Abstract
Glycerol metabolism provides a central link between sugar and fatty acid catabolism. In most bacteria, glycerol kinase plays a crucial role in regulating channel/facilitator-independent uptake of glycerol into the cell. In the firmicute Enterococcus casseliflavus, this enzyme’s activity is enhanced by phosphorylation of the histidine residue (His232) located in its activation loop, approximately 25 Å away from its catalytic cleft. We reported earlier that some mutations of His232 altered enzyme activities; we present here the crystal structures of these mutant GlpK enzymes. The structure of a mutant enzyme with enhanced enzymatic activity, His232Arg, reveals that residues at the catalytic cleft are more optimally aligned to bind ATP and mediate phosphoryl transfer. Specifically, the position of Arg18 in His232Arg shifts by ~1 Å when compared to its position in WT, His232Ala, and His232Glu enzymes. This new conformation of Arg18 is more optimally positioned at the presumed γ-phosphate location of ATP, close to the glycerol substrate. In addition to structural changes exhibited at the active site, the conformational stability of the activation loop is decreased, as reflected by ~35% increase in B-factors (“thermal factors”) in a mutant enzyme displaying diminished activity, His232Glu. Correlating conformational changes to alteration of enzymatic activities in the mutant enzymes identifies distinct localized regions that can have profound effects on intramolecular signal transduction. Alterations of pairwise interactions across the dimer interface can communicate phosphorylation states over 25 Å from the activation loop to the catalytic cleft, positioning Arg18 to form favourable interactions at the β,γ-bridging position to ATP. This would offset loss of the hydrogen bonds at the γ-phosphate of ATP during phosphoryl transfer to glycerol, suggesting that appropriate alignment of the second substrate of glycerol kinase, the ATP molecule, may largely determine the rate of glycerol-3-phosphate production.
In most bacteria, the uptake of glycerol is catalyzed in an energy-independent manner by a membrane channel protein, the glycerol facilitator (GlpF). The motive force for net glycerol diffusion results from the imbalance of intra- and extra-cellular glycerol concentrations caused by the metabolism of glycerol. Frequently, the enzyme glycerol kinase (GlpK) catalyzes the first step of glycerol metabolism by transforming the triol into glycerol-3-P (G3P). Because G3P is not recognized as substrate by GlpF, GlpK plays an essential role in trapping glycerol intracellularly.
In firmicutes (Gram positive) as well as enterobacteriaceae (Gram negative) GlpK synthesis is controlled by carbon catabolite repression and the enzyme activity is regulated by the sugar phosphotransferase system (PTS). The PTS is a complex carbohydrate transport and phosphorylation system and four of its five proteins (or domains) form a phosphorylation cascade using phosphoenolpyruvate (PEP) as phosphoryl donor (1). Mutations which affect one of the first two proteins of the PTS phosphorylation cascade, enzyme I (EI) or HPr, prevent the utilisation of glycerol (2-6). In in vitro studies, PEP, EI and HPr were found to specifically phosphorylate a conserved histidyl residue in GlpK of firmicutes (7-9), which is usually surrounded by three aromatic amino acids that are all present on a loop which we designate as the ‘activation loop’. In in vivo studies, the presence of PTS sugars, which leads to poor phosphorylation of the PTS proteins by a mechanism involving HPr kinase/phosphorylase (10-12), was shown to also drastically lower the extent of GlpK phosphorylation and activation in glycerol-grown cells (12). The near absence of GlpK activation in firmicutes during efficient PTS carbohydrate transport slows the formation of the inducer G3P(6) and therefore serves as an inducer exclusion mechanism (9).
Surprisingly, the mechanisms which control GlpK activity in firmicutes and enterobacteriaceae turned out to be completely different (1), although these enzymes usually exhibit between 50 and 60% sequence identity. In enterobacteriaceae such as E. coli, unphosphorylated EIIAGlc interacts with GlpK and inhibits its activity (13), thus preventing the biosynthesis of the inducer G3P. The interaction of EIIAGlc with GlpK occurs in the C-terminal domain about 30 Å away from the glycerol binding site (14) and is stimulated by the presence of Zn2+ ions (15). In GlpK of firmicutes, including Enterococcus casseliflavus, the activation loop is also located distal from the active site, but in a region opposite to the EIIAGlc interaction site in GlpK of enterobacteriaceae, as shown by the crystal structure (16).
Several spontaneous mutations in B. subtilis resulted in the ability to utilize glycerol in the absence of a functional PTS (17); these were found to be clustered to the glpK gene. GlpK of B. subtilis becomes phosphorylated at His-230 and one of the above mutations led to the replacement of His-230 with an arginine and another to the exchange of Phe-232 with a serine. These findings suggested that the above mutations and GlpK phosphorylation cause similar structural changes, leading to activation of the enzyme. Indeed, purified mutant GlpKs from E. casseliflavus (8) and B. subtilis (9) exhibited between 7- to 19-fold higher activity than the unphosphorylated wild-type (WT) enzyme.
We have previously determined the structures of E. casseliflavus WT GlpK with and without glycerol bound to the active site (16). Unfortunately, the lability of the P~nitrogen bond in phosphorylated GlpK (7) complicates direct structural analysis of the activated state of GlpK. Nevertheless, we expected that the structural changes in some of the above-mentioned GlpK mutants would resemble the structural changes caused by phosphorylation. Structural characterization of these GlpK mutants may yield information about the conformational changes triggered by phosphorylation of GlpK enzyme, leading to increased activity. Of particular interest is the His232Arg mutant GlpK from E. casseliflavus, which is altered at the regulatory phosphorylation site and exhibits the highest Vmax among the mutant proteins (8).
In previous studies of the WT GlpK apo and glycerol-complexed structures, binding of substrate induced structural heterogeneity in the dimer, with certain regions of the kinase exhibiting rmsd of up to 2.4 Å (16). It was proposed that these changes, in conjunction with the conformational differences of the activation loops, may represent possible structural adjustments that occur upon activation for intramolecular signal transduction, whereby phosphorylation of His232 is transmitted over 25 Å from the activation loop to the active site. Additionally, the WT structures suggested that dynamic conformational sampling likely occurs within the ternary complex of GlpK, glycerol, and ATP, until the two substrates are appropriately aligned for phosphoryl-transfer to occur. In this paper, we propose that phosphorylation of GlpK results in two major structural changes, with Arg18 playing a major role in substrate alignment. First, phosphorylation of His232 initiates conformational rearrangement of active site residues into a configuration that more optimally aligns the two substrates, glycerol and ATP, to enhance the kinetics of product formation and, second, the structural changes that lead to activation concomitantly limit the conformational dynamics of the enzyme. The structural results reported here are consistent with this proposed model of activation; in particular, the structure of His232Arg GlpK, a ‘hyperactive’ mutant, substantiates a mechanism involving components of structural alignment coupled to stabilization of distinct oligomeric states to activate the enzyme.
MATERIALS AND METHODS
Bacterial strains and plasmids
WT glpK was cloned, mutagenized, and subcloned as previously described (8). The pOXO4 (18) chimeras were transformed into JM109(DE3) (Promega AG, Mannheim, Germany) for expression. To clone the His232Arg glpK allele into the His-tag expression vector pQE30 (Qiagen, France), it was amplified by using the corresponding pOXO4 plasmid as template and the oligos glpKEcaHisF (5′-GGAGGATCCATGGCAGAAAAAAATTATG-3′) and glpKEcaHisR 5′-CCAAAGCTTTTACTCGCCTTCTTTCTTC-3′ as primers. The PCR product was cut with BamHI and HindIII and cloned into pQE30 cut with the same enzymes thus providing plasmid pQEglpKEcasHis232Arg. Transformation of strain NM522 (Stratagene Europe, The Netherlands) with this plasmid allowed the synthesis of His-tagged His232Arg mutant GlpK (hereon referred to as “His232Arg-B”).
Purification of E. casseliflavus GlpK mutants
All proteins used for crystallization were purified as previously described (8), except His-tagged His232Arg mutant GlpK, which was purified by metal chelate chromatography in the presence of glycerol 15% (v/v) by following the method described in (11). For all other proteins, separation on a mono Q column with an ÄKTA explorer system resulted in a single peak eluting at approximately 0.6 M NaCl. Using Amicon Ultra 15-30K concentrators, each enzyme was desalted, concentrated, and the buffer gradually exchanged with 50 mM Tris·HCl, pH 7.5 in three cycles. To complex with glycerol, 10% (w/v) of glycerol was added at each cycle of exchange. To obtain ethylene glycol (EG) complexed crystals, mutant enzymes were purified as above but omitting glycerol.
