Summary
In this report, we describe a novel method for producing mature and biologically active mono-biotinylated nerve growth factors (mBtNGF) that can be used for single molecule studies of real-time movement of neurotrophins within axons of neurons. We inserted an AviTag sequence into the C-terminal of the full length mouse preproNGF cDNA and cloned the fusion construct into a pcDNA3.1 mammalian expression vector. We also subcloned the E. coli biotin ligase, BirA, into a pcDNA3.1 vector. These two plasmids were then transiently co-expressed in HEK293FT cells. As a result, the AviTag located in the C-terminal of preproNGF was selectively ligated to a single biotin by BirA. The prepro sequence of NGF was subsequently cleaved within the cell. Mature mono-biotinylated NGF (mBtNGF) was secreted into cell culture media and was purified using Ni resin. We carried out activity assays and our results showed that mBtNGF retained biological activities that were comparable to normal NGF purified from mouse sub maxillary glands. We further verified the biotinylation efficiency of mBtNGF and the level of non-biotinylated NGF was virtually undetectable in the final preparation. Finally, by conjugating to quantum-dot streptavidin, mBtNGF was successfully used for single molecule study of axonal NGF trafficking in neurons.
Introduction
Neurotrophic factors (NTs) are a family of small protein ligands that play an important role in regulating many aspects of neuronal function including survival, phenotypic maintenance, and synaptic plasticity(Chao, 2003; Glebova and Ginty, 2005; Huang and Reichardt, 2001, 2003; Sofroniew et al., 2001). The NT family includes nerve growth factor (NGF), brain-derived neurotrophic factor (BDNF), neurotrophin-3 (NT-3) and neurotrophin-4 (NT-4) (Chao, 2003; Glebova and Ginty, 2005; Sofroniew et al., 2001). Each NT exerts its biological function through a specific tyrosine receptor kinase (Trk): TrkA for NGF, TrkB for BDNF, TrkC for NT-3 and NT-4(Chao, 2003; Huang and Reichardt, 2003). Mechanistically, NTs are released in-vivo from the target tissues which neurons innervate. These secreted NTs bind to and activate their respective surface receptors to initiate neurotrophic signaling at the neuronal axon terminus. Once bound, the ligand/receptor signaling complex becomes rapidly endocytosed via receptor mediated internalization. The ligand/receptor complex is then retrogradely transported from the axonal terminus to the soma to effect nuclear gene expression(Ginty and Segal, 2002; Ibanez, 2007; Reichardt and Mobley, 2004; Sofroniew et al., 2001; Zweifel et al., 2005). Increasing evidence demonstrates that defect/deficiency in axonally transported NTs is linked to neuronal atrophy in a spectrum of neurodegenerative disorders including Alzheimer’s disease (AD), Down syndrome (DS), prion disease, amyotrophic lateral sclerosis (ALS), and Huntington’s disease (HD) (Chao et al., 2006; Cooper et al., 2001; Megarbane et al., 2009; Wu et al., 2009). To gain further insight into the pathological mechanisms of these and other neurodegenerative diseases, it is imperative to develop tools/reagents that will afford the capability of detecting the molecular and cellular sequences of axonal transport of NTs with sensitive spatial and temporal resolutions.
Fluorescent single molecule detection methods offer a power tool to probe cellular processes such as axonal transport with spatial and temporal sensitivities that are unattainable using radioactive labels or conventional fluorescent proteins(Hohlbein et al., 2010; Pons and Mattoussi, 2009; Raj and van Oudenaarden, 2009). Indeed, impressive advance has been made using the techniques in understanding the molecular details of NGF endocytosis and its ensuing retrograde transport within axons. These studies have demonstrated that under physiological conditions, a single NGF dimer is internalized at the growth cone of chicken dorsal root ganglion neurons (Tani et al., 2005) and that a single NGF dimer is carried in an endosome that is retrogradely transported within axons of rat E16 dorsal root ganglion neurons (Cui et al., 2007). To examine axonal transport of NTs using single molecule methods, it is critical to explore different ways to tag NTs with suitable fluorescent labels. Ideally, the labeling method will help to produce a molecule with the following characteristics: 1) it can be labeled at selective site(s) that will not interfere with NTs’ biological activity; 2) Each NT molecule can be tagged with only one label to facilitate single molecule studies; 3) the fluorescent label itself is ultra-bright so that one fluorescent molecule can be visualized under cellular backgrounds; and 4) the label must have excellent photo stability i.e. resistance to photo bleaching, a desired feature for tracking the movement of the same molecules over an extended time period within the long axons of neurons.
To date, both direct and indirect labeling methods have been developed to tag NTs. One direct labeling technique involves over expressing, in a mammalian cell line, of fluorescent chimeras consisting of the full length NT cDNA construct with its C-terminus fused to a fluorescent protein such as GFP or mCherry e.g. BDNF-GFP (Adachi et al., 2005; Hartmann et al., 2001; Haubensak et al., 1998), or BDNF-mCherry(Her and Goldstein, 2008). Since the chimera proteins, expressed within neurons, must undergo the process of intra-neuronal and intra-axonal targeting, axonal movement of these fluorescent chimeras may not faithfully reflect the transport behavior of neurotrophic signals derived from the axonal targets. Further, current technologies do not permit the detection of single GFP or mCherry molecules under cellular backgrounds, hence each fluorescent signal detected may represent many fluorescent protein molecules i.e. aggregates. We tried both NGF-GFP and NGF-Cy3, neither could be detected in axonal transport assays using cultured rat dorsal root ganglion neurons by live imaging (data not shown). These limitations will hinder efforts for performing studies like those showing a single NGF dimer is internalized at the growth cone (Tani et al., 2005) and transported within axon (Cui et al., 2007). To directly label NGF a fluorescent dye like Cy3 was crosslinked to purified NGF (Tani et al., 2005). Fluorescent dyes have two distinct advantages over fluorescent proteins in that they are much smaller, and have a higher quantum yield (i.e. brighter). Unfortunately, Cy3 and other fluorescent dyes, while useful for tracking NGF movement at the growth cone, are sensitive to photo bleaching and are thus not suitable for long term tracking of molecule movement within axons of neurons. Similarly, NGF was crosslinked to biotin and conjugated to Alexa-488 streptavidin(Bronfman et al., 2003) or quantum dot-streptavidin(Cui et al., 2007). These conjugates were used to study intracellular receptor trafficking(Bronfman et al., 2003) or axonal transport(Cui et al., 2007). Unfortunately, chemical crosslinking may occur at multiple sites and potentially result in inactivation of NT molecules. For instance, following crosslinking to biotin, each NGF monomer was found to carry 5-9 biotins (Bronfman et al., 2003; Cui et al., 2007). The presence of multiple biotin moieties spreading across the entire NGF molecule may yield a heterogeneously biotinylated NGF preparation. Although the preparation, as a whole, may retain bioactivity, the fundamental problem with such samples is that some NGF molecules, due to different numbers and sites of biotinylation, may have different binding, signaling and trafficking properties from other molecules. As such, it is impossible to compare these heterogeneously labeled individual NGF molecules for their binding, trafficking and signaling.
