Abstract
Transforming growth factor-beta (TGFβ) superfamily ligands are produced by and act upon testicular cells to control testis morphogenesis and adult fertility. Ligand production changes during testis development and dysregulated signaling affects the number of cells comprising each lineage and their development, with several components of this diverse signaling pathway linked to male infertility. To test the hypothesis that TGFβ superfamily signaling regulators are differentially expressed during mouse testis development, we surveyed expression of Hgs, Zfyve9, Smurf1 and Net25 by northern blot and in situ hybridization and SMURF2 and MAN1 by western blot and immunohistochemistry. Expression of these genes is highly regulated and differs between the first spermatogenic wave and adult spermatogenesis. Zfyve9 transcripts were first detected in Sertoli cells and spermatogonia at 5 days post partum (dpp) whereas Hgs mRNA was first detected in pachytene spermatocytes at 15 dpp. Smurf1 mRNA was broadly expressed at 0 and 5 dpp but restricted to spermatogonia and early spermatocytes at 15 dpp and spermatogonia, spermatocytes and round spermatids in adults. SMURF2 was limited to gonocyte nuclei at birth but was nuclear in all cells at 5 dpp. SMURF2 was absent from 15 dpp differentiating spermatogonia and early spermatocytes but readily detected in adult pachytene spermatocytes and round spermatids. MAN1 and Net25 also had different expression profiles, with MAN1 undetectable at 5 dpp. Differential synthesis of signaling modulators explains how Sertoli cells and spermatogenic cells, which all possess TGFβ superfamily signaling machinery and reside within the same microenvironment, respond differently to the same ligand.
Keywords: activin, Sertoli, spermatogenesis, smad, smurf, SARA, MAN1
Introduction
Quantitatively normal spermatogenesis requires the appropriate specification, proliferation and maturation of testicular somatic and germ cell lineages. Initiated early in embryogenesis, these processes continue during fetal and juvenile postnatal life to establish a functional adult testis. In the adult, cycling of the adult seminiferous epithelium by the periodic entry of spermatogonial stem cells into the differentiation pathway enables ongoing sperm production throughout life.
Testis development and the maintenance of adult spermatogenesis are tightly controlled by the endocrine system as well as by hormones and growth factors produced within the testis. Ligands of the transforming growth factor-beta (TGFβ) superfamily, which includes the prototypical TGFβs, activins, bone morphogenetic proteins (BMPs), growth and differentiation factors (GDFs) and glial cell line-derived neurotrophic factor (GDNF), are key regulators of testis development and spermatogenesis (reviewed in ref. 1). Synthesis of these ligands within the testis changes during development2–4 and their dysregulated production has significant effects on the number of cells comprising each lineage, the timing of developmental events and the capacity of cells to mature. For example, spermatogonial stem cells are depleted in mice with reduced GDNF production whereas spermatogonia overproliferate and fail to differentiate when GDNF is ovexpressed.5 In mice lacking inhibin, and which therefore have excessive activin signaling, uncontrolled proliferation and failure of Sertoli cells to mature leads to the development of Sertoli cell tumours.6 Mice with reduced levels of bioactive activin have fewer Sertoli cells7 and display features of delayed Sertoli cell maturation8 whereas analysis of germ cell differentiation markers indicates the first wave of spermatogenesis is advanced.9 Conversely, mice unable to produce activin A have fewer Sertoli cells but double the normal number of gonocytes at birth.10 TGFβ superfamily responsiveness within the developing and adult testis must therefore be precisely regulated to ensure appropriate organ development and optimal fertility in adulthood.
TGFβ superfamily ligands initiate intracellular signaling pathways upon binding to cell surface receptor complexes. Ligand-bound receptors recruit and phosphorylate receptor-activated SMAD (R-SMAD) proteins which complex with Co-SMAD4, accumulate in the nucleus and regulate target gene transcription. TGFβs, activins, GDF3 and GDF9 signals are transduced by SMAD2 and SMAD3 whereas BMPs, GDF6 and GDF7 signal via SMAD1, SMAD5 and SMAD8.11 TGFβ superfamily ligands also activate non-canonical pathways, including the mitogen activated protein kinases (MAPKs), ERK1/2, p38 and JNK.12
Distinctly different effects of TGFβ superfamily ligands on the proliferation and maturation of somatic and germ cells indicate that although they reside in the same microenvironment and possess appropriate receptors and intracellular signal transduction machinery, adjacent cells have different capacities to transduce these signals and their responses differ. In investigating this, our laboratory has uncovered remarkable regulation of TGFβ superfamily signal transducers and signaling modulators in the developing and adult testis. Inhibitory (I-) SMAD6, which downregulates TGFβ superfamily signaling,13,14 is readily detected in gonocytes of the neonatal mouse testis and in spermatogonia at 5 dpp yet undetectable in spermatogonia at 15 dpp.15 Expression of I-SMAD7 is ubiquitous in the developing testis but in adulthood is restricted to spermatogonia, spermatocytes and round spermatids.15 Similarly, ubiquitous expression of the BMP-responsive Smad1, Smad5 and Smad8 transcripts in the developing testis contrasts with restricted distribution of these transcripts in adult germ cells.15 In addition, we have described the potential for cellular responses to activin and TGFβ to be modulated by the regulated production of SnoN, a transcriptional repressor which interacts with SMAD2 and SMAD3,16,17 and of the kinase-deficient pseudoreceptor BAMBI (BMP and Activin Membrane Bound Inhibitor), which blocks signal transduction.18,19
Based on these findings, we hypothesized that the expression of other TGFβ superfamily signaling regulators would also be highly modulated to effect cell-specific ligand responses. We selected six modulators, three functionally related pairs, for which pre-existing data indicated they are expressed in the developing mouse testis (Fig. 1). These were Hgs (hepatocyte growth factor-regulated tyrosine kinase substrate), Zfyve9 (murine homologue of human SARA [Smad Anchor for Receptor Activation]), Smurf1 (SMad Ubiquitination-Related Factor-1), SMURF2, Net25 (Nuclear Envelope Transmembrane protein 25) and MAN1. Hgs and Zfyve9 encode endosome-localized FYVE domain containing proteins that facilitate signal transduction by promoting SMAD2/SMAD3 association with receptor complexes to increase C-terminal SMAD phosphorylation and transcriptional activity.20,21 Smurf1 and SMURF2 are members of the HECT family of E3 ubiquitin ligases which target phosphorylated R-SMADs22 and activated receptor complexes for proteasomal degradation.23 MAN1 (Lemd3), a component of the inner nuclear membrane, downregulates TGFβ and BMP-mediated SMAD signaling by sequestering R-SMADs away from chromatin and by abrogating MAPK activity.24–26 NET25 (Lemd2), which is similar to MAN1 but lacks the SMAD-binding RRM domain, is a potent inhibitor of MAPK activity.27
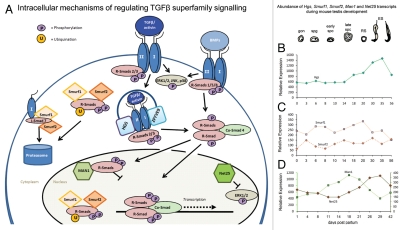
Figure 1.
