Abstract
Prostate cancer (PCa) is a major age-related malignancy as increasing age correlates with increased risk for developing this neoplasm. Similarly, alterations in circadian rhythms have also been associated with the aging population and cancer risk. The pineal hormone melatonin is known to regulate circadian rhythms, which are under the control of a core set of genes: Period 1, 2, 3 (Per 1 – 3); Cryptochrome 1, 2 (Cry 1, 2); Clock, and Bmal 1, 2. Melatonin levels have been shown to decrease in cancer patients and exogenous melatonin exhibits anti-proliferative effects against certain cancers. In this study, we challenged the hypothesis that melatonin imparts anti-proliferative effects in prostate cancer via resynchronization of deregulated core clock circuitry. We found that Clock and Per2 protein levels were downregulated whereas Bmal1 protein levels were upregulated in PCa cells, compared to normal prostate cells. Additionally, employing automated quantitative analysis of a microarray containing human tissues, we found that compared to benign tissues, Clock and Per2 levels were downregulated whereas Bmal1 levels were upregulated in PCa and other proliferative prostatic conditions. Overexpression of Per2 was found to result in a significant loss of PCa cell growth and viability. Interestingly, melatonin treatment resulted in an increase in Per2 and Clock and a reduction in Bmal1 in PCa cells. Further, melatonin treatment resulted in a resynchronization of oscillatory circadian rhythm genes (Dbp and Per2). Our data support our hypothesis and suggest that melatonin should be thoroughly investigated as an agent for the management of PCa and other age-related malignancies.
Keywords: circadian rhythm, prostate cancer, melatonin
Introduction
Prostate cancer (PCa) is an age-related disease with over 75% of cases diagnosed in men aged 65 and older. Additionally, it was estimated that in 2009 there would be 192,280 new cases of PCa diagnosed and 27,360 deaths will be attributed to this disease [1]. With the continuing shift of the American demographic pattern toward an older population, a further increase in the PCa cases and associated deaths are expected in the future. Therefore, PCa drug development efforts must focus towards identifying links between aging and PCa. Alterations in circadian rhythms as well as production of the pineal hormone melatonin (N-acetyl-5-methoxytryptamine) have been linked to aging and cancer [2–6].
Circadian rhythms obey intrinsic clocks to function and direct a variety of physiological and metabolic functions, such as core body temperature and sleep-wake cycles. These intrinsic clocks include the central pacemaker located in the suprachiasmatic nuclei (SCN), as well as several peripheral clocks. Interestingly, like PCa, changes in circadian rhythms as well as in the period of the circadian pacemaker controlling these rhythms have been linked to aging (reviewed in [7]). Disruption of rhythms has been shown to lead to a variety of conditions including insomnia, depression, heart disease, neurodegenerative disorders, and cancer [8]. Mechanistically, the circadian clock is composed of interlocked cycles of transcription and translation. The basic-helix-loop-helix (bHLH)-PAS transcription factors, Bmal1 and Clock, form a heterodimer complex and bind to E-box (CACGTC) promoters of various clock controlled genes such as Period 1, 2, 3 (Per 1 – 3) and Cryptochrome 1, 2 (Cry 1, 2) to induce their transcription [9]. Upon translation, the Per and Cry proteins inhibit Clock-Bmal1-mediated transcription in a negative feedback manner [10]. It has been proposed that circadian rhythm factors may play a role in carcinogenesis as there expression profiles have been shown to be altered in cancers [11–14].
In all mammals including humans, melatonin is synthesized from tryptophan in a circadian rhythmic fashion, under the control of various enzymes that are inhibited by light and stimulated at night in the pineal gland [3,4]. Light-at-night (LAN), due to night shift work or frequent changes in time zones, has been shown to cause decreases in melatonin levels with an increased risk for certain cancers such as breast, endometrial, colorectal, non-Hodgkin lymphoma and PCa [3,15–19]. The role of melatonin on circadian rhythm clock components is not well-studied and there are only a few reports that have investigated the role melatonin might play with the circadian rhythm clock components [20–22]. In the current study, we challenged the hypothesis that melatonin will impart anti-proliferative effects in PCa via resynchronization of the deregulated core clock circuitry [2]. Our data demonstrated that core clock components are indeed deregulated in human PCa and melatonin causes a resynchronization of the various clock genes, while having no effect on non-circadian rhythm genes, in PCa cells.
Methods and Materials
Cell Culture
The human prostate carcinoma cell lines viz. LNCaP, 22Rν1, DU145, and PC3 (obtained from American Type Culture Collection, ATCC, VA) were maintained in RPMI-1640, MEM, and F12K media (ATCC, VA) supplemented with FBS and antibiotics (penicillin/streptomycin). Normal human prostate epithelial cells PrEC (Cambrex, NJ) were maintained at standard cell culture conditions in PrEBM media with growth factors and supplements as recommended by the vendors (Cambrex, NJ). All cells were maintained at standard cell culture conditions (37°C, 5% CO2 in a humidified incubator) as recommended by the vendors.
Treatment of Cells with Melatonin
Cells were grown to 60% confluency and then treated with 100 μM, 1mM or 2mM melatonin (Enzo Life Sciences, PA; dissolved in a few drops of 100% ethanol and brought to desired concentration with PBS) for 24–48 hours.
Preparation of Protein Lysates and Western Blot Analysis
PCa cells were washed with ice-cold PBS, trypsinized and collected by centrifugation. Cell lysates were prepared using 1X RIPA lysis buffer, with freshly added PMSF and protease inhibitor cocktail (Cell Signaling, CA) and protein concentration was determined with BCA Protein Assay (Pierce, IL). For immunoblot analysis, 30–40 μg protein was subjected to SDS-PAGE and transferred onto a nitrocellulose membrane. Immunoblot analysis was performed using a variety of primary antibodies: anti-cleaved PARP (Cell Signaling, MA), anti-Clock, anti-β-actin, anti-Per2, and anti-Bmal1 (Santa Cruz, CA) and variety of secondary antibodies: goat anti-rabbit and goat anti-mouse HRP-conjugated antibodies (Upstate, MA), donkey anti-goat HRP-conjugated antibody (Santa Cruz, CA) followed by chemiluminescent detection. The quantification of protein was performed by a digital analyses of protein bands (TIFF images) using UN-SCAN-IT software.
