Abstract
Moving nuclei to specific intracellular locations is central to many cell and developmental processes. However, the molecular mechanisms of nuclear migration are poorly understood. We took advantage of the ability to film nuclear migration events in Caenorhabditis elegans embryos to gain insights into the mechanisms of nuclear migration. Mutations in unc-83 blocked the initiation of nuclear migration. UNC-83 recruits kinesin-1 and dynein to the nuclear envelope. Live imaging of mutants showed that kinein-1 provides the major force to move nuclei. Dynein is responsible to move nuclei backwards or to mediate nuclear rolling to by pass cellular roadblocks that impede efficient migration. Live imaging was also used to analyze the microtubule network, which is highly polarized and dynamic. This detailed mechanism of nuclear migration may be applicable to nuclear migration in other systems and for the movement of other large cellular cargo.
Key words: nuclear envelope, KASH, SUN, nuclear migration, nuclear envelope bridge, UNC-83, UNC-84
Introduction
“You can observe a lot just by watching.” - Yogi Berra
A wide variety of cellular processes, including fertilization, cell division, cell migration, establishment of cell polarity, differentiation and the organization of syncytia, depend on moving and anchoring nuclei to a specific location in the cell.1,2 Not surprisingly, defects in nuclear positioning disrupt neurological and muscular development and lead to a variety of diseases.1,2 Genetic screens for mutants with misplaced nuclei in yeast, filamentous fungi, worms and flies have identified many of the players involved in nuclear positioning. For example, analyses of the nud (nuclear distribution) mutants in Aspergillus implicate dynein and its regulatory components in nuclear migration.3 Genetic screens for mutants with misplaced nuclei in worms and flies originally established the importance of KASH and SUN proteins in nuclear migration.4–6 KASH and SUN proteins form a bridge across the nuclear envelope to connect the cytoskeleton to structural components inside the nucleus.1,2 This bridge is central to most characterized mechanisms of nuclear positioning. Many additional studies on SUN and KASH prtoeins have been done in mammalian tissue culture cells.7–11 However, these genetic and cellular approaches mostly fail to observe actual nuclear migration events; the genetic studies focused on terminal phenotypes and the tissue culture systems do not undergo long-distance nuclear migrations. A notable exception is the filming of short-range nuclear migrations in polarizing fibroblasts in culture.12,13 Heidi Fridolfsson from my lab set out to directly watch normal and mutant nuclear migration events in the developing Caenorhabditis elegans embryo.14 True to the Yogi-ism at the top of this section, she observed a lot.
A Model System for Nuclear Migration
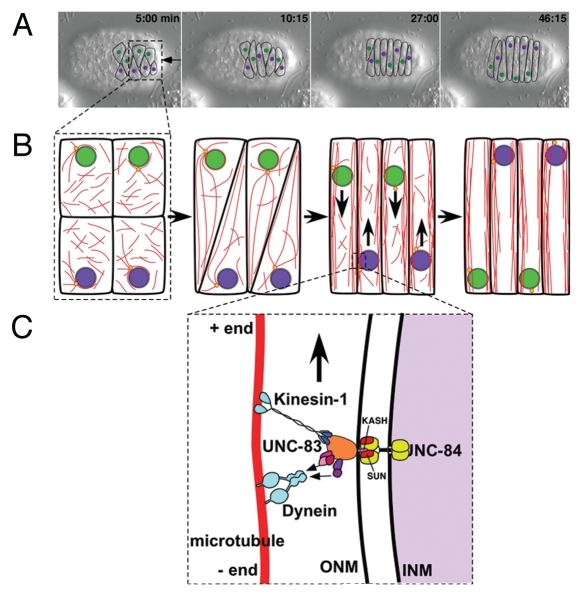
My lab uses C. elegans hypodermal cells as a model system to study the mechanisms of nuclear migration. An advantage of this system is the optical transparency of the animal; nuclei can be easily observed in live animals using DIC optics. More importantly, C. elegans has an invariant cell lineage. The Nobel-prize-winning studies of John Sulston and colleagues determined the position of every nucleus throughout development, including striking nuclear migrations in embryonic and larval hypodermal cells.15,16 On the dorsal surface of the pre-elongation embryo, fifteen hyp7 precursor cells align in two rows on either side of the dorsal midline, intercalate across the midline to form a row of column-shaped epithelial cells and undergo contralateral nuclear migrations across the dorsal midline (Fig. 1).16,17 The cells then fuse to form the dorsal hyp7 syncytium that secretes the dorsal cuticle. Midway through the first larval stage, twelve P-cell nuclei migrate from lateral positions to the ventral cord. Cells derived from P-cells form the vulva, contribute to the hypodermis and some become motor neurons.15
Figure 1.
