Summary
Infant leukaemia is an embryonal disease in which the underlying MLL translocations initiate in utero. Zebrafish offer unique potential to understand how MLL impacts haematopoiesis from the earliest embryonic timepoints and how translocations cause leukaemia as an embryonal process. In this study, a zebrafish mll cDNA syntenic to human MLL spanning the 5’ to 3’UTRs, was cloned from embryos, and mll expression was characterized over the zebrafish lifespan. The protein encoded by the 35-exon ORF exhibited 46.4% overall identity to human MLL and 68–100% conservation in functional domains (AT-hooks, SNL, CXXC, PHD, bromodomain, FYRN, taspase1 sites, FYRC, SET). Maternally supplied transcripts were detected at 0–2 hpf. Strong ubiquitous early zygotic expression progressed to a cephalo-caudal gradient during later embryogenesis. mll was expressed in the intermediate cell mass (ICM) where primitive erythrocytes are produced and in the kidney where definitive haematopoiesis occurs in adults. mll exhibits high cross species conservation, is developmentally regulated in haematopoietic and other tissues and is expressed from the earliest embryonic timepoints throughout the zebrafish lifespan. Haematopoietic tissue expression validates using zebrafish for MLL haematopoiesis and leukaemia models.
Keywords: zebrafish, Mll, molecular cloning, primitive haematopoiesis, definitive haematopoiesis
Introduction
Infant acute leukaemias are embryonal cancers in which the characteristic translocations leading to disruption of the MLL transcription factor that give rise to transformation initiate in utero (Ford, et al 1993). MLL translocations also are archetypal of secondary leukaemias after chemotherapeutic topoisomerase II poisons (Rowley and Olney 2002), and occur less often in childhood and adult acute leukaemias (Liedtke and Cleary 2009). Fusions of 5’ MLL with >60 partner genes, many of which encode transcriptional regulatory or cell signaling proteins (Meyer, et al 2009), generate diverse chimeric proteins believed to play key roles in leukaemogenesis by altering transcription (Liedtke and Cleary 2009). Patients with leukaemias marked by particular MLL translocations are at high risk for a poor outcome (Balgobind, et al 2009, Hilden, et al 2006, Pieters, et al 2007).
Despite substantial progress, how MLL affects normal haematopoiesis and how MLL translocations lead to leukemogenesis are incompletely understood. From murine models it is known that MLL is essential for primitive yolk sac (Hess, et al 1997, Yu, et al 1995) and definitive fetal liver (Yagi, et al 1998) haematopoiesis and is expressed in adult myeloid and lymphoid cells (Jude, et al 2007), though its precise roles remain enigmatic. Zebrafish are an attractive model to better understand MLL because of the embryonal origin of infant leukaemia (Ford, et al 1993), and the unique in vivo access to the earliest developmental timepoints inaccessible in mammals but possible with abundant, transparent, rapidly and externally growing zebrafish embryos (Payne and Look 2009).
MLL SNL (speckled nuclear localization), PHD (plant homeodomain) and SET (Su (var)3–9, Enhancer of zeste, trx) domains were highly conserved through evolution from Drosophila trx, pufferfish mll, murine Mll and the human ortholog (Caldas, et al 1998, Djabali, et al 1992, Tkachuk, et al 1992), predicting a similar zebrafish protein. Drosophila trx is a transcriptional maintenance factor for homeobox gene expression, which is antagonized by transcriptional repressive effects of Polycomb group proteins (Liedtke and Cleary 2009). Homeobox gene expression regulation by trx is critical for cell fate specification during development, a function conserved through evolution as evidenced by defective body patterning and lethality with Mll deficiency in mice, where Mll maintains homeobox gene expression during skeletal, neuronal, craniofacial and haematopoietic development (Hess, et al 1997, Yu, et al 1998, Yu, et al 1995).
The MLL oncoprotein undergoes taspase1 proteolysis into amino and carboxyl fragments with transcriptional repression and activation properties, which reassociate in a multiprotein complex (Hsieh, et al 2003, Yokoyama, et al 2002) that orchestrates epigenetic nucleosome and histone modifications (Milne, et al 2002). Its amino terminus contains AT-hooks that promote p21 and p27 upregulation, cell cycle arrest and monocyte differentiation (Caslini, et al 2000a), SNL motifs that direct subnuclear localization (Caslini, et al 2000b) and a CXXC region characteristic of chromatin associated proteins, which binds to non-methylated CpG dinucleotides (Allen, et al 2006, Caslini, et al 2000a, Xia, et al 2003, Yokoyama, et al 2002). Central PHD zinc fingers mediate homodimerization and protein interactions including binding to a nuclear cyclophilin, which modulates target gene expression (Fair, et al 2001). The carboxyl SET domain with histone H3K4-specific methyltransferase activity interacts with the SWI/SNF chromatin remodeling complex (Milne, et al 2002). Taspase1 proteolysis is critical for proper MLL nuclear sublocalization and homeobox gene regulation (Hsieh, et al 2003). Taspase1 proteolysis (Takeda, et al 2006) and bimodal MLL degradation by specific E3 ligases (Liu, et al 2007) affect cell cycle progression.
Until now, zebrafish Mll has not been characterized even though zebrafish orthologs of numerous mammalian haematopoietic genes (Chen and Zon 2009) and various leukaemia-associated oncogenes, tumor-suppressor genes and MLL partner genes (Song, et al 2004) have been identified and other zebrafish leukaemia models are emerging (Payne and Look 2009). Interestingly, mll recently was identified as a maternally supplied transcript in a genome-wide screen for SET domains with potential developmental roles in early histone programming (Sun, et al 2008). The purposes of this work were to clone the full-length zebrafish mll cDNA and define spatio-temporal mll expression throughout the zebrafish lifespan as a critical foundation for utilizing zebrafish for further investigations of MLL in developmental haematopoiesis and disease.
Methods and Materials
Gene prediction tools
The existence of a zebrafish mll gene and relationship to human MLL were queried with BLASTP (www.ncbi.nlm.nih.gov/BLAST/). Corresponding transcripts were studied using BLAST and ENSEMBL (www.ensembl.org). GNOMON was used to align the predicted transcript and protein to the genome (www.ncbi.nlm.nih.gov/genome/). CDART was employed to find relationships between the zebrafish and human proteins (www.ncbi.nlm.nih.gov/Structure/lexington/lexington.cgi). Synteny was analysed using the ENSEMBL genome assembly (Zv7) database.
Zebrafish maintenance
Wild-type zebrafish were raised and maintained under standard conditions and utilized according to US Institutional Animal Care and Use Committee (IACUC) guidelines.