Automated quantitative analysis (AQUA)
The tissue micro-array (TMA) was constructed using formalin-fixed, paraffin-embedded prostate specimens from the University of Wisconsin-Madison Pathology archive described elsewhere [23]. Tissues in the TMA were PCa local (N=59), PCa met (N=18), metastatic PCa (Met, N=18), benign prostate hyperplasia (BPH, N=24), high-grade intraepithelial neoplasia (HGPIN, N=19), and benign prostate tissues (N=48). For a quantitative analysis of Clock, Per2, and Bmal1, we employed immunofluorescence staining followed by AQUA analysis as previously described in great detail [23]. Briefly, TMA slide deparaffinization and antigen retrieval was performed following standard protocol. Prostate epithelium was masked using anti-cytokeratin and anti-E-cadherin cocktail and visualized with Alexa Fluor 555 (green) conjugated secondary antibody. 4, 6-diamidino-2-phenylindole (DAPI) was used to identify the nuclear compartment within the epithelial mask (blue). The target antigen was detected with specific antibody and visualized with Alexa Fluor 647 (red) conjugated secondary antibody. The resultant AQUA score is directly proportional to the number of molecules per unit area.
Transfection with human Per2
Human Per2 overexpression vector (hPer2), with a pCDNA backbone, was a kind gift from Nils Thoennissen (Division of Hematology and Oncology, Cedars-Sinai Medical Center, University of California-Los Angeles School of Medicine). Transfections with either hPer2 or pCDNA (control vector) were done using Lipofectamine 2000 according to the vendor’s protocol (Invitrogen, CA). After 48 hours, cells were collected for subsequent experiments.
Trypan Blue Exclusion Assay
Following transfections, cells were trypsinized and collected in a 1.5mL Eppendorf tube. The cells were pelleted by centrifugation and re-suspended in PBS (120 μL). Trypan blue (0.4% in PBS; 10 μL) was added to a smaller aliquot (10 μL) of cell suspension, and the number of cells (viable-unstained and non-viable-blue) were counted.
MTT Assay
The effect of the transfections on cell growth was assessed by MTT-based colorimetric assay employing the cell-proliferation kit (Roche Molecular Biochemicals, IN). The cells were subjected to transfections with pCDNA or hPer2 as described above and cell growth was evaluated as described in the manufacturer’s protocol.
Quantitative Real Time Reverse Transcriptase-PCR
RNA was isolated using Trizol reagent (Promega, WI) according to the vendor’s protocol. RNA was treated with DNAse (Promega, WI) and first strand cDNA was transcribed with 300ng random primers, 10mM dNTPs and 200 units of M-MLV reverse transcriptase (Promega, WI). Quantitative RT-PCR (qRT-PCR) was performed in triplicate with Platinum SYBR Green qPCR SuperMix-UDG (Takara, Japan) with 50ng first strand cDNA, 0.2 μM each forward and reverse primers. Human primer sequences used are as follows: Per2 Forward (5′-ACAGCTTTGGCTTCTGGTGT-3′), Per2 Reverse (5′-TATTGGCCATCATGGTCTGA-3″), Dbp Forward (5′-TAGAAGGAGCGCCTTGAGTC-3′), Dbp Reverse (5′-GCAACCCTCCAGTATCCAGA-3′), Clock Forward (5′-CAGAGCACCTTCCCTCAGTC-3′), Clock Reverse (5′-TTTCCCTCCTTTCCTCAGGT-3′), Bmal1 Forward (5′-TTCTCCAGGAGGCAAGAAGA-3′), Bmal1 Reverse (5′-TGCTGCCTCATCATTACTGG-3′), GAPDH Forward (5′-GAAGGTGAAGGTCGGAGTC-3′), GAPDH Reverse (5′ GAAGATGGTGATGGGATTTC-3′). The samples were cycled once for 50°C for 2 minutes for UDG incubation followed by 94°C for 2 minutes then 45 cycles of 95°C; 15 seconds, 60°C; 30 seconds each. Relative target mRNA was calculated using the ΔΔCT comparative method using GAPDH as an endogenous control. Purity of product was checked by dissociation curve analysis as well as running the samples on 3% agarose gel.
Circadian Variation Analysis
To test the circadian rhythmicity of various genes, 22Rν1 and PC3 cells were grown to 60% confluency and then synchronized via serum starvation for 48 hours. Cells were then treated with vehicle control or 1mM melatonin as described previously. Every four hours, for 36 hours, cells from each treatment group were trypsinized and collected. The pH of the media was measured at every time point to ensure proper and consistent growth conditions. RNA was isolated and qRT-PCR was performed as described previously. Expression values for each gene were tested for circadian rhythmicity using a cosine wave-fitting algorithm, COSOPT, as described elsewhere [24]. Briefly, COSOPT imports data and calculates the mean expression intensity and its corresponding standard deviation using period length parameters of 12–28 hours. Multiple-measures corrections of BETA (pMMC-β) values describe the goodness of fit. A significance threshold for pMMC-β was estimated based on known circadian clock genes and found to be <0.05.
Statistical Analysis
Statistical analyses were performed with the Student’s t test for independent samples and the data are expressed as means ± SD. Statistically significant p-values are provided for each individual figure.