Here we report a novel approach to produce mono-biotinylated NGF molecules. We incorporated the biotin acceptor peptide, AP-tag (also known as AviTag, www.avidity.com) (Chen et al., 2005; Howarth et al., 2005; Howarth and Ting, 2008) into the 3′-end of a full length NGF cDNA to generate the NGF-Avi construct. AP is a 15-17 amino acid recognition sequence containing a lysine residue that can be specifically ligated to one biotin by the Escherichia coli biotin ligase BirA (Chen et al., 2005). By co-expressing NGF-Avi and BirA in HEK293 cells, NGF-Avi was efficiently biotinylated and we were able to purify mature mono-biotinylated NGF (mBtNGF herein) to apparent homogeneity. We found the presence of the AviTag did not interfere with normal biological activities of NGF in activating TrkA-mediated signaling pathways. Finally, we mixed streptavidin-quantum dots (QD) with mBtNGF to produce ultra-bright and photo-bleaching resistant NGF conjugates that were used to track axonal movement of NGF by single molecule imaging in cultured rat dorsal root ganglion (DRG) neurons. The main advantages over previous approach are that the level of non-biotinylated NGF is less than 1% and that each protein has the same number of tags and at the same location, thus guaranteeing a homogenous preparation with uniform bioactivities and trafficking properties.
Materials and Methods
Cloning
Mouse preproNGF was amplified by PCR from a NGF-GFP plasmid (a generous gift from Professor Lessmann, Mainz, Germany). The forward primer sequence was: 5′-acgaattccaccatgtccatgttgttctacactctgatcactgcg-3′ and the reverse primer sequence was: 5′-gatggatccttcgtgccattcgattttctgagcctcgaagatgtcgttcagaccgccaccgacctccacggcggtggc-3′. The reverse primer contains a sequence coding for the 17 amino acid AviTag: GGGLNDIFEAQKIEWHE. The sequence was based on the #85 AviTag peptide sequence described previously (Schatz, 1993). One glutamic acid residue was added to the C-terminal AviTag based on a finding by Avidity (www.avidity.com) that it greatly enhanced the biotinylation rate of the AviTag (Beckett et al., 1999). Platinum pfx DNA polymerase (Invitrogen, Cat#11708021) was used following the manufacture’s instruction. The 50 μl of reaction was denatured at 94°C for 4min, followed by 25 cycles of amplification (30 sec at 94°C; 30 sec at 50°C; 90sec at 68°C). An additional extension was performed at 68°C for 4 min. The PCR product was purified and digested with EcoRI (Fermentas, Cat# FD0274) and BamHI (Fermentas, Cat# FD0054). It was ligated in-frame into the pcDNA3.1-myc-His vector that was predigested with EcoRI/BamHI. The resulting construct was designated as pcDNA3.1-NGFavi. BirA was amplified by PCR from pET21a-BirA (purchased from www.Addgene.com; Plasmid# 20857) (Howarth et al., 2005) using primers (forward primer: 5′-gtgaac atg gctagcatgact-3′) and reverse primer (5′-ggtgctcgagtcatgcggccgcaagct-3′ (containing an XhoI site). The PCR reaction was carried out using Pfx as described above. The PCR product was digested with XhoI (Fermentas, Cat# FD0694) and was subcloned into pcDNA3.1 myc.his (+) vector (Invitrogen) that was pre-cut with EcoRV (Fermentas, Cat# FD0303) and XhoI. The resulting plasmid was designated as pcDNA3.1-BirA. All primers were purchased from Elim Biopharmaceuticals, Inc (Hayward, CA). All constructs were verified by sequencing (Elim Biopharmaceuticals, Inc).
Cell culture and transfection
PC12 cells or a subclone of PC12 cells, PC12M were cultured as described previously (Wu et al., 2001; Wu et al., 2007). To induce neurite outgrowth, the culture media was changed to serum-free, DMEM-high glucose media (4.5 g/L glucose, Mediatech, Cat#10-013-CV) supplemented with either vehicle (0), or 10, 25, 100 ng/ml of NGF. HEK293FT cells (Invitrogen, Cat# R70007) cells were cultured in DMEM, 4.5 g/l glucose, 10% FBS, 1% penicillin/streptomycin. For transfection, HEK293FT cells were grown in 15 cm plates to ~70% confluency. Cells were changed to 25ml of DMEM-high glucose, serum-free media that was supplemented with 50 μM d-biotin (Sigma, Cat# B4639). 15-21 μg pcDNA3.1-NGFavi plasmids DNA plus 15-21 μg pcDNA3.1-BirA plasmids DNA were mixed with 1ml of DMEM-high glucose media and 60 μl of Turbofect (Fermentas, Cat# R0531). The mixture was incubated at room temperature for 15 min and was then added into the media by drop-wise method. Transfected HEK293FT cells were incubated at 37°C, 5% CO2. 72 hrs post transfection, media were collected for protein purification.