Molecular analyses of predicted zebrafish mll ortholog
The most highly conserved regions in human MLL, mouse Mll and pufferfish mll CXXC and SET domains were identified using ClustalW (www.ebi.ac.uk/clustalw/). Degenerate cross-species primer mixtures for degenerate reverse transcriptase polymerase chain reaction (dRT-PCR) (TABLE SI) were designed from these regions, accounting for fold degeneracy after examining codons with a mismatched base (boneslab.bio.ntnu.no/degpcrshortguide.htm). Eliminating sequence wobble in some codons reduced primer degeneracy, even if there was a mismatched base. Random hexamer primed first strand cDNA, prepared from total RNA from a whole wild-type adult zebrafish using SuperScript™ III reverse transcriptase (Invitrogen, Carlsbad, CA), was amplified with Expand High Fidelity Taq polymerase (Roche, Indianapolis, IN).
Cross-species Southern blot analysis was performed using the B859 human MLL (ALL-1) breakpoint cluster region (bcr) cDNA as probe (Gu, et al 1992). To predict restriction fragments that would be detected, restriction maps of a projected genomic sequence corresponding to a zebrafish mll cDNA derived with gene prediction tools (Entrez Gene 557048), were simulated in the region of highest homology to the probe. Genomic DNA (20 µg) from a whole wild-type adult zebrafish extracted using DNeasy (Qiagen, Valencia, CA), was digested with BamHI, BglII, NheI, SacI, XbaI or HindIII. Conditions were those employed for human DNAs (Felix, et al 1997); BamHI-digested human lymphocyte DNA (10 µg) was the control.
Molecular cloning of zebrafish mll cDNAs
TABLE SI contains details of the molecular cloning experiments including target amplicons. RT-PCR primers were designed from reference sequences derived with gene prediction tools, or sequences of partial cDNAs derived herein.
Total RNAs were isolated from whole wild-type adult zebrafish or 24 hpf (hours post fertilization) embryos (~25–50 per experiment) using TRIzol® reagent (Invitrogen) or RNeasy (Qiagen) (TABLE SI) and treated with DNase I (Roche); 1–5 µg of total RNA was used to prepare cDNAs. All RT-PCRs were replicated at least twice.
RT-PCR analysis of a predicted single transcript spanning two proximal chromosome 15 “similar to MLL” cDNAs (XM_680024, XM_679940) was performed by synthesizing oligo(dT) primed first strand cDNA with SuperScript™ II reverse transcriptase and performing PCR with Expand High Fidelity Taq Polymerase (Roche).
To generate cDNAs spanning exons 1–35 including all but 199 bases of the 5’ end and 46 bases of the 3’ end of the ORF by long-range RT-PCR, AccuPrime™ High Fidelity Taq DNA Polymerase (Invitrogen) and primers in TABLE SI were used to amplify either oligo-dT primed first strand cDNA from adults, or a mixture of oligo-dT primed and random hexamer primed first strand cDNA from 24 hpf embryos synthesized with a SuperScript™ II First Strand Synthesis kit (Invitrogen). Products were subcloned into pCR-XL-TOPO (Invitrogen) and a sequence contig generated.
The 5’ and 3’UTRs were cloned by RACE (Rapid Amplification of cDNA ends) with primers in TABLE SI using 5’ and 3’ RACE Systems (Invitrogen) according to manufacturer’s instructions for first strand cDNA synthesis, and the Expand High Fidelity PCR System (Roche) for PCRs. The products were subcloned into pCR 2.1-TOPO (Invitrogen); their sequences informed primers for further RT-PCRs.
The same first strand cDNAs used above to clone exons 1–35, and High Fidelity Taq Polymerase (Invitrogen) were employed to generate a cDNA with an engineered 5’ NotI site comprising the 5’UTR and spanning the BsrGI site in exon 3, and another cDNA containing exons 33–35 encompassing the ClaI site in exon 35 with an engineered 3’ KpnI site. A full-length ORF cDNA subclone from 24 hpf embryos was obtained by restriction enzyme cleavage of the 5’UTR-exon 3 PCR product with NotI and BsrGI, of the larger central exon 1–35 subclone with BsrGI and ClaI, and of the exon 33–35 PCR product with ClaI and KpnI, followed by ligation of the 5’ and 3’ fragments to the BsrGI or ClaI sites in the larger central fragment and to NotI and KpnI sites in pCR-XL-TOPO (Invitrogen).
A cDNA spanning the 5’UTR through the 3’UTR was generated from random hexamer primed cDNAs by long-distance RT-PCR using AccuPrime™ High Fidelity Taq DNA Polymerase (Invitrogen), subcloned into pCR-XL-TOPO (Invitrogen) and sequenced.
Bioinformatic analyses of cloned zebrafish mll cDNA and predicted protein
The ORF finder algorithm (www.ncbi.nlm.nih.gov/gorf/gorf.html) was used to identify the ATG codon consistent with the ORF and the Genome Browser Gateway (genome.ucsc.edu) to define exon/intron boundaries. The 3’ RACE PCR product was examined for polyadenylation signals and regulatory elements.
Protein domain alignment with human MLL was achieved using SMART (Simple Modular Architecture Research Tool, smart.embl-heidelberg.de/) and NCBI BLAST programs. ClustalW2 (www.ebi.ac.uk/Tools/clustalw2/) and the neighbor-joining method within the MEGA4 program (Tamura, et al 2007) was employed to place the protein predicted by the cloned cDNA in a consensus phylogram tree with MLL proteins of other species; amino acid identity to human was analysed using the EMBOSS Pairwise Alignment Algorithm Needle (www.ebi.ac.uk/emboss/align/).
Temporal RT-PCR analysis
First strand cDNAs synthesized from RNAs from pooled staged wild-type embryos and a whole wild-type adult using random hexamers (Applied Biosystems, Foster City, CA), were amplified using High Fidelity Taq Polymerase (Roche) and primers for zebrafish mll or tuba1 (TABLE SI). Where possible, amplicons spanned exon junctions. Amplification of zebrafish mll was confirmed by direct sequencing.
Temporal and tissue specific quantitative real-time PCR (Q-RT-PCR) analyses
mll expression was analysed in whole wild-type embryos, larvae and adults over a developmental timecourse, and in dissected adult tissues, compared to whole adult tissue. Random hexamer primed first strand cDNAs were amplified by Q-RT-PCR with the ABI Prism 7900HT sequence detection system (Applied Biosystems), Platinum SYBR Green SuperMix (Invitrogen) as per manufacturer's instructions, and primers targeting exons 1–3 (TABLE SI).
The temporal Q-RT-PCR experiment was repeated 4 times on entirely independent animals using 3 replicates per probeset per experiment.
To quantify mll expression in tissues, respective organs microdissected from three wild-type adults using standard methods were homogenized together, total RNAs were prepared and quadruplicate experiments (three replicates per probeset apiece) were performed on each sample. The same procedure was repeated on RNAs from pooled tissues from 3 more adults. The control sample for all 8 experiments was prepared by extracting total RNA after homogenizing two each whole wild-type adult males and females altogether.
Relative mll expression over the developmental timecourse, or in specific dissected adult tissues compared to whole wild-type adult, was determined by calculating 2−ΔCT values (Livak and Schmittgen 2001) after normalization to bactin1.