Results
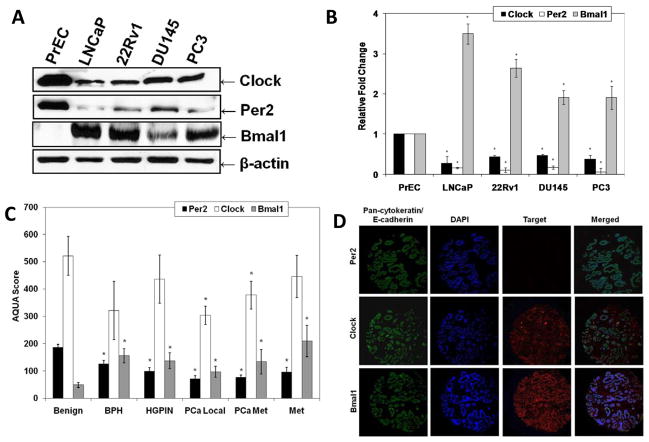
Studies have shown that certain circadian genes are altered in certain cancer [11–14]. However, the expression patterns of clock genes have not been studied in PCa. Therefore, we first evaluated the protein expression levels of core clock genes Clock, Bmal1 and Per2 in human PCa cells LNCaP, 22Rν1, DU145 and PC3, compared to normal prostate epithelial PrEC cells. As shown in figure 1, we found that endogenous Clock and Per2 protein levels were decreased and whereas Bmal1 protein levels were increased in PCa cells compared to normal PrEC cells (Fig. 1A and 1B), suggesting that the expression patterns of circadian rhythm genes are altered in PCa cells compared to normal prostate cells. Further, as shown in the supplementary figure, our data also demonstrated binding between Clock and Bmal1 in all PCa cells (Supplemental Fig. 1A and B), suggesting that despite of altered expression profile of the core circadian rhythm genes in human PCa, they are still able to complex with their counterparts.
Fig. 1.
Endogenous expression levels of Clock, Per2 and Bmal1 protein in PCa versus normal prostate specimens. A) Western blot analysis of Clock, Per2, and Bmal1: Clock, Per2, and Bmal1 protein levels were determined by Western blot analysis. Equal loading was confirmed by reprobing the blot for β-actin; B) Quantitation of Clock, Per2, and Bmal1 protein levels: Western blot analysis was quantitated by densitometric analysis of protein bands. The data are normalized to β-actin. C) Automated quantitative analysis (AQUA) of Clock, Per2, and Bmal1 in a human prostate tissue micro-array (TMA). The levels of Per2, Clock and Bmal1 were determined in a human TMA using AQUA. D) Representative immunofluorescence staining for Per2, Clock and Bmal1 in a human prostate TMA: Prostate epithelium was tagged with Alexa Fluor 555 (green). DAPI identified the nuclear compartment (blue). The target markers, Per2, Clock, and Bmal1, were visualized with Alexa Fluor 647 (red). Composite images demonstrate the targets and their localization within the tissue core. The data are expressed as mean ± SD of three experiments (*p ≤ 0.01 compared to appropriate controls). Details of the experiments are given in “Materials and Methods.
We next assessed the expression patterns of Clock, Bmal1 and Per2 genes in human prostate tissues using AQUA. The AQUA system quantitates protein expression within sub-cellular compartments in tissue sections. This multi-tissue proteomic analysis platform combines fluorescence-based image analysis with automated microscopy and high-throughput tissue microarray technologies [25,26]. For this purpose, we utilized a custom tissue micro-array (TMA) containing 336 spots which were composed of benign prostate, benign prostate hyperplasia (BPH), high-grade intraepithelial neoplasia (HGPIN), localized prostate cancer (PCa local), aggressive prostate cancer with metastasis (PCa met), and metastatic PCa from either lymph nodes, colon, or brain (Met) tissues. Validation of the TMA and AQUA system have been performed previously and are described elsewhere [23].
As shown by AQUA, we found that the expression levels of Per2 were significantly lower in all proliferative prostate diseases (BPH, HGPIN, PCa local, PCa met, Met), (AQUA scores, 126.6 ± 11.7, 9.92 ± 12.9, 71.4 ± 12.0, 77.0 ± 7.99, 96.0 ± 17.99, respectively) compared with benign prostate tissue (AQUA score, 186.8 ± 10.8) (Fig. 1C). Additionally expression levels of Clock were also found to be decreased in all proliferative prostate diseases (BPH, HGPIN, PCa local, PCa met, Met; AQUA scores, 321.7 ± 106.6, 436.2 ± 88.2, 304.5 ± 33.4, 378.1 ± 51.2, 446.2 ± 77.5, respectively) compared with benign prostate tissue (AQUA score, 521.7 ± 71.1) (Fig. 1C). Further, expression levels of Bmal1 were found to be significantly upregulated in all proliferative prostate diseases (BPH, HGPIN, PCa local, PCa met, Met; AQUA scores, 156.3 ± 26.0, 138.4 ± 28.8, 97.2 ± 19.8, 134.3 ± 44.7, 209.8 ± 56.7, respectively) compared with benign prostate tissue (AQUA scores, 49.1 ± 9.1) (Fig. 1C). The prostate epithelium was distinguished from stroma with a pan-cytokeratin and E-cadherin antibody cocktail tagged with Alexa Fluor 555 (green). The nuclear compartment within the epithelial mask was visualized with DAPI (blue). The targets (Per2, Clock, and Bmal1) were visualized with Alexa Fluor 647 (red). Composite merged images demonstrate the targets in the membrane/cytoplasmic compartment or nuclear compartment. Representative pictures were taken to confirm appropriate staining (Fig. 1D). These results obtained in human prostate tissues confirmed the results observed in human PCa cells and again suggest that circadian rhythm genes, Per2, Clock and Bmal1, are altered in PCa samples compared to normal prostate samples. These results coupled with the observations in human PCa cells (Fig. 1A and 1B) suggest that circadian rhythm genes, Per2, Clock and Bmal1, are altered in PCa samples compared to normal prostate samples.