(A) Intracellular modulators of the SMAD signaling pathway. The binding of a dimeric TGFβ superfamily ligand (e.g., TGFβ, activin or BMP) to its respective type I and type II receptors at the cell surface initiates phosphorylation of R-SMADs (SMAD2 and SMAD3 in the case of activins and TGFβ and SMAD1, SMAD5 and SMAD8 in response to BMPs). Phosphorylated R-SMADs complex with Co-SMAD4, accumulate in the nucleus and, in association with other transcriptional co-regulators, drive or repress target gene expression. TGFβ superfamily members can also activate the mitogen-activated protein kinases ERK1/2, p38 and JNK. SMAD2/3-mediated signaling is enhanced by the endosomal proteins HGS (hepatocyte growth factor-regulated tyrosine kinase substrate) and ZFYVE9 zinc finger FYVE domain containing 9 protein (the murine homologue of human SARA). SMURF1 and SMURF2 are E3 ubiquitin ligases that cause proteosomal degradation of activated receptors (in conjunction with the inhibitory SMAD7) and of SMAD2 and SMAD3, thereby downregulating activin/TGFβ signaling. MAN1 and NET25 localize to the inner nuclear membrane. MAN1 (Lemd3) antagonizes activin/TGFβ/BMP signaling by sequestering SMAD2/3 and SMAD1/5 away from chromatin binding. NET25 (Lemd2) is a truncated form of MAN1 that is unable to bind SMAD proteins but which is a potent inhibitor of mitogen activated protein kinases (MAPKs). Production of one or more of these modulators has the potential to selectively influence the capacity of a cell to respond to one or more TGFβ superfamily ligands. (B–D) Differential expression of genes encoding related TGFβ superfamily signaling modulators during mouse testis development, as determined by Affymetrix microarray. (B) Transcript levels of the endosome-localized HGS, (C) Smurf1 and Smurf2 and (D) Man1 and Net25 exhibit distinct profiles corresponding to progressive changes in the cellular populations throughout the first wave of spermatogenesis. Man1 transcript levels are highest between days 18 and 21, concordant with the prevalence of spermatocytes while Hgs and Net25 levels peak around day 26–35, when round spermatids predominate. Data are compiled from Schultz et al. 2003 and Shima et al. 2004 (Hgs: GDS605; Smurf1 and Smurf2: GDS606; Man1 and Net25: GDS409, available at www.ncbi.nlm.nih.gov/geo). Time of first appearance of differentiating germ cell types are depicted above graphs; gon: gonocyte, spg: spermatogonia; early spc: early spermatocytes; late spc: late spermatocytes; RS: round spermatids; ES: elongating spermatids.
We demonstrate that the expression of Hgs, Zfyve9, Smurf1 and Net25 mRNAs and the production and localization of SMURF2 and MAN1 proteins are highly regulated in somatic cells and germ cells in the developing and adult mouse testis. Our findings suggest that the specific functions of each enables cell-specific fine-tuning of cellular responses to TGFβ superfamily ligands and suggest a possible mechanism by which cells within the same microenvironment respond differently to surrounding cues.
Results
Hgs, Zfyve9, Smurf1, SMURF2, Net25 and MAN1 are expressed in the immature and adult mouse testis.
To identify whether regulators of TGFβ superfamily signaling have distinctive expression profiles during murine testis development, we initially surveyed existing GEO Profile datasets (www.ncbi.nlm.nih.gov/geo28) corresponding to Affymetrix microarray analysis of testis RNA from mice spanning birth through adulthood.29 The Hgs transcript level increased two-fold by 35 dpp relative to levels in 0–14 dpp testes and then reduced by half in the adult testis (day 56) (Fig. 1B). No probe set existed for Zfyve9. Smurf1 and Smurf2 transcripts did not change remarkably during postnatal testis development (Fig. 1C). An inverse relationship between Net25 and Man1 transcript profiles was apparent. Man1 transcripts peaked around 18 dpp but by maturity, levels had reduced to those measured in the newborn testis. Net25 levels peaked later, around 29 dpp (Fig. 1D). These initial data indicated the potential for differential production of TGFβ superfamily regulators with related functions. We pursued these observations by investigating the presence of Hgs, Zfyve9, Smurf1 and Net25 mRNAs in testes of immature (5–15 dpp) and adult mice by northern blot and in situ hybridization and examined expression of SMURF2 and MAN1 proteins, for which specific antibodies were available, by western blot and immunohistochemistry.
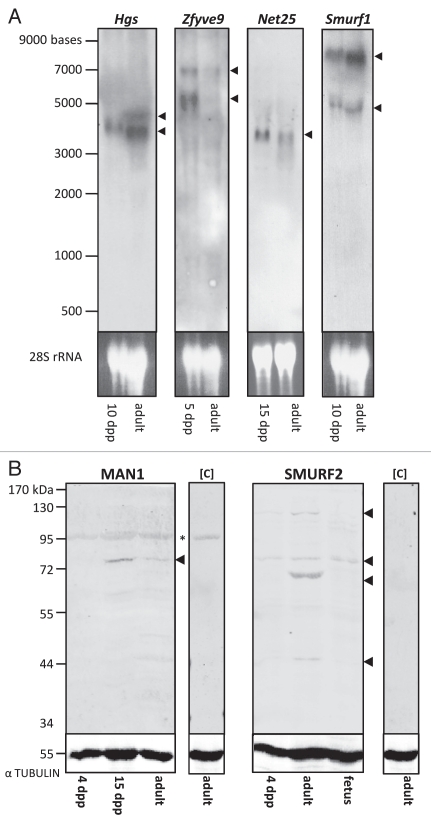
Northern blot analysis (Fig. 2A) identified a single transcript for Hgs of approximately 4 kb in 10 dpp testis and two transcripts in adult testis, one of 4 kb and a second transcript of apparently lesser abundance at 4.5 kb. Two Zfyve9 transcripts of approximately 5 and 7 kb were detected, with the smaller species present at relatively greater levels in the immature (5 dpp) compared to the adult sample. One distinct 3.5 kb Net25 transcript was detected in both immature (15 dpp) and adult testis samples. Two Smurf1 transcripts were detected in immature (10 dpp) and adult mouse testis, one at 7 kb and a second, of lesser abundance, at 5.3 kb.
Figure 2.
Hgs, Zfyve9, Net25 and Smurf1 mRNAs and MAN1 and SMURF2 proteins are expressed in immature and adult testes. (A) Northern blot analysis of Hgs, Zfyve9, Net25 and Smurf1 transcripts in total RNA isolated from immature and adult mouse testes, detected using digoxigenin-labeled antisense riboprobes and indicated by black arrowheads. Transcript sizes were determined using an RNA ladder (left). Ethidium bromide stained gel (lower part) indicates the amount of RNA loaded per lane. (B) Western blot analysis of immature and adult mouse testis lysates detected bands of the expected sizes for MAN1 and SMURF2 proteins in whole testis lysates at the indicated ages. Asterisk indicates non-specific binding of secondary antibody used for MAN1 detection, as determined by blotting adult testis lysate in the absence of primary antibody (control, [C]). SMURF2 protein was detected at the expected size in 4 dpp mouse testis lysates and lysates prepared from whole 12.5 dpc fetus. Additional bands detected in adult testis lysates at 44, 72 and 130 kDa suggest the existence of different SMURF2 isoforms. No band was detected in adult testis lysates in the absence of anti-SMURF2 primary antibody [control, (C)]. Detection of α-TUBULIN (lower part) indicates amount of protein present in each lane.