Whole mount in situ hybridization (WISH) analysis of mll in zebrafish embryos
Probes were created using 662 bp 5’ (5’UTR-exon 3) and 447 bp 3’ (exon 33–35) plasmid subclones corresponding to positions −46 to 616 and 12181 to 12657 in cDNA clone EU179544. Plasmids were linearized and anti-sense and sense probes were generated using the DIG RNA labeling kit SP6/T7 (Roche). In situ hybridization was performed as previously described (Panzer, et al 2005). Images were acquired with a compound microscope (DMR, Leica, Allendale, NJ) equipped with a digital camera (DC500, Leica).
Results
Prediction of zebrafish mll ortholog using bioinformatics
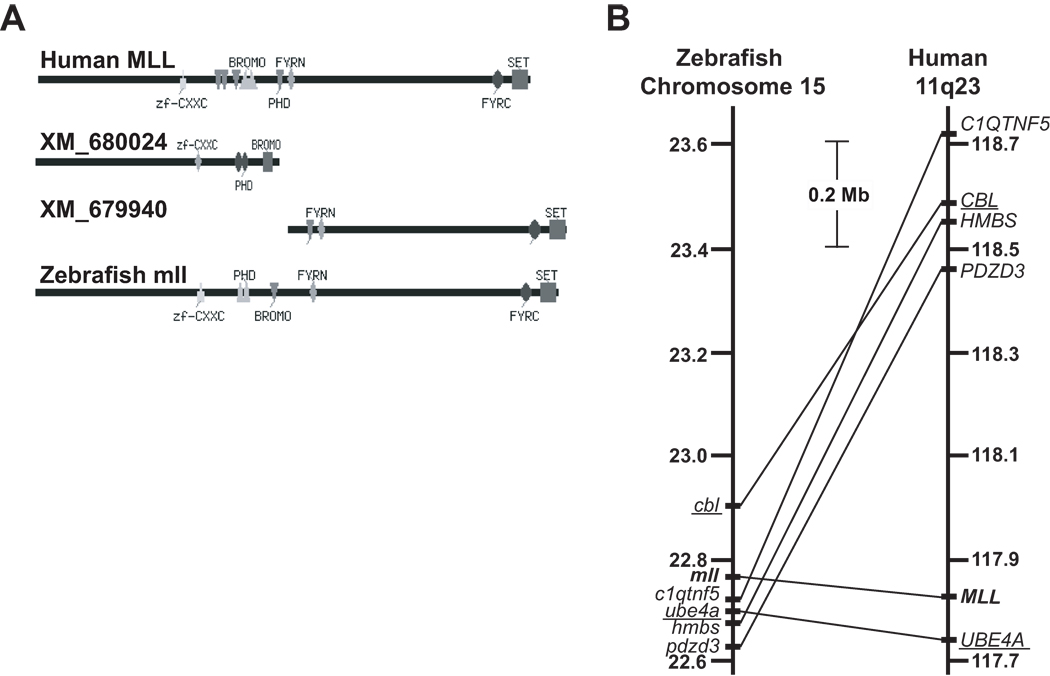
At the outset of this work several gene and protein prediction methods supported the existence of a single zebrafish mll gene with functional similarity to human MLL. BLASTP searching using human MLL (Accession no. NP_005924) as the reference, identified two putative zebrafish “similar to MLL proteins” (Accession nos. XP_685032, XP_685116), for which there were corresponding GenBank entries for predicted 5’ and 3’ 17- and 18-exon transcripts in close proximity on zebrafish chromosome 15 (Accession nos. XM_680024, XM_679940). The two transcripts were predicted by ENSEMBL and GNOMON, respectively, to comprise a single gene (Accession no. NW_633640) coding for one transcript (Entrez Gene 557048). CDART analysis (Geer, et al 2002) indicated conservation of the important human MLL domains in the predicted protein from this composite transcript (FIG 1A).
Fig 1.
Bioinformatic predictions of zebrafish mll ortholog. (A) CDART analysis of human MLL protein and zebrafish “similar to MLL proteins.” Conserved domains of human MLL, putative zebrafish “similar to MLL” proteins, and hypothetical single zebrafish Mll protein obtained by joining of the two zebrafish “similar to MLL” protein sequences are shown. Abbreviation: zf, zinc finger. (B) Syntenic relationship between human MLL and zebrafish mll genes determined using ENSEMBL genome assembly (Zv7) database. Genes in same map order and similar uninterrupted segments (UBE4A/ube4a and CBL/cbl) are underlined. Human MLL and zebrafish mll are bold. Distances are to scale.
Human and zebrafish genes can have one-to-many or, due to duplication during evolution, many-to-many relationships (Tatusov, et al 1997); these predictions of a single gene, transcript and protein were most consistent with a one-to-one relationship. Importantly, a conserved block of syntenic chromosome band 11q23 genes surrounding human MLL and the predicted mll on zebrafish chromosome 15 was identified (FIG 1B), suggesting that zebrafish mll is a functionally similar ortholog (Barbazuk, et al 2000) to human MLL.