Protein purification
Media were harvested and adjusted to 30 mM phosphate buffer, pH8.0, 500 mM NaCl, 20 mM imidazole and a cocktail of protease inhibitors (1 mM PMSF from Sigma Cat# P7626, and 1 μl/ml aprotinin from Sigma, Cat#A6279). After incubation on ice for 15 min, media were cleared by centrifugation at 18, 000 rpm for 30 min using a Beckman JA-20 rotor. Ni-NTA resins (Qiagen, Cat# 30250) were rinsed with the washing buffer (30 mM phosphate buffer, pH8.0, 500 mM NaCl, 20 mM imidazole and a cocktail of protease inhibitors from Sigma, Cat# S8820). Ni-NTA resins were added to the media at a concentration of 0.3 ml Ni-NTA/100 ml media and incubated overnight with rotation at 4°C. The media/Ni-NTA slurry were loaded onto a column and the captured Ni-NTA resins were washed with 10ml wash buffer and eluted with 5ml elution buffer (30 mM phosphate buffer, pH8.0, 500 mM NaCl, 300 mM imidazole, protease inhibitors). Every 500 μl volume of eluction was collected. The purity and concentration of NGF was assessed by SDS-PAGE using a silver staining kit (Fast silver, G-Biosciences, Cat# 786-30, Maryland Heights, MO). Known quantities of NGF purified from mouse sub maxillary glands were used as standards. The first two eluted fraction contained most of purified protein.
DRG culture, live cell imaging and data analysis
Embryonic dorsal root ganglia (DRGs E15-16) were isolated from Sprague-Dawley as described in (Cui et al., 2007; Wu et al., 2007) with some minor modifications. Briefly, whole dorsal root ganglia from E16 rat embryos were extracted and cultured in vitro for 6-8 days in culturing media that were alternated between growth media (Neurobasal media containing 10% FBS, 2mM L-glutamax, 1% penicillin/streptomycin and 100ng/ml of NGF) for two days and selection media: Neurobasal media containing 2mM L-glutamax (Invitrogen: Cat#:35050-061), 5 μM of cytosine β-D-arabinofuranoside (C1768 from Signma) and 100 ng/ml NGF for two days. DRGs were then harvested and dissociated in 0.25% Trypsin (Invitrogen) prior to plating into the microfluidic chambers (Cui et al., 2007; Taylor et al., 2006). When cultured for over 7-10 days in vitro, cell bodies of dissociated DRG neurons tend to aggregate and axons bundle up. This makes it hard to distinguish individual axons from each other. By harvesting, triturating and replating DRG cultures at day 7. The neurons were cultured for an additional 3-4 day. We were able to separate DRG neurons into single cells with individual axons that can be easily identified. By doing so, we found that it was much easier to track signals in individual axons. The microfluidic chambers, manufactured in house, were laid onto 24 × 48 mm glass coverslips that were pre-coated with poly-L-Lysine (Sigma, Cat# P8920) as described (Taylor et al., 2006). Dissociated DRG neurons were plated into the cell body chamber. The growth/selection scheme outlined above was repeated. Axons from the DRG neurons started to cross the microgrooves in ~3 day and reached the axonal chamber in another 7-8 days.
Prior to live imaging of axonal transport of NGF, all compartments (cell body and axonal chambers) of the DRG neurons were thoroughly rinsed and were depleted of NGF in NGF-free, serum-free Neurobasal media for 3 hrs. To prepare the mBtNGF-Quantum dot conjugates (mBtNGF-QD), 100 nM of mBtNGF dimer were mixed with 100 nM QD655-streptavidin conjugates (Invitrogen, Cat# Q10121MP) in Neurobasal media and were incubated on ice for 30 min. mBtNGF-QD655 were added to a final concentration of 0.2 nM to the axonal chambers for 2hrs at 37°C. Live cell imaging of mBtNGF-QD655 transport with the proximal axons was carried out using a Leica DMI6000B inverted microscope. The scope was equipped with an environmental chamber that maintained a constant temperature (37°C) and CO2 (5%) during live imaging. A set of Texas red excitation/emission filter cubes was used to visualize the QD655 signal. Time lapse images were acquired at the speed of 1 frame/ second and were captured using a CCD camera (Rolera-MGi Fast 1397 from Qimaging). All data were processed and analyzed using NIH ImageJ.
SDS-PAGE/blotting
Standard protocols were followed for SDS-PAGE and blotting. Rabbit IgGs against NGF were from Santa Cruz Biotechnology Inc (Santa Cruz, CA, Cat# sc-549). Rabbit IgGs against Trk-pTyr, pErk1/2, total Erk1/2 were from Cell Signaling Technology (Danvers, MA, Cat #9141, 9101, 9102 respectively). Rabbit monoclonal antibody against pAkt was from Epitomics Inc (Cat#2214-1). Rabbit IgGs against AviTag were purchased from Genscript (Piscataway, NJ, Cat# A00674). HRP-streptavidin was from Chemicon (San Diego, CA, Cat# 18-152). Streptavidin-agarose beads were from Invitrogen (Carlsbad, CA, Cat# SA 100-04) or Pierce (Rockland, IL, Cat# 20349). For precipitation of proteins with trichloroacetic acid (TCA from Sigma, Cat# T6399), 100% ice-cold TCA was added to the supernatant to a final concentration of 5-7% and incubated on ice for 20 min. The sample was centrifuged for 20 min at 14,000 rpm in a tabletop microfuge. The pellet was washed three times with acetone, air-dried and boiled in SDS loading buffer.
