Abstract
Purpose
We examined the feasibility of using CYP2D6 genotyping to determine optimal tamoxifen dose and investigated whether the key active tamoxifen metabolite, endoxifen, could be increased by genotype-guided tamoxifen dosing in patients with intermediate CYP2D6 metabolism.
Patients and Methods
One hundred nineteen patients on tamoxifen 20 mg daily ≥ 4 months and not on any strong CYP2D6 inhibiting medications were assayed for CYP2D6 genotype and plasma tamoxifen metabolite concentrations. Patients found to be CYP2D6 extensive metabolizers (EM) remained on 20 mg and those found to be intermediate (IM) or poor (PM) metabolizers were increased to 40 mg daily. Eighty-nine evaluable patients had tamoxifen metabolite measurements repeated 4 months later.
Results
As expected, the median baseline endoxifen concentration was higher in EM (34.3 ng/mL) compared with either IM (18.5 ng/mL; P = .0045) or PM (4.2 ng/mL; P < .001). When the dose was increased from 20 mg to 40 mg in IM and PM patients, the endoxifen concentration rose significantly; in IM there was a median intrapatient change from baseline of +7.6 ng/mL (−0.6 to 23.9; P < .001), and in PM there was a change of +6.1 ng/mL (2.6 to 12.5; P = .020). After the dose increase, there was no longer a significant difference in endoxifen concentrations between EM and IM patients (P = .84); however, the PM endoxifen concentration was still significantly lower.
Conclusion
This study demonstrates the feasibility of genotype-driven tamoxifen dosing and demonstrates that doubling the tamoxifen dose can increase endoxifen concentrations in IM and PM patients.
INTRODUCTION
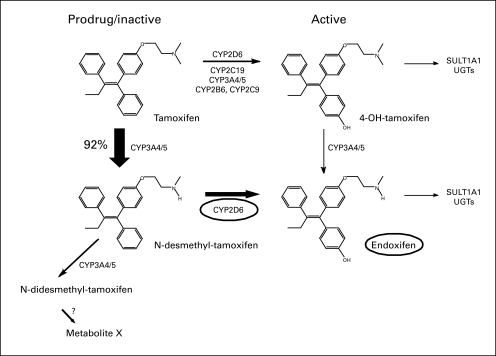
Approximately 80% of the 182,000 invasive breast cancers diagnosed this year in the United States will be endocrine receptor–positive appropriate for tamoxifen treatment.1 Tamoxifen is a prodrug, and may need to be metabolized to be active; up to half of those taking it may not receive the full benefit because of genetic differences that limit this metabolism.2 Tamoxifen is metabolized to the antiestrogen 4-hydroxytamoxifen by several enzymes. However, a preferred route is the formation of the intermediate metabolite N-desmethyltamoxifen by cytochrome P450 3A4/5 with cytochrome P450 2D6 (CYP2D6) further metabolizing N- desmethyltamoxifen to the most abundant active metabolite, endoxifen (Fig 2).3–5 Endoxifen and 4-hydroxytamoxifen have a 50-fold higher affinity for the estrogen receptor (ER) than tamoxifen, and the plasma concentration of endoxifen is 5 to 10 times higher than 4-hydroxytamoxifen; for this reason endoxifen is considered the major active metabolite.6,7 Multiple studies examining endoxifen in ER binding and inhibition of estrogen-induced cell proliferation demonstrate that endoxifen is a highly active metabolite of tamoxifen.6,8,9
Fig 2.
Pathways of tamoxifen and the main CYP enzymes. Concept adapted from Goetz et al.5
Over 75 reduced-activity alleles of CYP2D6 have been detected that vary in ability to convert tamoxifen to endoxifen. In previous CYP2D6 studies, approximately 50% of women were extensive metabolizers (EM; active CYP2D6), 43% were intermediate metabolizers (IM; reduced but not inactive CYP2D6), and 7% were poor metabolizers (PM, inactive CYP2D6).10 CYP2D6 genetic variation affects endoxifen concentrations.7,11
Some, but not all,12–19 of the initial studies showed worse disease-free survival (DFS) for tamoxifen-treated patients with breast cancer with CYP2D6 dysfunctional alleles,20–32 or those who were taking CYP2D6-interacting medications.24,27,33,34 However, two large studies showed no association between CYP2D6 genotype and outcome, which has raised concern about CYP2D6 as a tamoxifen efficacy biomarker.35,36
Tamoxifen is US Food and Drug Administration approved at both 20 mg and 40 mg per day, and has been used safely and effectively at both doses.37,38 It is unknown what effect changing tamoxifen dose might have on endoxifen concentrations in a dysfunctional allele population.
Modern technologic advances aiding in therapeutic decision making include pharmacogenomic assays to identify patient-based factors in drug metabolism. In addition to using these assays to choose among drugs, it is possible that they can help optimize drug dosing. In this study, we examined the feasibility and impact of using CYP2D6 genotyping to determine tamoxifen dose and demonstrated that endoxifen concentrations can be safely increased by individualized tamoxifen dosing.
PATIENTS AND METHODS
Study Population
Women (≥ 18 years old) were eligible if they were on tamoxifen 20 mg daily for at least 4 months (ensuring steady-state concentrations), nonpregnant/nonlactating, had an Eastern Cooperative Oncology Group performance status of 0 to 2, normal kidney, liver, and bone marrow function. Patients were excluded if they had a history of thromboembolic disease, prior liver or bone marrow transplant, or had received blood product transfusions within the past 3 months. Concurrent medication therapy with medications known to inhibit CYP2D6 was not permitted (ie, buproprion, amiodarone, haloperidol, indinavir, ritonavir, quinidine, duloxetine, paroxetine, or fluoxetine). The protocol was approved at each respective institutional review board and all participants provided written informed consent.
CYP2D6 Genotyping
CYP2D6 genotyping was performed in a Clinical Laboratory Improved Amendments–certified clinical laboratory using the AmpliChip CYP450 test (Roche Diagnostics, Indianapolis, IN) for the major CYP2D6 alleles (Data Supplement). The turnaround time for the CYP2D6 genotyping was 1 to 2 weeks to patient and physician notification of their genotype group.