Cross species molecular analyses detect highly conserved regions of zebrafish mll
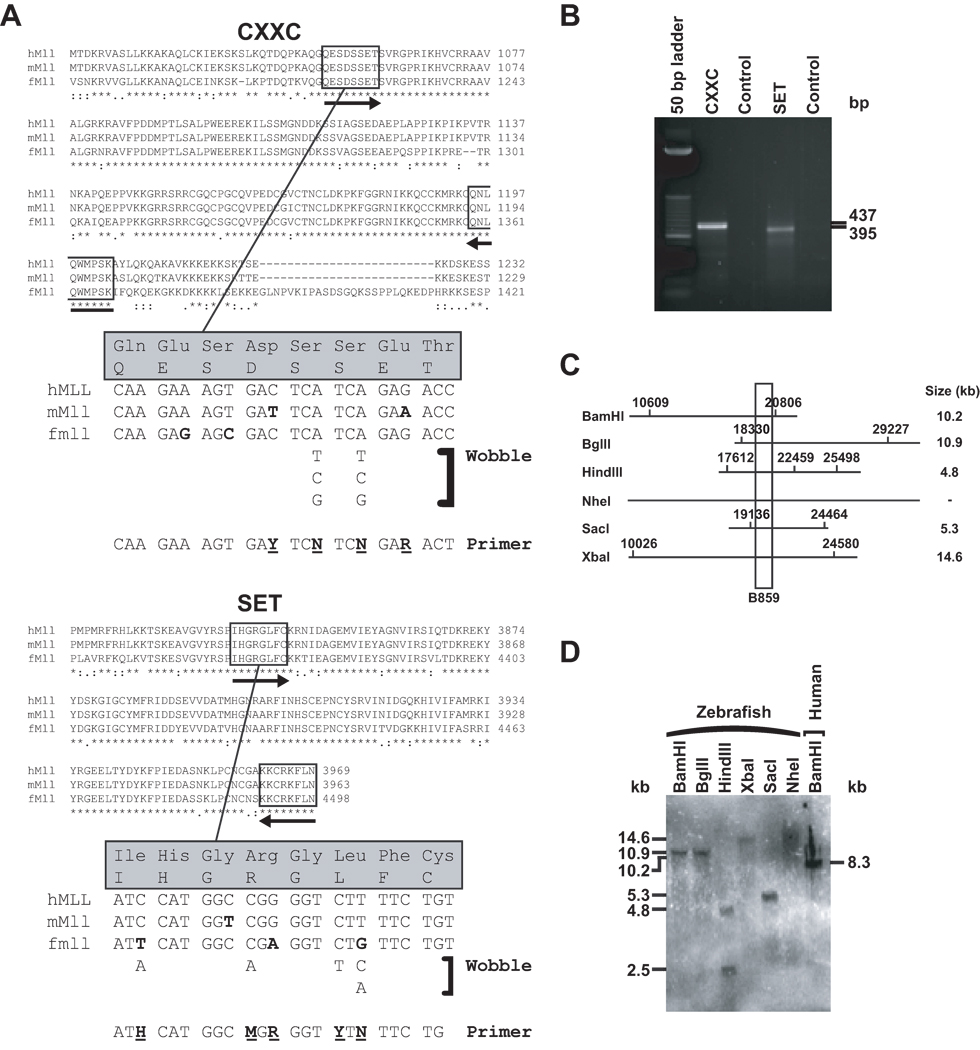
RT-PCR experiments on total RNA from a zebrafish adult using degenerate primers matching MLL CXXC and SET domain amino acids (FIG 2A) that are highly conserved between human, murine and pufferfish, yielded products (FIG 2B) with 100% identity to corresponding regions in the predicted “similar to MLL” transcripts (Accession nos. XM_680024, XM_679940). These results indicated that transcript regions encoding MLL functional domains were highly conserved through evolution and gave the first experimental evidence that the transcript suggested by gene prediction tools was bona fide zebrafish mll.
Fig 2.
Molecular identification of predicted mll ortholog in zebrafish. (A) Amino acid sequences from highly conserved CXXC and SET domains in human MLL and mouse and pufferfish (fugu) proteins corresponding to amplicons in degenerate reverse transcriptase (dRT)-PCR. Boxed insets indicate amino acids used to design sense (forward arrows) and antisense (reverse arrows) primers. Sense primer designs are shown in detail at bottom. Bold indicates mismatched bases across species. Bold underlined indicates degenerate bases used in primers (Y, C/T; R, A/G; M, A/C; H, A/C/T; N, A/C/G/T). Reverse primers were designed using similar considerations. (B) Products of dRT-PCR analysis of whole wild-type adult zebrafish RNA with degenerate cross-species primers, sequences of which were 100% identical to corresponding regions in predicted XM_680024 and XM_679940 cDNAs. (C) Simulated restriction maps of predicted 36,662 bp zebrafish mll genomic sequence (Entrez Gene ID 557048) in region most homologous to B859 human MLL bcr cDNA (Gu, et al 1992); approximately 90 bases of predicted zebrafish mll cDNA matched the probe exactly. (D) Cross-species Southern blot analysis of whole wild-type adult zebrafish genomic DNAs with indicated restriction enzymes using B859 human MLL bcr cDNA (Gu, et al 1992) as probe and BamHI digested normal human peripheral blood lymphocyte DNA as the control.
Similarly, cross-species Southern blot analysis demonstrated that the human MLL cDNA probe (Gu, et al 1992) was able to detect zebrafish mll. FIG 2C summarizes the simulated restriction mapping of the projected 36,662 bp zebrafish mll genomic sequence (Entrez Gene 557048) and projected restriction enzyme fragments in the region of highest homology to the probe. The sizes and numbers of BamHI, BglII, XbaI, SacI and NheI fragments found by actual Southern blot analysis exactly matched those that were predicted (FIG 2D). However, a single HindIII fragment was predicted and two fragments were detected, probably due to generation of the zebrafish mll genomic sequence with a gene prediction tool.
Molecular cloning of full-length zebrafish mll ORF and flanking UTRs
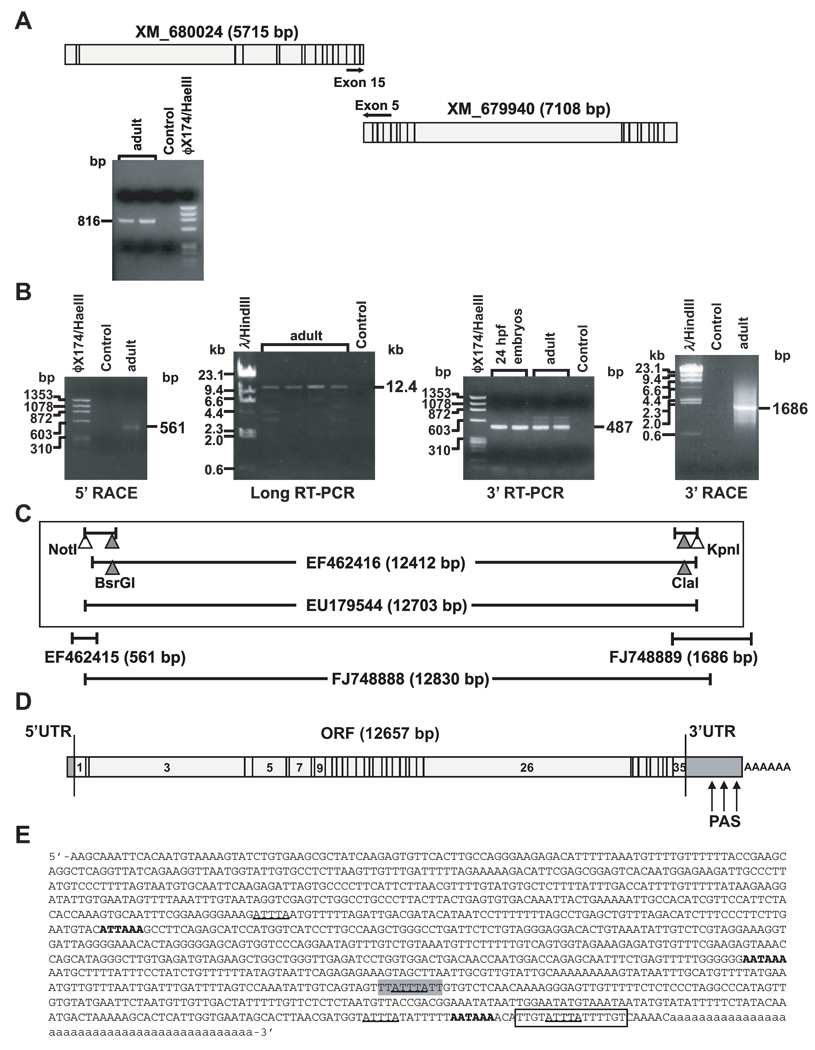
RT-PCR analysis of whole wild-type adult RNA amplified a single transcript spanning the predicted cDNAs for both “similar to MLL proteins”, giving evidence that the two cDNAs were from the same gene (FIG 3A); a similar partial cDNA cloned from zebrafish kidney marrow (Accession no. DQ355790) supported these results. Nonetheless, it was important to clone the full-length cDNA because cDNAs derived with gene prediction tools are imprecise, especially at exon boundaries.