Results
1. Expression and purification of mBtNGF
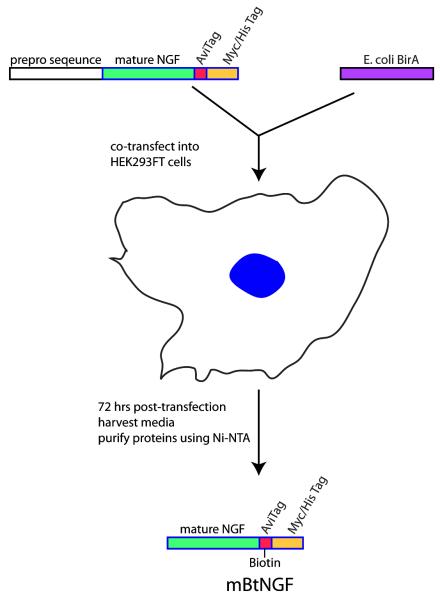
As described in the Materials and Methods section, we introduced by PCR an AviTag to the carboxyl terminal of the mouse preproNGF sequence to generate the pcDNA3.1-NGFavi construct. The PCR product was inserted immediately to the upstream of the myc-his tags of the pcDNA3.1 myc-His expression vector. The molecular mass of the full length fusion protein was predicted to be ~32.2 kDa (http://ca.expasy.org/tools/pi_tool.html). Similarly, the E.coli BirA sequence was also cloned into a pcDNA 3.1 vector by PCR to make the pcDNA3.1-BirA expression vector. Our strategy involved co-transfecting the two expression plasmids into 293FT cells (Fig 1). The BirA biotin ligase, once expressed in the cell, was expected to ligate a biotin molecule to the specific lysine site (K12) contained within the AviTag (GGGLNDIFEAQKIEWHE) of the NGFavi construct. The prepro sequence was cleaved prior to the fusion protein exiting the cell, thus only the mature NGF tagged with a single-biotin moiety together with the myc-his tags (mBtNGF) was secreted into the media (Fig 1). The media was collected and mBtNGF with a predicted molecular mass of 18.7 kDa (http://ca.expasy.org/tools/pi_tool.html) was purified using Ni-NTA resin following a standard protocol (Fig 1).
Figure 1.
Experimental designs for cloning, expression and purification of mBtNGF. As described in Materials and Methods, a mouse cDNA sequence encoding the full length of prepro NGF with a C-terminal AviTag is co-transfected with an expression vector for the E.coli biotin ligase (BirA) into 293FT cells. The specific “Lysine” residue within the AviTag is ligated to a biotin by BirA. Upon or during secretion, the N-terminal prepro sequence is cleaved and the mature protein with the biotinylated AviTag is released into the media. Mono-biotinylated NGF is secreted into the media and can be recovered from the media by Ni-NTA purification.
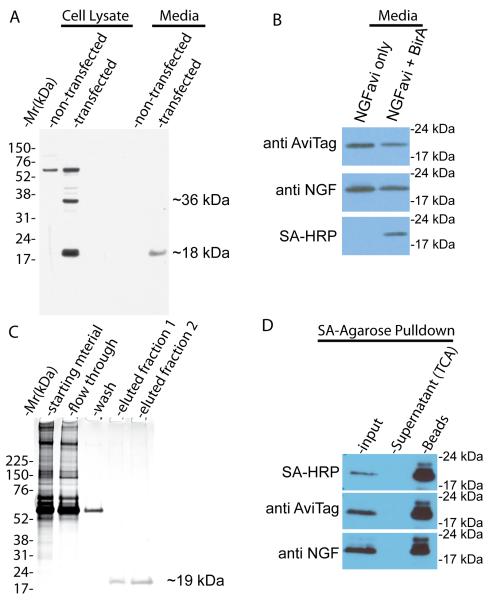
To validate our strategy, the pcDNA3.1-NGFavi plasmid was transfected into HEK293FT cells for 72 hrs. Cells were maintained in serum-free DMEM-high glucose media supplemented with 50μM of D-biotin. Both cell lysate and culture media were collected from transfected cells as well as from untransfected cells. The samples were analyzed by SDS-PAGE/immunoblotting with an antibody specific to the AviTag. As shown in Fig 2A, the anti-AviTag antibody recognized a nonspecific band with a molecular mass of ~60 kDa in both the transfected and non-transfected HEK293FT cell lysates. In cells transfected with the plasmid, the anti-AviTag antibody recognized two specific bands with apparent molecular mass of ~35 kDa and ~19 kDa, which are consistent with the predicted molecular mass of the full length preproNGFavi fusion protein and the processed mature NGFavi fusion protein (18.7 kDa), respectively (Fig 2A). In the media, the antibody detected no signal for the ~35 kDa protein. The ~19 kDa processed mature NGFavi was the only band that was detected with the antibody (Fig 2A). No signal was detected in the media from untransfected cells (Fig 2A). These results demonstrated that mature mBtNGF, but not the unprocessed form, is the predominant form secreted into the media. To investigate if mature NGFavi was biotinylated by BirA, we transfected HEK293FT cells with either pcDNA3.1-NGFavi alone or with pcDNA3.1-NGFavi and pcDNA3.1-BirA. Media were collected and analyzed by SDS-PAGE/immunoblotting. Both anti-AviTag and anti-NGF antibodies recognized the ~19 kDa band in media from both the single and double-transfected cells (Fig 2B). However, only the double tranfected sample gave rise to a positive signal when the PVDF membrane was probed with streptavidin-HRP (Fig 2B), that specifically recognizes biotinylated proteins species. These results confirm that the presence of BirA is necessary and sufficient in biotinylation of NGFavi as designed. We designate the mono-biotinylated NGFavi product as mBtNGF herein.
Figure 2.
Purification and biochemical characterization of mature mBtNGF. A: Detection of the mature form, but not the immature form, of mBtNGF in the culture media of 293FT cells following transfection. Cell lysates and media collected from transfected and non-transfected 293FT cells were separated by SDS-PAGE and blotted with anti-AviTag antibodies. The unprocessed full-length protein (~35 kDa) and the mature protein (~19 kDa) are marked. B: BirA-dependent biotinylation. Media from cells that were double-transfected with the NGFavi construct and the BirA construct (NGF + BirA) or with the NGFavi construct alone (NGF only) were separated by SDS-PAGE and blotted with indicated antibodies. The biotinylated forms were detected only with streptavidin-HRP (SA-HRP). C: Purification of mature mBtNGF to apparent homogeneity. Starting materials, flow-through, wash and two eluted fractions (25 μl each) from Ni-NTA column purification were separated by SDS-PAGE followed by silver staining. The eluted fractions contains only the ~19kDa species that corresponds to the mature mBtNGF. D: Assay for biotinylation efficiency of mBtNGF. Purified mBtNGF was incubated with streptavidin-agarose beads for 2hr at 4°C. The beads were washed and boiled in SDS sample buffer. The supernatants were collected and precipitated with TCA. These samples along with input were analyzed by SDS-PAGE and immunoblotting. The blot was probed with SA-HRP, with anti AviTag and anti NGF antibodies. All signals were detected in the SA-bead sample and no signal was seen in the supernatant.