Nuclear migration in embryonic C. elegans hypodermal cells. (A) DIC time-lapse images of nuclear migration. The dorsal surface of a pre-elongation embryo is shown with the anterior to the left. The dorsal mid-line is marked with an arrow. Cell-cell boundaries of hyp7 precursors are drawn on top in black. Nuclei that migrate down (from the right to the left of the embryo) are marked in green and nuclei migrating up (from left to right of the embryo) are in purple. (B) Cartoon showing intercalation, nuclear migration and microtubule organization in hyp7 precursors. Cells first align in two rows either side of the dorsal midline (left) and then intercalate (second from left). Microtubules (red) reorganize during intercalation to eventually form bundles polarized in the direction of nuclear migration (right). Centrosomes (yellow circles) remain associated with nuclei, but do not nucleate the vast majority of microtubules. (C) A model for how UNC-83 interacts with microtubule motors at the nuclear envelope. UNC-84 (yellow) is in the inner nuclear membrane (INM). The SUN domain of UNC-84 (red) interacts with the KASH domain of UNC-83 (pink) to span the nuclear envelope. UNC-83 spans the outer nuclear membrane (ONM) and its cytoplasmic domain (orange) interacts with proteins that regulate microtubule motors kinesin-1 and dynein. Kinesin-1 then provides the major force to move nuclei toward the plus ends of microtubules. Dynein moves nuclei backwards and causes nuclei to roll to bypass cellular roadblocks. See text for full details.
Genetic screens for lineage mutants by Sulston and Horvitz found mutations in two genes, unc-83 and unc-84, that disrupted both P-cell and hyp7 nuclear migration.18,19 Defects in nuclear migration lead to P-cell death, which disrupts neuronal and vulval development, resulting in animals that are uncoordinated and unable to lay eggs. Defects in hyp7 nuclear migration lead to mis-positioned nuclei in the dorsal cord rather than the contralateral side of the animal. Normally, there are no nuclei in the dorsal cord, but in unc-83 and unc-84 mutants, about 12 nuclei (85–90%) are mis-positioned in each dorsal cord.4,20
UNC-83, UNC-84 and Microtubule Motors Mediate Nuclear Migration
Molecular characterization of unc-84 and unc-83 showed that they both encode proteins of the nuclear envelope.4,20 UNC-84 is a founding member of the SUN (Sad1 and UNC-84) family of proteins that are conserved across eukaryotes.4 Like other SUN proteins, UNC-84 is an integral component of the inner nuclear membrane with its conserved SUN domain in the perinuclear space of the nuclear envelope.21,22 UNC-83 is a component of the outer nuclear membrane with a small C-terminal KASH domain inserted into the perinuclear space and a large cytosolic domain without homology to any domains of known function.20,22 The SUN domain of UNC-84 directly interacts with the KASH domain of UNC-83 to anchor UNC-83 in the outer nuclear membrane.22 Thus, UNC-84 and UNC-83 form a bridge across the nuclear envelope (Fig. 1C). In general, KASH-SUN bridges across the nuclear envelope are used to connect structural elements of the nucleus to components of the cytoskeleton.1,2