Fig 3.
Molecular cloning of zebrafish mll cDNA. (A) Reverse transcriptase (RT)-PCR design and products identifying single transcript spanning predicted cDNAs for both “similar to MLL proteins” in wild-type adult zebrafish total RNA. (B) Products of 5’ RACE containing 5’UTR (left), central ORF fragment obtained by long-range RT-PCR with gene-specific primers from predicted exons 1 and 35 (middle left, Accession no. EF462416), products of RT-PCR with primers from exon 33 and the predicted exon 35 containing the 3’ end of ORF to stop codon (middle right) and products of 3’ RACE PCR (right). (C) Strategy for subcloning cDNA spanning flanking sequence in 5’UTR and entire ORF through stop codon (Accession no. EU179544) into pCR-XL-TOPO vector (Top, boxed). Restriction sites in overlapping partial cDNAs (filled triangles) and engineered restriction sites (open triangles) that were used are indicated. cDNA subclones of 5’UTR (Accession no. EF462415), or spanning 3’ORF, 3’UTR and poly-A tail (Accession no. FJ748889) (middle), and cDNA subclone of entire ORF and partial flanking UTRs from single long-range PCR (Accession no. FJ748888) (bottom), are diagramed with respective GenBank entries. (D) Structure of zebrafish mll cDNA including UTRs. PAS indicates polyadenylation signals. (E) Sequence of zebrafish mll 3’UTR. AUUUA pentamers, underlined; UUAUUUAUU nonamer, grey highlight; polyadenylation signals corresponding to (D), bold; U-rich element, boxed; poly-A tail, lowercase.
5’ RACE, long-distance RT-PCR, conventional RT-PCR, and 3’ RACE, respectively, yielded cDNAs comprising the 5’UTR, a ~12.4 kb central fragment of the ORF, the 3’ ORF to the stop codon, and the entire 3’UTR through the poly-A tail, from whole wild-type zebrafish adults and 24 hpf embryos (FIG 3B). Further RT-PCR amplification of the 5’UTR with an engineered NotI site, and ligation of overlapping fragments, enabled subcloning of an mll cDNA spanning the 5’UTR through the stop codon from 24 hpf embryos (FIG 3C, boxed) (Accession no. EU179544). The 5’UTR and 3’UTR sequences (FIG 3C, middle) informed additional long-range RT-PCR and subcloning of a 12.8 kb cDNA from 24 hpf embryos spanning the entire ORF and flanking UTRs (Accession no. FJ748888) (FIG 3C, bottom).
FIG 3D summarizes the structure of the zebrafish mll cDNA. Four potential ATG start codons were identified, only one of which was consistent with the ORF, indicating that 152 bases of subclone EF462415 (FIG 3C, middle) were from the 5’UTR. The ORF in zebrafish mll (Accession no. EU179544) contained 12657 bases, compared to the 11910 base human ortholog (Accession no. NM_005933). Comparison with the UCSC zebrafish genome database to define exon/intron boundaries indicated that zebrafish mll spans 36.7 kb and has 35 exons (FIG 3D) of similar lengths to those in human MLL. Whereas the first eight exons all align, zebrafish mll exon 9 resembles human MLL exons 9–10, suggesting splicing as one unit; zebrafish mll exons 10–35 then maintain similarity to human MLL exons 11–36 (TABLE I).
Table I.
Lengths and nucleotide identity in exons of zebrafish mll cDNA (Accession No. EU179544) and human MLL (Accession No. L04284)
| Exon Number | Exon Length (bp) | |||
|---|---|---|---|---|
| Zebrafish mll | Human MLL | Zebrafish mll | Human MLL | Nucleotide Identity (%) |
| 1 | 1 | 246 | 432 | 41.5 |
| 2* | 2* | 70 | 70 | 48.2 |
| 3 | 3 | 3344 | 2654 | 47.2 |
| 4* | 4* | 178 | 178 | 79.2 |
| 5 | 5 | 226 | 235 | 68.9 |
| 6 | 6 | 62 | 65 | 69.6 |
| 7 | 7 | 480 | 378 | 45.6 |
| 8 | 8 | 71 | 74 | 54.5 |
| 9 | 237 | 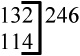 |
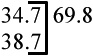 |
|
| 10* | 11* | 147 | 147 | 69.6 |
| 11* | 12* | 96 | 96 | 88.5 |
| 12* | 13* | 121 | 121 | 81.1 |
| 13* | 14* | 114 | 114 | 78.3 |
| 14* | 15* | 185 | 185 | 70.4 |
| 15* | 16* | 174 | 174 | 61.5 |
| 16* | 17* | 111 | 111 | 82.9 |
| 17* | 18* | 74 | 74 | 74.3 |
| 18 | 19 | 203 | 194 | 63.8 |
| 19 | 20 | 101 | 107 | 60.4 |
| 20* | 21* | 138 | 138 | 76.8 |
| 21* | 22* | 159 | 159 | 70.3 |
| 22* | 23* | 118 | 118 | 72.8 |
| 23* | 24* | 79 | 79 | 78.8 |
| 24* | 25* | 161 | 161 | 65.7 |
| 25 | 26 | 147 | 186 | 41.0 |
| 26 | 27 | 4447 | 4249 | 49.1 |
| 27 | 28 | 69 | 81 | 42.6 |
| 28 | 29 | 83 | 65 | 42.9 |
| 29 | 30 | 168 | 171 | 59.1 |
| 30* | 31* | 75 | 75 | 70.9 |
| 31* | 32* | 175 | 175 | 71.5 |
| 32* | 33* | 108 | 108 | 80.6 |
| 33* | 34* | 84 | 84 | 69.3 |
| 34* | 35* | 130 | 130 | 73.8 |
| 35* | 36* | 276 | 276 | 79.1 |
| Totals | 12657 | 11910 | 54.5 | |
Legend: Asterisk indicates exon of exact same length. Note that zebrafish mll exon 9 is more identical to human MLL exons 9–10 than exon 9; this alignment is supported by high conservation of sizes and nucleotide identity in ensuing 3’ exons.