Crystallization
Crystallization conditions for each GlpK mutant are reported in Table 1. Protein stock concentrations used for crystallization were 12 mg/mL; all crystals were obtained at 20°C via the vapour diffusion method with hanging-drops set ups in 24-well VDX plates (Hampton Research). For the His232Arg, His232Ala, and His232Glu proteins, each sample drop contained 2 μl protein solution at 10 mg/mL concentration and 2 μl crystallization solution. For the His232Arg-B, each sample drop contained 3 μl of protein solution at 8 mg/ml and 1μl of crystallization solution; all hanging drop setups were suspended over 1 mL of well solution. To complex with EG, crystals were grown without glycerol from proteins purified in the absence of glycerol. These crystals were soaked for 5 minutes in a solution containing the artificial mother liquor and ethylene glycol (EG) at a final concentration of 23-25% (Table 1) and subsequently flash-cooled at 100 K by plunging these into liquid nitrogen or under cryo-cooled nitrogen stream. His232Arg-B glycerol complex crystals were soaked in paraffin oil for 30 seconds to remove aqueous solution from crystal surfaces, preventing ice formation. His232Ala crystals were soaked in mother liquor supplemented with an additional 4% PEG 400, to a final concentration of 33% PEG 400 as a cryoprotectant. For His232Arg-EG and His232Glu-EG crystals, the EG is both a glycerol-analogue and a cryoprotectant.
Table 1.
Crystallization Conditions for GlpK Mutant Enzymes
| Enzyme | Crystallization conditions | Glycerol added | Space Group |
|---|---|---|---|
| His232Arg + glycerol | 0.05M KH2PO4, 17% (w/v) PEG 8000 |
10% (Co-crystallized) |
P21212 |
| His232Arg + ethylene glycol |
0.05M KH2PO4, 17% (w/v) PEG 8000 |
0% Soaked with 23% ethylene glycol |
P21 |
| His232Arg-B + glycerol | 2.0M (NH4)2SO4, 5% (v/v) isopropanol |
0% 15% present during purification (Co-crystallized) |
P21212 |
| His232Glu + glycerol | 0.1M HEPES pH 6.5, 10mM ZnSO4, 25% (w/v) PEG 550 MME |
10% (Co-crystallized) |
P21212 |
| His232Glu + ethylene glycol |
0.05M KH2PO4, 20% (w/v) PEG 8000 |
0% Soaked with 25% ethylene glycol |
P21 |
| His232Ala + glycerola |
29% PEG 400, 0.1 M sodium acetate pH 4.5, 0.1 M calcium acetate |
10% (Co-crystallized) |
P21212 |
Data collection
Data sets were collected at a wavelength of 0.97 Å at the Argonne National Laboratory, on beamline 22 on a MAR225 CCD for the His232Glu glycerol complex crystals and on beamline 23 on a MAR300 CCD for the His232Arg EG complex crystals. For the His232Arg glycerol and His232Glu EG complex crystals, data were collected at the Swiss Light Source, on beamline X10SA on a MAR225 CCD at a wavelength 0.98 Å. The His232Ala glycerol complex crystal data were collected at National Synchrotron Light Source-Brookhaven National Laboratory beamline X6A at a wavelength of 0.97 Å, as previously described (19). Data for the His232Arg-B glycerol complex crystals were collected on the European Synchrotron Radiation Facility in Grenoble, France, beamline ID14-3.
Data for His232Arg were processed using XDS (20) to 1.7 Å; data for the His232Glu and for His232Arg-B complex were processed with HKL2000 and scaled with SCALEPACK (21) to 1.75 Å and 1.85 Å, respectively. The data for the His232Arg-EG and His232Glu-EG were processed with d*TREK (22) and scaled to 2.60 Å and 2.50 Å, respectively. Statistics for all data sets are shown in Table 2.
Table 2.
Data Statistics
| Data set | His232Arg | His232Arg | His232Arg-B | His232Glu | His232Glu | His232Ala |
|---|---|---|---|---|---|---|
| glycerol complex |
ethylene glycol |
glycerol complex |
glycerol complex |
ethylene glycol |
glycerol complex |
|
| Data collection | ||||||
| Space group | P21212 | P21 | P21212 | P21212 | P21 | P21212 |
| Total reflections | 930616 | 180270 | 1589192 | 891523 | 215415 | 173577 |
| Independent reflections | 115245 | 92379 | 87047 | 100197 | 73411 | 63284 |
| Unit cell (Å) | ||||||
| a | 96.8 | 98.8 | 92.6 | 96.7 | 98.6 | 95.6 |
| b | 200.5 | 104.3 | 192.2 | 199.6 | 105.2 | 200.0 |
| c | 56.5 | 114.2 | 57.8 | 56.8 | 114.3 | 56.4 |
| β | 114.1 | 114.6 | ||||
| Resolution (Å) | 50-1.7 | 50-2.30 | 30 - 1.85 | 50-1.75 | 50-2.5 | 50-2.03 |
| Completeness (%) | 99.7 (100) a | 98.5 (97.0) | 97.8 (97.8) | 97.6 (81.4) | 99.8(99.8) | 88.8 (78.1) |
| I/σ(I) | 9.3(2.8) | 11.1 (2.09) | 16.2 (2.15) | 8.4(2.5) | 6.5 (1.70) | 11.8 (2.35) |
| Rsym b (%) | 8.7 (49.8) a | 8.2(52.8) | 7.3 (59.8) | 9.5 (47.4) | 9.0(36.8) | 9.1 (38.7) |
| Refinement statistics | ||||||
| Rcryst c (%) | 21.74 | 18.65 | 21.5 | 22.11 | 23.40 | 20.21 |
| Rfree d (%) | 23.61 | 23.64 | 24.4 | 23.89 | 27.86 | 22.28 |
| Average B | 37.03 | 54.6 | 24.5 | 35.3 | 43.64 | 25.8 |
| Ramachandran Statistics | ||||||
| R.m.s.d. | ||||||
| bond lengths (Å) | 0.0051 | 0.0065 | 0.0053 | 0.0057 | 0.0067 | 0.0056 |
| bond angles (°) | 1.27 | 1.31 | 1.31 | 1.26 | 1.33 | 1.26 |
| Most favored | 91.1 | 87.0 | 90.2 | 90.5 | 86.3 | 88.5 |
| Additionally allowed | 8.1 | 11.9 | 9.1 | 9.0 | 12.5 | 10.4 |
| Generously allowed | 0.3 | 0.6 | 0.3 | 0.2 | 0.7 | 0.6 |
| Disallowed | 0.5 | 0.5 | 0.3 | 0.2 | 0.5 | 0.6 |
Numbers in parentheses represent values in the highest resolution shell (last of 10 shells).
Rsym = ΣhΣi ∣I(h,i) - <I(h)>∣ / ΣhΣi I(h,i) where I(h,i) is the intensity value of the i-th measurement of h and <I(h)> is the corresponding mean value of I(h) for all i measurements.
Rcryst = Σ ∣∣Fobs∣ - ∣Fcalc∣∣ / Σ ∣Fobs∣, where ∣Fobs∣ and ∣Fcalc∣ are the observed and calculated structure factors, respectively.
Rfree with a 10% subset of all reflections ever used in crystallographic refinement.
Phasing, Refinement, and Structural Analysis
All GlpK mutants complexed with glycerol crystallized in the orthorhombic space group P21212. His232Arg-EG and His232Glu-EG crystallized in space group P21. For all structures except His232Arg-B, phasing information were obtained by using the X subunit of His232Ala complex as an initial search model (16) for molecular replacement in Phaser (23). His232Arg-B complex was solved by molecular replacement, initially to 3.5 Å resolution with MOLREP (24) using molecule O of E. casseliflavus free enzyme (pdb accession number 1r59) (16).
Phaser was used for rotational and translational searches to phase all other mutant data. In the His232Glu and His232Arg complex, the asymmetric units contain a X-O dimer, which was previously identified as the likely physiological oligomer (16). Difference Fourier (Fo-Fc) maps confirmed the presence of one glycerol molecule per subunit in the mutant complexes. In both His232Glu-EG and His232Arg-EG GlpK, the asymmetric units contain two dimers. Models were modified to their respective residue at His232 for each mutant and refined in Refmac5 (25), part of the CCP4 suite (26), and CNS (27), initially applying non-crystallographic symmetry (NCS) restraints. Simulated annealing followed by validation using omit and composite omit maps generated in CNS (27) were used at each refinement cycle to confirm correctness of model building. The final cycles of refinement were done without application of NCS restraints. Non-protein densities (>2.5σ) corresponding to water molecules were found using Arp/wArp in CCP4 (28); N- and C-terminal residues were added in electron densities (>3σ). The His232Arg-B complex structure was refined in CNS (27). A clear electron density corresponding to a glycerol molecule was observed in the active site of both monomers O and X. As with the structures described above, the free R factor(29, 30) was used to monitor each cycle of refinement. Refinement initially included simulated annealing runs and application of NCS constraints, followed by individual B factor refinement. In the final steps, water molecules were placed in residual densities (>2.5σ).