In order to elucidate the molecular mechanisms used by UNC-83 to move nuclei, Marina Meyerzon and Heidi Fridolfsson in my lab identified binding partners of the novel, cytosolic domain of UNC-83. The kinesin-1 light chain KLC-2 and three dynein regulators, the dynein LC8 light chain DLC-1, the Bicaudal-D homolog BICD-1 and the NudE homolog NUD-2, were found to interact with UNC-83.23,24 Genetic analysis showed that mutations in kinesin-1 components caused severe nuclear migration defects where most hyp7 nuclei were mis-positioned to the dorsal cord.24 However, mutations in dynein or its regulatory components caused only very minor nuclear migration phenotypes where only 2–15% of hyp7 nuclei were found in the dorsal cord.23 These results support a model where UNC-83 functions as a cargo adaptor to recruit both kinesin-1 and dynein to the surface of the nucleus. Kinesin-1 provides the major force to move nuclei and dynein plays a relatively minor role to ensure the fidelity of the migration. However, these studies are limited by the fact that only terminal phenotypes, presumed to represent failures in nuclear migration, were analyzed. Furthermore, these reports failed to determine how microtubule motors of opposite polarity are coordinated to move nuclei.
Watching Nuclear Migration
To better understand how kinesin-1 and dynein interact to move nuclei, we employed time-lapse, differential interference contrast (DIC) microscopy to observe actual nuclear migration events in wild type and mutant embryos.14 Normally, nuclear migration begins shortly after the completion of intercalation. Nuclei migrate slowly, about 13.6 µm/hour, taking 30–40 minutes to reach the contralateral side of the cell (Fig. 1). Nuclei move short distances with peak velocities about ten times the average velocity and have frequent pauses. Furthermore, nuclear migration is bidirectional with short, backwards movements. Nuclei are nearly the width of the cell. Therefore, dramatic rearrangements of the cellular architecture must occur during migration. As a nucleus moves forward, a large build-up of cytoplasmic granules accumulates in front of the migrating nucleus. When the nucleus reaches about half way across the cell, the build-up of granules must be resolved. Often, this is accomplished by a nuclear rolling event where the nucleus rolls more than 360° in a span of a few minutes. As the nucleus rolls, the granules squeeze by, allowing nuclear migration to continue.14
In unc-83 or unc-84 null mutant backgrounds, nuclear migration completely fails.14 Nuclei do not move in the first 10 minutes after intercalation is complete, and frequently do not migrate at all in the entire 40 minutes that we can film before the embryo reorients within the eggshell. Our data suggest that 30–60 minutes after they were supposed to migrate, nuclei are pushed to the dorsal cord by the migration of underlying body-wall muscles. Nuclei that do not end up in the dorsal cord remain in their original lateral positions. Thus, the hyp7 nuclear migration defect in unc-83 or unc-84 mutants is 100% penetrant.14
Our model predicts that kinesin-1 is the major force producer for migration because mutations in klc-2 lead to most hyp7 nuclei being mis-positioned in the dorsal cord.24 When filmed, about 83% of nuclei fail to migrate in klc-2(km11) mutant embryos.14 These failed migration events look just like unc-83 mutants. However, about 17% of nuclei migrate normally with bi-directional movements and occasional rolling events. klc-2(km11) is a loss-of-function, but not a null, allele,24 which likely explains why the phenotype is weaker than unc-83 mutant embryos.