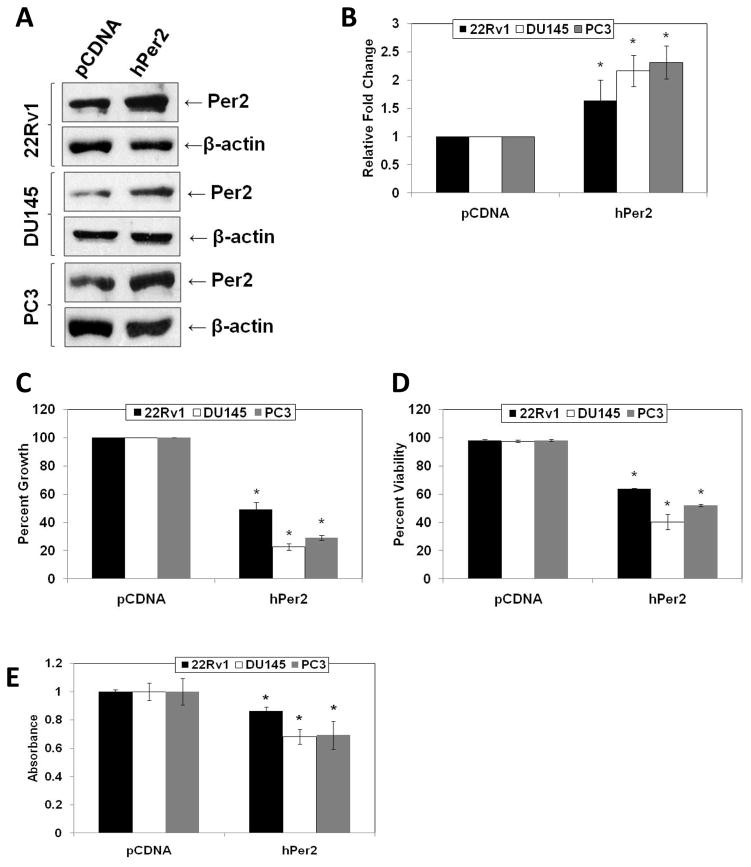
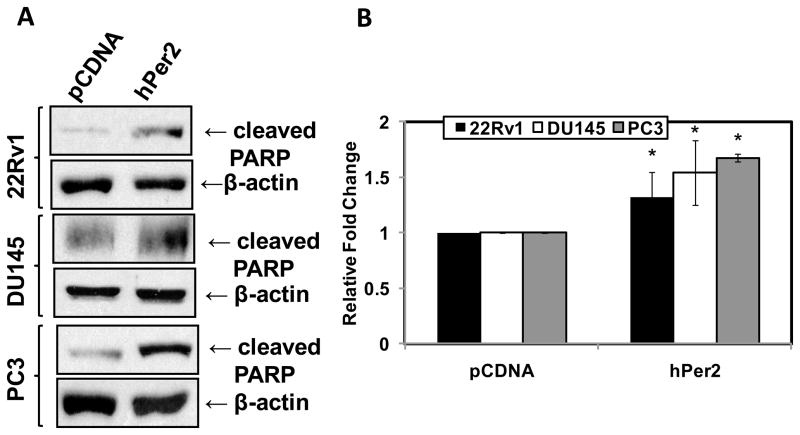
Because we observed different expression profile of Per2 in PCa cells compared to normal prostate cells, we investigated the effect of a forced overexpression of human Per2 in human PCa cells. We successfully transfected 22Rν1, DU145 and PC3 cells with either human Period2 (hPer2) or pCDNA control vector (Fig. 2A and B). Further, we used trypan blue assay to assess the effect of hPer2 overexpression cell growth and viability. We observed a significant inhibition of PCa cell growth and viability with hPer2 overexpression in 22Rv1, DU145 and PC3 cells (Fig. 2C and 2D). This was further confirmed by measuring a decrease in cell growth with a MTT assay (Fig. 2E). Further, apoptosis was measured by evaluating the expression of cleaved poly ADP ribose polymerase (PARP). Compared to vector controls, hPer2 transfected PCa cells showed a significant increase in cleaved PARP compared to pCDNA transfected cells (Fig. 3A and 3B), suggesting an induction of apoptosis of PCa cells as a result of Per2 overexpression.
Fig. 2.
Overexpression of Per2 in PCa cells. A) Effect of overexpression on Per2 protein levels: Following Lipofectamine 2000 mediated transfections of hPer2 or pCDNA in 22Rν1, DU145 and PC3 cells, Per2 protein levels were detected by Western blot analysis. Equal loading was confirmed by re-probing the blot for β-actin; B) Quantitation of Per2 protein levels: Western blot data was quantitated by a densitometric analysis of protein bands. The data were normalized to β-actin. C) Effect of Per2 overexpression on PCa cell growth as assessed by Trypan Blue analysis: 22Rν1, DU145 and PC3 cells transfected with hPer2 or pCDNA were analyzed by Trypan Blue assay to assess cell growth. Cell growth is expressed as percent growth (from total number of cells); D) Effect of Per2 overexpression on PCa cell viability as assessed by Trypan Blue analysis: 22Rν1, DU145 and PC3 cells transfected with hPer2 or pCDNA were analyzed by Trypan Blue assay to assess cell viability. Cell viability is expressed as the percent viable cells out of the total number of cells. E) Effect of Per2 overexpression on cell growth in PCa cells as assessed by MTT assay: The effect of hPer2 or pCDNA transfections on 22Rν1, DU145 and PC3 cell growth was assessed by MTT-based colorimetric assay. The cells were subjected to transfections with pCDNA or hPer2 as described and cell growth was evaluated as described in the manufacturer’s protocol. The data are expressed as the mean ± SD of three experiments (*p ≤ 0.01). Details of the experiments are given in “Materials and Methods.”
Fig. 3.
Effect of Per2 overexpression on apoptosis. A) Effect of Per2 over-expression on cleaved PARP protein levels: Following lipofectamine mediated transfection of hPer2 or pCDNA in 22Rν1, DU145 and PC3 cells, cleaved PARP protein levels were detected by Western blot analysis. Equal loading was confirmed by re-probing the blot for β-actin; B) Quantitation of cleaved PARP protein levels: Western blot data was quantitated by a densitometric analysis of protein bands. The data (relative density normalized to β-actin) are expressed as mean ± SD of three experiments (p-value ≤ 0.01*).