We next carried out protein purification using Ni-NTA resin following a standard protocol as described in the Materials and Methods. Aliquots from different stages of the purification protocol were saved. These samples were separated on a 4-12% SDS-PAGE followed by silver staining. As shown in Fig 2C, a single band with an apparent molecular mass of ~19 kDa was specifically eluted in fraction 1 and 2. It was the only protein species that was detected by silver staining as compared to the starting materials. We estimate that we routinely recovered ~50-60 μg mBtNGF proteins from 50 ml of culture media.
To assay for the efficiency of biotinylation, we performed protein purification from media collected from double transfected cells using Ni-NTA as described above. The purified proteins were re-incubated with streptavidin-agarose beads (SA-agarose). The supernatants were collected and were precipitated with trichloroacetic acid. The beads were washed and boiled in SDS sample buffer. All protein samples from the supernatant, beads along with input were analyzed by SDS-PAGE and probed with either SA-HRP, or anti-AviTag or anti-NGF antibodies. The results showed that all signals were pulled down with the SA-agarose beads and no signals were detected in the supernatant (Fig 2D). According to a densitometer analysis of Fig 2D, we estimated that the maximal percentage of non-biotinylated NGF is less than 1%. Based on these results, we conclude that the NGFavi product was efficiently biotinylated by BirA within HEK293FT cells.
2. Activity Assays of mBtNGF
To test if mBtNGF retained NGF’s biological activity in stimulating the NGF/TrkA signaling pathways, we investigated its effect in PC12 cells. PC12 is a model cell line that expresses both TrkA and p75 (Wu et al., 2001). NGF treatment of PC12 cells leads to activation and endocytosis of the TrkA receptor. Among the many downstream signaling pathways that are initiated by the NGF/TrkA signaling pathways, a sustained activation of the Erk1/2 mitogen-activated protein kinase signaling pathway is believed to play a necessary role for NGF-induced differentiation of PC12 cells i.e. neurite outgrowth. Activated Akt signaling pathway suppresses apoptosis by targeting pro-apoptotic proteins such as Bad, which is also crucial in neuronal survival.
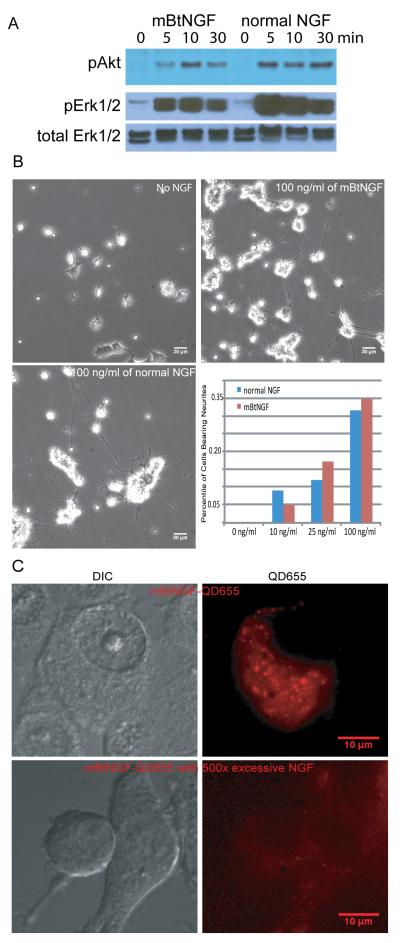
To examine if mBtNGF was able to induce the activation of the Akt and Erk1/2 signaling pathways in PC12 cells, 50-60% confluent PC12 cells were deprived of serum and NGF for 4 hrs, followed by treatment with either 100ng/ml of normal NGF or mBtNGF at 37°C for 0, 5, 10, 30 min. Cells were rinsed and lysed in RIPA buffer. Lysate proteins were analyzed by SDS-PAGE/immunoblotting. Activation of Akt and Erk ½ i.e. phosphorylation, was detected with antibodies that specifically recognize the phoshporylated form of Akt and Erk1/2, but not the non-phosphorylated forms. As shown in Fig 3A, similar to normal NGF, mBtNGF was also capable of inducing the phosphorylation of Akt and Erk1/2 at 5, 10 and 30 min of treatment. We re-probed the blot with an antibody that recognizes total Erk1/2 (phosphorylated and nonphosphorylated) as a loading control (Fig 3A). We thus conclude mBtNGF, like NGF, was able to activate the Akt and Erk1/2 signaling pathways.
Figure 3.
Analysis of bioactivities of mBtNGF using PC12 cells. A: Activation of Akt and Erk1/2 signaling pathways by mBtNGF. PC12 cells were serum-starved and were stimulated for 0, 5, 10 and 30 min with either 100ng/ml mBtNGF or 100 ng/ml normal NGF. Cell lysates were analyzed by SDS-PAGE/immunoblotting with indicated antibodies. B: Neurite outgrowth assay. PC12 cells were treated with vehicle only (0), or 10, 25 or 100ng/ml mBtNGF or normal NGF at the same concentrations for 6 days. The percentage of cells bearing neurites longer than three times of cell body was presented. Typical micrographs of PC12 cells treated with the vehicle (no NGF), 100 ng/ml mBtNGF or normal NGF were shown. C: Assay for internalization of mBtNGF. PC12 cells after incubating with either 0.2 nM of mBtNGF-QD655 or with 0.2 nM mBtNGF-QD655 together with 100 nM of normal NGF. Both DIC and corresponding fluorescence images were captured and shown.