The small number of hyp7 nuclei in the dorsal cord of dynein or dynein-regulatory-component mutants suggests that dynein plays an unknown regulatory role in nuclear migration.23 RNAi against dynein heavy chain leads to early embryonic lethality,25 making filming of nuclear migration in later embryogenesis impossible. Thus, most of our live filming was done in the nud-2(ok949); bicd-1(RNAi) background—the strongest, viable defect of the dynein-regulatory genes. Nuclear migration initiates normally in nud-2(ok949); bicd-1(RNAi) hyp7 precursors; nuclei move at wildtype velocities to the middle of the cell, where cytoplasmic granules begin to accumulate.14 Two-thirds of the time, mutant nuclei overcome the blockage and complete their migrations normally. The other third of the time, mutant nuclei are unable to resolve the blockage of cytoplasmic granules and fail to complete their migrations. Upon closer examination, nud-2(ok949); bicd-1(RNAi) nuclei fail to undergo any backwards movements and migration failure is correlated with a failure to roll.14 To completely eliminate the role of dynein, unc-83 null animals were rescued by expression of KLC-2 fused to a KASH domain. In these transgenic animals, kinesin is recruited to the nuclear envelope in the absence of UNC-83 and the unc-83 nuclear positioning defect is approximately 50% rescued.24 When filmed, none of the KLC-2::KASH transgenic nuclei that migrate ever move backwards or roll.14 We conclude that dynein is used to mediate short backwards movements of nuclei and to roll nuclei. Most of the time, kinesin-1 can resolve the cellular roadblocks without dynein, but dynein is important to resolve some blockages of cytoplasmic granules and to ensure efficient nuclear migration.
Looking at Microtubules
If the plus-end directed microtubule motor kinesin-1 is responsible for the forward movement of nuclei and the minus-end directed motor dynein mediates backwards movements, the microtubule network must be somehow polarized. It was previously predicted that microtubules form stable polarized bundles during intercalation that are used as tracks during nuclear migration.17,24 We showed that γ-tubulin rearranges from primarily centrosome localization during the rapid cellular divisions to a distributed pattern along the cellular cortex just before the beginning of hyp7 precursor intercalation.14 This suggests that in hyp7 precursors, the microtubule network is centrosome independent, which could explain why the centrosome apparently plays no role in hyp7 nuclear migration.14,20 To examine the microtubule network in live embryos, we expressed tubulin and the plus-end tip tracker EB1 as GFP fusion proteins specifically in embryonic hypodermal cells.14 Surprisingly, bundles of microtubules in hyp7 precursors during nuclear migration are dynamic; FRAP studies showed t1/2 for recovery of 7.4 seconds.14 However, microtubules are polarized, as shown by tracking EB1::GFP comets. Over 90% of microtubules grow in the direction of nuclear migration.14 These data nicely fit our model where the plus-end directed motor kinesin-1 is the major force producer and dynein functions to roll nuclei or move them backwards (towards the minus ends of microtubules) to bypass cellular roadblocks and ensure efficient nuclear migration.
Coordination of Kinesin-1 and Dynein for Bi-Directional Movements
A single cellular cargo will often bind both plus and minus-end-directed microtubule motors, but how the relative outputs of such motors are coordinated is poorly understood. Three non-mutually exclusive models could explain why both kinesin-1 and dynein are recruited to many cellular cargos. In the first, kinesin-1 and dynein are interdependent; a mutation in one disrupts movements in both directions.26 Alternatively, dynein could function as a drag on kinesin-1 in a tug-of-war model.27 Our data for nuclear migration favors a third model, where the motors mediate bidirectional movements that could allow the cargo to switch microtubule tracks, avoid obstacles or correct errors as previously suggested.28,29 As the largest cellular cargo, the stress of moving the nucleus requires coordinating many motors to generate forces for processive movement. UNC-83 interacts with both the LC8 light chain DLC-1 and the C-terminus of NudE.23 The C-terminus of NudE also binds DLC-1 to recruit LIS-1 and make dynein more processive.30 Thus it is interesting to speculate that UNC-83 regulates dynein activation through DLC-1, which binds UNC-83, NUD-2, EGAL-1 and dynein IC. The LC8 light chain has been proposed to act as a dimerization hub in a variety of networks;31 the regulation of these interactions therefore warrant future studies. Our data show that KLC-2 binds UNC-83 in a different region than the dynein regulators.23 Thus, UNC-83 could act as both a cargo adaptor to recruit kinesin-1 and dynein to the surface of nuclei and as an integrator of dynein and kinesin-1 activities. Similar roles have been proposed for the Drosophila KASH protein Klarsicht.32
Other Mechanisms to Move Nuclei
The kinesin-1-dependent mechanism to move C. elegans embryonic hyp7 nuclei that is the focus of our study is just one of many emerging mechanisms to move nuclei. In C. elegans, there are at least two other mechanisms. The forces to move pronuclei toward one another in newly fertilized embryos are provided by dynein, but unlike in hyp7 cells, this is a centrosome-dependent process.25 Similar to UNC-83's role in hyp7 cells, the KASH protein ZYG-12 is required for dynein localization to the surface of pronuclei; ZYG-12 is also required for attachment of centrosomes to male pronuclei.33 Second, in C. elegans larval P-cells, unc-83 null mutations are temperature sensitive, suggesting the existence of an alternative pathway.20 My lab has unpublished genetic data that shows a mechanism independent of UNC-83 functions to move P-cell nuclei at the unc-83 permissive temperature.