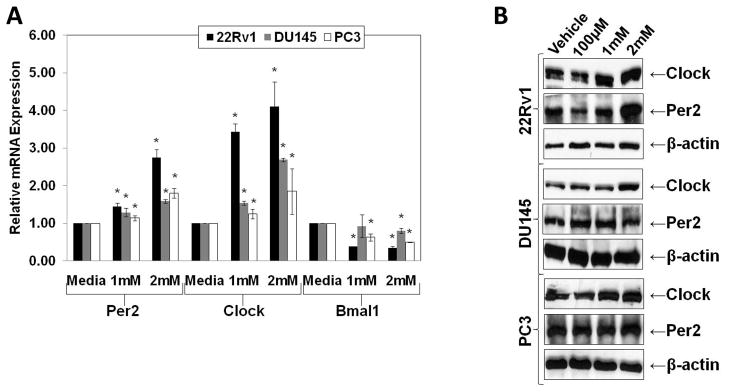
We demonstrated that some core circadian rhythm genes are altered in PCa cells and tissues. In the next series of experiments, we evaluated the effects of circadian rhythm related hormone melatonin on clock genes in human PCa cells. Our data demonstrated that melatonin treatment (1mM and 2mM; 24 hours) caused a significant upregulation in mRNA levels of Clock and Per2 and a significant downregulation of Bmal1 mRNA in 22Rν1, DU145 and PC3 cells (Fig. 4A). Further, using Western blot analysis, we found that Clock and Per2 protein levels were also increased in a pattern similar to that observed at the mRNA level, with melatonin treatment (100 μM, 1mM, and 2mM; 48 hours) in PCa cells (Fig. 4B). This suggested that melatonin significantly alters the expression of core clock genes.
Fig. 4.
Effect of melatonin on Per2, Clock and Bmal1 mRNA and protein levels in human PCa cells. A) Effect of melatonin on Per2, Clock, and Bmal1 mRNA levels: The relative expression levels of Per2, Clock, and Bmal1 transcripts in PCa cell lines treated with melatonin (1mM or 2mM; 24 hours) were determined by qRT-PCR. The data (relative density normalized to GAPDH) is expressed as mean ± SD of three experiments (*p ≤ 0.01); B) Effect of melatonin on Per2 and Clock protein levels: The protein levels of Clock and Per2 were determined in PCa cells were treated with melatonin (100 μM, 1mM or 2mM; 48 hours) by Western blot analysis. Equal loading was confirmed by reprobing the blot for β-actin. Details of the experiments are given in “Materials and Methods.”
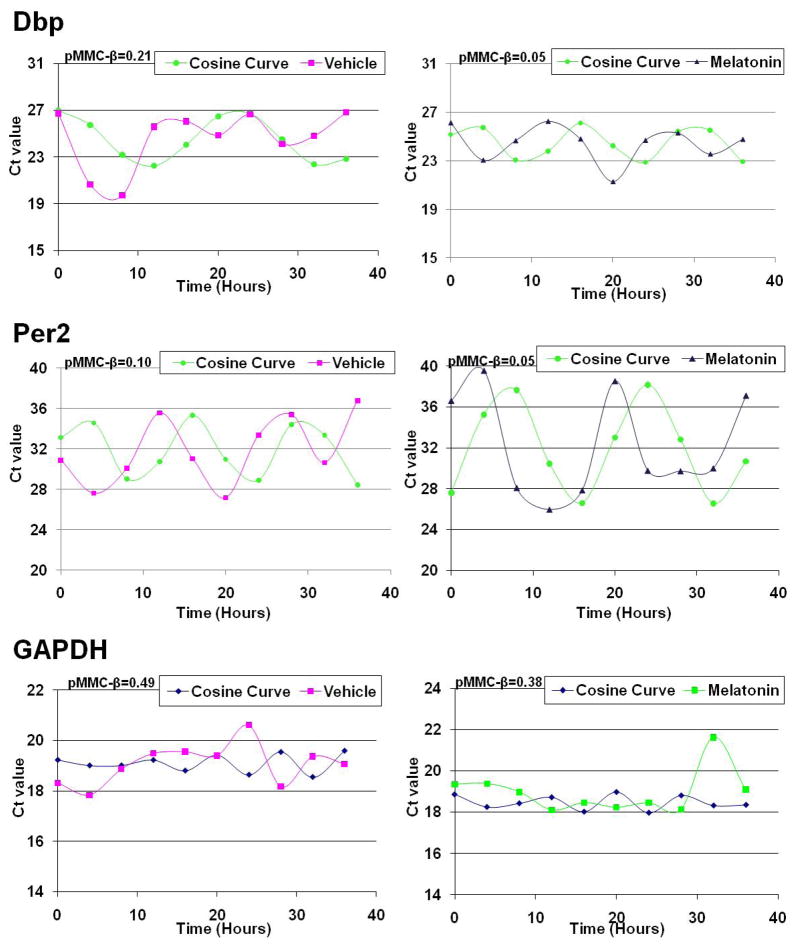
Next we further investigated if melatonin affects in vitro rhythmicity of circadian-regulated genes. To accomplish this, we first synchronized PCa cells with a 48 hour serum starvation, which was confirmed via cell cycle analysis to cause a G0/G1 cell cycle arrest (data not shown). The cells were then released and treated with either vehicle control or 1mM melatonin followed by a periodic collection every four hours for 36 hours. From these cells RNA was isolated and qRT-PCR was performed. To ensure that cells were in optimal growing conditions, the pH of the media was checked and found to be consistent with a variance of only ± 0.2 in each treatment (data not shown). Rhythmicity of genes was identified using the COSOPT algorithm with a pMMC-β cut-off of 0.05 and a period length restriction of 12–28 hours. As shown by the data, the well established cycling genes, Dbp and Per2 (Fig. 5) were found to have rhythmic oscillation in PC3 PCa cells following melatonin treatment. Interestingly, the oscillations were not seen in vehicle treated cells. We observed pMMC-β values of 0.05 for both Dbp and Per2 with melatonin treatment and pMMC-β values of 0.21 and 0.10 for Dbp and Per2 with vehicle treatment, respectively. We also assessed the oscillation pattern of GAPDH, as a non-oscillating housekeeping gene. Our data demonstrated that melatonin did not induce any sort of rhythmicity in GAPDH (Fig. 5) in PC3 cells, as the pMMC-β values were 0.49 for vehicle and 0.38 for melatonin treatment. Additionally, we also found that other non-oscillating genes such as Rap/Gap, PacS1 and Neud4 had pMMC-β values about 0.05 with melatonin treatment in PC3 cells (data not shown). This data suggested that melatonin has the ability to “re-synchronize” the core clock genes in PCa cells.