The portion of the 3’ RACE PCR product non-overlapping with the 3’ ORF indicated that the 3’UTR is 1193 bases long; a 44 base poly-A sequence followed (FIG 3E). Two AAUAAA and one AUUAAA canonical cis-elements, which serve as polyadenylation signals (PAS) (Hu, et al 2005), were identified. A U-rich element indicative of a strong PAS occurred between the more 3’ AAUAAA and the poly-A tail (Hu, et al 2005). There were four AUUUA AU-rich elements (AREs) one of which was embedded in a UUAUUUAUU nonamer, which has been implicated in mRNA destabilization (Barreau, et al 2005, Zubiaga, et al 1995).
Zebrafish mll ortholog contains all critical functional domains of human MLL
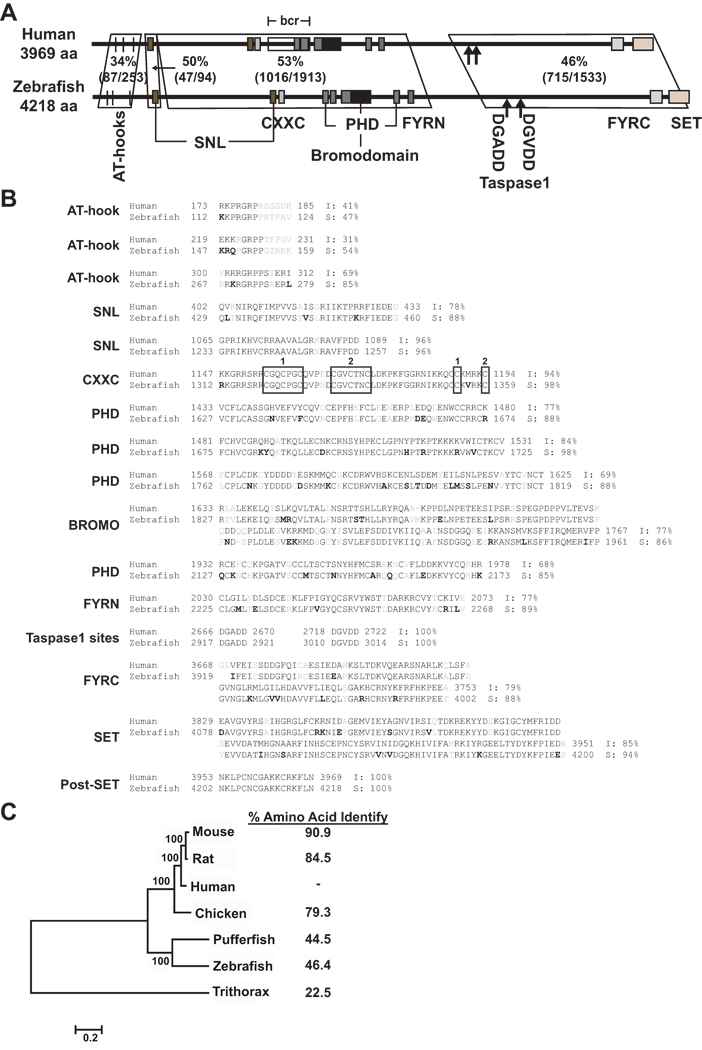
Alignments of the protein encoded by the cloned cDNAs identified all of the important functional domains of human MLL including AT-hooks, SNL, CXXC, PHD, bromodomain, FYRN, taspase1 sites, FYRC, and SET. There are 4218 amino acids in zebrafish Mll with 46.4% overall sequence identity to the 3969 amino acid human protein. Regionally, the highest amino acid identity (53%) is in the central portion of the protein containing the PHDs, bromodomain and FYRN, but high identity (50%) also was detected in a less well defined amino terminal region, and in the more carboxyl terminal region where the taspase1 sites, FYRC and SET domain are located (FIG 4A). However, there is much higher (68–100%) sequence similarity and, in some instances, identity within the functional domains (FIG 4B). As expected, phylogram tree analysis revealed closer relationships between human and mammalian MLL proteins relative to the other non-mammalian vertebrates zebrafish and pufferfish and the more distant fly (FIG 4C). Still, there was substantial conservation of all of the critical functional domains from zebrafish to human.
Fig 4.
Comparisons of protein encoded by cloned zebrafish mll cDNA to human MLL. (A) Conserved domains found by SMART (smart.embl-heidelberg.de/) are indicated. The percentages indicate amino acid identity determined by NCBI BLAST. (B) Protein domain alignment found by SMART with percentage of amino acid identity and similarity within domains determined by EMBOSS Pairwise Alignment Algorithm Needle (www.ebi.ac.uk/emboss/align/). Black indicates amino acid identity; gray, non-conservative substitution; bold, conservative substitution; numbered boxes, CGXCXXC motifs and distal cysteine residues in CXXC domain. (C) Consensus phylogram tree of MLL proteins produced by ClustalW and the neighbor-joining method within the MEGA4 program (Tamura, et al 2007). Note node values (100) indicating 100% bootstrap confidence derived from 1000 replicates. Branch lengths represent evolutionary change. Numbers at right represent the percentage of amino acid identity to human determined using EMBOSS Pairwise Alignment Algorithm Needle (www.ebi.ac.uk/emboss/align/). The accession numbers for this tree analysis are: NP_005924, Human MLL; NP_001101609, Rat Mll, NP_001074518, Mouse Mll; XP_417896.2, Chicken Mll; AF036382.1, pufferfish mll; EU179544, zebrafish Mll; NP_599109, trithorax.
Whereas the predicted single zebrafish Mll protein derived from a composite transcript from the two proximal predicted similar to mll transcripts using CDART contained only two PHDs (FIG 1A), all four PHDs are represented in the protein predicted by the cloned cDNA (FIG 4A,B), further underscoring the merit of molecular cloning of the full-length ORF.
mll mRNA is maternally supplied, expressed throughout the zebrafish lifespan and in haematopoietic tissues
From other studies it is well established that haematopoiesis in zebrafish progresses through primitive and definitive waves and generates similar blood cell lineages as in mammals, although in different anatomic sites. Whereas in mammals primitive haematopoiesis takes place in the extraembryonic yolk sac, primitive haematopoiesis in zebrafish is intraembryonic. First, primitive macrophages are produced in early gastrula in the anterior lateral mesoderm (ALM) (Herbomel, et al 1999). Then primitive erythroid cells are detected as early as the 2 somite stage (10–11 hpf) in the posterior stripes (posterior lateral mesoderm; PLM) that, at ~18 hpf, converge to form the indermediate cell mass (ICM) (Detrich, et al 1995, Thompson, et al 1998). A transient wave of haematopoiesis generating definitive erythroid progenitors recently was suggested in the murine yolk sac (Palis 2008) and definitive haematopoiesis in mammals also occurs transiently in the placenta (Rhodes, et al 2008, Zeigler, et al 2006). Likewise, in zebrafish, once circulation is established (~24 hpf) there is a transient definitive wave of haematopoiesis in the posterior blood island (PBI) producing erythromyeloid progenitors between 24–36 hpf (Bertrand, et al 2007, Rhodes, et al 2005). Intraembryonic definitive haematopoiesis producing haematopoietic stem cells (HSCs) in mammals first occurs in the aorto-gonad-mesonephros (AGM), then in fetal liver and finally in the bone marrow (Chen and Zon 2009). Between ~30 hpf to 3 days post fertilization (dpf) in zebrafish there is migration of definitive HSCs from the AGM first to the caudal haematopoietic tissue (CHT), which is remodeled from the PBI and considered the zebrafish equivalent of mammalian fetal liver (Murayama, et al 2006), as well as to the thymus and, then ~4 dpf, to the kidney marrow, which is the zebrafish equivalent of the bone marrow in mammals where adult HSCs are produced (Bertrand, et al 2007, Chen and Zon 2009).