Root mean square deviations were determined after superposition of specified regions using R_fit in CNS. Difference distance matrices were calculated with DDMP (31, 32). Average B-values were calculated in CNS and solvent accessibilities were calculated using “accessible surface areas” in CCP4. Structural analysis and verification were done through Procheck (33) and SFcheck (34) in CCP4, errat (35), and verify3D (36). For structural analysis of His232Arg-B complex, models were superimposed and rmsd for defined Cα atoms were calculated in O using default parameters (37). Summary of the refinement and model statistics is tabulated in Table 2.
Results and Discussion
General Structural Features
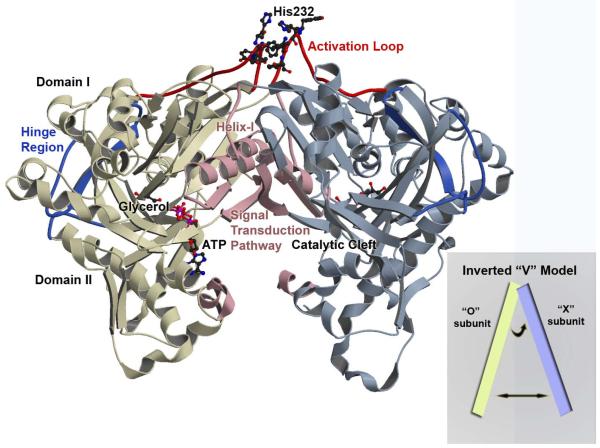
Each subunit of the homodimeric E. casseliflavus GlpK is composed of two major domains, I and II (16). Previously defined as the most likely physiologically-relevant oligomeric state, the O-X dimer (16), is composed of two monomeric subunits related by an axis of NCS 2-fold symmetry of 178-179°. This dimeric arrangement whereby the monomer subunits are related by a NCS 2-fold symmetry axis, parallel to the major molecular axis of the protein, has also been recently found in another structure of a glycerol-metabolism enzyme, the glycerol-3-phosphate dehydrogenase, a monotopic membrane protein (38). Asymmetry of monomer subunits within an O-X dimer was additionally observed in previous and current structures of both E. coli and E. casseliflavus GlpK (14, 16) and are consistent with half-of-the-sites reactivity and negative cooperativity with respect to ATP concentration reported for the E. coli GlpK enzyme (39, 40).
The catalytic cleft, containing the glycerol and ATP binding sites, is located at the interface of domains I and II in each monomer. A “hinge region” serves as a linker between domains I and II, at the periphery of the binding cleft, opposite of the entrance to the active site. Regulation of E. casseliflavus GlpK activity is through phosphorylation of a conserved histidine, His232, in a region referred to as the “activation loop” (225-240) and ~25 Å from the catalytic cleft, at the top of domain I (16) (Figure 1). Structural changes caused by phosphorylation of His232 appear to be transduced through the molecule to the active site since the rate of glycerol phosphorylation is enhanced. His232 mutations modify the measurable WT GlpK activity: the His232Arg enzyme displays activity levels comparable to that of the P~GlpK, His232Ala exhibits WT dephospho GlpK activity, while His232Glu results in less than half of WT activity (8).
Figure 1. Overview of the Enterococcus casseliflavus GlpK O-X Dimer.
A ribbon depiction through Cα-backbone positions showing the overall topology and fold of the wild-type GlpK with the O-subunit colored in yellow and X-subunit in blue. Each subunit is composed of domain I and domain II with the catalytic cleft located at the interface of the two domains. The hinge region is colored in deep blue and delineates the boundary of the two domains and the location of the substrate binding site, with a molecule of glycerol depicted in ball-in-stick. The ATP-binding site is at the entrance of the catalytic cleft and ATP is modeled in ball-in-stick; its position is based on the E. coli ADP-complexed structure (PDB accession number 1GLB; 14) with the γ-phosphate position defined by the phosphate and sulfate ion location from the His232Arg and His232Glu GlpK structures, respectively. The activation loop is colored in red, with His232 and adjacent aromatic residues shown in ball-and-stick. Structural changes initiated by phosphorylation of His232 is transduced to the catalytic cleft along a pathway that is established by helix-I and the O-X dimer interface, colored in pink. The inset shows a schematic of the “inverted-V” topology, described in the text.
As in the crystal structures of the WT GlpK, all mutants form a pseudotetramer, generated through application of crystallographic symmetry operations. In this manner, two equivalent dimers, O-X and Y-Z, are formed. As the O subunit is equivalent to the Y and the X subunit is equivalent to the Z in all GlpK structures, all discussions of the O-X dimer apply equally to the Y-Z dimer. Consequently, the contacts that occur between the alternative dimeric form, the X-Z dimer (and O-Y dimer), are also similar to the crystal contacts made in the WT crystals.
Catalytic Site
Binding of glycerol to GlpK has been proposed to be an ordered reaction, with glycerol binding first followed by ATP (39-41). Although the majority of studies have been conducted on the E. coli GlpK enzyme, sequence conservation of catalytic cleft residues are found across organisms, in contrast to greater sequence diversity found at regulatory regions of the GlpKs. Consequently, active site configuration and catalytic mechanism are likely to be similar amongst the various GlpK enzymes.
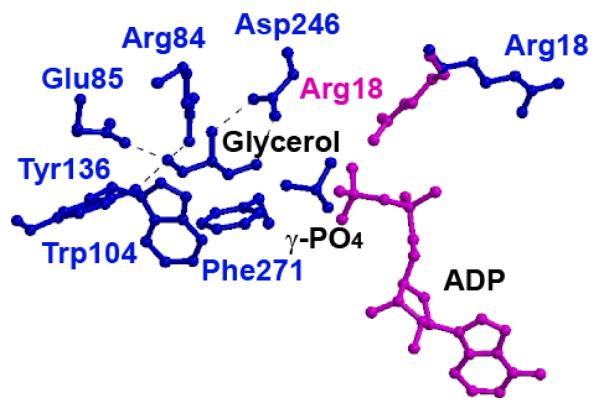
In the complex structures, glycerol is bound deep in the catalytic cleft, held in place by hydrogen bonds to multiple residues (Figure 2). The presence of a single glycerol molecule per subunit is supported by strong electron density apparent in Fo-Fc maps of the catalytic clefts of the glycerol-bound His232Arg, His232Arg-B, His232Glu, and His232Ala structures. The His232Arg, His232Glu and His232Ala active sites have strong similarity around the glycerol binding site, with the hydrogen bonding distances between the glycerol and amino acids in the active site differing by less than 0.2 Å. At the active site, Glu85 assists Arg84, Asp 246, and Tyr136 in anchoring and aligning the bound glycerol. In all glycerol-bound structures, Glu85 has clearly defined electron density; simulated-annealing omit-map analysis confirms its orientation although its phi/psi values place Glu85 in an unallowed region on the Ramachandran plot leading to the speculation that Glu85 may be ‘energetically primed’ to mediate binding of glycerol and release of the G3P product as its conformation is invariant across all the GlpK complex structures. Residues Trp104 and Phe271 provide hydrophobic surfaces that interact with the glycerol carbons, further aligning glycerol (Figure 2). The similarities between the orientations of active site residues across all GlpK structures suggest that the differences in activity are not due to direct changes in glycerol binding at the catalytic cleft.
Figure 2. Glycerol Binding Site in His232Arg and His232Glu GlpK Mutants.
The glycerol binding site from the His232Arg GlpK enzyme, with ADP modeled in according to the E. coli structure (PDB accession number 1GLB; 14). In the His232Glu GlpK structure (blue), Arg18 points away from the glycerol, whereas in the His232Arg GlpK (pink), Arg18 points toward the substrate, glycerol. In this conformation, Arg18 bridges between the γ,β-phosphate group of ATP and glycerol, enhancing G3P formation, as detailed in the text. Binding of glycerol is predominantly mediated through residues Trp104 and Phe271, which provide large hydrophobic surfaces to align the glycerol substrate. Hydrogen bond distances between glycerol hydroxyl oxygens to active site residues converge within 0.2 Å in all the structures; these include Arg84 (2.7 - 2.8 Å), Glu85 (2.5 - 2.7 Å), Tyr136 (2.6 - 2.8 Å), and Asp246 (2.5 - 2.7 Å).