Fig. 5.
Effect of melatonin on the rhythmic gene expression of Dbp, Per2, and GAPDH in PC3 cells. PC3 cells were synchronized via serum starvation and then collected every four hours for a total of 36 hours following treatment with melatonin. The relative expression levels of Dpb, Per2 and GAPDH transcripts were determined by qRT-PCR. Expression values for each gene were tested for circadian variation using COSOPT. After analysis, the relative Ct values and the generated cosine curve versus time (hours) were plotted and pMMC-β value was calculated for vehicle and melatonin treated PC3 cells. Data represents three experiments with similar results. Details of the experiments are given in “Materials and Methods.”
Discussion
Studies have suggested an association between aging and the risk of developing PCa. However, the mechanism of this association is not well understood. This study was designed to determine the possible role of circadian rhythm machinery in PCa. Circadian rhythms are rhythmic oscillations in various biological processes that are regulated by an endogenous clock (reviewed in [27]). It has been reported that aging alters certain rhythmic cellular functions which have biological outcomes such as changes in sleeping patterns, alterations in melatonin synthesis and release, and disruptions in the levels of a variety of circulating hormones (reviewed in [7]).
Normally, the core clock components, Clock, Bmal1, Per 1, 2, and 3 and Cry 1 and 2 genes function in concert to maintain a tightly-regulated 24 hour rhythmic period, which if altered can led to a number of different disease states, including cancer. The circadian rhythm machinery has been shown to be functioning normally in many disease-free models such as in normal prostate tissues [28], but interestingly, a number of those same circadian rhythm players have been shown to have altered expression levels in various cancers [11–14,19,29–35]. Interestingly, not only are the expression profiles of the circadian rhythm components altered in cancers, but they also have been reported to cause a lack of synchronization of circadian rhythms [12,36,37]. For example, You et al. [12] reported that in tumor cells mPer1 and mPer2 gene expression patterns failed to cycle with the expected daily rhythms. We therefore suggest that not only are the altered expression patterns of clock genes important in cancer specimens, but the rhythmic oscillation alterations which they cause are additionally important to maintain proper cell growth, metabolism and hormone release.
Interestingly, low levels of melatonin have been associated with cancer and melatonin has also been shown to control the circadian rhythm machinery [19,22,38–41]. The benefits of directly or indirectly quantifying melatonin levels and using them as a measurement of circadian dysregulation and/or chronodisruption have been reported [42–44]. Studies have shown that exogenous melatonin can have anti-proliferative and anti-cancer effects. Thus, there appears to be an association between melatonin, circadian rhythms and various core circadian rhythm genes.
In this study, we demonstrated that several important core clock components (Per2, Clock and Bmal1) are deregulated in PCa cells as well as in human tissue samples. We also demonstrated that melatonin is able to alter the levels of various circadian rhythm genes by re-synchronizing a rhythmic pattern of gene expression. We suggest that the altered expression patterns as well as the proper rhythmicity of important clock components may be responsible for an ‘out of sync’ state, thereby causing defects in various mechanistic controls responsible for the control of cell growth/cell death, metabolism and hormone release, ultimately leading to cancer development. It is possible that the dysregulation of circadian rhythms observed in PCa cells may be due to decreased in vivo levels of melatonin. Therefore, exogenous melatonin may be able to resynchronize the circadian rhythm that may be helpful in providing novel avenues in PCa management. The concentrations of melatonin used in this study are high that may raise a concern regarding relevance of our findings. It has been shown that the uptake of melatonin by PCa cells, under in vitro conditions, is poor and therefore higher concentrations in the media do not necessarily correlate with internal concentrations [45]. Further, the uptake of melatonin was found to be inconsistent in PCa cell [45], therefore, a higher concentration of melatonin in the media is needed to ensure equal intracellular melatonin concentration in PCa cells. Indeed, more in-depth mechanistic studies both in appropriate in vitro and in vivo models are needed to confirm and substantiate our findings, especially with low and chronic melatonin exposure.
Supplementary Material
Acknowledgments
This work was partly supported by National Institutes of Health (T32ES00715–30 and F31 AT005393-01). Additionally, we would like to thank Nils Thoennissen (Division of Hematology and Oncology, Cedars-Sinai Medical Center, University of California-Los Angeles School of Medicine) for generously providing us with the hPer2 vector as well as Thomas Pier (Department of Pathology and Laboratory Medicine, University of Wisconsin-Madison) for his help with the AQUA system.