In mammals, Nesprin-4 plays a role analogous to UNC-83 in recruiting the light chain of kinesin-1 to the surface of the nucleus to mediate nuclear movement towards the plus ends of microtubules in highly polarized, secretory epithelial cells.34 In HeLa cells, BicD is recruited to nuclear pore complexes in G2. BicD then recruits dynein and regulates kinesin-1 at the nuclear envelope to position nuclei and centrosomes prior to mitosis.35 In wounded mouse fibroblast tissue culture cells, the KASH protein Syne/Nesprin-1 moves nuclei by tethering the nuclear envelope to moving actin fibers.9 Progenitor cells in developing vertebrate neuroepithelia undergo cell cycle dependent interkinetic nuclear migrations.36 In some neuroepithelia, kinesin-3 moves nuclei away from centrosomes and the apical surface during G1 of the cell cycle and dynein moves nuclei back to the apical surface in G2.37 Actinmyosin contraction moves nuclei in other neuroepithelia.38 KASH proteins likely mediate these nuclear migrations;27,39,40 Syne/Nesprin-2 has been shown to interact with both dynein and kinesin-1 and is required for multiple nuclear migration events in developing neurons.40
These examples show that a toolbox of motors and structural elements are available to move nuclei. At different times in development and in different systems, various combinations of KASH-SUN bridges, centrosomes, dynein, kinesin, actin flow and actin-myosin contraction are used to mediate nuclear migration. Thus, future studies should focus both on careful elucidation of individual mechanisms in well-studied systems and on characterizing more poorly understood nuclear migration events.
Summary and Future Directions
In summary, our live analysis of wild type and mutant nuclear migration events in embryonic C. elegans hypodermal cells has led to one of the most completely understood molecular mechanisms to move nuclei (Fig. 1). Kinesin-1 is recruited to the outer surface of the nuclear envelope by the KASH protein UNC-83 and provides the major force to propel nuclei forward along a dynamic and polarized microtubule network. Dynein, also recruited to the nuclear envelope by UNC-83, functions to roll nuclei or to move them backwards in order to bypass cellular roadblocks of cytoplasmic granules.
Many questions about hyp7 nuclear migration remain: How is the non-centrosomal microtubule network organized and how is polarity of microtubules maintained? Can a single molecule of UNC-83 bind both kinesin-1 and dynein components simultaneously? If so, how are the relative outputs of these opposing motors regulated? Once nuclei migrate, they must become anchored in place, which uses the KASH protein ANC-1.6 How does the UNC-84-UNC-83 bridge for migration switch to an UNC-84-ANC-1 bridge for anchorage? For that matter, how do KASH-SUN bridges form in the first place? And finally, what role does the structural integrity of the nuclear envelope and lamina play in migration? Such questions will keep my group and others busy for the foreseeable future.