Previous structures of GlpK in the absence and in complex with glycerol revealed that substrate binding results in closure of the catalytic cleft, bringing domains I and II into closer proximity (14, 16, 42). This change is characterized by a rotation of ~6° degrees with a translational component of ~0.2-0.5 Å about an axis orthogonal to the catalytic cleft normal. While glycerol binds deep in the catalytic cleft, the putative ATP binding site is at the entrance of the catalytic cleft, as defined by structures complexed with ADP and ATP-analogues (42, 43). Whereas glycerol is readily identified in the active site, additional significant positive residual electron densities indicative of a tetragonal molecule are found close to glycerol in the His232Arg, His232Arg-B, and the His232Glu difference density maps, in a position that bridges glycerol and the ATP binding site (14). Based on the crystallization conditions for each protein, a phosphate ion was modelled in His232Arg density while a sulfate ion was placed in His232Glu and His232Arg-B. After refinement, the phosphate group is 4.4 Å from the glycerol O3 atom while the sulfate is 4.8 Å (His232Glu) and 3.5 Å (His232Arg-B) (Figure 2).
Using the phosphate/sulfate ion to position the γ-phosphate group of an ATP molecule aligns the γ -phosphate for an in-line transfer, with reasonable distances and geometry. Chiral phosphate studies on the E. coli GlpK enzyme demonstrated an inversion of stereochemical configuration of the γ-phosphoryl group of ATP, upon transfer to glycerol by GlpK (44). For the E. casseliflavus enzyme, less is known regarding inversion of the stereochemical configuration although the high sequence homology at the active sites supports conservation of reaction mechanism.
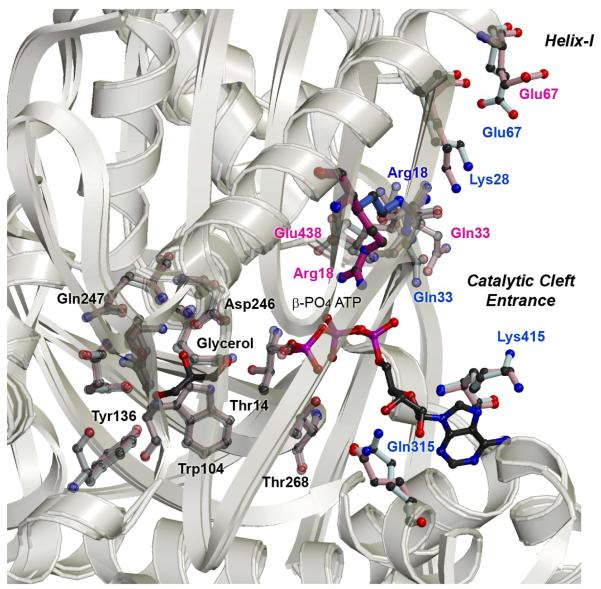
Previous structural results identified Arg18 (Arg17 in E. coli) in playing a role in mediating the phosphoryl transfer reaction from ATP to glycerol (16). In the His232Glu and WT structures, Arg18 forms hydrogen bonds to Glu438, altering its rotameric configuration so it no longer points toward the ATP binding site (Figure 3). In contrast, Arg18 in the His232Arg structure is positioned to interact with two of the phosphoryl oxygens at the β-position of the modelled ATP at distances of 3.4 Å and 2.8 Å. In this conformation, Arg18 forms favourable interactions at the β,γ-bridging position to offset the loss of the hydrogen bonds at the γ-phosphate of ATP, which might explain why His232Arg mutant exhibits higher activity. Accordingly, in the His232Arg mutant GlpK, the frequency with which the γ-phosphate position of ATP would be productively bound and appropriately positioned for catalysis after binding of both substrates is increased, due to the altered conformation of Arg18.
Figure 3. Superposition of His232Arg and His232Glu Active Sites and O-X Interface Regions.
The most significant structural changes are found at the O-X dimer interface that links the activation loop to the active site. As visualized from superpositioning these regions in the His232Arg and His232Glu GlpK mutant enzymes, the conformations of specific residues are altered and these changes ultimately repositions Arg18 closer to the ATP substrate. Most of the structural differences at the active site are clustered around the entrance of the catalytic cleft, at the nucleotide-binding site, whereas the conformation of residues involved in glycerol binding are maintained in all of the mutant structures. Structural differences are found to be distinctly clustered into three major regions – the activation loop, helix-I, and ATP binding site – whereas, overall, the monomer subunits superimposes well, as seen by the ribbon depiction showing an overlay of the Cα-positions of His232Arg and His232Glu structures (in yellow, for clarity, only the O-subunit is shown).
Correlated to the repositioning of Arg18 in the His232Arg mutant GlpK, Glu438 conformation also differs when compared to the His232Ala and His232Glu structures. In His232Arg mutant GlpK Glu438 shifts to a conformation in which it points toward the ATP binding site; this coordinated, pairwise change in Arg18-Glu438 configuration specifically positions active site residues into proximity of the nucleotide binding site and suggests that Glu438 may be involved in aligning Arg18, stabilizing its conformation for ATP interactions.
Further conformational changes are clustered around residues located at the entrance of the catalytic cleft, the nucleotide binding site. In the His232Arg structure, residues Glu331 and Lys 415 rotameric conformations are in closer proximity to the N6 atom of adenine at 3.05 Å and 3.25 Å, respectively, compared to 4 Å and 5.1 Å in the His232Glu mutant GlpK. Although cleft closure upon binding of glycerol limits active site access after the first substrate is bound, the ATP binding site is at the entrance of the catalytic cleft and binding of ADP results in minor structural changes (40, 42). This suggests that ATP may diffuse in and out of the nucleotide binding site until ATP is aligned for productive phosphoryl transfer. Thus, in the His232Arg mutant, repositioning of residues Glu331 and Lys415 may serve to increase binding affinity and frequency of productive alignment, augmenting Arg18 and Glu438 interactions, so that ATP association is increased relative to dissociation, enhancing the rate of G3P production.
To further dissect whether glycerol binding leads to structural rearrangements that result in stabilization of the activation loop, a glycerol analogue, ethylene glycerol (EG), was complexed to mutant GlpKs His232Arg and His232Glu (denoted as His232Arg-EG and His232Glu-EG). The EG is bound at the same region as glycerol, with C1 and C2 of EG superimposable with C1 and C2 of glycerol. As described below, binding of EG results in catalytic cleft closure but the conformation of the activation loop and interface residues differs from that found in the glycerol-complexed structures.
Hinge region
The hinge region includes a linker portion comprised of residues 277-287, a turn, residues 398-405, and three β-strands encompassing residues 269-277, 288-295, and 298-308 (Figure 1). Two of the β-strands, 269-277 and 298-308, continue beyond the hinge into the catalytic cleft. Thus, the hinge region may be affected by structural changes that occur in the activation loop and the catalytic site. For both His232Glu and His232Arg, the linker region between domains I and II have two segments with comparatively higher B factor values and less well defined electron densities. The first segment is comprised of residues 277-287 and the second segment is comprised of residues 399-403. In His232Arg, the average B value for these two segments is 44.3 Å2 versus 25.6 Å2 for the overall protein. In His232Glu, the average B value for the combined segments is 37.1 Å2 compared to the overall B value for the structure of 25.6 Å2 and in the His232Ala, the values are 26.6 Å2 and 41.1 Å2 for the overall and combined segments, respectively. The relatively higher B values indicate that these regions have greater dynamics and motion that permits the catalytic cleft to open and close.
Activation Loop and Dimer Interface: Pathway of Intramolecular Signal Transduction
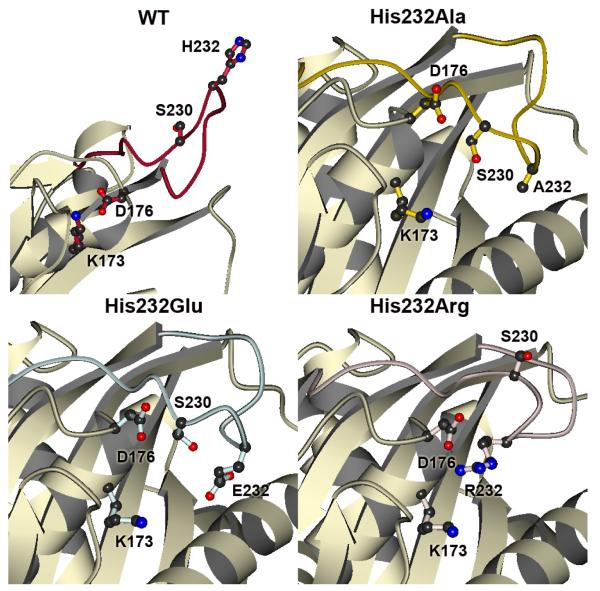
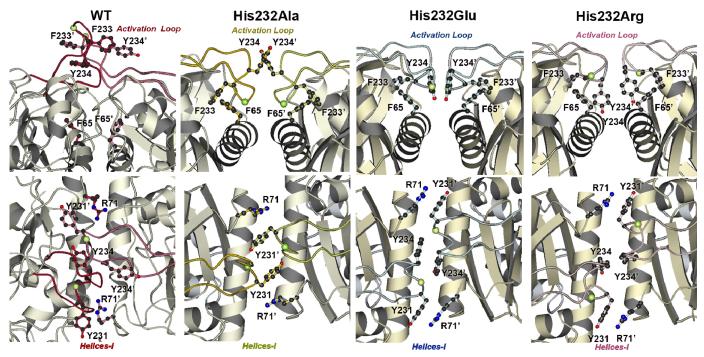
The activation loop encompassing residues 225-240 of the WT GlpK complex protrudes from the surface of domain I (Figure 1; Figure 4). As previously discussed, the extended conformation may allow the docking of HPr, the protein that phosphorylates GlpK at His232 to activate the enzyme. The extended WT loop conformation is stabilized through interactions between His232 from each monomer subunit, in an edge to face orientation. Additionally, immediate neighboring hydrophobic residues surrounding residue 232 -- Tyr231, Phe233, Tyr234 -- form a distinct cluster at the base of the loop. In contrast to the extended conformation of the WT GlpK, the activation loops of the His232 mutant enzymes are all folded down to the surface of domain I. Comparisons of the activation loops of His232Ala, His232Glu, and His232Arg proteins reveal differences that are mainly clustered into two distinct regions, the dimer interface and the periphery of the catalytic cleft.