References
- 1.JEMAL A, SIEGEL R, WARD E, et al. Cancer statistics, 2009. CA Cancer J Clin. 2009;59:225–249. doi: 10.3322/caac.20006. [DOI] [PubMed] [Google Scholar]
- 2.JUNG-HYNES B, AHMAD N. SIRT1 controls circadian clock circuitry and promotes cell survival: a connection with age-related neoplasms. FASEB J. 2009;9:2803–2809. doi: 10.1096/fj.09-129148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.JUNG B, AHMAD N. Melatonin in cancer management: progress and promise. Cancer Res. 2006;66:9789–9793. doi: 10.1158/0008-5472.CAN-06-1776. [DOI] [PubMed] [Google Scholar]
- 4.REITER RJ. Melatonin: the chemical expression of darkness. Mol Cell Endocrinol. 1991;79:C153–C158. doi: 10.1016/0303-7207(91)90087-9. [DOI] [PubMed] [Google Scholar]
- 5.PARK JW, HWANG MS, SUH SI, et al. Melatonin down-regulates HIF-1 alpha expression through inhibition of protein translation in prostate cancer cells. J Pineal Res. 2009;46:415–421. doi: 10.1111/j.1600-079X.2009.00678.x. [DOI] [PubMed] [Google Scholar]
- 6.JOO SS, YOO YM. Melatonin induces apoptotic death in LNCaP cells via p38 and JNK pathways: therapeutic implications for prostate cancer. J Pineal Res. 2009;47:8–14. doi: 10.1111/j.1600-079X.2009.00682.x. [DOI] [PubMed] [Google Scholar]
- 7.PANDI-PERUMAL SR, SEILS LK, KAYUMOV L, et al. Senescence, sleep, and circadian rhythms. Ageing Res Rev. 2002;1:559–604. doi: 10.1016/s1568-1637(02)00014-4. [DOI] [PubMed] [Google Scholar]
- 8.GRIMALDI B, NAKAHATA Y, KALUZOVA M, et al. Chromatin remodeling, metabolism and circadian clocks: the interplay of CLOCK and SIRT1. Int J Biochem Cell Biol. 2009;41:81–86. doi: 10.1016/j.biocel.2008.08.035. [DOI] [PubMed] [Google Scholar]
- 9.REPPERT SM, WEAVER DR. Coordination of circadian timing in mammals. Nature. 2002;418:935–941. doi: 10.1038/nature00965. [DOI] [PubMed] [Google Scholar]
- 10.LEE C, ETCHEGARAY JP, CAGAMPANG FR, et al. Posttranslational mechanisms regulate the mammalian circadian clock. Cell. 2001;107:855–867. doi: 10.1016/s0092-8674(01)00610-9. [DOI] [PubMed] [Google Scholar]
- 11.FU L, PELICANO H, LIU J, et al. The circadian gene Period2 plays an important role in tumor suppression and DNA damage response in vivo. Cell. 2002;111:41–50. doi: 10.1016/s0092-8674(02)00961-3. [DOI] [PubMed] [Google Scholar]
- 12.YOU S, WOOD PA, XIONG Y, et al. Daily coordination of cancer growth and circadian clock gene expression. Breast Cancer Res Treat. 2005;91:47–60. doi: 10.1007/s10549-004-6603-z. [DOI] [PubMed] [Google Scholar]
- 13.CHEN ST, CHOO KB, HOU MF, et al. Deregulated expression of the PER1, PER2 and PER3 genes in breast cancers. Carcinogenesis. 2005;26:1241–1246. doi: 10.1093/carcin/bgi075. [DOI] [PubMed] [Google Scholar]
- 14.HUA H, WANG Y, WAN C, et al. Circadian gene mPer2 overexpression induces cancer cell apoptosis. Cancer Sci. 2006;97:589–596. doi: 10.1111/j.1349-7006.2006.00225.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.BLASK DE, SAUER LA, DAUCHY RT. Melatonin as a chronobiotic/anticancer agent: cellular, biochemical, and molecular mechanisms of action and their implications for circadian-based cancer therapy. Curr Top Med Chem. 2002;2:113–132. doi: 10.2174/1568026023394407. [DOI] [PubMed] [Google Scholar]
- 16.KUBO T, OZASA K, MIKAMI K, et al. Prospective cohort study of the risk of prostate cancer among rotating-shift workers: findings from the Japan collaborative cohort study. Am J Epidemiol. 2006;164:549–555. doi: 10.1093/aje/kwj232. [DOI] [PubMed] [Google Scholar]
- 17.VISWANATHAN AN, HANKINSON SE, SCHERNHAMMER ES. Night shift work and the risk of endometrial cancer. Cancer Res. 2007;67:10618–10622. doi: 10.1158/0008-5472.CAN-07-2485. [DOI] [PubMed] [Google Scholar]
- 18.VISWANATHAN AN, SCHERNHAMMER ES. Circulating melatonin and the risk of breast and endometrial cancer in women. Cancer Lett. 2009;281:1–7. doi: 10.1016/j.canlet.2008.11.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.REITER RJ, TAN DX, KORKMAZ A, et al. Light at night, chronodisruption, melatonin suppression, and cancer risk: a review. Crit Rev Oncog. 2007;13:303–328. doi: 10.1615/critrevoncog.v13.i4.30. [DOI] [PubMed] [Google Scholar]
- 20.TORRES-FARFAN C, SERON-FERRE M, DINET V, et al. Immunocytochemical demonstration of day/night changes of clock gene protein levels in the murine adrenal gland: differences between melatonin-proficient (C3H) and melatonin-deficient (C57BL) mice. J Pineal Res. 2006;40:64–70. doi: 10.1111/j.1600-079X.2005.00279.x. [DOI] [PubMed] [Google Scholar]
- 21.TORRES-FARFAN C, ROCCO V, MONSO C, et al. Maternal melatonin effects on clock gene expression in a nonhuman primate fetus. Endocrinology. 2006;147:4618–4626. doi: 10.1210/en.2006-0628. [DOI] [PubMed] [Google Scholar]
- 22.DUPRE SM, BURT DW, TALBOT R, et al. Identification of melatonin-regulated genes in the ovine pituitary pars tuberalis, a target site for seasonal hormone control. Endocrinology. 2008;149:5527–5539. doi: 10.1210/en.2008-0834. [DOI] [PubMed] [Google Scholar]
- 23.WARREN M, TWOHIG M, PIER T, et al. Protein expression of matriptase and its cognate inhibitor HAI-1 in human prostate cancer: a tissue microarray and automated quantitative analysis. Appl Immunohistochem Mol Morphol. 2009;17:23–30. doi: 10.1097/PAI.0b013e31817c3334. [DOI] [PubMed] [Google Scholar]