To test if long-term treatment of PC12 cells with mBtNGF induced neurite outgrowth in these cells, we added 10, 25, 100 ng/ml of mBtNGF or 10, 25, 100 ng/ml of normal NGF into the culture media for 6 days. In a parallel experiment, a well of cells that were treated with the NGF vehicle only was also prepared as a control. As shown in Fig 3B, mBtNGF promoted neurite outgrowth in PC12 cells and the length of these neurites were comparable to that induced with normal NGF (most have the length > three times of the cell body). The percentile of cells with neurites that were three time of the cell body in length was counted and shown (Fig 3B). We did notice that cells, treated with the vehicle only, also appeared to have processes, but the length of these processes were much shorter than those cells treated with either mBtNGF or normal NGF.
To see if mBtNGF was internalized into PC12 cells following conjugation to QD655 streptavidin, we conjugated mBtNGF dimer to QD655-streptavidin at a 1:1 ratio as described. We chose PC12M cells for these experiments, because PC12M is a cell line that is more suitable for cell biology studies due to its large size and flat morphology. Cells were deprived of serum and NGF for 2 hrs and 0.2 nM of the mBtNGF-QD655 conjugate (final concentration) was applied to the media for 30 min. Cells were rinsed and were immediately imaged by live imaging microscopy for QD655 signals. Both the DIC and QD655 images of a typical cell were presented in Fig 3C (top panel). These images showed that QD655 was present in the cytoplasm of the cell (Fig 3C, top panel). To ensure that internalization of these signals was receptor-mediated, we treated PC12M cells with 0.2nM of mBtNGF-QD655 in the presence of 100 nM of normal NGF prior to imaging (Fig 3C, bottom panel). Our results showed that very little fluorescence was detected inside the cell under these conditions. We thus conclude that mBtNGF specifically binds to its surface receptors and is internalized through receptor-mediated endocytosis.
3. Live Imaging of Axonal Transport of mBtNGF
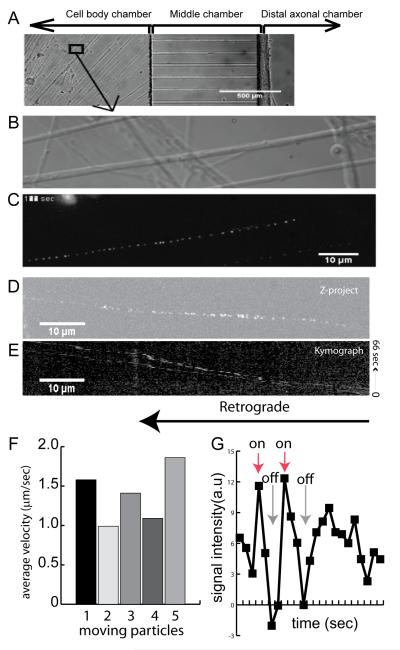
To determine whether mBtNGF could be used for studies of axonal transport, we elected to use rat E15-16 DRG neurons. Neurons were cultured in microfluidic chambers that were divided into three compartments; cell body, middle and distal axonal chambers (Fig 4A). After axon crossed the microgrooves, all three compartments of chambers were washed and starved for serum and NGF for 3 hrs. Only the distal axonal chamber was supplied with 0.2 nM of mBtNGF-QD655 for 2 hrs at 37°C. Axonal movement of QD655 signals within the proximal axon near the cell body was imaged as described in Materials and Methods. Time-lapse images were recorded at 1 frame/sec. Corresponding DIC images were also captured. Results from a typical experiment were presented in Fig 4B-E. Axonal movement of a single mBtNGF-QD655 signal within the proximal axons (DIC image in Fig 4B) was processed using NIH ImageJ. The time-lapse trajectory shows that the signal was moving in a processive manner towards the cell body (retrograde) (Fig 4C). Examination of time-lapse movies revealed that most of the moving signal displayed “blinking”, an intrinsic characteristic of a single quantum dot (Supplemental Movies). The “blinking” property of a single QD was further demonstrated in Fig 4G. We measured the signal intensity of a moving particle, the result showed that the signal was either “on” or “off” for a period of time (Fig 4G). Unlike an out of focus signal that showed a gradual decrease in signal intensity, the signal intensity for a single QD suddenly disappeared and re-appeared (Fig 4G). We also performed kymograph analysis of the time-lapse images. The image of Z-project of max intensity and the kymograph of two moving particles from Movie 2 (Supplemental) are presented (Fig 4D-E). Our results confirmed that the retrograde movement of mBtNGF-QD655 showed a typical “stop and go” pattern. The average transport speed within axons was ~1.4μm/sec (Fig 4F). This is in excellent agreement with our previously published results (Cui et al., 2007).
Figure 4.
Single molecule imaging of transport of mBtNGF-QD655 in axons of dorsal root ganglion (DRG). A: A phase contrast image of DRG neurons that were cultured in the microfluidic chambers. Following addition of 0.2 nM mBtNGF-QD655 to the distal axons, real time imaging of transport of mBtNGF-QD655 within the proximal axons was carried out using a 100x oil objective lens. A high resolution DIC image of proximal axons of DRG neurons is shown in B. Time-lapse trajectory of mBtNGF-QD655 movement within the corresponding axons is shown in C. Kymograph analysis of Movie 2 (supplemental materials) was also performed using ImageJ. The result of Z-project is shown in D and the resulting kymograph is shown in E. F: A histogram shows the average velocities of retrogradely transported endosomes. G: A line graph shows the blinking properties of single QDs. Fluorescence intensity of a moving particle was measured frame by frame. Following background subtraction, the signal intensity displays a pattern of either complete “on” or complete “off”, characteristic of blinking of a single QD, whereas an out-of-focus signal should show a slow drop in fluorescence intensity instead.