The purpose of the next experiments was to characterize developmental mll expression over the zebrafish lifespan, tissue-specific mll expression in adults, and spatio-temporal mll expression in embryos, all in relation to the timepoints when, and tissues where, haematopoiesis occurs in zebrafish.
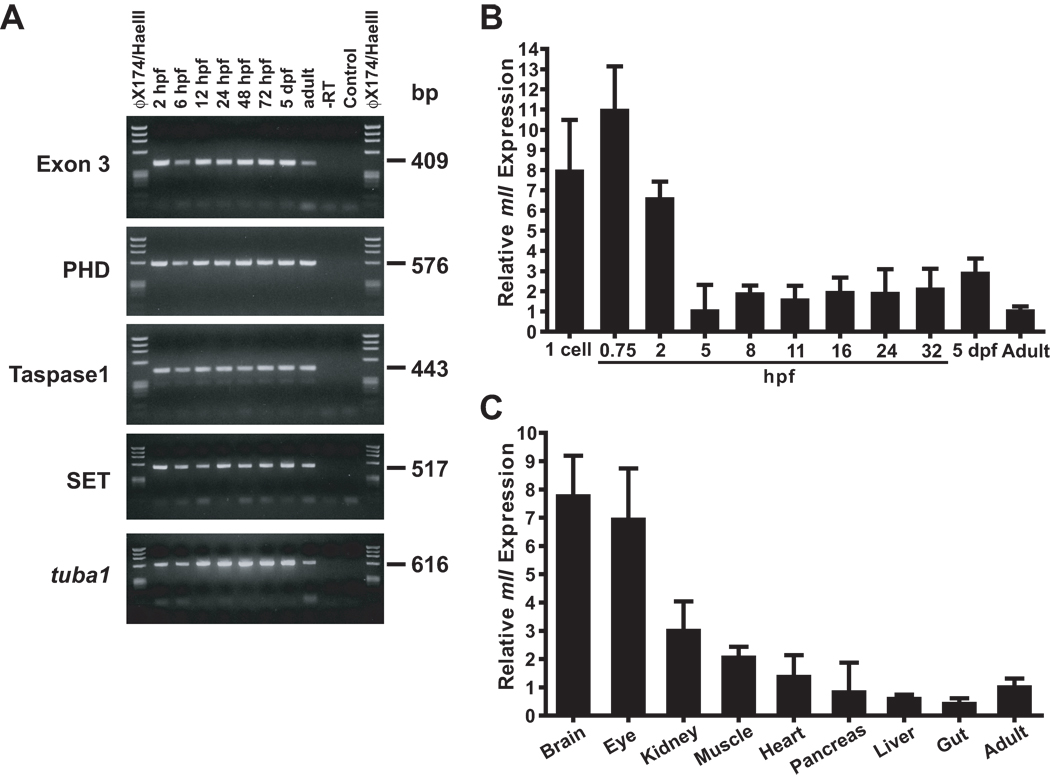
First mll expression was examined in whole wild-type zebrafish embryos, larvae and adults over a developmental timecourse. Analysis of four different regions of the transcript (TABLE SI) by RT-RCR and analysis of exons 1–3 by Q-RT-PCR (TABLE SI) in whole wild-type zebrafish, demonstrated mll expression throughout embryonic development and in the adult. RT-PCR yielded products at all embryonic timepoints tested (2, 6, 12, 24, 48, 72 hpf) and in 5 dpf larvae and adults (FIG 5A). Similarly, Q-RT-PCR yielded products at as early as 0 and 0.75 hpf, throughout embryonic development as well as in adults (FIG 5B). Given that zebrafish zygotic gene expression does not begin until 3 hpf and most maternal transcripts are degraded by 5 hpf, after which most transcripts are zygotic (Chatterjee, et al 2005, Christie, et al 2004), these results suggest the presence of maternally supplied mll transcripts at the single cell stage through the earliest developmental stages in the embryo. Furthermore, mll transcript levels were highest at these timepoints (0–2 hpf) (FIG 5B). The next Q-RT-PCR experiments comparing mll transcript abundance in dissected wild-type adult zebrafish tissues to that in whole adults showed enriched expression in the kidney (FIG 5C). Consistent with what was expected from mammals (Yu, et al 1998), mll was expressed abundantly in the brain. As the kidney marrow is the site of definitive haematopoiesis in adults (Chen and Zon 2009, Weinstein, et al 1996), these results demonstrated zebrafish mll expression in a haematopoietic tissue.
Fig 5.
Analysis of mll expression in whole zebrafish and demonstration of enrichment of zebrafish mll expression in haematopoietic tissues. (A) Temporal RT-PCR analysis of zebrafish mll mRNA expression. Developmental timepoints are indicated above each lane; target amplicons are at left. Sequencing confirmed respective transcripts. (B) Q-RT-PCR analysis of developmental mll expression. mll amplicon spanned exons 1–3; bactin1 housekeeping gene target amplicon was exons 4–5. Expression normalized to bactin1 is shown for each timepoint relative to whole adult. Data were analysed using comparative Ct method (Livak and Schmittgen 2001). Results represent 4 experiments per timepoint with 3 replicates per probeset per experiment; error bars represent standard deviation. Abbreviations: hpf, hours post-fertilization; dpf, days post-fertilization. (C) Q-RT-PCR analysis of mll expression in dissected wild-type zebrafish adult tissues. Results represent quadruplicate experiments on RNAs from respective pooled dissected tissues of 3 wild-type adults, and independent quadruplicate experiments on RNAs from respective pooled dissected tissues of 3 more wild-type adults (8 experiments per tissue total). The same probesets were used as in (B). Normalized expression is shown relative to the RNA sample prepared from two whole wild-type adult males and females each. Data were analysed using comparative Ct method (Livak and Schmittgen 2001). Error bars represent standard deviation of all 8 experiments. Note enriched mll expression in the adult kidney.