Intervention
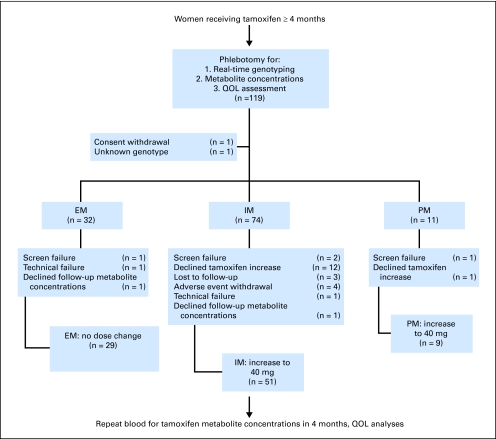
After enrollment, participants underwent phlebotomy to obtain specimens for CYP2D6 genotype and tamoxifen metabolite assessment (Fig 1). For dosing purposes, patients were classified as EM, IM, or PM based on their respective genotypes. Patients with two CYP2D6 alleles with normal metabolic activity (EM/EM) were considered to be EM and were instructed to remain on their dose of tamoxifen 20 mg daily, as was one patient with an ultrarapid metabolism allele. Patients with one or more CYP2D6 alleles with reduced enzymatic activity or heterozygous for an inactive allele were considered IM (EM/IM, EM/PM, IM/IM, IM/PM); those with two inactive alleles were considered PM (PM/PM). Patients with IM or PM genotypes were instructed to increase their tamoxifen to 40 mg daily (given as 20 mg twice daily). Adherence to therapy was recorded via a self-reported pill diary. Quality of life (QOL) assessment (including hot flashes) was performed using the Functional Assessment of Cancer Therapy Endocrine Subscale (FACT-B [es]) and the Breast Cancer Prevention Trial Menopausal Symptom Scale (BCPT-MSS) at time of study consent.39,40 After 4 months ± 4 weeks of genotype directed therapy, tamoxifen metabolite concentrations were repeated and toxicity and QOL measures were assessed using the National Cancer Institute Common Toxicity Criteria for Adverse Events, version 3.0 and the FACT- B (es) and BCPT-MSS. At the end of protocol-directed therapy, the nature of further endocrine therapy was at the discretion of the patient and treating physician.
Fig 1.
CONSORT diagram. EM, extensive metabolizers; IM, intermediate metabolizers; PM, poor metabolizers; QOL, quality of life.
Tamoxifen and Metabolite Measurement in Plasma
Tamoxifen and its metabolites were measured by liquid chromatography tandem mass spectrometry (LC-MS/MS). Standard curves and quality controls were run with each sample batch. The intraday coefficient of variance was lower than 10% for all compounds, while the interday was lower than 15%. An intrapatient assay was not run for each sample set, but repeated measurement was performed in a subset of samples. The results show that the intrasample variability was lower than 10%. The Data Supplement details a full description of this method.
Statistical Methods
The primary objective was to evaluate change in plasma endoxifen concentrations after an increase in tamoxifen dose from 20 mg to 40 mg among patients with IM CYP2D6 genotypes. Endoxifen concentrations were measured at baseline and after 4 months on tamoxifen. A one-sample Wilcoxon test was used to test the null hypothesis that the change in endoxifen concentration from baseline equals zero. Pair-wise two-sample Wilcoxon rank sum test was used to test the difference in endoxifen concentration among the three genotype groups (EM, IM, and PM). We posited that a potentially clinically meaningful end point would be to achieve the same endoxifen concentration in IM patients on a higher tamoxifen dose as EM patients on 20 mg per day. Assuming that endoxifen concentrations are 40% higher in EM than IM patients,7,11 12% nonevaluability, a two-sided significance level of .05, and a sample size of 40 patients with IM CYP2D6 genotypes, 100 patients would provide 84% power.41 Actual enrollment was 119 because at the time of accrual completion 19 patients were in equal stages of screening. The tests were repeated for tamoxifen, N-desmethyltamoxifen, 4-hydroxytamoxifen and for the endoxifen/N-desmethlytamoxifen ratio. The Data Supplement details the generalized estimating equation (GEE) method.
Patient responses to the survey questions in the FACT-B (es) and BCPT-MSS at baseline and 4 months after the treatment were reported. A proportional odds model being fit with the GEE method42 was used to evaluate the changes in response over the two time points and for the three genotype groups. The outcome scores used are in an ordinal scale, ranging from 0 “not at all” to 4 “very much.”
Unless noted, Fisher's exact test was used to evaluate baseline characteristic differences between genotype groups. All analyses were performed using SAS statistical software, version 9.2 (SAS Institute Inc, Cary, NC).
RESULTS
Patients and Genotyping
A total of 119 patients were enrolled in this study; one withdrew consent immediately after registration, one had an allele of unknown significance (CYP2D6*25) and was excluded from endoxifen concentration analyses, and 28 withdrew before having their 4-month metabolite concentrations drawn, leaving 89 patients for the analysis of the tamoxifen dose intervention (Table 1; Fig 1). Table 2 presents the CYP2D6 allele frequencies from our population. A higher than expected proportion of patients (72%) had at least one PM or IM allele. The odds of an African American having a CYP2D6 allele consistent with reduced metabolism were about two times higher than those in other (mostly white) patients (odds ratio [OR], 2.26; 95% CI, 1.17 to 4.37).
Table 1.