The antibody to MAN1 detected a protein at the expected size of 82 kDa30 by western blot in lysates from 15 dpp and adult mouse testes, but not in testis lysates from 4 dpp mice (Fig. 2B). A band of 86 kDa, the predicted size of SMURF2 (http://www.genecards.org/cgi-bin/carddisp.pl?gene=SMURF2), was detected in testis lysates from 4 dpp and adult mice as well as lysates prepared from whole 12.5 dpc fetus which was used as a positive control for protein size (Fig. 2B). The presence of additional bands at 44, 72 and 130 kDa in adult testis lysates, but which were not detected in fetal lysates, suggests the possibility that different SMURF2 isoforms exist in the testis.
Each member of the three functional pairs of TGFβ superfamily signaling regulators are differentially expressed in developing and adult mouse testes.
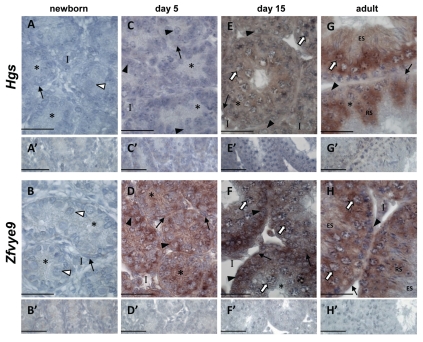
In the newborn testis, neither Hgs nor Zfyve9 mRNAs were detected (Fig. 3A and B). While absence of Hgs persisted at 5 dpp, Zfyve9 expression was readily detected in Sertoli cells, peritubular cells and spermatogonia at this age (Fig. 3C and D). By 15 dpp, a low level of signal indicated the presence of Hgs transcripts in spermatocytes (Fig. 3E). Zfyve9 transcripts were present in peritubular myoid, interstitial and germ cells, with signal more intense in spermatogonia relative to spermatocytes, but apparently absent from Sertoli cells (Fig. 3F). In the adult testis, Hgs mRNA was detected in spermatocytes, round spermatids and elongating spermatids (Fig. 3G) whereas Zfyve9 was most apparent in spermatogonia, spermatocytes and round spermatids (Fig. 3H).
Figure 3.
Hgs and Zfyve9 are differentially expressed in the developing postnatal and adult mouse testis. In situ hybridisation using DIG-labeled riboprobes localizes Hgs and Zfyve9 mRNAs (purple staining) in mouse testis sections at the indicated ages, counterstained with Harris Haematoxylin (blue) to visualize chromatin. In all cases, no signal was detected when sections were incubated with sense probe (A′–H′). White arrowhead: gonocytes; black arrow head: spermatogonia; white arrow: spermatocytes; RS: round spermatids; ES: elongating spermatids; asterisk: Sertoli cell cytoplasm; black arrow: peritubular myoid cells; I: interstitium. Scale bars = 50 µm.
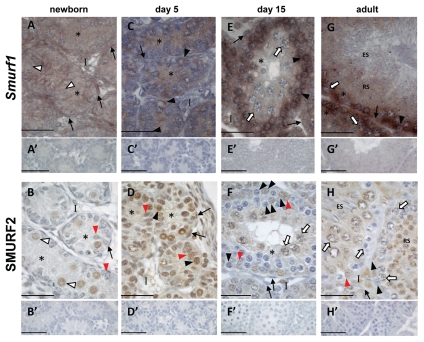
At birth, Smurf1 mRNA was readily detected in all cells, whereas SMURF2 protein was restricted to gonocyte nuclei (Fig. 4A and B). In the 5 dpp testis, Smurf1 expression was limited to Sertoli cells and spermatogonia, contrasting with the detection of SMURF2 in the nuclei of all cells at this age (Fig. 4C and D). At 15 dpp, Smurf1 was detected in all cells, with signal relatively stronger in spermatogonia and early meiotic cells, compared to late pachytene spermatocytes (Fig. 4E). SMURF2 protein was prominent in Sertoli cell nuclei, the cytoplasm of some, but not all, interstitial cells and both nucleus and cytoplasm of pachytene spermatocytes, a pattern distinctly different to that of Smurf1 transcripts. No protein was detected in B-type spermatogonia, preleptotene-leptotene spermatocytes or peritubular myoid cells (Fig. 4F). In the adult seminiferous epithelium, Smurf1 mRNA was present in Sertoli cells, spermatogonia and spermatocytes, with faint signal in round spermatids and no signal detected in and elongating spermatids (Fig. 4G). SMURF2 protein was readily detected in the nucleus and cytoplasm of Sertoli cells, spermatogonia, late pachytene spermatocytes and round spermatids but was absent from early spermatocytes and elongating spermatids (Fig. 4H).
Figure 4.
Smurf1 and SMURF2 expression is dynamic in immature and adult mouse testes. Localization of Smurf1 mRNA by in situ hybridisation using a DIG-labeled riboprobe (purple staining) and immunohistochemistry localizing SMURF2 protein (brown staining) in mouse testis sections at the indicated ages. Chromatin is visualized by counterstaining with Harris Haematoxylin (blue). In all cases, no signal was detected when sections were incubated with sense probe (A′, C′, E′ and G′) or in the absence of primary antibody (B′, D′, F′ and H′). White arrowhead, gonocytes; black arrow head, spermatogonia; white arrow, spermatocytes; RS, round spermatids; ES, elongating spermatids; asterisk, Sertoli cell cytoplasm; red arrowhead, Sertoli cell nucleus; black arrow, peritubular myoid cells; I, interstitium. Scale bars = 50 µm.
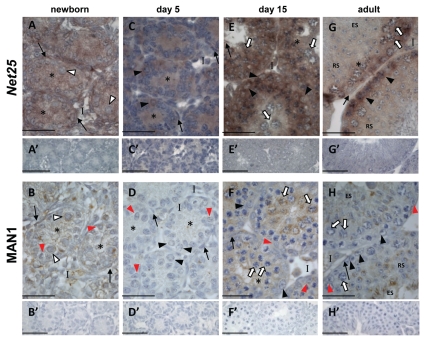
At birth, both Net25 (Fig. 5A) and MAN1 (Fig. 5B) were evident in all testicular cell types. Net25 mRNA continued to be detected in all cells of the 5 dpp testis (Fig. 5C) whereas MAN1 protein appeared absent (Fig. 5D), consistent with the inability to detect MAN1 protein in 4 dpp testis lysates by western Blot (Fig. 2B). At 15 dpp, both Net25 (Fig. 5E) and MAN1 (Fig. 5F) were readily detected in all cells, with intense MAN1 signal in pachytene spermatocyte cytoplasm. In the adult testis, Net25 mRNA was readily detected in Sertoli cells, spermatogonia and spermatocytes with signal intensity reduced in round spermatids and faint to absent in elongating spermatids (Fig. 5G). MAN1 protein was limited to the acrosomal region of round and elongating spermatids (Fig. 5H). The inability to detect MAN1 in pachytene spermatocytes of the adult testis was in stark contrast to the intense cytoplasmic signal observed in pachytene spermatocytes at 15 dpp.