Acknowledgements
Foremost, I thank Heidi Fridolfsson for her significant effort, patience and dedication toward filming nuclear migration and her comments on this manuscript. I thank all the other past and present members of the Starr lab for elucidating so many interesting aspects of UNC-84 and UNC-83 mechanisms of nuclear migration. Finally, I thank those individuals who initiated investigation of unc-83 and unc-84 that made our studies possible, especially John Sulston, Robert Horvitz, William Fixsen, Min Han and Chris Malone. Studies in the Starr lab are supported by grant R01GM073874 from the NIH.
References
- 1.Starr DA. A nuclear-envelope bridge positions nuclei and moves chromosomes. J Cell Sci. 2009;122:577–586. doi: 10.1242/jcs.037622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Starr DA, Fridolfsson HN. Interactions Between Nuclei and the Cytoskeleton Are Mediated by SUNKASH Nuclear-Envelope Bridges. Annu Rev Cell Dev Bio. 2010 doi: 10.1146/annurev-cellbio-100109-104037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Xiang X, Fischer R. Nuclear migration and positioning in filamentous fungi. Fungal Genet Biol. 2004;41:411–419. doi: 10.1016/j.fgb.2003.11.010. [DOI] [PubMed] [Google Scholar]
- 4.Malone CJ, Fixsen WD, Horvitz HR, Han M. UNC-84 localizes to the nuclear envelope and is required for nuclear migration and anchoring during C. elegans development. Development. 1999;126:3171–3181. doi: 10.1242/dev.126.14.3171. [DOI] [PubMed] [Google Scholar]
- 5.Mosley-Bishop KL, Li Q, Patterson L, Fischer JA. Molecular analysis of the klarsicht gene and its role in nuclear migration within differentiating cells of the Drosophila eye. Curr Biol. 1999;9:1211–1220. doi: 10.1016/s0960-9822(99)80501-6. [DOI] [PubMed] [Google Scholar]
- 6.Starr DA, Han M. Role of ANC-1 in tethering nuclei to the actin cytoskeleton. Science. 2002;298:406–409. doi: 10.1126/science.1075119. [DOI] [PubMed] [Google Scholar]
- 7.Crisp M, Burke B. The nuclear envelope as an integrator of nuclear and cytoplasmic architecture. FEBS Lett. 2008;582:2023–2032. doi: 10.1016/j.febslet.2008.05.001. [DOI] [PubMed] [Google Scholar]
- 8.Haque F, Mazzeo D, Patel JT, Smallwood DT, Ellis JA, Shannahan CM, et al. Mammalian SUN protein networks at the inner nuclear membrane and their role in laminopathy disease processes. J Biol Chem. 2010;285:3487–3498. doi: 10.1074/jbc.M109.071910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Luxton GWG, Gomes ER, Folker ES, Vintinner E, Gundersen GG. Linear arrays of nuclear envelope proteins harness retrograde actin flow for nuclear migration. Science. 2010;329:956–959. doi: 10.1126/science.1189072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ostlund C, Folker ES, Choi JC, Gomes ER, Gundersen GG, Worman HJ. Dynamics and molecular interactions of linker of nucleoskeleton and cytoskeleton (LINC) complex proteins. J Cell Sci. 2009;122:4099–4108. doi: 10.1242/jcs.057075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wilhelmsen K, Litjens SH, Kuikman I, Tshimbalanga N, Janssen H, van den Bout I, et al. Nesprin-3, a novel outer nuclear membrane protein, associates with the cytoskeletal linker protein plectin. J Cell Biol. 2005;171:799–810. doi: 10.1083/jcb.200506083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gomes ER, Jani S, Gundersen GG. Nuclear movement regulated by Cdc42, MRCK, myosin and actin flow establishes MTOC polarization in migrating cells. Cell. 2005;121:451–463. doi: 10.1016/j.cell.2005.02.022. [DOI] [PubMed] [Google Scholar]