Figure 4. The Activation Loops of WT, His232Ala, His232Arg, and His232Glu GlpK.
The activation loop in WT (red) is fully extended, with His232 pointing away from domain I (16). In His232Ala (yellow), His232Glu (light blue), and His232Arg (pink) GlpK structures, the loops are folded down towards domain I, although specific interactions are altered in these mutant structures. As comparative references between these structures, three residues (Lys173, Asp176, Ser230) that map the conformational space around the mutation site are depicted in ball-and-stick. In the His232Arg GlpK, the arginine is part of a charge cluster located at the top of domain I and involves multiple arginines from both O and X subunits, converging on Asp176 at the nexus of this cluster. These interactions form an extensive hydrogen-bonding network. Correlating these results to a phosphorylated histidine moiety in the activated WT, stabilization may be achieved through electrostatic interactions between an anionic, phosphorylated His232 and Lys173, which is 4.0-4.6 Å away from the Arg232 of His232Arg GlpK. Extensive interactions between Glu232 and surrounding residues are notably absent and likely related to the increased thermal factors of the activation loop region in the His232Glu GlpK.
In all the structures -- WT, His232Ala, His232Glu, and His232Arg -- the O-X dimer interface is stabilized predominantly by hydrophobic interactions between tryptophan (Trp54), tyrosines (Arg231,234), phenylalanines (Phe65, 233), isoleucines (Ile61, 66), and alanine (Ala62). Additional electrostatic interactions augmenting these involve acidic, basic, and polar residues, including serines (Ser59, 68, 230), aspartate (Asp176), glutamine (Gln58), glutamate (Glu), arginines (Arg71, 229), lysine (Lys173), and asparagine (Asn55). In contrast to the E. coli GlpK (14) the O-X dimer was previously proposed (16) as the likely physiologically relevant oligomeric form. Modifications of specific, correlated interactions along the dimeric interface are apparent when the WT and mutant GlpK structures are compared. These changes mediate the conformational changes initiated at the activation loop, centered at residue 232, to the catalytic cleft through the interface helix (‘helix-I”), comprised of residues 49-69 (Figure 1). In particular, the hydrophobic cluster formed by aromatic amino acids that surround residue 232 shows the largest conformational changes, forming new interface surfaces that may function as the trigger for communicating the phosphorylation state of His232. These changes are transduced along helix-I to the active site, exemplified by the significant differences in interactions between Arg71, Tyr231, Phe233, and Tyr234 between the WT and mutant GlpKs (Figures 4 and 5). In particular, conformation of Tyr234 dramatically changes, swinging almost 180° and rotating nearly 90° from pointing toward the top of the dimer in WT GlpK to pointing down and into the respective monomers in the His232Arg mutant (Figure 5).
Figure 5. The O-X Dimer Interface Regions of WT, His232Ala, His232Arg, and His232Glu GlpK.
The loop regions of the His232 mutants are similarly folded over domain I but the pairwise interactions across the dimer interface are changed among these structures. The interface perspectives shown in B are rotated approximately 90° from those shown in A. The Cα carbon of residue 232, the site of mutation, is denoted by a yellow-green ball. In the His232Glu, Tyr234 (O) phenolic ring forms face-to-face interactions to Tyr234 (X), with the π–π interactions permitting greater conformational flexibility. In contrast, in addition to changes in the aromatic interactions around Tyr234, an electrostatic network formed by Arg232 further stabilizes the O-X dimer interactions. In the His232Ala structure, additional conformational changes are presented which may also lead to enhanced stabilization of the O-X dimer interactions compared to those found in the His232Glu structure.
In the WT structure, Phe233 and Tyr234 form π-stacking interactions at the loop, stabilizing the extended loop conformation and allowing His232 to be presented for phosphorylation. In contrast, in all mutant GlpK structures, hydrophobic interactions between Phe233 and Phe65 ‘lock’ the loop conformation in such a way that it is folded against domain I, driven predominantly through modification of interactions involving the hydrophobic and polar residues defined above.
In the His232Arg mutant, the activation loops are well-defined with an average B factor of 27.8 Å2, close to the overall average B-factor of 25.6 Å2 for the main protein chain. Arg232 is part of a charge cluster at the top of domain I, at the dimer interface, with Arg 229 (O), Arg232 (O) and Arg 71 (X) converging with Asp176 (O) and Tyr231 (O) at the nexus of this cluster to form an extensive hydrogen-bonding network (O and X in parentheses denote the subunits from which the residues derive). This cluster is reproduced at the other subunit and additional interactions are formed as a result of the His232Arg mutation, stabilizing the dimer by extensive, albeit highly localized, electrostatic interactions at the outermost region of domain I. The tyrosyl hydroxyl of residue 231 then displaces the helix-I, shifting it down towards the catalytic cleft. Concomitantly, shifts in the β-strand containing residues 30-40, located at the exterior strand of a 3-stranded β-sheet at the catalytic cleft, trigger the movement of Arg18 at the active site, located on the middle strand of this sheet, toward the substrate binding sites. Overall, these changes result in further closing the catalytic cleft, positioning active site residues Glu438 and Arg18 to more optimally bridge the β-phosphate of ATP, as described earlier.
The average B value for the loop region of the His232Arg-EG is 42.7 Å2 with an average B factor of 36.9 Å2 for the main protein chain; in comparison, in His232Arg GlpK complexed with glycerol, the average B factor of the loop region is 27.8 Å2. Despite the higher thermal factors, the electrostatic cluster at the activation loop is also found in the His232Arg-EG. While the overall conformation of the activation loop of the His232Arg mutant GlpK is maintained in both the glycerol and EG complexed structures, greater differences are found between the glycerol and EG complex in the His232Glu mutant. These differences suggest that glycerol binding is uncoupled from activation and regulation, defining two distinct stages involved in catalysis.
The loop region of His232Glu GlpK is not as well defined as the His232Arg complex. The average B factor in this region in His232Glu glycerol complex is 42.2 Å2, comparatively higher than the average B factor of 25.6 Å2 for the main protein chain. Interactions of Glu232 differ from those found for Arg232, particularly at the interface between the X and O subunits (Figure 4). Lack of contacts at the Glu232 residue is striking, except for a lone hydrogen bonding interaction between Oε1 of Glu232 and the backbone O of Ser230 at 2.6 Å, in contrast to the network of electrostatic connections formed by Arg232. As in the His232Arg structure, hydrophobic residues flanking residue 232 on the activation loop are inserted at the dimer interface of His232Glu GlpK; however, the specific conformations of these residues are altered; the Tyr234 (O) phenolic ring forms face-to-face interactions to Tyr234 (X) at distances of 3.6-6 Å between atoms pairs (Figure 5). Alignment of their respective aromatic rings in a parallel orientation permits greater conformational motion as distances up to 10 Å between ring centroids can be accommodated (45). Overall, lack of specific interactions at Glu232 residue along with altered conformation of O-X interface aromatic residues lead to a greater flexibility and dynamics of the activation loop in the His232Glu mutant GlpK. The importance of the aromatic residues neighbouring residue 232 has also been found in B. subtilis GlpK, whereby a Phe232Ser mutation (equivalent to residue Tyr234 in E. casseliflavus GlpK) activated the enzyme (17).