Discussion
We described here a novel strategy for producing mono-biotinylated NGF (mBtNGF) that retained NGF’s full biological activity. By introducing the AviTag at the carboxyl terminal of NGF, each NGF molecule was ensured to be biotinylated at the specific lysine residue contained within the AviTag. We demonstrated that co-expression of BirA with the NGFavi construct in 293FT cells resulted in highly efficient biotinylation of the NGF fusion protein. The amount of non-biotinylated NGF fusion proteins was not detectable by immunoblotting. This is particularly important for studying if or not NGF is co-transported in axons with its receptors by live imaging. The presence of non-labeled NGF such as in the randomly labeled NGF preparation will not allow one to reach a conclusion. Furthermore, mBtNGF was purified to apparent homogeneity by one step Ni-NTA purification. We have also confirmed that mBtNGF was biologically active in activating the Akt and Erk1/2 pathway and inducing neurite outgrowth in PC12 cells. Finally, when applied to compartmentalized DRG neuron cultures, we were able to image and quantitate axonal retrograde transport of mBtNGF labeled with QD at the single molecule level.
Site-specific biotinylation of NGF represents a significant advance in labeling NGF. Previously NGF was biotinylated using chemical crosslinker (Bronfman et al., 2003). It frequently resulted in labeling of a single NGF molecule with 5-9 biotins that spread across the entire NGF molecule (Bronfman et al., 2003; Cui et al., 2007). As a result, it is difficult to direct the binding of QD to a specific site. It is entirely possible that attachment of QD to the different biotin sites will differentially affect NGF’s ability to bind and to signal. Therefore, even when mixed at 1:1 ratio (NGF dimer:QD), the resulting mixture will give rise to mostly one QD per NGF dimer. Yet, the QDs may bind to different biotin sites for different NGF dimers. Characterizing the effect of QD binding to these 5-9 biotin sites on NGF’s activity will be very challenging. Our method eliminates these uncertainties. By introducing the AviTag to the carboxyl terminal of NGF, we ensured that each NGF molecule was only ligated to a single biotin specifically by BirA and at the designated site (K12 with the AivTag). As a result, when mBtNGF dimer was mixed with QDs at 1:1 ratio, the reaction will yield a highly uniform labeling mixture i.e. each NGF dimer will have one QD label and importantly all QDs will bind to exactly the same site of the NGF dimer molecule. We believe that mBtNGF will serve as a powerful tool for single molecule study of NGF signaling and trafficking.
The other advantage our method offers is the fact that by co-expressing BirA with the pcDNA3.1-NGFavi construct, we found that NGFavi was biotinylated with an excellent efficiency. Non-biotinylated NGFavi was detected by immunoblotting. Previously by our estimate, ~5-10% of NGF was left non-biotinylated following the reaction with the chemical crosslinking method (Bronfman et al., 2003; Cui et al., 2007). A similar problem also existed when we tried to perform in a test tube biotinylation of purified NGFavi proteins from singly transfected cell media using a BirA ligation kit purchased from Avidity (data not shown). Purified BirA may not be stable and the efficiency of in vitro biotinylation varied greatly from experiment to experiment in our experience. In addition, these extra steps will invariably result in some loss of NGF’s biological activity.
We further demonstrated that mBtNGF, when conjugated to QD-streptavidin, can be used successfully to track axonal movement of NGF in rat DRG neurons. We did notice that the majority of QD signals that was moving with the axon, displayed “blinking”, a characteristic of a single QD. These signals were retrogradedly transport along the axons with an average speed of 1.4 μm/sec. These observations are all in excellent agreement with our previously published results(Cui et al., 2007).
In summary, our method offers a fast (one step purification) and efficient way of producing mono-biotinylated and biologically active NGF (mBtNGF). Utilizing the same procedure we have also successfully produced two NGF mutant proteins (data not shown). We believe that our method can also be applied to produce other neurotrophins such as BDNF, NT3, NT4 in a similar fashion for imaging trafficking of these neurotrophic factors within live neurons.
Supplementary Material
Acknowledgement
We would like to thank Dr W.C. Mobley for his support and Sadie Bartholomew for her help with DRG dissection and culture. These studies are supported by NIH grants (2PN2EY16525, NS24054, NS055371), and grants from the Hillblom Foundation and the Down Syndrome Research and Treatment Foundation.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Adachi N, Kohara K, Tsumoto T. Difference in trafficking of brain-derived neurotrophic factor between axons and dendrites of cortical neurons, revealed by live-cell imaging. BMC Neurosci. 2005;6:42. doi: 10.1186/1471-2202-6-42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beckett D, Kovaleva E, Schatz PJ. A minimal peptide substrate in biotin holoenzyme synthetase-catalyzed biotinylation. Protein Sci. 1999;8:921–9. doi: 10.1110/ps.8.4.921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bronfman FC, Tcherpakov M, Jovin TM, Fainzilber M. Ligand-induced internalization of the p75 neurotrophin receptor: a slow route to the signaling endosome. J Neurosci. 2003;23:3209–20. doi: 10.1523/JNEUROSCI.23-08-03209.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chao MV. Neurotrophins and their receptors: a convergence point for many signalling pathways. Nat Rev Neurosci. 2003;4:299–309. doi: 10.1038/nrn1078. [DOI] [PubMed] [Google Scholar]
- Chao MV, Rajagopal R, Lee FS. Neurotrophin signalling in health and disease. Clin Sci (Lond) 2006;110:167–73. doi: 10.1042/CS20050163. [DOI] [PubMed] [Google Scholar]