Figure 5.
Net25 and MAN1 are differentially expressed in the developing and adult mouse testis. Localization of Net25 mRNA by in situ hybridisation using a DIG-labelled riboprobe (purple staining) and immunohistochemistry localizing MAN1 protein (brown staining) in mouse testis sections at indicated ages. Chromatin is visualized by counterstaining with Harris Haematoxylin (blue). In all cases, no signal was detected when sections were incubated with sense probe (A′, C′, E′ and G′) or in the absence of primary antibody (B′, D′, F′ and H′). White arrowhead, gonocytes; black arrow head, spermatogonia; white arrow, spermatocytes; RS, round spermatids; ES, elongating spermatids; asterisk, Sertoli cell cytoplasm; red arrowhead, Sertoli cell nucleus; black arrow, peritubular myoid cells; I, interstitium. Scale bars = 50 µm.
Discussion
Here we report that positive and negative modulators of TGFβ superfamily signaling display dynamic expression patterns and subcellular localization in the seminiferous epithelium of the developing and adult mouse testis. These data extend previous findings from our laboratory of highly regulated testicular expression of the inhibitory SMAD6 and SMAD7,15 the transcriptional repressor SnoN16 and the pseudoreceptor BAMBI18 and are consistent with current knowledge of TGFβ superfamily regulation of testis development and adult fertility.
The functional pairs of regulators studied here, Hgs and Zfyve9, Smurf1 and SMURF2 and Net25 and MAN1, are not co-regulated in somatic and germ cells of the developing or adult mouse testis. Based on the capacity of these related gene products to exert similar as well as unique effects on SMAD and MAPK activity, their regulated synthesis may enable discrete switches in cellular responses to TGFβ superfamily ligand stimulation. Furthermore, their distinctly different expression patterns in the first wave of spermatogenesis compared to the cycling adult seminiferous epithelium highlights the growing understanding that both germ cells and somatic cells respond differently to ligand stimulation in the juvenile versus mature testis.
Regulated production of signal-promoting and signal-inhibiting factors may direct germ cell responses to activin and BMPs at the onset of spermatogenesis.
In the neonatal testis, gonocyte re-entry into the cell cycle, migration to the basement membrane and transition into spermatogonia occur in the presence of high activin levels.4 Activin increases gonocyte numbers and impairs their differentiation into spermatogonia31 yet later promotes spermatogonial proliferation,32 illustrating the necessity for tightly regulated germ cell responses to activin at the time when the spermatogonial stem cell population is being established and the first spermatogonia enter the differentiation pathway. Our finding of a shift from the expression of signal-inhibitory factors (SMURF2, MAN1) to expression of a signal-promoting factor (Zfyve9) as gonocytes differentiate into spermatogonia suggests that regulated expression of signaling modulators may influence the change in the germ cell response to activin during this time.
BMP ligands also have distinct effects on mouse germ cells and Sertoli cells at the onset of the first wave of spermatogenesis around 5 dpp. BMP2 and BMP7 enhance spermatogonial and Sertoli cell proliferation, respectively,33 whereas BMP4 activates SMAD5, promoting spermatogonial proliferation and upregulating production of the survival and differentiation factor c-kit.34 Importantly, as activin opposes BMP4 actions at this age by downregulating c-kit synthesis,9 it is essential to differentially regulate spermatogonial responses to activin and BMP. As HGS interacts with SMAD5 to repress BMP-induced transcription in human chondrocytes35 and MAN1 abrogates SMAD1- and SMAD5-mediated BMP signaling,36 the absence of Hgs transcripts and MAN1 protein in 5 dpp spermatogonia may reflect a signaling status in germ cells that is permissive to BMP actions as they begin to differentiate.
A SMAD3-selective response of developing sertoli cells to activin corresponds to regulated expression of Zfyve9 and Hgs.
High activin levels in the neonatal testis also correlate with the most active period of postnatal Sertoli cell proliferation.37,38 Our inability to detect Hgs and Zfyve9 in the newborn testis, and the substantially delayed onset of Hgs expression relative to Zfyve9 during testis development, may be accounted for by the differential effects of SARA (human homologue of murine ZFYVE9) and HGS on activation of SMAD2 and SMAD3. Both SARA and HGS interact with internalized activin and TGFβ receptors at the early endosome to maximize SMAD activation.21,39–41 Although SARA interacts efficiently with both SMAD2 and SMAD3,39 SARA is necessary for maximal SMAD2 phosphorylation and transcriptional activity42 but is dispensible for efficient SMAD3-mediated signaling.43 HGS promotes activation of both SMAD2 and SMAD3,20 and whereas SMAD2 activation is increased when HGS and SARA are co-expressed,20 HGS can actually inhibit SMAD3-mediated signaling.43 We have previously described that activin signals via SMAD3 but not SMAD2, in immature (6 dpp) Sertoli cells.8 Our findings that Zfyve9 is absent from Sertoli cells at birth and that Hgs expression is not detected in immature Sertoli cells are consistent with conditions which selectively permit SMAD3-mediated but not SMAD2-mediated signaling and may represent the mechanism underlying preferential utilization of SMAD3 in response to activin.
Partitioning the regulation of the canonical versus non-canonical signaling response.
Dissimilar expression of MAN1 and Net25 may reflect distinct mechanisms by which TGFβ superfamily signaling blockades are segregated into broad ablation of SMAD and MAPK activity by MAN1 compared to select downregulation of MAPK activity by NET25. This is particularly relevant to spermatogonial stem cells, which in addition to interpreting activin and BMP inputs must also respond appropriately to GDNF, which signals via the non-canonical MAPK pathway. Selective blockade of MAPK activity may be influenced by the presence of Net25 transcripts in the absence of MAN1 protein in germ cells at 5 dpp, representing a means to regulate the response of spermatogonial stem cells to diverse signaling inputs. Later, in meiotic germ cells, the prominent, unexpected localization of MAN1 in the cytoplasm of pachytene spermatocytes at 15 dpp is identical to the localization of SMAD3 in pachytene spermatocytes at this age,44 suggesting the possibility that MAN1 may also function in the cytoplasm of these cells to regulate SMAD3 activity.
In Sertoli cells, TGFβ activates p38MAPK to regulate cyclic formation and breakdown of the blood-testis barrier (reviewed in ref. 45) which is essential for progression of germ cell differentiation through meiosis and spermiogenesis. Appropriate activin signals are also essential for normal Sertoli cell function.6,8 Differential Net25 and MAN1 production in developing and adult Sertoli cells may therefore selectively modulate SMAD and MAPK responses of Sertoli cells to TGFβ superfamily ligands.