To ascertain whether activation loop conformation and O-X dimer interface interactions are affected by glycerol binding, these regions in the glycerol and EG complexed structures were compared. The average B factor of the loop of His232Glu-EG is 52.0 Å2 and thus significantly higher than the average B value of 35.9 Å2 for the main protein chain and of 42.2 Å2 for the loop region of the His232Glu-glycerol complex. Overall, the differences observed in the conformation of the His232Glu loops and O-X dimer interface suggest that there are greater dynamics present in these regions. This conformational flexibility is independent of substrate binding, as indicated when the glycerol and EG complexes of His232Arg and His232Glu GlpK structures are compared.
As in the other His232 GlpK mutant structures, the activation loop of His232Ala is folded down against the surface of domain I. In this conformation, the conserved aromatic residues located in this loop are repositioned to insert into the interface region. However, residues Tyr231, Phe233, and Tyr234 have distinctly different interactions from those exhibited in the His232Glu protein. The smaller size of Ala232 along with the lack of charge allows Tyr234 of each subunit to flip out of the dimer interface, leading to changes in positioning of adjacent residues at the activation loop and slight opening of the ‘top’ of domain I, relative to the His232Arg and His232Glu GlpK structures. To visualize, if the GlpK dimer is represented as an inverted “V” with the O and X subunits represented by each leg of the V, the distance between the two legs at the bottom of the V, opposite of the top of domain I, is closest in the His232Ala, as a result of Tyr234 flipping out of the O-X dimer interface (Figure 5). The average B factor for the activation loop of His232Ala is 29.7 Å2, comparable to the average B factor of 26.6 Å2 for the main protein chain. This suggests that the His232Ala activation loop is stabilized in a conformation that constrains alternative O-X dimer interface modifications that are needed to position residues at the catalytic cleft to mediate phosphorylation of the glycerol substrate. The conformation of the His232Ala is ‘locked’ in a stable but inactive conformation, consistent with the activity of the His232Ala GlpK mutant, which cannot be activated, and is thus similar in activity to the unphosphorylated GlpK.
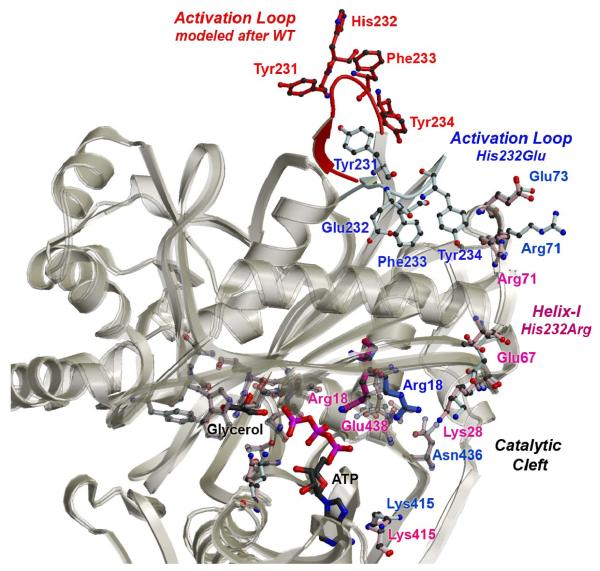
Along helix-I, residues clustered at Arg71, at the top of domain I and close to the activation loop, have altered conformations in the His232Arg and His232Glu mutants. Located at one end of helix-I, residue Glu67 also exhibit modified conformation. The location of Glu67, at the junction between the O-X interface and active site, suggests that changes at the interface are propagated to active site residues involved in ATP and glycerol binding through Glu67 (Figure 6). An intramolecular signal transduction pathway along helix-I, which links the activation loop to the active site, is supported by mutation of Glu65Gly, which led to altered activity in the B. subtilis GlpK enzyme (17). Although Glu65 (corresponding to Glu67 in E. casseliflavus GlpK) is not directly involved in binding of either substrates, our current results indicate that perturbation of the intramolecular signal transduction cascade can be rationally linked to modification of enzyme activity. Consequently, helix-I defines the signal transduction pathway initiated by His232 phosphorylation to the active site in GlpK (Figures 1, 6).
Figure 6. Model of a Pathway of Intramolecular Signal Transduction.
Residues exhibiting the greatest conformational changes in the His232Arg (in pink) and His232Glu (in blue) GlpK structures are depicted in ball-and-stick and labeled accordingly. To model the conformational changes that may occur upon phosphorylation of His232, the WT activation loop (in red) was grafted onto the His232Arg structure (in pink). As shown by a superposition of the Cα positions of the His232Arg and His232Glu O-subunits, represented as white ribbons, rmsd deviations are small at the glycerol binding site and domain conformations are mostly maintained. However, conformational changes initiated by phosphorylation of His232 at the activation loop can be propagated to the active site by modifying interactions at helix-I, the O-X dimer interface, and residues involved in nucleotide binding. Consequently, conformational changes can be propagated >25 A from the activation loop to the active site by coordinated changes initiated by His232 phosphorylation, ultimately resulting in increasing G3P production by enhancing ATP binding. Except for the WT activation loop encompassing residues 230-236, the His232Arg structure was used to model a possible activated conformation of the GlpK enzyme.
In summary, comparative analysis between the His232Ala, His232Glu, and His232Arg mutant enzyme structures and to the WT GlpK identifies two key regions that are involved in modulating the activity of the enterococcal GlpK enzyme. The conformation of the activation loop, at the top of domain I, influences the conformation of hydrophobic residues surrounding residue 232 and alterations can transduce an activation signal through helix-I, which bridges the regulatory site and the catalytic cleft. Consequently, intramolecular signal transduction from the activation loops to the active sites can be transmitted distally over 25 Å by subtle but correlated structural adjustments of hydrophobic residues at the O-X dimer interface and these changes are initiated through conformational changes at residue 232.
Conclusions
Our structural studies on GlpK provide insights into the mechanism of activation via phosphorylation and intramolecular signal transduction. We have examined mutant GlpKs to characterize the structural changes that lead to activation or inactivation of the enzyme. Of particular interest is the structure of the His232Arg GlpK, which reflects an activated structural state of the enzyme. The collective structural changes characterized in the WT and His232 mutants lead to a model of activation in E. casseliflavus GlpK enzyme.
In the WT structures, the region around helix-I exhibited the greatest structural difference between the O and X subunits and these conformational changes were proposed to be ‘snapshots’ of possible structural modifications leading to repositioning of residues at the active site. The current results support such a model, which is based on the following structural changes identified in the His232Arg structures: i) conformation of the activation loop is directed by interactions centered at residue 232; alterations of the loop structure lead to formation of new interaction motifs, including an electrostatic cluster at the O-X dimer interface that stabilizes the overall His232Arg structure; ii) changes in the activation loop conformation are transmitted via modified pairwise interactions across the O-X dimer interface region and transduced to the catalytic cleft through helix-I; iii) correlated changes in configuration of active site residues involve Glu458 and aligns Arg18 at the nucleotide binding site to bridge the β-phosphate group of a modelled ATP molecule; iv) conformational dynamics likely play a key role in regulation; v) conformational changes can be transmitted over 25 Å by subtle and localized rearrangement of two distinct regions of GlpK enzyme, the activation loop and the O-X dimer interface.
The fact that GlpK is activated by Arg232 mutation and not by Glu232 may seem counterintuitive; however, this can be rationalized from the overall structural changes observed in the His232Arg enzyme. Rather than charge as the predominantly driving force, it is the size of the side chain of the residue at position 232 and, specifically, whether this residue can initiate new O-X interface contacts. The new interactions formed in the His232Arg structure are highly localized, focused at the topmost periphery of domain I. Contacts formed at the electrostatic cluster result in the largest angular spread between the O and X subunits of the dimer. Using the inverted V description presented in the His232Ala structure, the distance between the two legs at the bottom of the V is greatest in the His232Arg mutant structure, followed by the His232Glu, with His232Ala having the closest distance between the two legs (Figure 1). Quantitatively, the His232Arg O subunit is ~1 Å further apart from the X subunit, with a rotation of 2° about the 2-fold NCS symmetry axis when compared to the His232Ala O and X subunits. Presuming that the conformation and interactions observed in the His232Arg reflects that of the phosphorylated state of the GlpK enzyme (or represents one of a subset of activated conformational states), stabilization of an activated form can be achieved through electrostatic contacts, as observed in the His232Arg structure. Accordingly, interactions between a phosphorylated histidine and a basic residue, such as Arg71 or Lys173 is plausible, as the NZ atom of Lys173 is 4.0 Å and 4.6 Å away from the NH2 atom of Arg232 in the X and O subunits, respectively.