Baseline Characteristics (n = 89), Grouped by CYP2D6 Metabolism Phenotype, and No. of Patients With Each CYP2D6 Combination Grouped by CYP2D6 Metabolism Phenotype (n = 89)
| Characteristic | EM (n = 29) | IM (n = 51) | PM (n = 9) | P |
|---|---|---|---|---|
| Median age, years | 49 | 49 | 47 | .587* |
| Range | 35-77 | 35-72 | 36-56 | |
| Race/ethnicity | .738 | |||
| White | 22 | 40 | 6 | |
| African American | 4 | 9 | 3 | |
| Asian | 1 | 2 | — | |
| Hispanic | 2 | — | — | |
| Median tamoxifen duration, years | 0.7 | 1.3 | 1.0 | .055* |
| Range | 0.3-4.4 | 0.3-5.0 | 0.5-3.0 | |
| Menopause | .892 | |||
| Pre/peri | 16 | 30 | 4 | |
| Post | 13 | 21 | 5 | |
| SSRI/SNRI use | .489 | |||
| Total | 5 | 16 | 1 | |
| Citalopram | 3 | 4 | — | |
| Venlafaxine | 1 | 9 | 1 | |
| Escitalopram | 1 | 3 | — | |
| Reason on tamoxifen | .07 | |||
| Invasive carcinoma | 28 | 41 | 9 | |
| DCIS | 1 | 10 | — | |
| Prior chemotherapy | .041 | |||
| Yes | 22 | 27 | 8 | |
| No | 7 | 24 | 1 | |
| EM | EM/UM = 1; 1.1% | |||
| EM/EM = 28; 31.5% | ||||
| IM | EM/IM = 20; 22.5% | |||
| EM/PM = 19; 21.3% | ||||
| IM/IM = 4; 4.5% | ||||
| IM/PM = 8; 9.0% | ||||
| PM | PM/PM = 9; 10.1% |
Abbreviations: DCIS, ductal carcinoma in situ; EM, extensive metabolizer; IM, intermediate metabolizer; PM, poor metabolizer; SNRI, serotonin-norepinephrine reuptake inhibitor; SSRI, selective serotonin reuptake inhibitor.
The Kruskal-Wallis test was used to test the shift of location from the median value.
Table 2.
CYP2D6 Allele Frequencies (n = 118)
| Allele | EM |
PM *3-6 | IM |
Other | |||||
|---|---|---|---|---|---|---|---|---|---|
| *1, *2 | *35 | *9 | *10 | *17 | *29 | *41 | |||
| Total patients (n = 118) | 0.48 | 0.02 | 0.25 | 0.02 | 0.03 | 0.04 | 0.02 | 0.13 | .02 |
| African American patients (n = 25) | 0.32 | 0 | 0.22 | 0 | 0 | 0.18 | 0.08 | 0.14 | .06 |
| Non-African American patients (n = 93) | 0.53 | 0.02 | 0.25 | 0.02 | 0.03 | 0.01 | 0 | 0.13 | .01 |
Abbreviations: EM, extensive metabolizer; IM, intermediate metabolizer; PM, poor metabolizer.
Adherence to Protocol
For the patients completing protocol-directed therapy, 51% (45 of 89) did not report missing any tamoxifen doses. Of those that reported missed doses, 66% (29 of 44) reported missing five or fewer doses. Overall, 93% (83 of 89) reported missing 10 or fewer doses over the 4-month period. Six patients (six of 89; 7%) reported missing 12 to 27 doses. The Data Supplement shows reported adherence by genotype group.
Endoxifen Concentrations
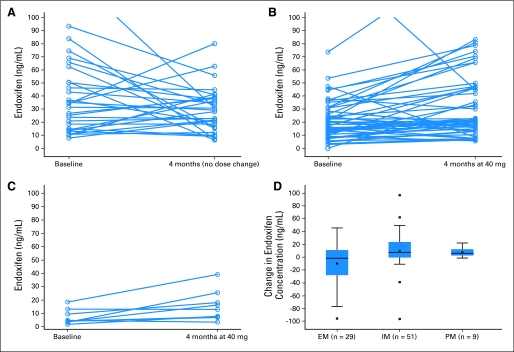
The results are summarized in Table 3. In the univariate analysis the median baseline endoxifen concentration was significantly higher in the EM patients compared with either IM (46% higher; P = .0045), or PM (88% higher; P < .001) patients, and the median endoxifen concentration in PM was 77% lower than in IM (P = .0006). Thus, as expected, there was a statistically significant baseline difference among the underlying distributions in the three genotype groups. There was no statistical difference between baseline endoxifen concentrations and concentrations 4 months later among the EM group (tamoxifen 20 mg/day; P = .25). The tamoxifen dose increase from 20 mg to 40 mg resulted in a significant rise in endoxifen concentrations in both IM (P < .001) and PM (P = .020) patients. After 4 months, there was no significant difference in median endoxifen concentrations between EM taking 20 mg and IM taking 40 mg (P = .84) even though the baseline median endoxifen concentration was almost twice as great for the EM than for the IM group. The median endoxifen concentration in the PM group was still significantly lower than in the EM (P = .016) and IM (P = .019) groups (Figs 3A and 3B, 3C, 3D). Based on three multiple comparisons, the Bonferroni P value is .017. The Data Supplement shows the results of the IM patients split into EM/IM, IM/IM, IM/PM, and EM/PM. Median endoxifen concentrations increased within all IM subsets (significant for EM/IM and EM/PM). Splitting these groups did not explain the interpatient variability seen in response to the tamoxifen dose increase, although the numbers in each subset are small.
Table 3.
Median Endoxifen Concentrations for the Three CYP2D6 Genotype Groups and Change Over Time (EM patients no change in dose; IM and PM patients increased tamoxifen dose)
| Genotype Category | No. Evaluable at Baseline | Median Endoxifen Baseline* (ng/mL) | No. Completing Protocol-Driven Therapy | Median Endoxifen Baseline (ng/mL) | Median Endoxifen 4 Months Later† (ng/mL) | Intrapatient Change From Baseline (ng/mL) |
P‡ | |
|---|---|---|---|---|---|---|---|---|
| Median | Interquartile Range | |||||||
| EM | 32 | 34.9 | 29 | 34.3 | 29.2 | −1.5 | −28 to 11.2 | .25 |
| IM | 74 | 19.8 | 51 | 18.5 | 21.8 | +7.6 | −0.6 to 23.9 | < .001 |
| PM | 11 | 4.6 | 9 | 4.2 | 12.9 | +6.1 | 2.6 to 12.5 | .020 |
Abbreviations: EM, extensive metabolizer; IM, intermediate metabolizer; PM, poor metabolizer.
Medians of the three groups are significantly different from each other with EM v IM: P = .0054; EM v PM: P < .001; IM v PM: P < .001.
Represents aggregate data within genotype groups, not intrapatient data.
Relates to the median intrapatient change from baseline.
Fig 3.