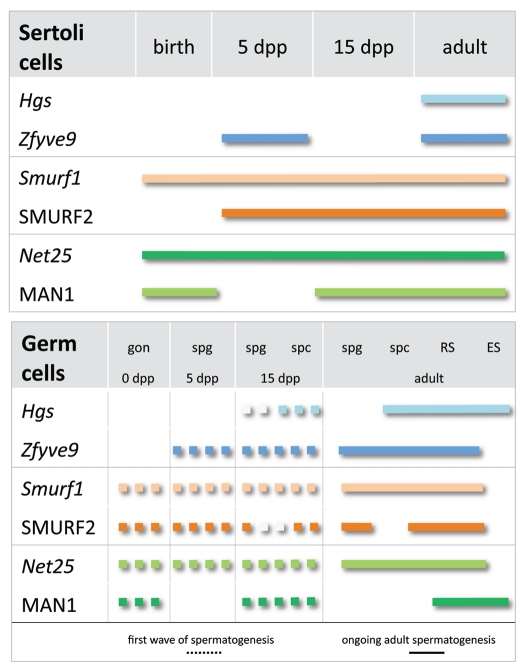
In summary, we describe striking differences in the expression of the related Hgs and Zfyve9, Smurf1 and SMURF2 and Net25 and MAN1 in germ cells and somatic cells during the first wave of spermatogenesis and in the adult testis, consistent with existing knowledge of TGFβ superfamily regulation of testis development and adult spermatogenesis (Fig. 6). Selective production of positive (HGS, ZFYVE9) and negative signaling (SMURF1, SMURF2, NET25, MAN1) regulators provide evidence of cell autonomous regulation of TGFβ superfamily signaling, contributing valuable knowledge to understanding how neighbouring cells, which each possess signaling machinery and are within the same microenvironment, respond differently to TGFβ superfamily signals.
Figure 6.
Intracellular modulators of TGFβ superfamily signaling are differentially expressed in germ cells of the developing and adult mouse testis. Striking differences in the expression of the related Hgs and Zfyve9, Smurf1 and SMURF2 and Net25 and MAN1 were detected within germ and Sertoli cells between the first wave of spermatogenesis and in the cycling adult seminiferous epithelium. Selective modulation of responses to TGFβ superfamily ligands during differentiation may be endowed by regulated production of positive (HGS, ZFYVE9) and negative signaling (SMURF1, SMURF2, NET25, MAN1) regulators. Due to their individual capacities to modulate signal transduction, differential expression of these functionally related pairs may confer important switches in the cellular responses to TGFβ superfamily ligands. Lower panel: dotted line indicates expression pattern in germ cells during the first wave of spermatogenesis; solid line indicates expression in germ cells of the adult testis. Gon, gonocyte; spg,spermatogonium; spc, spermatocyte; RS, round spermatid; ES, elongating spermatid.
Materials and Methods
Geo profiles.
The expression graphs for Hgs, Smurf1, Smurf2, Net25 and Man1 were generated by downloading publically available data records from the NCBI website (www.ncbi.nlm.nih.gov/geo), generated as described in reference 28 and 29. Expression values for each gene in GEO Datasets GDS409, GDS605 and GDS606 were chosen from probe sets that yielded values above 50 and graphed using Microsoft Excel.
Experimental animals and tissues.
Newborn, 5 dpp, 15 dpp and adult outbred mice (C56Bl/6 × CBA) were obtained from Monash University Central Animal Services. Juvenile animals were killed by decapitation and adult animals were asphyxiated with CO2 followed by cervical dislocation before tissue removal. All investigations conformed to the NHMRC/CSIRO/AAC Code of Practice for the Care and Use of Animals for Experimental Purposes and were approved by the Monash University Standing Committee on Ethics in Animal Experimentation. Testes for RNA and protein extraction were snap frozen on dry ice and either processed immediately or stored at −80°C until required. Intact tissue samples for in situ hybridization and immunohistochemistry were placed in Bouin's fixative for 5 hrs immediately after collection then dehydrated through a graded ethanol series and embedded in paraffin. Sections of 3–5 µm were placed on Superfrost Plus II slides (Lomb Scientific #4758).
RNA isolation, cDNA synthesis, northern blot analysis and in situ hybridization.
RNA was prepared from testis tissue using TRIzol reagent (Invitrogen #15596026) and contaminating genomic DNA was eliminating using DNAfree (Applied Biosystems, #AM1906) according to the manufacturer's guidelines. cDNA synthesis was performed by reverse transcribing 1 µg of total RNA using 100 U Superscript III reverse transcriptase (Invitrogen #18080093) with 2.5 µM random hexamer oligonucleotides (Roche, #58002113-1) according to manufacturer's guidelines. Primer sequences, accession numbers of genes from which primers were designed and region amplified are listed in Table 1. Amplification parameters were 95°C for 4 mins, 40 cycles of 95°C (30 s), 60°C (60 s) and 72°C (30 s) using 1 µl cDNA.
Table 1.
Oligonucleotide primer sequences used to amplify Hgs, Zfyve9, Smurf1 and Net25
| Gene | Accession number | Forward primer (5′-3′) | Reverse primer (5′-3′) | Region amplified |
| Hgs | NM_001159328 | TGT GTG AGC CCT GCT ATG AG | CTGCCTTGGATGTGCTGTA | 724–770 bp |
| Zfyve9 | NM_183300 | CAG GAA CTC TGG CTG TGC A | ACC AGT GAG ATG AGG ATG G | 456–628 bp |
| Smurf1 | NM_001038627 | CCC TTG CCA AGA TTG TTG T | CCA CTA TTT GGC CAC GAA CT | 390–688 bp |
| Net25 | NM_146075 | GAG GCG CAG GAG TAC ATA GC | GGC ACC AGA AGA AGA GTG AGC | 954–1203 bp |
Probes for northern blot and in situ hybridization were derived from RTPCR products which were cloned into pGEM T-Easy (Promega Corp., #A1360) following the manufacturer's instructions and sequenced for verification (Big Dye Terminator v3.1 Cycle Sequencing Kit, ABIPRISM 377 DNA Sequencer, Applied Biosystems) by the Gandel Charitable Trust Sequencing Centre, Monash Institute of Medical Research, Clayton, VIC, Australia. PCR amplification of these plasmids using M13 forward and reverse primers produced products that included T7 and SP6 RNA polymerase binding sites which were used as templates for in vitro transcription to yield sense and antisense cRNAs using digoxigenin-(DIG-) labeled dNTPs (Roche #1277073).
Northern blots were performed to assess the specificity of probe target recognition and to establish transcript sizes. Twenty to twentyfive µg of total RNA isolated from immature (5, 10 or 15 dpp) and adult mouse testes were separated on 1.1% agarose/formaldehyde gels and transferred to Hybond N membranes (Amersham Biosciences, #RPN303N). Membranes were prehybridized at 68°C with Ultrahyb (Ambion, AM8669) for 1–2 hrs then hybridized with Ultrahyb containing 25 ng/ml antisense probe at 68°C overnight. Membranes were then washed to a stringency of 0.1× standard saline citrate (SSC) and 0.1% sodium dodecyl sulfate (SDS) at 68°C. Bound DIG-labeled riboprobe was detected using an anti-DIG antibody (Roche, #11093274910). Chemiluminescent signal generated by CDP-Star (Roche, #11685672001) substrate was detected by exposure of membranes to Kodak Hyperfilm (Amersham Biosciences, #28-9068-36). Northern blots were performed twice.