Modifications of nucleotide binding as a result of domain motion have been observed in actin complexes and in E. coli GlpK complexed with various nucleoside triphosphate analogues (42, 46). Structural changes through rigid-body rotation of domains leading to alterations of binding affinities and/or ligand coordination were described as ‘bond racheting’, originally observed in the actin complex structures (46, 47). In the E. casseliflavus GlpK structures described here, the changes observed at the nucleoside-binding site of the catalytic cleft have similarities to the bond racheting mechanism although the conformational changes are more subtle in magnitude. Rearrangement of active site residues arise from changes in interaction across the O-X dimer interface, most notably focused around aromatic residues immediately adjacent to residue 232. Correlated conformational changes are initiated by structural changes originating at the activation loop and propagated along a pathway defined by the interface helices (residues 49-69) of the O-X dimer, ultimately changing the configuration of residues involved in nucleotide binding. These changes are consistent with an earlier proposal whereby coupling of small-magnitude domain motions with correct positioning of the triphosphate chain of ATP were described as energetically-plausible structural changes and readily explains changes in Vmax of G3P production (42). In this model, regulation of enzyme activity is partly through alteration of the frequency with which the γ-phosphate position of ATP is productively bound and appropriately positioned for catalysis after the binding of both substrates and rationally links structural changes to modulation of enzymatic activity.
As described for the His232Glu structure, conformational instability precludes defined structural interactions along the activation loop and the O-X dimer interface, the proposed pathway of intramolecular signal transduction that communicates the phosphorylation state of His232. Whether the conformation of the ‘overactive’ His232Arg GlpK captures the structure of the phosphorylated, activated GlpK enzyme is unknown. Only in few cases has it been possible to obtain the structure of a His-phosphorylated protein. The formation of specific ion-pair hydrogen bonds between the second, non-phosphorylated nitrogen atom of the histidine imidazole ring and an acidic amino acid side chain seems to contribute to the stabilization of the phospho-histidine in these proteins (48). Such stabilization does not seem to be operative in bacterial GlpKs. Nonetheless, it is likely that the correlated changes found in the His232Arg mutant depict a possible regulatory mechanism in E. casseliflavus GlpK.
The conformational changes observed in the WT and mutant structures of GlpK reported here provide additional evidence that domain motion is correlated to modulation of catalytic activity. This mechanism of regulation appears to control both E. coli and E. casseliflavus GlpK enzymes, despite differences in how these structural changes are initiated. Whether structural reorganization arises from protein-protein interactions through formation of EIIAGlc-GlpK complex in E. coli or via phosphorylation of a histidyl residue in E. casseliflavus GlpK, the result of these regulatory interactions converge in both cases to modulating the binding of ATP.
Acknowledgments
This work was supported in part by grants from the National Institutes of Health (GM66466, JIY) and the Pennsylvania Department of Health (JIY); the Department specifically disclaims responsibility for any analyses, interpretations or conclusions. Additional support is provided by the CNRS, the INRA, and the AgroParisTech (JD, PB). We thank the staff at the General Medicine and Cancer Institutes Collaborative Access Team (GM/CA-CAT) beamline 23-ID and the Southeast Regional Collaborative Access Team (SER-CAT) beamline 22-ID, both at the Advanced Photon Source, Argonne National Laboratory, for access and technical assistance. We thank the staff from ESRF for their attentive technical assistance. GM/CA CAT has been funded in whole or in part with Federal funds from the National Cancer Institute (Y1-CO-1020) and the National Institute of General Medical Science (Y1-GM-1104). Use of the Advanced Photon Source is supported by the U. S. Department of Energy, Office of Science, Office of Basic Energy Sciences, under Contract No. W-31-109-Eng-38.
References
- (1).Deutscher J, Franke C, Postma PW. How phosphotransferase system-related protein phosphorylation regulates carbohydrate metabolism in bacteria. Microbiol. Mol. Biology. Rev. 2006;70 doi: 10.1128/MMBR.00024-06. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (2).Niaudet B, Gay P, Dedonder R. Identification of the structural gene of the PEP-phosphotransferase Enzyme I in Bacillus subtilis Marburg. Mol. Gen. Genet. 1975;136:337–349. doi: 10.1007/BF00341718. [DOI] [PubMed] [Google Scholar]
- (3).Reizer J, Novotny MJ, Stuiver I, Saier MH., Jr. Regulation of glycerol uptake by the phosphoenolpyruvate-sugar phosphotransferase system in Bacillus subtilis. J. Bacteriol. 1984;159:243–250. doi: 10.1128/jb.159.1.243-250.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (4).Romano AH, Saier MH, Jr., Harriott OT, Reizer J. Physiological studies on regulation of glycerol utilization by the phosphoenolpyruvate:sugar phosphotransferase system in Enterococcus faecalis. J. Bacteriol. 1990;172:6741–6748. doi: 10.1128/jb.172.12.6741-6748.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (5).Gonzy-Treboul G, de Waard JH, Zagorec M, Postma PW. The glucose permease of the phosphotransferase system of Bacillus subtilis: evidence for IIGlc and IIIGlc domains. Mol. Microbiol. 1991;5:1241–1249. doi: 10.1111/j.1365-2958.1991.tb01898.x. [DOI] [PubMed] [Google Scholar]
- (6).Beijer L, Rutberg L. Utilisation of glycerol and glycerol 3-phosphate is differently affected by the phosphotransferase system in Bacillus subtilis. FEMS Microbiol. Lett. 1992;79:217–220. doi: 10.1111/j.1574-6968.1992.tb14043.x. [DOI] [PubMed] [Google Scholar]
- (7).Deutscher J, Sauerwald H. Stimulation of dihydroxyacetone and glycerol kinase activity in Streptococcus faecalis by phosphoenolpyruvate-dependent phosphorylation catalyzed by enzyme I and HPr of the phosphotransferase system. J. Bacteriol. 1986;166:829–836. doi: 10.1128/jb.166.3.829-836.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (8).Charrier V, Buckley E, Parsonage D, Galinier A, Darbon E, Jaquinod M, Forest E, Deutscher J, Claiborne A. Cloning and sequencing of two enterococcal glpK genes and regulation of the encoded glycerol kinases by phosphoenolpyruvate-dependent, phosphotransferase system-catalyzed phosphorylation of a single histidyl residue. J. Biol. Chem. 1997;272:14166–14174. doi: 10.1074/jbc.272.22.14166. [DOI] [PubMed] [Google Scholar]
- (9).Darbon E, Servant P, Poncet S, Deutscher J. Antitermination by GlpP, catabolite repression via CcpA, and inducer exclusion elicited by P~GlpK dephosphorylation control Bacillus subtilis glpFK expression. Mol. Microbiol. 2001;43:1039–1052. doi: 10.1046/j.1365-2958.2002.02800.x. [DOI] [PubMed] [Google Scholar]
- (10).Deutscher J, Saier MH., Jr ATP-dependent protein kinase-catalyzed phosphorylation of a seryl residue in HPr, a phosphate carrier protein of the phosphotransferase system in Streptococcus pyogenes. Proc. Natl. Acad. Sci. USA. 1983;80:6790–6794. doi: 10.1073/pnas.80.22.6790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (11).Mijakovic I, Poncet S, Galinier A, Monedero V, Fieulaine S, Janin J, Nessler S, Marquez JA, Scheffzek K, Hasenbein S, Hengstenberg W, Deutscher J. Pyrophosphate-producing protein dephosphorylation by HPr kinase/phosphorylase: a relic of early life? Proc. Natl. Acad. Sci. USA. 2002;99:13442–13447. doi: 10.1073/pnas.212410399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (12).Deutscher J, Bauer B, Sauerwald H. Regulation of glycerol metabolism in Enterococcus faecalis by phosphoenolpyruvate-dependent phosphorylation of glycerol kinase catalyzed by enzyme I and HPr of the phosphotransferase system. J. Bacteriol. 1993;175:3730–3733. doi: 10.1128/jb.175.12.3730-3733.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (13).Postma PW, Epstein W, Schuitema ARJ, Nelson SO. Interaction between IIIGlc of the phosphoenolpyruvate:sugar phosphotransferase system and glycerol kinase of Salmonella typhimurium. J. Bacteriol. 1984;158:351–353. doi: 10.1128/jb.158.1.351-353.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (14).Hurley JH, Faber HR, Worthylake D, Meadow ND, Roseman S, Pettigrew DW, Remington SJ. Structure of the regulatory complex of Escherichia coli IIIGlc with glycerol kinase. Science. 1993;259:673–677. [PubMed] [Google Scholar]
