley K, Gannavarapu A, Chintagumpala MM, Levy ML, Plon SE. Intron-size constraint as a mutational mechanism in Rothmund-Thomson syndrome. Am J Hum Genet. 2002;71:165–167. doi: 10.1086/341234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weterman MAJ, van Ruissen F, de Wissel M, Bordewijk L, Samijn JPA, van der Pol WL, Meggouh F, Baas F. Copy number variation upstream of PMP22 in Charcot-Marie-Tooth disease. Eur J Hum Genet. 2010;18:421–428. doi: 10.1038/ejhg.2009.186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wong L-JC, Dimmock D, Geraghty MT, Quan R, Lichter-Konecki U, Wang J, Brundage EK, Scaglia F, Chinault AC. Utility of oligonucleotide array-based comparative genomic hybridization for detection of target gene deletions. Clin Chem. 2008;54:1141–1148. doi: 10.1373/clinchem.2008.103721. [DOI] [PubMed] [Google Scholar]
- Yang TP, Patel PI, Chinault AC, Stout JT, Jackson LG, Hildebrand BM, Caskey CT. Molecular evidence for new mutation at the hprt locus in Lesch-Nyhan patients. Nature. 1984;310:412–414. doi: 10.1038/310412a0. [DOI] [PubMed] [Google Scholar]
- Yatsenko AN, Shroyer NF, Lewis RA, Lupski JR. An ABCA4 genomic deletion in patients with Stargardt disease. Hum Mutat. 2003;21:636–644. doi: 10.1002/humu.10219. [DOI] [PubMed] [Google Scholar]
- Yeo GW, Xu X, Liang TY, Muotri AR, Carson CT, Coufal NG, Gage FH. Alternative splicing events identified in human embryonic stem cells and neural progenitors. PLoS Comput Biol. 2007;3:e196. doi: 10.1371/journal.pcbi.0030196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang F, Carvalho CMB, Lupski JR. Complex human chromosomal and genomic rearrangements. Trends Genet. 2009a;25:298–307. doi: 10.1016/j.tig.2009.05.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang F, Khajavi M, Connolly AM, Towne CF, Batish SD, Lupski JR. The DNA replication FoSTeS/MMBIR mechanism can generate genomic, genic and exonic complex rearrangements in humans. Nat Genet. 2009b;41:849–853. doi: 10.1038/ng.399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang F, Potocki L, Sampson JB, Liu P, Sanchez-Valle A, Robbins-Furman P, Navarro AD, Wheeler PG, Spence JE, Brasington CK, Withers MA, Lupski JR. Identification of uncommon recurrent Potocki-Lupski syndrome-associated duplications and the distribution of rearrangement types and mechanisms in PTLS. Am J Hum Genet. 2010a;86:462–470. doi: 10.1016/j.ajhg.2010.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang F, Seeman P, Liu P, Weterman MA, Gonzaga-Jauregui C, Towne CF, Batish SD, De Vriendt E, De Jonghe P, Rautenstrauss B, Krause K-H, Khajavi M, Posadka J, Vandenberghe A, Palau F, Van Maldergem L, Baas F, Timmerman V, Lupski JR. Mechanisms for nonrecurrent genomic rearrangements associated with CMT1A or HNPP: Rare CNVs as a cause for missing heritability. Am J Hum Genet. 2010b;86:892–903. doi: 10.1016/j.ajhg.2010.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou X-P, Hampel H, Thiele H, Gorlin RJ, Hennekam RCM, Parisi M, Winter RM, Eng C. Association of germline mutation in the PTEN tumour suppressor gene and Proteus and Proteus-like syndromes. Lancet. 2001;358:210–211. doi: 10.1016/s0140-6736(01)05412-5. [DOI] [PubMed] [Google Scholar]
- Zhuang J, Tromp G, Kuivaniemi H, Nakayasu K, Prockop DJ. Deletion of 19 base pairs in intron 13 of the gene for the proα2(I) chain of type-I procollagen (COL1A2) causes exon skipping in a proband with type-I osteogenesis imperfecta. Hum Genet. 1993;91:210–216. doi: 10.1007/BF00218258. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.