- 13.Luxton GW, Gomes ER, Folker ES, Vintinner E, Gundersen GG. Linear arrays of nuclear envelope proteins harness retrograde actin flow for nuclear movement. Science. 2010;329:956–959. doi: 10.1126/science.1189072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Fridolfsson HN, Starr DA. Kinesin-1 and dynein at the nuclear envelope mediate the bidirectional migrations of nuclei. J Cell Biol. 2010;191:115–128. doi: 10.1083/jcb.201004118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sulston JE, Horvitz HR. Post-embryonic cell lineages of the nematode, Caenorhabditis elegans. Dev Biol. 1977;56:110–156. doi: 10.1016/0012-1606(77)90158-0. [DOI] [PubMed] [Google Scholar]
- 16.Sulston JE, Schierenberg E, White JG, Thomson JN. The embryonic cell lineage of the nematode Caenorhabditis elegans. Dev Biol. 1983;100:64–119. doi: 10.1016/0012-1606(83)90201-4. [DOI] [PubMed] [Google Scholar]
- 17.Williams-Masson EM, Heid PJ, Lavin CA, Hardin J. The cellular mechanism of epithelial rearrangement during morphogenesis of the Caenorhabditis elegans dorsal hypodermis. Dev Biol. 1998;204:263–276. doi: 10.1006/dbio.1998.9048. [DOI] [PubMed] [Google Scholar]
- 18.Horvitz HR, Sulston JE. Isolation and genetic characterization of cell-lineage mutants of the nematode Caenorhabditis elegans. Genetics. 1980;96:435–454. doi: 10.1093/genetics/96.2.435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sulston JE, Horvitz HR. Abnormal cell lineages in mutants of the nematode Caenorhabditis elegans. Dev Biol. 1981;82:41–55. doi: 10.1016/0012-1606(81)90427-9. [DOI] [PubMed] [Google Scholar]
- 20.Starr DA, Hermann GJ, Malone CJ, Fixsen W, Priess JR, Horvitz HR, et al. unc-83 encodes a novel component of the nuclear envelope and is essential for proper nuclear migration. Development. 2001;128:5039–5050. doi: 10.1242/dev.128.24.5039. [DOI] [PubMed] [Google Scholar]
- 21.Lee KK, Starr DA, Cohen M, Liu J, Han M, Wilson KL, et al. Lamin-dependent localization of UNC-84, a protein required for nuclear migration in C. elegans. Mol Biol Cell. 2002;13:892–901. doi: 10.1091/mbc.01-06-0294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.McGee MD, Rillo R, Anderson AS, Starr DA. UNC-83 Is a KASH Protein Required for Nuclear Migration and Is Recruited to the Outer Nuclear Membrane by a Physical Interaction with the SUN Protein UNC-84. Mol Biol Cell. 2006;17:1790–1801. doi: 10.1091/mbc.E05-09-0894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Fridolfsson HN, Ly N, Meyerzon M, Starr DA. UNC-83 coordinates kinesin-1 and dynein activities at the nuclear envelope during nuclear migration. Dev Biol. 2010;338:237–250. doi: 10.1016/j.ydbio.2009.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Meyerzon M, Fridolfsson HN, Ly N, McNally FJ, Starr DA. UNC-83 is a nuclear-specific cargo adaptor for kinesin-1-mediated nuclear migration. Development. 2009;136:2725–2733. doi: 10.1242/dev.038596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gonczy P, Pichler S, Kirkham M, Hyman AA. Cytoplasmic dynein is required for distinct aspects of MTOC positioning, including centrosome separation, in the one cell stage Caenorhabditis elegans embryo. J Cell Biol. 1999;147:135–150. doi: 10.1083/jcb.147.1.135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ally S, Larson AG, Barlan K, Rice SE, Gelfand VI. Opposite-polarity motors activate one another to trigger cargo transport in live cells. J Cell Biol. 2009;187:1071–1082. doi: 10.1083/jcb.200908075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Del Bene F, Wehman AM, Link BA, Baier H. Regulation of neurogenesis by interkinetic nuclear migration through an apical-basal notch gradient. Cell. 2008;134:1055–1065. doi: 10.1016/j.cell.2008.07.