Endoxifen concentration (ng/mL) change over time, demonstrating (A) no significant difference in extensive metabolizers (EM; P = .25; no dose change); (B) a significant increase (P < .001) in endoxifen after 4 months of increased tamoxifen in intermediate metabolizers (IM); and (C) a significant increase (P = .020) in endoxifen after 4 months of increased tamoxifen in poor metabolizers (PM), although still lower than other groups. (D) Box and whisker plot of change over time of endoxifen concentration by genotype group (EM, IM, PM), demonstrating an increase in endoxifen concentration in the groups with increased dose (IM, PM; P < .001 and P = .020, respectively). The two ends of the boxes indicate the upper and lower quartiles, the median is the middle line, the mean is the hollow square within the boxes, and the whiskers represent the upper and lower fence of the data. Observations outside the fences are identified as outliers with a square. The EM/ultrarapid metabolizer (UM) patient's value lies within the ends of the boxes (between the upper and lower quartiles) for the EM group (change of −17 ng/mL). (A) Value outside the figure for baseline = 126 ng/mL. The EM/UM patient had a baseline endoxifen concentration of 37 ng/mL and a 4-month concentration of 20 ng/mL. (B) Value outside range of figure for 4 months = 170 ng/mL and value outside figure for baseline = 141 ng/mL.
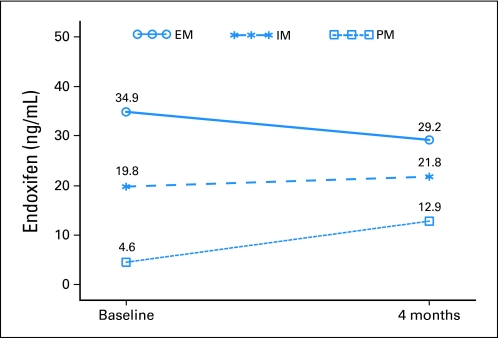
In order to address the pattern of change in median endoxifen concentration from baseline, the GEE approach was used for the repeated measurements analyses. The results suggested that the IM group had a significant increase (+7.6 ng/mL; P = .0062) in endoxifen concentration from baseline to 4 months later when compared to the EM group (Fig 4). There was also a convergence of IM and EM with the former increasing significantly and a nonsignificant but numerical decrease in the latter. While the majority of EM patients had no notable change in endoxifen concentration, eight patients had a marked decrease that is poorly understood (all reported complete adherence). Eight EM patients had 4-month endoxifen concentrations lower than 20 ng/mL (comparable to PM, six reporting complete adherence). Five EM patients had baseline endoxifen concentrations lower than 20 ng/mL that increased at 4 months (no prestudy adherence data was obtained). As anticipated, an increase in dose for the PM resulted in a significant pattern of change (P = .0035) in median endoxifen concentration when compared to EM, but not between IM and PM. The data for tamoxifen, N-desmethyltamoxifen, 4-hydroxytamoxifen concentrations, and the endoxifen/N-desmethyltamoxifen ratio are located in the Data Supplement.
Fig 4.
Median endoxifen concentration by genotype group for the generalized estimating equation shown to address the pattern of change over time for the three genotype groups. The intermediate metabolizers (IM) group had a significant increase in endoxifen concentration when compared to the extensive metabolizers (EM) group (P = .0062). The poor metabolizers (PM) group also had a significant pattern of change (P = .0035) when compared to the EM group.
Toxicity/QOL
No thromboembolic events were seen. There was one episode of grade 3 vaginal bleeding (IM patient, 1 month into the double dose, endometrial biopsy negative for carcinoma). All other adverse events were lower than grade 3, including the three other reasons for withdrawal (ie, nausea, cramping, joint aches). There was no evidence of a difference of mean hot flash scores among groups at baseline (EM, 2.3; IM, 1.9; PM, 2.5; P = .18), or at 4 months (EM, 1.9; IM, 1.9; PM, 2.3; P = .71), or over time irrespective of dose (Data Supplement). The other measured endocrine symptoms for the FACT-B (es) and the BCPT-MSS are described in the Data Supplement, and with few exceptions, do not show a difference between genotype groups or between patients who received different doses of tamoxifen (based on 180 multiple comparisons, the Bonferroni P value is .00028). There were no associations between hot flashes and endoxifen concentrations at baseline (P = .97) or at 4 months (P = .14; Data Supplement).
DISCUSSION
This study demonstrates the feasibility of genotype-driven dosing of tamoxifen for the substantial proportion of women who are intermediate metabolizers of the agent. Like others, we noted good correlation between baseline plasma endoxifen concentrations and CYP2D6 genotype. Our data suggest that doubling the dose of tamoxifen can raise endoxifen concentrations in IM patients to the extent that the differences between EM and IM concentrations are no longer statistically significant. While the endoxifen concentrations in PM patients also increased, these increases were more modest than in the IM group, and the final endoxifen concentration in these patients remained significantly lower than in the other two groups. That the endoxifen concentrations increased at all in PM patients is interesting, and may reflect metabolism by other enzymes in the pathway. This question will be addressed in a recently completed 500 patient expansion study.
These data address one aspect of the clinical utility of genotype-guided dosing, namely that in the large group of patients who possess intermediate metabolizing genes, it is possible to at least partly compensate for inherited inadequate metabolism by changing the dose. We found that the 40 mg tamoxifen dose is safe, US Food and Drug Administration approved, and efficacious. If further studies of tamoxifen dosing are pursued, far higher doses may be considered,43 however, extrapolating from our data in the PM group and assuming a linear relationship, increasing the endoxifen concentrations in the PM group to those in the EM group may take a dose more than 100 mg to achieve. It should be noted that at high tamoxifen doses, an increase in retinopathy has been documented.44 However, this study was not designed to demonstrate that actively managing endoxifen concentrations will improve outcomes. It is not clear that CYP2D6 by itself can predict outcomes; two recent studies in postmenopausal women did not find an association.35,36 Tamoxifen and the non-CYP2D6 metabolites produce greater than 99% ER saturation in postmenopausal women45 possibly making endoxifen activity less relevant. Whether study of additional enzymes or a focus on premenopausal women will demonstrate relevance for outcome is yet unknown. While some initial studies did demonstrate worse outcomes (DFS or time to progression) for those who are CYP2D6 IM or PM on tamoxifen compared to EM,20–33 this finding has never been consistent.12–19,35,36 Evidence that supports the effect of tamoxifen metabolism on breast cancer outcomes comes from drug-interaction studies that demonstrate concurrent use of medications (especially selective-serotonin reuptake inhibitors) that inhibit CYP2D6 reduce DFS,24,27,33,34 although this is also somewhat controversial.35 Given that the impact of these CYP2D6/drug interactions is to decrease endoxifen concentrations, our findings may have implications for patients on CYP2D6-interacting drugs who cannot change drugs and are appropriate for tamoxifen; however the ability to alter endoxifen concentrations in the setting of CYP2D6 inhibitors is not established here.