In situ hybridization was used to localize Hgs, Zfyve9, Smurf1 and Net25 transcripts in mouse testis sections. Hybridization was performed with 100–400 ng probe per slide at 50–60°C with stringency washes to 0.1× SSC at the hybridization temperature. Bound DIG-labeled riboprobe was detected using an anti-DIG antibody (Roche, #11093274910) and visualized by purple staining using 5-Bromo-4chloro-3-indoyl phosphate/nitroblue tetrazolium (BCIP/NBT) substrate (Thermo Scientific, #34042). Sections were counterstained with Harris haematoxylin to visualize chromatin and mounted in GVA aqueous mounting solution (Invitrogen #00-800). Both antisense and sense (negative control) probes were used at the same concentration on each sample, in every experiment, for each set of conditions tested. In situ hybridization was performed at least three times for each age using tissues from at least three different animals. Images were captured using a Leica DMR microscope with a Leica DC200 digital camera (Leica).
Western blot and immunohistochemistry.
Western blots were performed using lysates from 4 dpp, 15 dpp or adult mouse testes and from whole fetus at embryonic day 12.5. Samples were homogenized at 4°C in RIPA buffer (150 mM sodium chloride, 1% Nonidet P-40, 0.5% Tween-20, 0.1% SDS, 1 mM ethylene-diaminetetraacetic acid) in the presence of protease inhibitors (Protease inhibitor cocktail set III, Sigma Aldrich, #P8340). Samples were incubated on ice for 10 mins then centrifuged at 13,000 rpm for 10 mins. Supernatant was recovered and lysate concentration was determined using the Bio-Rad DC protein assay (Bio-Rad, #500-0116). Thirty µg of protein per lane was separated by electrophoresis in a 10% SDS polyacrylamide gel against protein size standards (Fermentas, #SM0671). Lysates were diluted 1:1 in SDS reducing buffer (0.17 M Tris-HCl pH 6.8, 34% glycerol, 0.1% β-mercaptoethanol, 2% SDS, 0.01% bromophenol blue), incubated at 95°C for 10 mins then placed on ice before loading into gel. Samples underwent electrophoresis at 35 mA for 1.5 hrs in running buffer consisting of 3 g/l Tris base, 14.4 g/l glycine, 1 g/l SDS, pH 8.3. Following electrophoresis, proteins were transferred to Hybond C nitrocellulose membrane (Amersham Biosciences, #RPN203E) for 1.5 hrs in transfer buffer (3 g/l Tris base, 14.4 g/l glycine, 10% methanol) at 80 V. Membranes were air dried, prewet with TBS then blocked for 1 hr in 2:1 TBS:Odyssey blocking buffer (LICOR Biosciences #927-40000). Primary antibody incubation was carried out overnight at 4°C in blocking buffer plus 0.1% Tween. Anti-SMURF2 (Abcam #ab38543) was used at 250 ng/ml and anti-MAN1 (Santa Cruz #sc-19785) was used at 200 ng/ml. Anti-alpha-TUBULIN (Sigma Aldrich, #T5168) was used as a loading control at a dilution of 1:6,000. Unbound primary antibody was washed off by 4 × 5 minute washes in 1× TBS plus 0.1% Tween. Bound primary antibodies were detected using donkey-anti-rabbit AlexaFluor 680 (Invitrogen, #A10043) (SMURF2), donkey-anti-goat IR-800 (Rockland Immunochemicals #605-732-125) (MAN1) or rabbit-anti-mouse IR-800 (Rockland Immunochemicals, #310-432-002) (alpha-TUBULIN) at 1:10,000 dilution in blocking solution with 0.1% Tween and 0.01% SDS for 1 hr at room temp then washed 4 × 5 mins in TBS plus 0.1% Tween. Bound antibody was detected with the LICOR Odyssey System (John Morris Scientific). Western blots were performed once and negative control blots were performed for each experiment using adult mouse testis lysate in the absence of primary antibody to assess background signal.
Immunohistochemistry was performed to localize SMURF2 and MAN1 in Bouins fixed testis sections. Briefly, sections were dewaxed, rehydrated and treated with 0.3% hydrogen peroxide [5 min, room temperature (RT)] to quench endogenous peroxidases. To detect SMURF2 and MAN1, antigen retrieval was performed by heating in 50 mM glycine pH 3.5 and maintaining temperature at 90°C for 10 mins using 800 W microwave oven then left to cool for 20 minutes. Slides were washed 3 × 5 min at RT in tris-buffered saline (50 mM Tris, 150 mM NaCl, pH 7.5) (TBS) between all subsequent incubations. Blocking solution and antibody diluent consisted of 5% normal serum diluted in TBS/0.1% BSA (Sigma Aldrich, #A7906) and performed for at least 20 mins at RT in a humid chamber. Sections were incubated with primary antibody overnight at RT in a humid chamber. Anti-SMURF2 (Abcam, #ab38543) was used at 10 ng/ml and anti-MAN1 (Santa Cruz, #sc-19785) at 2 ng/ml. Bound anti-SMURF2 was detected using biotinylated anti-rabbit antibody (Chemicon, #AP322B) and anti-MAN1 was detected with biotinylated anti-goat (DAKO, #E0466). Signal was amplified with Vectastain Elite ABC kit reagents according to the manufacturer's instructions (Vector Laboratories, #PK-6100) followed by detection with DAB (3,3-diaminobenzidine tetrahydrochloride, DAKO, #K3468) to produce a brown precipitate. Harris haemotoxylin was used as a counterstain to enable visualization of chromatin. Sections were dehydrated in an ethanol series and mounted under DPX (Sigma Aldrich, #44581). Immunohistochemistry was performed at least three times for each age using tissues from at least three different animals. For each antibody in each experiment, the negative control to detect non-specific binding of secondary and tertiary reagents consisted of identical treatments with the exception that the primary antibody was omitted and in all cases, no signal was observed. Images were captured using a Leica DMR microscope with a Leica DC200 digital camera.
Acknowledgements
Support from the NHMRC of Australia (#545917 and #545916 to K.L.), the Australian Research Council (#348239 to K.L.), Monash University (Postgraduate Scholarship and Bridging Postdoctoral Fellowship to C.I.) is hereby acknowledged.