- Chen I, Howarth M, Lin W, Ting AY. Site-specific labeling of cell surface proteins with biophysical probes using biotin ligase. Nat Methods. 2005;2:99–104. doi: 10.1038/nmeth735. [DOI] [PubMed] [Google Scholar]
- Cooper JD, Salehi A, Delcroix JD, Howe CL, Belichenko PV, Chua-Couzens J, Kilbridge JF, Carlson EJ, Epstein CJ, Mobley WC. Failed retrograde transport of NGF in a mouse model of Down’s syndrome: reversal of cholinergic neurodegenerative phenotypes following NGF infusion. Proc Natl Acad Sci U S A. 2001;98:10439–44. doi: 10.1073/pnas.181219298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cui B, Wu C, Chen L, Ramirez A, Bearer EL, Li WP, Mobley WC, Chu S. One at a time, live tracking of NGF axonal transport using quantum dots. Proc Natl Acad Sci U S A. 2007;104:13666–71. doi: 10.1073/pnas.0706192104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ginty DD, Segal RA. Retrograde neurotrophin signaling: Trk-ing along the axon. Curr Opin Neurobiol. 2002;12:268–74. doi: 10.1016/s0959-4388(02)00326-4. [DOI] [PubMed] [Google Scholar]
- Glebova NO, Ginty DD. Growth and survival signals controlling sympathetic nervous system development. Annu Rev Neurosci. 2005;28:191–222. doi: 10.1146/annurev.neuro.28.061604.135659. [DOI] [PubMed] [Google Scholar]
- Hartmann M, Heumann R, Lessmann V. Synaptic secretion of BDNF after high-frequency stimulation of glutamatergic synapses. Embo J. 2001;20:5887–97. doi: 10.1093/emboj/20.21.5887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haubensak W, Narz F, Heumann R, Lessmann V. BDNF-GFP containing secretory granules are localized in the vicinity of synaptic junctions of cultured cortical neurons. J Cell Sci. 1998;111(Pt 11):1483–93. doi: 10.1242/jcs.111.11.1483. [DOI] [PubMed] [Google Scholar]
- Her LS, Goldstein LS. Enhanced sensitivity of striatal neurons to axonal transport defects induced by mutant huntingtin. J Neurosci. 2008;28:13662–72. doi: 10.1523/JNEUROSCI.4144-08.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hohlbein J, Gryte K, Heilemann M, Kapanidis AN. Surfing on a new wave of single-molecule fluorescence methods. Phys Biol. 2010;7:031001. doi: 10.1088/1478-3975/7/3/031001. [DOI] [PubMed] [Google Scholar]
- Howarth M, Takao K, Hayashi Y, Ting AY. Targeting quantum dots to surface proteins in living cells with biotin ligase. Proc Natl Acad Sci U S A. 2005;102:7583–8. doi: 10.1073/pnas.0503125102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Howarth M, Ting AY. Imaging proteins in live mammalian cells with biotin ligase and monovalent streptavidin. Nat Protoc. 2008;3:534–45. doi: 10.1038/nprot.2008.20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang EJ, Reichardt LF. Neurotrophins: roles in neuronal development and function. Annu Rev Neurosci. 2001;24:677–736. doi: 10.1146/annurev.neuro.24.1.677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang EJ, Reichardt LF. Trk receptors: roles in neuronal signal transduction. Annu Rev Biochem. 2003;72:609–42. doi: 10.1146/annurev.biochem.72.121801.161629. [DOI] [PubMed] [Google Scholar]
- Ibanez CF. Message in a bottle: long-range retrograde signaling in the nervous system. Trends Cell Biol. 2007;17:519–28. doi: 10.1016/j.tcb.2007.09.003. [DOI] [PubMed] [Google Scholar]
- Megarbane A, Ravel A, Mircher C, Sturtz F, Grattau Y, Rethore MO, Delabar JM, Mobley WC. The 50th anniversary of the discovery of trisomy 21: the past, present, and future of research and treatment of Down syndrome. Genet Med. 2009;11:611–6. doi: 10.1097/GIM.0b013e3181b2e34c. [DOI] [PubMed] [Google Scholar]
- Pons T, Mattoussi H. Investigating biological processes at the single molecule level using luminescent quantum dots. Ann Biomed Eng. 2009;37:1934–59. doi: 10.1007/s10439-009-9715-0. [DOI] [PubMed] [Google Scholar]
- Raj A, van Oudenaarden A. Single-molecule approaches to stochastic gene expression. Annu Rev Biophys. 2009;38:255–70. doi: 10.1146/annurev.biophys.37.032807.125928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reichardt LF, Mobley WC. Going the distance, or not, with neurotrophin signals. Cell. 2004;118:141–3. doi: 10.1016/j.cell.2004.07.008. [DOI] [PubMed] [Google Scholar]
- Schatz PJ. Use of peptide libraries to map the substrate specificity of a peptide-modifying enzyme: a 13 residue consensus peptide specifies biotinylation in Escherichia coli. Biotechnology (N Y) 1993;11:1138–43. doi: 10.1038/nbt1093-1138. [DOI] [PubMed] [Google Scholar]
- Sofroniew MV, Howe CL, Mobley WC. Nerve growth factor signaling, neuroprotection, and neural repair. Annu Rev Neurosci. 2001;24:1217–81. doi: 10.1146/annurev.neuro.24.1.1217. [DOI] [PubMed] [Google Scholar]
- Tani T, Miyamoto Y, Fujimori KE, Taguchi T, Yanagida T, Sako Y, Harada Y. Trafficking of a ligand-receptor complex on the growth cones as an essential step for the uptake of nerve growth factor at the distal end of the axon: a single-molecule analysis. J Neurosci. 2005;25:2181–91. doi: 10.1523/JNEUROSCI.4570-04.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor AM, Rhee SW, Jeon NL. Microfluidic chambers for cell migration and neuroscience research. Methods Mol Biol. 2006;321:167–77. doi: 10.1385/1-59259-997-4:167. [DOI] [PubMed] [Google Scholar]
- Wu C, Cui B, He L, Chen L, Mobley WC. The coming of age of axonal neurotrophin signaling endosomes. J Proteomics. 2009;72:46–55. doi: 10.1016/j.jprot.2008.10.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu C, Lai CF, Mobley WC. Nerve growth factor activates persistent Rap1 signaling in endosomes. J Neurosci. 2001;21:5406–16. doi: 10.1523/JNEUROSCI.21-15-05406.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu C, Ramirez A, Cui B, Ding J, Delcroix JD, Valletta JS, Liu JJ, Yang Y, Chu S, Mobley WC. A functional dynein-microtubule network is required for NGF signaling through the Rap1/MAPK pathway. Traffic. 2007;8:1503–20. doi: 10.1111/j.1600-0854.2007.00636.x. [DOI] [PubMed] [Google Scholar]
- Zweifel LS, Kuruvilla R, Ginty DD. Functions and mechanisms of retrograde neurotrophin signalling. Nat Rev Neurosci. 2005;6:615–25. doi: 10.1038/nrn1727. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.