Our study is one of two tamoxifen CYP2D6 studies15 that included a significant percentage of African American patients. We found that the allele frequency of reduced metabolism alleles was greater in African Americans than in other patients with the IM allele *17 being common in African Americans. This may partially explain the disproportionally worse outcome suffered by premenopausal African American women with breast cancer.46
We found no association between endoxifen concentrations and hot flashes, nor between genotype group or dose of tamoxifen and hot flashes. However, a study size of 500 would be required to identify a 10% difference in hot flash scores, larger than the study presentedhere. Although some have suggested that endocrine symptoms, especially hot flashes, can be used as a proxy for endoxifen concentrations,47,48 we would recommend caution until further data becomes available in using vasomotor symptoms to determine the efficacy of tamoxifen. In general, physicians should be cautious when discussing emerging genomic markers with patients. In 2009, a community-based survey suggested that nearly one in three oncologists had ordered CYP2D6 genotyping.49 Our data suggest that participants in pharmacogenomic trials have a high expectation of benefit despite emphasis in informed consent documents of the scientific purpose of the research and the uncertainty of any direct benefit.50
A clear limitation of this study is the lack of clinical outcome data. To demonstrate a clinical impact of genotype-driven dosing of tamoxifen, a prospective, randomized controlled trial of different doses of tamoxifen for IMs and possibly PMs with clinical end points would be needed. While our study was predicated on genotype-driven dosing, trying to dose to a particular endoxifen concentration might be more efficacious. However, the precise clinically relevant endoxifen concentration is unknown, and efficacy may not be solely mediated via endoxifen. While genotype-driven therapy may impact outcome, the impact is likely in the increase of the active metabolite concentration and thus in the end, the directed therapy may be the endoxifen concentration.
While the majority of EM patients had no notable change in endoxifen concentration, eight patients had a marked decrease and eight had PM-like concentrations that are poorly understood. Adherence was self-reported, which has clear limitations,51 and up to 68% of tamoxifen patients are nonadherent.52 Also, large increases were seen in about one third of the IM patients, while marginal increases were seen in others, a phenomenon that is also poorly understood. Standard curves and quality controls were run with each sample batch with very good agreement between these runs. Although we did not run an intrapatient assay for each sample, the assay is robust and in a subset of samples the results showed low intrasample variability. To obtain a complete picture of tamoxifen metabolism and for individualized dosing to be relevant, one needs to examine the effect of alterations in other genes that metabolize tamoxifen (or eliminate endoxifen such as the UDP-glucuronosyltranferases) as these genes may also be impacting endoxifen concentrations, and may be responsible for the low endoxifen concentrations seen in some EM patients.
It is possible that the lack of statistical significance in endoxifen concentration between the IM group at 40 mg and the EM group is due to the sample size. This study had 80% power to detect a 40% difference in endoxifen concentrations, making the lack of an adequate sample size an unlikely explanation for this finding. The expansion study mentioned above will have power higher than 99%.
Endoxifen is a highly active tamoxifen metabolite, and this study suggests that pharmacogenomic knowledge can be successfully used to manage drug concentrations. CYP2D6 testing does not currently meet evidence for clinical use in that endoxifen concentration may not affect outcome, or it may matter in some as-yet unidentified subsets but not in others.
Supplementary Material
Footnotes
See accompanying editorial on page 3206 and article on page 3240; listen to the podcast by Dr Leyland-Jones at www.jco.org/podcasts
Supported by Grant No. CA58223 from the National Cancer Institute Specialized Programs of Research Excellence, North Carolina University Cancer Research Fund, University of North Carolina at Chapel Hill Investments for the Future Grant No. 6231, Laboratory Corporation of America, Roche Diagnostics, American Society of Clinical Oncology Foundation and Breast Cancer Research Foundation, National Institute of General Medical Sciences Pharmacogenomics Research Network Award (U-01GM061373) to the Consortium on Breast Cancer Pharmacogenomics.
Presented in part in poster format at the 45th Annual Meeting of the American Society of Clinical Oncology, Orlando, FL, May 29-June 2, 2009; the 2009 Breast Cancer Symposium, San Francisco, CA, October 9-10, 2009; and the San Antonio Breast Cancer Symposium, San Antonio, TX, December 9-13, 2009.
Authors' disclosures of potential conflicts of interest and author contributions are found at the end of this article.
Clinical trial information can be found for the following: NCT00764322.
AUTHORS' DISCLOSURES OF POTENTIAL CONFLICTS OF INTEREST
Although all authors completed the disclosure declaration, the following author(s) indicated a financial or other interest that is relevant to the subject matter under consideration in this article. Certain relationships marked with a “U” are those for which no compensation was received; those relationships marked with a “C” were compensated. For a detailed description of the disclosure categories, or for more information about ASCO's conflict of interest policy, please refer to the Author Disclosure Declaration and the Disclosures of Potential Conflicts of Interest section in Information for Contributors.