Abbreviations
- BCIP/NBT
5-bromo-4chloro-3-indoyl phosphate/nitroblue tetrazolium
- BMP
bone morphogenetic protein
- BSA
bovine serum albumin
- dpp
days post partum
- DAB
3,3-diaminobenzidine tetrahydrochloride
- DIG
digoxigenin
- ES
elongating spermatid
- GDF
growth and differentiation factor
- GDNF
glial cell-line derived neurotrophic factor
- GEO
gene expression omnibus
- gon
gonocyte
- GVA
glycerol vinyl alcohol
- MAPK
mitogen activated protein kinase
- NCBI
national center for biotechonology information
- NHMRC
national health and medical research council
- RS
round spermatid
- RT
room temperature
- RTPCR
reverse transcription polymerase chain reaction
- SARA
smad anchor for receptor activation
- SDS
sodium dodecyl sulphate
- spc
spermatocyte
- spg
spermatogonium
- TBS
tris-buffered saline
- TGFβ
transforming growth factor beta
References
- 1.Itman C, Mendis S, Barakat B, Loveland KL. All in the family: TGF-beta family action in testis development. Reproduction. 2006;132:233–246. doi: 10.1530/rep.1.01075. [DOI] [PubMed] [Google Scholar]
- 2.Mullaney BP, Skinner MK. Transforming growth factor-beta (beta 1, beta 2, and beta 3) gene expression and action during pubertal development of the seminiferous tubule: potential role at the onset of spermatogenesis. Mol Endocrinol. 1993;7:67–76. doi: 10.1210/mend.7.1.8446109. [DOI] [PubMed] [Google Scholar]
- 3.Buzzard JJ, Loveland KL, O'Bryan MK, O'Connor AE, Bakker M, Hayashi T, et al. Changes in circulating and testicular levels of inhibin A and B and activin A during postnatal development in the rat. Endocrinology. 2004;145:3532–3541. doi: 10.1210/en.2003-1036. [DOI] [PubMed] [Google Scholar]
- 4.Barakat B, O'Connor AE, Gold E, de Kretser DM, Loveland KL. Inhibin, activin, follistatin and follicle stimulating hormone serum levels and testicular production are highly modulated during the first spermatogenic wave in mice. Reproduction. 2008;136:345–359. doi: 10.1530/REP-08-0140. [DOI] [PubMed] [Google Scholar]
- 5.Meng XM, Lindahl M, Hyvonen ME, Parvinen M, de Rooij DG, Hess MW, et al. Regulation of cell fate decision of undifferentiated spermatogonia by GDNF. Science. 2000;287:1489–1493. doi: 10.1126/science.287.5457.1489. [DOI] [PubMed] [Google Scholar]
- 6.Matzuk MM, Finegold MJ, Su JG, Hsueh AJ, Bradley A. Alpha-inhibin is a tumour-suppressor gene with gonadal specificity in mice. Nature. 1992;360:313–319. doi: 10.1038/360313a0. [DOI] [PubMed] [Google Scholar]
- 7.Brown CW, Houston-Hawkins DE, Woodruff TK, Matzuk MM. Insertion of Inhbb into the Inhba locus rescues the Inhba-null phenotype and reveals new activin functions. Nat Genet. 2000;25:453–457. doi: 10.1038/78161. [DOI] [PubMed] [Google Scholar]
- 8.Itman C, Small C, Griswold MD, Nagaraja AK, Matzuk MM, Brown C, et al. Developmentally regulated SMAD2 and SMAD3 utilization directs activin signalling outcomes. Dev Dyn. 2009;238:1688–700. doi: 10.1002/dvdy.21995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Mithraprabhu S, Mendis S, Meachem S, Tubino L, Matzuk MM, Brown CW, et al. Activin bioactivity affects germ cell differentiation in the postnatal mouse testis in vivo. Biol Reprod. 2010;82:980–990. doi: 10.1095/biolreprod.109.079855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Mendis S, Meachem S, Sarraj M, Loveland KL. Activin A balances Sertoli and germ cell proliferation in the fetal mouse testis. Biol Reprod. 2011;84:379–391. doi: 10.1095/biolreprod.110.086231. [DOI] [PubMed] [Google Scholar]
- 11.Feng XH, Derynck R. Specificity and versatility in TGFbeta signaling through Smads. Ann Rev Cell Dev Biol. 2005;21:659–693. doi: 10.1146/annurev.cellbio.21.022404.142018. [DOI] [PubMed] [Google Scholar]
- 12.Moustakis A, Heldin CH. Non-Smad TGFbeta signals. J Cell Sci. 2005;118:3573–3584. doi: 10.1242/jcs.02554. [DOI] [PubMed] [Google Scholar]
- 13.Kavsak P, Rasmussen RK, Causing CG, Bonni S, Zhu H, Thomsen GH, et al. Smad7 binds to Smurf2 to form an E3 ubiquitin ligase that targets the TGFbeta receptor for degradation. Mol Cell. 2000;6:1365–1375. doi: 10.1016/s1097-2765(00)00134-9. [DOI] [PubMed] [Google Scholar]
- 14.Hata A, Lagna G, Massague J, Hemmati-Brivanlou A. Smad6 inhibits BMP/Smad1 signaling by specifically competing with the Smad4 tumour suppressor. Genes Dev. 1997;12:186–197. doi: 10.1101/gad.12.2.186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Itman C, Loveland KL. SMAD expression in the testis: an insight into BMP regulation of spermatogenesis. Dev Dyn. 2008;237:97–111. doi: 10.1002/dvdy.21401. [DOI] [PubMed] [Google Scholar]
- 16.Itman C, Whiley PA, Zhou W, Meistrich ML, Sahin Z, Loveland KL. Regulated production of SnoN2 is a feature of testicular differentiation. Microsc Res Tech. 2009;72:833–844. doi: 10.1002/jemt.20739. [DOI] [PubMed] [Google Scholar]
- 17.Stroschein SL, Wang W, Zhou S, Zhou Q, Luo K. Negative feedback regulation of TGFbeta signaling by the SnoN oncoprotein. Science. 1999;286:771–774. doi: 10.1126/science.286.5440.771. [DOI] [PubMed] [Google Scholar]
- 18.Loveland KL, Bakker M, Meehan T, Christy E, von Schonfeldt V, Drummond A, et al. Expression of Bambi is widespread in juvenile and adult rat tissues and is regulated in male germ cells. Endocrinology. 2003;144:4180–4186. doi: 10.1210/en.2002-0124. [DOI] [PubMed] [Google Scholar]
- 19.Onichtchouk D, Chen YG, Dosch R, Gawantka V, Delius H, Massague J, et al. Silencing of TGFbeta signalling by the pseudoreceptor BAMBI. Nature. 1999;401:480–485. doi: 10.1038/46794. [DOI] [PubMed] [Google Scholar]
- 20.Miura S, Takeshita T, Asao H, Kumura Y, Murata K, Sasaki Y, et al. Hgs (Hrs), a FYVE domain protein, is involved in Smad signaling through cooperation with SARA. Mol Cell Biol. 2000;20:9346–9355. doi: 10.1128/mcb.20.24.9346-9355.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Itoh F, Divecha N, Brocks L, Oomen L, Janssen HJC, et al. The FYVE domain in Smad anchor for receptor activation (SARA) is sufficient for localization of SARA in early endosomes and regulates TGFbeta/Smad signalling. Genes to Cells. 2002;7:321–331. doi: 10.1046/j.1365-2443.2002.00519.x. [DOI] [PubMed] [Google Scholar]