- 24.PANDA S, ANTOCH MP, MILLER BH, et al. Coordinated transcription of key pathways in the mouse by the circadian clock. Cell. 2002;109:307–320. doi: 10.1016/s0092-8674(02)00722-5. [DOI] [PubMed] [Google Scholar]
- 25.WARREN M, TWOHIG M, PIER T, et al. Protein expression of matriptase and its cognate inhibitor HAI-1 in human prostate cancer: a tissue microarray and automated quantitative analysis. Appl Immunohistochem Mol Morphol. 2009;17:23–30. doi: 10.1097/PAI.0b013e31817c3334. [DOI] [PubMed] [Google Scholar]
- 26.KOLESAR J, HUANG W, EICKHOFF J, et al. Evaluation of mRNA by Q-RTPCR and protein expression by AQUA of the M2 subunit of ribonucleotide reductase (RRM2) in human tumors. Cancer Chemother Pharmacol. 2009;64:79–86. doi: 10.1007/s00280-008-0845-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.FU L, LEE CC. The circadian clock: pacemaker and tumour suppressor. Nat Rev Cancer. 2003;3:350–361. doi: 10.1038/nrc1072. [DOI] [PubMed] [Google Scholar]
- 28.BEBAS P, GOODALL CP, MAJEWSKA M, et al. Circadian clock and output genes are rhythmically expressed in extratesticular ducts and accessory organs of mice. FASEB J. 2009;23:523–533. doi: 10.1096/fj.08-113191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.GERY S, KOMATSU N, KAWAMATA N, et al. Epigenetic silencing of the candidate tumor suppressor gene Per1 in non-small cell lung cancer. Clin Cancer Res. 2007;13:1399–1404. doi: 10.1158/1078-0432.CCR-06-1730. [DOI] [PubMed] [Google Scholar]
- 30.GERY S, GOMBART AF, YI WS, et al. Transcription profiling of C/EBP targets identifies Per2 as a gene implicated in myeloid leukemia. Blood. 2005;106:2827–2836. doi: 10.1182/blood-2005-01-0358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.CHU LW, ZHU Y, YU K, et al. Correlation between circadian gene variants and serum levels of sex steroids and insulin-like growth factor-I. Cancer Epidemiol Biomarkers Prev. 2008;17:3268–3273. doi: 10.1158/1055-9965.EPI-08-0073. [DOI] [PubMed] [Google Scholar]
- 32.CHU LW, ZHU Y, YU K, et al. Variants in circadian genes and prostate cancer risk: a population-based study in China. Prostate Cancer Prostatic Dis. 2008;11:342–348. doi: 10.1038/sj.pcan.4501024. [DOI] [PubMed] [Google Scholar]
- 33.POGUE-GEILE KL, LYONS-WEILER J, WHITCOMB DC. Molecular overlap of fly circadian rhythms and human pancreatic cancer. Cancer Lett. 2006;243:55–57. doi: 10.1016/j.canlet.2005.11.049. [DOI] [PubMed] [Google Scholar]
- 34.YEH KT, YANG MY, LIU TC, et al. Abnormal expression of period 1 (PER1) in endometrial carcinoma. J Pathol. 2005;206:111–120. doi: 10.1002/path.1756. [DOI] [PubMed] [Google Scholar]
- 35.ZHU Y, STEVENS RG, HOFFMAN AE, et al. Testing the circadian gene hypothesis in prostate cancer: A population-based case-control study. Cancer Res. 2009 doi: 10.1158/0008-5472.CAN-09-0648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.CAO Q, GERY S, DASHTI A, et al. A role for the clock gene Per1 in prostate cancer. Cancer Res. 2009 doi: 10.1158/0008-5472.CAN-08-4199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.TANIGUCHI H, FERNANDEZ AF, SETIEN F, et al. Epigenetic inactivation of the circadian clock gene BMAL1 in hematologic malignancies. Cancer Res. 2009;69:8447–8454. doi: 10.1158/0008-5472.CAN-09-0551. [DOI] [PubMed] [Google Scholar]
- 38.OTALORA BB, MADRID JA, ALVAREZ N, et al. Effects of exogenous melatonin and circadian synchronization on tumor progression in melanoma-bearing C57BL6 mice. J Pineal Res. 2008;44:307–315. doi: 10.1111/j.1600-079X.2007.00531.x. [DOI] [PubMed] [Google Scholar]
- 39.DOYLE SE, GRACE MS, MCIVOR W, et al. Circadian rhythms of dopamine in mouse retina: the role of melatonin. Vis Neurosci. 2002;19:593–601. doi: 10.1017/s0952523802195058. [DOI] [PubMed] [Google Scholar]
- 40.ZEMAN M, SZANTOOVA K, STEBELOVA K, et al. Effect of rhythmic melatonin administration on clock gene expression in the suprachiasmatic nucleus and the heart of hypertensive TGR(mRen2)27 rats. J Hypertens. 2009;27(Suppl 6):S21–6. S21–S26. doi: 10.1097/01.hjh.0000358833.41181.f6. [DOI] [PubMed] [Google Scholar]
- 41.ONSO-VALE MI, ANDREOTTI S, MUKAI PY, et al. Melatonin and the circadian entrainment of metabolic and hormonal activities in primary isolated adipocytes. J Pineal Res. 2008;45:422–429. doi: 10.1111/j.1600-079X.2008.00610.x. [DOI] [PubMed] [Google Scholar]
- 42.MIRICK DK, DAVIS S. Melatonin as a biomarker of circadian dysregulation. Cancer Epidemiol Biomarkers Prev. 2008;17:3306–3313. doi: 10.1158/1055-9965.EPI-08-0605. [DOI] [PubMed] [Google Scholar]
- 43.ERREN TC, REITER RJ, PIEKARSKI C. Light, timing of biological rhythms, and chronodisruption in man. Naturwissenschaften. 2003;90:485–494. doi: 10.1007/s00114-003-0468-6. [DOI] [PubMed] [Google Scholar]
- 44.ERREN TC, REITER RJ. Defining chronodisruption. J Pineal Res. 2009;46:245–247. doi: 10.1111/j.1600-079X.2009.00665.x. [DOI] [PubMed] [Google Scholar]
- 45.HEVIA D, SAINZ RM, BLANCO D, et al. Melatonin uptake in prostate cancer cells: intracellular transport versus simple passive diffusion. J Pineal Res. 2008;45:247–257. doi: 10.1111/j.1600-079X.2008.00581.x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.