Employment or Leadership Position: Steven M. Anderson, Laboratory Corporation of America (C); Kenneth J. Friedman, Laboratory Corporation of America (C) Consultant or Advisory Role: Karen E. Weck, Roche Diagnostics (C); Jeffrey M. Peppercorn, Novartis (C), Genetech (C); David A. Flockhart, Medco Health Solutions (C), Coriell (C); Howard L. McLeod, Myriad Genetics (C), Federal Drug Administration (C), Medco Health Solutions (C), Gentris (C) Stock Ownership: Steven M. Anderson, Laboratory Corporation of America Honoraria: Jeffrey M. Peppercorn, Novartis, Genetech Research Funding: Jeffrey M. Peppercorn, Novartis; David A. Flockhart, Pfizer, Novartis; Howard L. McLeod, Myriad Genetics Expert Testimony: None Other Remuneration: None
AUTHOR CONTRIBUTIONS
Conception and design: William J. Irvin Jr, Christine M. Walko, Karen E. Weck, Joseph G. Ibrahim, E. Claire Dees, Jeffrey M. Peppercorn, Steven M. Anderson, David A. Flockhart, Howard L. McLeod, James P. Evans, Lisa A. Carey
Financial support: Karen E. Weck, James P. Evans, Lisa A. Carey
Administrative support: William J. Irvin Jr, Steven M. Anderson
Provision of study materials or patients: William J. Irvin Jr, E. Claire Dees, Susan G. Moore, Oludamilola A. Olajide, Mark L. Graham, Sean T. Canale, Rachel E. Raab, Steven W. Corso, Lisa A. Carey
Collection and assembly of data: William J. Irvin Jr, Karen E. Weck, Joseph G. Ibrahim, Steven M. Anderson, Kenneth J. Friedman, Evan T. Ogburn, Zeruesenay Desta, Howard L. McLeod, Lisa A. Carey
Data analysis and interpretation: William J. Irvin Jr, Christine M. Walko, Karen E. Weck, Joseph G. Ibrahim, Wing K. Chiu, E. Claire Dees, Jeffrey M. Peppercorn, Steven M. Anderson, Kenneth J. Friedman, Zeruesenay Desta, David A. Flockhart, Howard L. McLeod, Lisa A. Carey
Manuscript writing: William J. Irvin Jr, Christine M. Walko, Karen E. Weck, Joseph G. Ibrahim, Wing K. Chiu, E. Claire Dees, Susan G. Moore, Oludamilola A. Olajide, Mark L. Graham, Sean T. Canale, Rachel E. Raab, Steven W. Corso, Jeffrey M. Peppercorn, Steven M. Anderson, Kenneth J. Friedman, Evan T. Ogburn, Zeruesenay Desta, David A. Flockhart, Howard L. McLeod, James P. Evans, Lisa A. Carey
Final approval of manuscript: William J. Irvin Jr, Christine M. Walko, Karen E. Weck, Joseph G. Ibrahim, Wing K. Chiu, E. Claire Dees, Susan G. Moore, Oludamilola A. Olajide, Mark L. Graham, Sean T. Canale, Rachel E. Raab, Steven W. Corso, Jeffrey M. Peppercorn, Steven M.Anderson, Kenneth J. Friedman, Evan T. Ogburn, Zeruesenay Desta, David A. Flockhart, Howard L. McLeod, James P. Evans, Lisa A. Carey
REFERENCES
- 1.Surveillance, Epidemiology, and End Results. Cancer of the Breast. In: Ries LAG, Melbert D, Krapcho M, et al., editors. SEER Stat Fact Sheets. Bethesda, MD: National Cancer Institute; 2008. [Google Scholar]
- 2.Hoskins JM, Carey LA, McLeod HL. CYP2D6 and tamoxifen: DNA matters in breast cancer. Nat Rev Cancer. 2009;9:576–586. doi: 10.1038/nrc2683. [DOI] [PubMed] [Google Scholar]
- 3.Lien EA, Solheim E, Kvinnsland S, et al. Identification of 4-hydroxy-N-desmethyltamoxifen as a metabolite of tamoxifen in human bile. Cancer Res. 1988;48:2304–2308. [PubMed] [Google Scholar]
- 4.Desta Z, Ward BA, Soukhova NV, et al. Comprehensive evaluation of tamoxifen sequential biotransformation by the human cytochrome P450 system in vitro: Prominent roles for CYP3A and CYP2D6. J Pharmacol Exp Ther. 2004;310:1062–1075. doi: 10.1124/jpet.104.065607. [DOI] [PubMed] [Google Scholar]
- 5.Goetz MP, Rae JM, Suman VJ, et al. Pharmacogenetics of tamoxifen biotransformation is associated with clinical outcomes of efficacy and hot flashes. J Clin Oncol. 2005;23:9312–9318. doi: 10.1200/JCO.2005.03.3266. [DOI] [PubMed] [Google Scholar]
- 6.Lim YC, Desta Z, Flockhart DA, et al. Endoxifen (4-hydroxy-N-desmethyl-tamoxifen) has anti-estrogenic effects in breast cancer cells with potency similar to 4-hydroxy-tamoxifen. Cancer Chemother Pharmacol. 2005;55:471–478. doi: 10.1007/s00280-004-0926-7. [DOI] [PubMed] [Google Scholar]
- 7.Stearns V, Johnson MD, Rae JM, et al. Active tamoxifen metabolite plasma concentrations after coadministration of tamoxifen and the selective serotonin reuptake inhibitor paroxetine. J Natl Cancer Inst. 2003;95:1758–1764. doi: 10.1093/jnci/djg108. [DOI] [PubMed] [Google Scholar]
- 8.Lim YC, Li L, Desta Z, et al. Endoxifen, a secondary metabolite of tamoxifen, and 4-OH-tamoxifen induce similar changes in global gene expression patterns in MCF-7 breast cancer cells. J Pharmacol Exp Ther. 2006;318:503–512. doi: 10.1124/jpet.105.100511. [DOI] [PubMed] [Google Scholar]
- 9.Johnson MD, Zuo H, Lee KH, et al. Pharmacological characterization of 4-hydroxy-N-desmethyl tamoxifen, a novel active metabolite of tamoxifen. Breast Cancer Res Treat. 2004;85:151–159. doi: 10.1023/B:BREA.0000025406.31193.e8. [DOI] [PubMed] [Google Scholar]
- 10.Bradford LD. CYP2D6 allele frequency in European Caucasians, Asians, Africans and their descendants. Pharmacogenomics. 2002;3:229–243. doi: 10.1517/14622416.3.2.229. [DOI] [PubMed] [Google Scholar]