- 22.Ebisawa T, Fukuchi M, Murakami G, Chiba T, Tanaka K, Imamura T, et al. Smurf1 interacts with transforming growth factor-beta type I receptor through Smad7 and induces receptor degradation. J Biol Chem. 2001;276:12477–12480. doi: 10.1074/jbc.C100008200. [DOI] [PubMed] [Google Scholar]
- 23.Lin X, Liang M, Feng XH. Smurf2 is a ubiquitin E3 ligase mediating proteasome-dependent degradation of Smad2 in TGFbeta signalling. J Biol Chem. 2000;275:36818–36822. doi: 10.1074/jbc.C000580200. [DOI] [PubMed] [Google Scholar]
- 24.Wagner N, Weyhersmuller A, Blauth A, Schuhmann T, Heckmann M, Krohne G, et al. The Drosophila LEM-domain protein MAN1 antagonizes BMP signaling at the neuromuscular junction and the wing crossveins. Dev Biol. 2010;339:1–13. doi: 10.1016/j.ydbio.2009.11.036. [DOI] [PubMed] [Google Scholar]
- 25.Ishimura A, Ng JK, Taira M, Young SG, Osada SI. Man1, an inner nuclear membrane protein, regulates vascular remodeling by modulating transforming growth factor beta signaling. Development. 2006;133:3919–3928. doi: 10.1242/dev.02538. [DOI] [PubMed] [Google Scholar]
- 26.Hellemans J, Preobrazhenska O, Willaert A, Debeer P, Verdonk PCM, Costa T, et al. Loss-of-function mutations in LEMD3 result in osteopoikilosis, Buschke-Ollendorff syndrome and melorheostosis. Nat Genet. 2004;36:1213–1218. doi: 10.1038/ng1453. [DOI] [PubMed] [Google Scholar]
- 27.Huber MD, Guan T, Gerace L. Overlapping functions of nuclear envelope proteins NET25 (Lem2) and Emerin in regulation of extracellular signal-regulated kinase signalling in myoblast differentiation. Mol Cell Biol. 2009;29:5718–5728. doi: 10.1128/MCB.00270-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Barrett T, Suzek TO, Broup DB, Wilhite SE, Ngau WC, Ledoux P, et al. NCBI GEO: mining millions of expression profiles—database and tools. Nucleic Acids Res. 2005;33:562–566. doi: 10.1093/nar/gki022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Shima JE, McLean DJ, McCarrey JR, Griswold MD. The murine testicular transcriptome: characterizing gene expression in the testis during the progression of spermatogenesis. Biol Reprod. 2004;71:319–330. doi: 10.1095/biolreprod.103.026880. [DOI] [PubMed] [Google Scholar]
- 30.Lin F, Blake DL, Callebaut I, skerjanc IS, Holmer L, McBurney MW, et al. MAN1, an inner nuclear membrane protein that shares the LEM domain with lamina-associated polypeptide 2 and emerin. J Biol Chem. 2000;275:4840–4847. doi: 10.1074/jbc.275.7.4840. [DOI] [PubMed] [Google Scholar]
- 31.Meehan T, Schlatt S, O'Bryan M, de Kretser DM, Loveland KL. Regulation of germ cell and Sertoli cell development by activin, follistatin and FSH. Dev Biol. 2000;220:225–237. doi: 10.1006/dbio.2000.9625. [DOI] [PubMed] [Google Scholar]
- 32.Mather JP, Attie KM, Woodruff TK, Rice GC, Phillips DM. Activin stimulates spermatogonial proliferation in germ-Sertoli cell cultures from immature rat testis. Endocrinology. 1990;127:3206–3214. doi: 10.1210/endo-127-6-3206. [DOI] [PubMed] [Google Scholar]
- 33.Puglisi R, Montanari M, Chiarella P, Stefanini M, Boitani C. Regulatory role of BMP2 and BMP7 in spermatogonia and Sertoli cell proliferation in the immature mouse. Eur J Endocrinol. 2004;151:511–520. doi: 10.1530/eje.0.1510511. [DOI] [PubMed] [Google Scholar]
- 34.Pellegrini M, Grimaldi P, Rossi P, Geremia R, Dolci S. Developmental expression of BMP4/ALK3/SMAD5 signalling pathway in the mouse testis: a potential role of BMP4 ini spermatogonia differentiation. J Cell Sci. 2003;116:3363–3372. doi: 10.1242/jcs.00650. [DOI] [PubMed] [Google Scholar]
- 35.Haag J, Chubinskaya S, Aigner T. Hgs physically interacts with Smad5 and attenuates BMP signalling. Exper Cell Res. 2006;312:1153–1163. doi: 10.1016/j.yexcr.2006.01.019. [DOI] [PubMed] [Google Scholar]
- 36.pan D, Estevez-Salmeron LD, Stroschein SL, Zhu X, He J, Zhou S, et al. The integral inner nuclear membrane protein MAN1 physically interacts with the R-Smad proteins to repress signalling by the transforming growth factor beta superfamily of cytokines. J Biol Chem. 2005;289:15992–16001. doi: 10.1074/jbc.M411234200. [DOI] [PubMed] [Google Scholar]
- 37.Boitani C, Stefanini M, Fragale A, Morena AR. Activin stimulates Sertoli cell proliferation in a defined period of rat testis development. Endocrinology. 1995;136:4538–4544. doi: 10.1210/endo.136.12.7588293. [DOI] [PubMed] [Google Scholar]
- 38.Buzzard JJ, Farnworth PG, de Kretser DM, E OCA, Wreford NG, Morrison JR. Proliferative phase Sertoli cells display a developmentally regulated response to activin in vitro. Endocrinology. 2003;144:474–483. doi: 10.1210/en.2002-220595. [DOI] [PubMed] [Google Scholar]
- 39.Tsukazaki T, Chiang T, Davison AF, Attisano L, Wrana JL. SARA, a FYVE domain protein that recruits Smad2 to the TGFβ receptor. Cell. 1998;95:779–791. doi: 10.1016/s0092-8674(00)81701-8. [DOI] [PubMed] [Google Scholar]
- 40.Hayes SA, Zarnegar M, Sharma M, Yang F, Peehl DM, ten Dijke P, et al. Smad3 represses androgen receptor-mediated transcription. Cancer Res. 2001;61:2112–2118. [PubMed] [Google Scholar]
- 41.Panopoulou E, Gillooly DJ, Wrana JL, Zerial M, Stenmark H, Murphy C, et al. Early endosomal regulation of Smad-dependent signaling in endothelial cells. J Biol Chem. 2002;227:18046–18052. doi: 10.1074/jbc.M107983200. [DOI] [PubMed] [Google Scholar]
- 42.Runyan CE, Schnaper HW, Poncelet AC. The role of internalization in transforming growth factor beta1-induced Smad2 association with Smad anchor for receptor activation (SARA) and Smad2-dependent signaling in human mesangial cells. J Biol Chem. 2005;289:8300–8308. doi: 10.1074/jbc.M407939200. [DOI] [PubMed] [Google Scholar]
- 43.Goto D, Nakajima N, Mori Y, Kurasawa K, Kitamura N, Iwamoto I. Interaction between Smad anchor for receptor activation and Smad3 is not essential for TGFbeta/Smad3 mediated signaling. Biochem Biophys Res Commun. 2001;281:1100–1105. doi: 10.1006/bbrc.2001.4489. [DOI] [PubMed] [Google Scholar]
- 44.Itman C, Wong C, Hunyadi B, Ernst M, Jans DA, Loveland KL. Smad3 regulates androgen responsiveness and influences maturation in the juvenile testis. Endocrinology. 2011 doi: 10.1210/en.2010-1453. [DOI] [PubMed] [Google Scholar]
- 45.Lui WY, Lee WM, Cheng CY. TGFbetas: their role in testicular function and Sertoli cell tight junction dynamics. Int J Androl. 2003;26:147–160. doi: 10.1046/j.1365-2605.2003.00410.x. [DOI] [PubMed] [Google Scholar]