- (15).Pettigrew DW, Feese M, Meadow ND, Remington SJ, Roseman S. Zn(II)-mediated protein interactions in Escherichia coli signal transduction: cation-promoted association of the phosphotransferase system regulatory protein IIIGlc with target protein glycerol kinase. In: Torriani-Gorini A, Yagil E, Silver S, editors. Phosphate in Microorganisms. Cellular and Molecular Biology. ASM Press; Washington: 1994. pp. 335–342. [Google Scholar]
- (16).Yeh JI, Charrier V, Paulo J, Hou L, Parsonage D, Darbon E, Claiborne A, Hol WGJ, Deutscher J. Structures of enterococcal glycerol kinase in the absence and presence of glycerol: Correlation of conformation to substrate binding and mechanism of activation by phosphorylation. Biochemistry. 2004;43:362–373. doi: 10.1021/bi034258o. [DOI] [PubMed] [Google Scholar]
- (17).Wehtje C, Beijer L, Nilsson RP, Rutberg B. Mutations in the glycerol kinase gene restore the ability of a ptsGHI mutant of Bacillus subtilis to grow on glycerol. Microbiology. 1995;141:1193–1198. doi: 10.1099/13500872-141-5-1193. [DOI] [PubMed] [Google Scholar]
- (18).Parsonage D, Miller H, Ross RP, Claiborne A. Purification and analysis of streptococcal NADH peroxidase expressed in Escherichia coli. J. Biol. Chem. 1993;268:3161–3167. [PubMed] [Google Scholar]
- (19).Vahedi-Faridi A, Stojanoff V, Yeh JI. The effects of flash-annealing on glycerol kinase crystals. Acta Cryst. 2005;D61:982–989. doi: 10.1107/S0907444905012746. [DOI] [PubMed] [Google Scholar]
- (20).Kabsch W. Automatic processing of rotation diffraction data from crystals of initially unknown symmetry and cell constant. J.Appl.Cryst. 1993;26:795–800. [Google Scholar]
- (21).Otwinowski Z, Minor W. Processing of X-ray Diffraction Data Collected in Oscillation Method. In: Carter CW, Sweet RM, editors. Macromolecular Crystallography Part A. 1997. [DOI] [PubMed] [Google Scholar]
- (22).Pflugrath JW. The finer things in X-ray diffraction data collection. Acta Cryst. 1999;D55:1718–1725. doi: 10.1107/s090744499900935x. [DOI] [PubMed] [Google Scholar]
- (23).McCoy AJ, Grosse-Kunstleve RW, Storoni LC, Read RJ. Likelihood-enhanced fast translation functions. Acta. Cryst. D. biol. Crystallogr. 2005;61:458–464. doi: 10.1107/S0907444905001617. [DOI] [PubMed] [Google Scholar]
- (24).Vagin A, Tepliakov A. MOLREP: an Automated Program for Molecular Replacement. J. Appl. Cryst. 1997;30:1022–1025. [Google Scholar]
- (25).Winn MD, Isupov MN, Murshudov GN. Use of TLS parameters to model anisotropic displacements in macromolecular refinement. Acta Cryst. 2001;D 57:122–133. doi: 10.1107/s0907444900014736. [DOI] [PubMed] [Google Scholar]
- (26).The CCP4 Suite: Programs for Protein Crystallography. Acta Cryst. 1994;D50:760–763. doi: 10.1107/S0907444994003112. 50. [DOI] [PubMed] [Google Scholar]
- (27).Brünger AT. X-PLOR. version 3.1 Yale University Press; New Haven and London: 1993. [Google Scholar]
- (28).Lamzin VS, Perrakis A, Wilson KS. ARP/wARP’s model-building algorithms. I. The main chain. Acta Cryst. 2002;D 57:1445–1450. doi: 10.1107/s0907444902005462. [DOI] [PubMed] [Google Scholar]
- (29).Brünger AT. Free R value: a novel statistical quantity for assessing the accuracy of crystal structures. Nature. 1992;355:472–475. doi: 10.1038/355472a0. [DOI] [PubMed] [Google Scholar]
- (30).Kleywegt GJ. Separating model optimization and model validation in statistical cross-validation as applied to crystallography. Acta Cryst. 2007;D 63:939–940. doi: 10.1107/S0907444907033458. [DOI] [PubMed] [Google Scholar]
- (31).Fleming P. DDMP Program. Center for Structural Biology at Yale University. Yale University; New Haven, CT: [Google Scholar]
- (32).Richards FM, Kundrot CE. Identification of structural motifs from protein coordinate data: secondary structure and first-level supersecondary structure. PROTEINS. 1988;3:71–84. doi: 10.1002/prot.340030202. [DOI] [PubMed] [Google Scholar]
- (33).Laskowski RA, Moss DS, Thornton JM. PROCHECK: a program to check the stereochemical quality of protein structures. J. Appl. Cryst. 1993;26:283–291. [Google Scholar]
- (34).Vaguine AA, Richelle J, Wodak SJ. SFCHECK: a unified set of procedures for evaluating the quality of macromolecular structure-factor data and their agreement with the atomic model. Acta Crystallogr D Biol Crystallogr. 1999;55:191–205. doi: 10.1107/S0907444998006684. [DOI] [PubMed] [Google Scholar]
- (35).Colovos C, Yeates TO. Verification of protein structures: patterns of nonbonded atomic interactions. Protein Sci. 1993;2:1511–1519. doi: 10.1002/pro.5560020916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (36).Eisenberg D, Lüthy R, Bowie JU. VERIFY3D: assessment of protein models with three-dimensional profiles. Methods Enzymol. 1997;277:396–404. doi: 10.1016/s0076-6879(97)77022-8. [DOI] [PubMed] [Google Scholar]
- (37).Harris M, Jones TA. Molray - a web interface between O and the POV-Ray ray tracer. Acta Cryst. 2001;D 57:1201–1203. doi: 10.1107/s0907444901007697. [DOI] [PubMed] [Google Scholar]
- (38).Yeh JI, Chinte U, Du S. Structure of glycerol-3-phosphate dehydrogenase, an essential monotopic membrane enzyme involved in respiration and metabolism. PNAS U.S.A. 2008;9 doi: 10.1073/pnas.0712331105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (39).Thorner JW, Paulus H. Catalytic and Allosteric Properties of Glycerol Kinase from Escherichia coli. Journal of Biological Chemistry. 1973;248:3922–3832. [PubMed] [Google Scholar]
- (40).Pettigrew DW, Yu G, Liu Y. Nucleotide regulation of Escherichia coli glycerol kinase: initial-velocity and substrate binding studies. Biochemistry. 1990;29:8620–8627. doi: 10.1021/bi00489a018. [DOI] [PubMed] [Google Scholar]
- (41).Bennett WS, Jr, Steitz TA. Structure of a complex between yeast hexokinase and glucose. II. Detailed comparison of conformation and active site configuration with the native hexokinase B monomer and dimer. J. Mol. Biol. 1980;140:211–230. doi: 10.1016/0022-2836(80)90103-5. [DOI] [PubMed] [Google Scholar]
- (42).Bystrom CE, Pettigrew DW, Branchaud BP, O’Brien P, Remington SJ. Crystal structures of Escherichia coli glycerol kinase variant S58 -> W in complex with nonhydrolyzable ATP analogues reveal a putative active conformation of the enzyme as a result of domain motion. Biochemistry. 1999;38:3508–3518. doi: 10.1021/bi982460z. [DOI] [PubMed] [Google Scholar]
- (43).Ormö M, Bystrom CE, Remington SJ. Crystal structure of a complex of Escherichia coli glycerol kinase and an allosteric effector fructose 1,6-bisphosphate. Biochemistry. 1998;37:16565–16572. doi: 10.1021/bi981616s. [DOI] [PubMed] [Google Scholar]
- (44).Blättler WA, Knowles JR. Stereochemical course of phosphokinases. The use of adenosine [.gamma.-(S)-16O,17O,18O]triphosphate and the mechanistic consequences for the reactions catalyzed by glycerol kinase, hexokinase, pyruvate kinase, and acetate kinase. Biochemistry. 1979;18:3927–3933. doi: 10.1021/bi00585a013. [DOI] [PubMed] [Google Scholar]
- (45).McGaughey GB, Gagnes M, Rappe AK. π-Stacking Interactions. Journal of Biological Chemistry. 1998;273:15458–15463. doi: 10.1074/jbc.273.25.15458. [DOI] [PubMed] [Google Scholar]
- (46).Schutt CE, Lindberg U, Lyslik J, Strauss N. The structure of crystalline profilin-beta-actin. Nature. 1993;365:810–816. doi: 10.1038/365810a0. [DOI] [PubMed] [Google Scholar]
- (47).Chik JK, Lindberg U, Schutt CE. The structure of an open state of beta-actin at 2.65 A resolution. J Mol Biol. 1996;263:607–623. doi: 10.1006/jmbi.1996.0602. [DOI] [PubMed] [Google Scholar]
- (48).Puttick J, B. E, Delbaere LT. Histidine phosphorylation in biological systems. Biochim Biophys Acta. 2007;1784:100–105. doi: 10.1016/j.bbapap.2007.07.008. [DOI] [PubMed] [Google Scholar]