- 11.Jin Y, Desta Z, Stearns V, et al. CYP2D6 genotype, antidepressant use, and tamoxifen metabolism during adjuvant breast cancer treatment. J Natl Cancer Inst. 2005;97:30–39. doi: 10.1093/jnci/dji005. [DOI] [PubMed] [Google Scholar]
- 12.Dezentje VO, van Blijderveen NJ, Gelderblom H, et al. Effect of concomitant CYP2D6 inhibitor use and tamoxifen adherence on breast cancer recurrence in early-stage breast cancer. J Clin Oncol. 2010;28:2423–2429. doi: 10.1200/JCO.2009.25.0894. [DOI] [PubMed] [Google Scholar]
- 13.Goetz MP, Berry DA, Klein TE. Adjuvant tamoxifen treatment outcome according to cytochrome P450 2D6 (CYP2D6) phenotype in early stage breast cancer: Findings from the International Tamoxifen Pharmacogenomics Consortium. Presented at the San Antonio Breast Cancer Symposium; December 9-13, 2009; San Antonio, TX. abstr 33. [Google Scholar]
- 14.Okishiro M, Taguchi T, Jin Kim S, et al. Genetic polymorphisms of CYP2D6 10 and CYP2C19 2, 3 are not associated with prognosis, endometrial thickness, or bone mineral density in Japanese breast cancer patients treated with adjuvant tamoxifen. Cancer. 2009;115:952–961. doi: 10.1002/cncr.24111. [DOI] [PubMed] [Google Scholar]
- 15.Nowell SA, Ahn J, Rae JM, et al. Association of genetic variation in tamoxifen-metabolizing enzymes with overall survival and recurrence of disease in breast cancer patients. Breast Cancer Res Treat. 2005;91:249–258. doi: 10.1007/s10549-004-7751-x. [DOI] [PubMed] [Google Scholar]
- 16.Wegman P, Elingarami S, Carstensen J, et al. Genetic variants of CYP3A5, CYP2D6, SULT1A1, UGT2B15 and tamoxifen response in postmenopausal patients with breast cancer. Breast Cancer Res. 2007;9:R7. doi: 10.1186/bcr1640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wegman P, Vainikka L, Stal O, et al. Genotype of metabolic enzymes and the benefit of tamoxifen in postmenopausal breast cancer patients. Breast Cancer Res. 2005;7:R284–90. doi: 10.1186/bcr993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Dezentje VO, Van Schaik RH, Vletter-Bogaatz JM, et al. Pharmacogenetics of tamoxifen in relation to disease-free survival in a Dutch cohort of the tamoxifen exemestane adjuvant multinational (TEAM) trial. J Clin Oncol. 2010;28:70s. abstr 510. [Google Scholar]
- 19.Ramon y Cajal T, Altes A, Pare L, et al. Impact of CYP2D6 polymorphisms in tamoxifen adjuvant breast cancer treatment. Breast Cancer Res Treat. 2009;119:33–38. doi: 10.1007/s10549-009-0328-y. [DOI] [PubMed] [Google Scholar]
- 20.Bijl MJ, van Schaik RH, Lammers LA, et al. The CYP2D6*4 polymorphism affects breast cancer survival in tamoxifen users. Breast Cancer Res Treat. 2009;118:125–130. doi: 10.1007/s10549-008-0272-2. [DOI] [PubMed] [Google Scholar]
- 21.Goetz MP, Suman V, Ames MM, et al. Tamoxifen pharmacogenetics of CYP2D6, CYP2C19, and SULT1A1: Long-term follow-up of the North Central Cancer Treatment Group 89-30-52 adjuvant trial. Presented at the San Antonio Breast Cancer Symposium; December 10-14, 2008; San Antonio, TX. abstr 6037. [Google Scholar]
- 22.Schroth W, Antoniadou L, Fritz P, et al. Breast cancer treatment outcome with adjuvant tamoxifen relative to patient CYP2D6 and CYP2C19 genotypes. J Clin Oncol. 2007;25:5187–5193. doi: 10.1200/JCO.2007.12.2705. [DOI] [PubMed] [Google Scholar]
- 23.Kiyotani K, Mushiroda T, Sasa M, et al. Impact of CYP2D6*10 on recurrence-free survival in breast cancer patients receiving adjuvant tamoxifen therapy. Cancer Sci. 2008;99:995–999. doi: 10.1111/j.1349-7006.2008.00780.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gonzalez-Santiago S, Zárate R, Haba-Rodriguez J, et al. CYP2D6*4 polymorphism as blood predictive biomarker of breast cancer relapse in patients receiving adjuvant tamoxifen. J Clin Oncol. 2007;25:25s. abstr 590. [Google Scholar]
- 25.Newman WG, Hadfield KD, Latif A, et al. Impaired tamoxifen metabolism reduces survival in familial breast cancer patients. Clin Cancer Res. 2008;14:5913–5918. doi: 10.1158/1078-0432.CCR-07-5235. [DOI] [PubMed] [Google Scholar]
- 26.Xu Y, Sun Y, Yao L, et al. Association between CYP2D6 *10 genotype and survival of breast cancer patients receiving tamoxifen treatment. Ann Oncol. 2008;19:1423–1429. doi: 10.1093/annonc/mdn155. [DOI] [PubMed] [Google Scholar]
- 27.Goetz MP, Knox SK, Suman VJ, et al. The impact of cytochrome P450 2D6 metabolism in women receiving adjuvant tamoxifen. Breast Cancer Res Treat. 2007;101:113–121. doi: 10.1007/s10549-006-9428-0. [DOI] [PubMed] [Google Scholar]
- 28.Goetz MP, Suman VJ, Couch FJ, et al. Cytochrome P450 2D6 and homeobox 13/interleukin-17B receptor: Combining inherited and tumor gene markers for prediction of tamoxifen resistance. Clin Cancer Res. 2008;14:5864–5868. doi: 10.1158/1078-0432.CCR-08-0619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kiyotani K, Mushiroda T, Imamura CK, et al. Significant effect of polymorphisms in CYP2D6 and ABCC2 on clinical outcomes of adjuvant tamoxifen therapy for breast cancer patients. J Clin Oncol. 2010;28:1287–1293. doi: 10.1200/JCO.2009.25.7246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lim HS, Ju Lee H, Seok Lee K, et al. Clinical implications of CYP2D6 genotypes predictive of tamoxifen pharmacokinetics in metastatic breast cancer. J Clin Oncol. 2007;25:3837–3845. doi: 10.1200/JCO.2007.11.4850. [DOI] [PubMed] [Google Scholar]