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ross JL, Shuman H, Holzbaur EL, Goldman YE. Kinesin and dynein-dynactin at intersecting microtubules: motor density affects dynein function. Biophys J. 2008;94:3115–3125. doi: 10.1529/biophysj.107.120014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Welte MA. Bidirectional transport along microtubules. Curr Biol. 2004;14:525–537. doi: 10.1016/j.cub.2004.06.045. [DOI] [PubMed] [Google Scholar]
- 30.McKenney RJ, Vershinin M, Kunwar A, Vallee RB, Gross SP. LIS1 and NudE induce a persistent dynein force-producing state. Cell. 2010;141:304–314. doi: 10.1016/j.cell.2010.02.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Barbar E. Dynein light chain LC8 is a dimerization hub essential in diverse protein networks. Biochemistry. 2008;47:503–508. doi: 10.1021/bi701995m. [DOI] [PubMed] [Google Scholar]
- 32.Shubeita GT, Tran SL, Xu J, Vershinin M, Cermelli S, Cotton SL, et al. Consequences of motor copy number on the intracellular transport of kinesin-1-driven lipid droplets. Cell. 2008;135:1098–1107. doi: 10.1016/j.cell.2008.10.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Malone CJ, Misner L, Le Bot N, Tsai MC, Campbell JM, Ahringer J, et al. The C. elegans hook protein, ZYG-12, mediates the essential attachment between the centrosome and nucleus. Cell. 2003;115:825–836. doi: 10.1016/s0092-8674(03)00985-1. [DOI] [PubMed] [Google Scholar]
- 34.Roux KJ, Crisp ML, Liu Q, Kim D, Kozlov S, Stewart CL, Burke B. Nesprin 4 is an outer nuclear membrane protein that can induce kinesin-mediated cell polarization. Proc Nat Acad Sci USA. 2009;106:2194–2199. doi: 10.1073/pnas.0808602106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Splinter D, Tanenbaum ME, Lindqvist A, Jaarsma D, Flotho A, Yu KL, et al. Bicaudal D2, dynein and kinesin-1 associate with nuclear pore complexes and regulate centrosome and nuclear positioning during mitotic entry. PLoS Biol. 2010;8:1000350. doi: 10.1371/journal.pbio.1000350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Taverna E, Huttner WB. Neural progenitor nuclei IN motion. Neuron. 2010;67:906–914. doi: 10.1016/j.neuron.2010.08.027. [DOI] [PubMed] [Google Scholar]
- 37.Tsai JW, Lian WN, Kemal S, Kriegstein AR, Vallee RB. Kinesin 3 and cytoplasmic dynein mediate interkinetic nuclear migration in neural stem cells. Nat Neurosci. 2010;13:1463–1471. doi: 10.1038/nn.2665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Norden C, Young S, Link BA, Harris WA. Actomyosin is the main driver of interkinetic nuclear migration in the retina. Cell. 2009;138:1195–1208. doi: 10.1016/j.cell.2009.06.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Tsujikawa M, Omori Y, Biyanwila J, Malicki J. Mechanism of positioning the cell nucleus in vertebrate photoreceptors. Proc Nat Acad Sci USA. 2007;104:14819–14824. doi: 10.1073/pnas.0700178104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Zhang X, Lei K, Yuan X, Wu X, Zhuang Y, Xu T, et al. SUN1/2 and Syne/Nesprin-1/2 complexes connect centrosome to the nucleus during neurogenesis and neuronal migration in mice. Neuron. 2009;64:173–187. doi: 10.1016/j.neuron.2009.08.018. [DOI] [PMC free article] [PubMed] [Google Scholar]