- 31.Thompson A, Quinlan P, Bray S, et al. CYP2D6 genotype affects outcome in post-menopausal breast cancer patients treated with tamoxifen monotherapy. Presented at the 2009 Breast Cancer Symposium; December 9-13, 2009; San Antonio, TX. abstr 35. [Google Scholar]
- 32.Schroth W, Goetz MP, Hamann U, et al. Association between CYP2D6 polymorphisms and outcomes among women with early stage breast cancer treated with tamoxifen. JAMA. 2009;302:1429–1436. doi: 10.1001/jama.2009.1420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Aubert RE, Stanek EJ, Yao J, et al. Risk of breast cancer recurrence in women initiating tamoxifen with CYP2D6 inhibitors. J Clin Oncol. 2009;27(suppl):9s. abstr CRA508. [Google Scholar]
- 34.Kelly CM, Juurlink DN, Gomes T, et al. Selective serotonin reuptake inhibitors and breast cancer mortality in women receiving tamoxifen: A population based cohort study. BMJ. 2010;340:c693. doi: 10.1136/bmj.c693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Rae JM, Drury S, Hayes DF, et al. Lack of correlation between gene variants in tamoxifen metabolizing enzymes with primary endpoints in the ATAC trial. Presented at the San Antonio Breast Cancer Symposium; December 8-12, 2010; San Antonio, TX. abstr S1-7. [Google Scholar]
- 36.Leyland-Jones B, Regan MM, Bouzyk M, et al. Outcome according to CYP2D6 genotype among postmenopausal women with endocrine-responsive early invasive breast cancer randomized in the BIG 1-98 trial. Presented at the San Antonio Breast Cancer Symposium Abstract; December 8-12, 2010; San Antonio, TX. #S1-8. [Google Scholar]
- 37.Bratherton DG, Brown CH, Buchanan R, et al. A comparison of two doses of tamoxifen (Nolvadex) in postmenopausal women with advanced breast cancer: 10 mg bd versus 20 mg bd. Br J Cancer. 1984;50:199–205. doi: 10.1038/bjc.1984.163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ward HW. Anti-oestrogen therapy for breast cancer: A trial of tamoxifen at two dose levels. BMJ. 1973;1:13–14. doi: 10.1136/bmj.1.5844.13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Fallowfield LJ, Leaity SK, Howell A, et al. Assessment of quality of life in women undergoing hormonal therapy for breast cancer: Validation of an endocrine symptom subscale for the FACT-B. Breast Cancer Res Treat. 1999;55:189–199. doi: 10.1023/a:1006263818115. [DOI] [PubMed] [Google Scholar]
- 40.Stanton AL, Bernaards CA, Ganz PA. The BCPT symptom scales: A measure of physical symptoms for women diagnosed with or at risk for breast cancer. J Natl Cancer Inst. 2005;97:448–456. doi: 10.1093/jnci/dji069. [DOI] [PubMed] [Google Scholar]
- 41.Noether GE. Sample size determination for some common nonparametric tests. J Am Stat Assoc. 1987;82:645–647. [Google Scholar]
- 42.Stokes ME, Davis CS, Kock G. ed 2. Cary, NC: SAS Institute Inc; 2000. Categorical Data Analysis Using the SAS System; pp. 514–518. [Google Scholar]
- 43.Stahl M, Wilke H, Schmoll HJ, et al. A phase II study of high dose tamoxifen in progressive, metastatic renal cell carcinoma. Ann Oncol. 1992;3:167–168. doi: 10.1093/oxfordjournals.annonc.a058136. [DOI] [PubMed] [Google Scholar]
- 44.Kaiser-Kupfer MI, Lippman ME. Tamoxifen retinopathy. Cancer Treat Rep. 1978;62:315–320. [PubMed] [Google Scholar]
- 45.Dowsett M, Haynes BP. Hormonal effects of aromatase inhibitors: Focus on premenopausal effects and interaction with tamoxifen. J Steroid Biochem Mol Biol. 2003;86:255–263. doi: 10.1016/s0960-0760(03)00365-0. [DOI] [PubMed] [Google Scholar]
- 46.Carey LA, Perou CM, Livasy CA, et al. Race, breast cancer subtypes, and survival in the Carolina Breast Cancer Study. JAMA. 2006;295:2492–2502. doi: 10.1001/jama.295.21.2492. [DOI] [PubMed] [Google Scholar]
- 47.Madlensky L, Flatt SW, Natarajan L, et al. Hot flashes are associated with CYP2D6 genotype in breast cancer survivors taking tamoxifen. Presented at the San Antonio Breast Cancer Symposium; December 10-14, 2008; San Antonio, TX. abstr 6045. [Google Scholar]
- 48.Mortimer JE, Flatt SW, Parker BA, et al. Tamoxifen, hot flashes and recurrence in breast cancer. Breast Cancer Res Treat. 2008;108:421–426. doi: 10.1007/s10549-007-9612-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Peppercorn J, Hamilton E, Qiu S, et al. Practice and attitudes amond US oncologists towards CYP2D6 testing for patients on tamoxifen. Presented at the San Antonio Breast Cancer Symposium; December 9-13, 2009; San Antonio, TX. abstr 1077. [Google Scholar]
- 50.Irvin WJ, Carey LA, Olajide O, et al. Patients' understanding of a CYP2D6 tamoxifen genotyping study. Presented at the San Antonio Breast Cancer Symposium; December 9-13, 2009; San Antonio, TX. abstr 608. [Google Scholar]
- 51.Potter L, Oakley D, de Leon-Wong E, et al. Measuring compliance among oral contraceptive users. Fam Plann Perspect. 1996;28:154–158. [PubMed] [Google Scholar]
- 52.Hershman DL, Kushi LH, Shao T, et al. Early discontinuation and nonadherence to adjuvant hormonal therapy in a cohort of 8,769 early-stage breast cancer patients. J Clin Oncol. 2010;28:4120–4128. doi: 10.1200/JCO.2009.25.9655. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.