Characterization of spatio-temporal mll expression in whole wild-type zebrafish using WISH
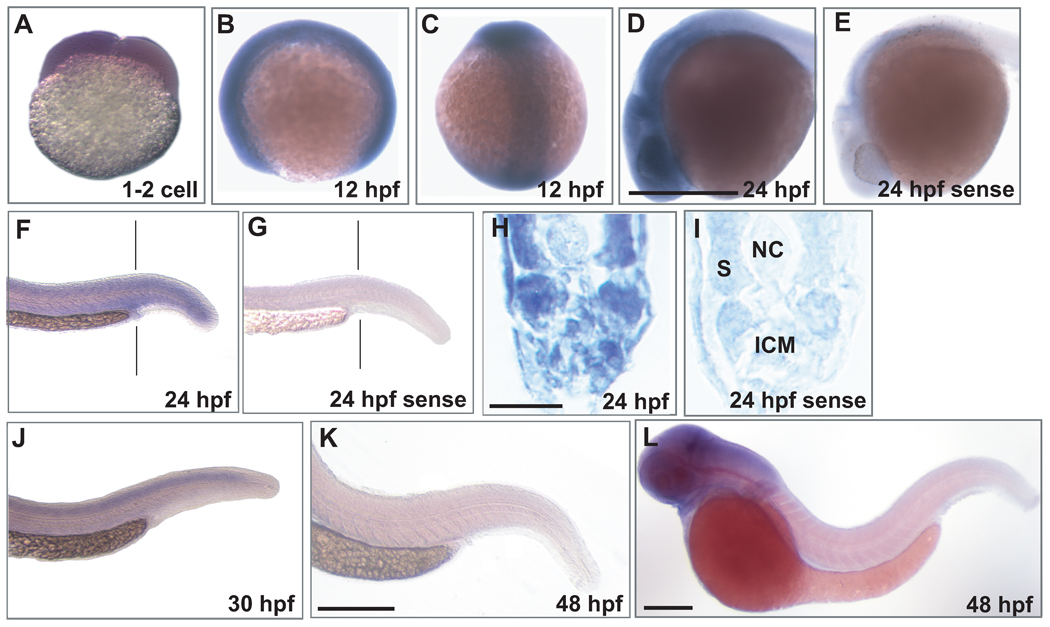
Next, WISH analyses of mll expression over a developmental timecourse were performed on zebrafish embryos. The results demonstrated abundant mll expression at the 1–2 cell stage (FIG 6A), consistent with the RT- and Q-RT-PCR results suggesting that mll transcripts are maternally supplied (FIG 5). The strong signal in the head and tailbud, among other tissues, suggested abundant zygotic mll transcripts throughout the 12 hpf embryo (FIG 6B,C). At 24 hpf, mll transcripts were highly expressed in the central nervous system (CNS) (FIG 6D) but expressed in peripheral tissues at lower levels including in the ICM, where primitive erythrocytes are produced (Bertrand, et al 2007, Chen and Zon 2009, Detrich, et al 1995) (FIG 6F–I). mll expression was absent over the yolk, which is not a site of primitive haematopoiesis in the zebrafish, but rather is a site to which primitive haematopoietic cells from the ALM and PLM migrate before circulation is established (Detrich, et al 1995, Herbomel, et al 1999) (FIG 6B,C,D). A prominent cephalo-caudal gradient was observed by 30–48 hpf. At 30 hpf low level mll mRNA expression was detected in the spinal cord, ICM and other caudal tissues (FIG 6J), whereas by 48 hpf mll mRNA expression was not detected by WISH in these tissues but remained high in the CNS (FIG 6K,L) consistent with the role of Mll as a modulator of neuronal development in mice (Yu, et al 1998). Sense controls, examples of which are shown at 24 hpf, did not show hybridization (FIG 6E,G,I).
Fig 6.
WISH analysis of mll mRNA expression in wild-type zebrafish embryos. Anti-sense (A–D,F,H, J–L) mll probe, and sense negative control probe (E, G, I) were used; examples of sense control are at 24 hpf. A. At the 1–2 cell stage, each cell is positive, suggesting that mll transcripts are maternally supplied. B, C. At 12 hpf, mll transcripts are ubiquitously expressed throughout the embryo. D. By 24 hpf, mll transcripts are highly expressed in the CNS. F. By 24 hpf, mll transcript expression is lower than at 12 hpf, but is present in peripheral tissues including the ICM (F, vertical line) at lower levels. H. Transverse section (vertical line in F) just posterior to yolk extension showing mll expression at 24 hpf in the posterior ICM and other peripheral tissues. ICM, intermediate cell mass; NC, notochord; S, somite. J. At 30 hpf, mll transcripts still remain highly expressed in the CNS (not shown) but are lower in the tail and in region of ICM. K, L. By 48 hpf, mll expression is not detected in peripheral tissues and is absent from the ICM, although transcripts are still expressed in the CNS. WISH probes from 2 regions of mll (662 base probe from 5’UTR-exon 3; 477 base probe from exon 33-exon 35) gave the same results. Scale bars: D, 500 µm (A–E); H, 20 µm (H, I); L, 500 µm (F, G, J, L); K, 500 µm (K).
Discussion
The first aspect of this work on molecular cloning established that there is a single mll gene in zebrafish syntenic to human MLL, which encodes not only all of the same functional domains in a structurally similar ORF, but also regulatory elements found in mammalian 3’UTRs controlling transcript turnover. The molecular cloning provided prerequisite cDNA sequences to investigate developmental and tissue specific expression. Elucidation of mll as a developmentally regulated gene expressed first as maternally supplied transcripts at the one cell stage and in the early embryo and, later, as zygotic transcripts in tissue-specific patterns throughout further embryogenesis into the adult, opens entirely new avenues to better understand Mll as an in vivo transcriptional modulator at whole organism and cellular levels throughout the zebrafish lifespan. Additionally, mll transcripts were detected in haematopoietic cells and tissues, providing rationale to elaborate roles of MLL in developmental haematopoiesis and leukemogenesis using zebrafish.
Pursuit of a zebrafish mll ortholog was strengthened by prior characterization of a single MLL-like gene with structural similarity and high overall sequence identity to human MLL in pufferfish (fugu), which is another teleost (Caldas, et al 1998). Because synteny is an important gauge of functional similarity (Barbazuk, et al 2000), the conserved block of syntenic genes suggested that the predicted single zebrafish mll on chromosome 15 was an ortholog with the same function as human MLL, rather than a paralog with a different function arising through gene duplication during evolution (Tatusov, et al 1997).
Our bioinformatics predictions of a single syntenic zebrafish mll were first borne out when cross-species dRT-PCR and Southern blot analyses with a human probe detected conserved regions of zebrafish mll. Amplification of a transcript spanning predicted chromosome 15 cDNAs for both “similar to MLL” proteins then gave experimental evidence that the two cDNAs comprised a single gene. The molecular cloning of the full length ORF and flanking UTRs that followed is significant because, until now, there were only partially cloned cDNAs of limited regions of the ORF, and the UTRs were totally uncharacterized.
At the genomic level, the 12657 bp ORF is a close replica of human MLL and, with only one exception, exhibits identical exon/intron structure. We also found all of the same functional domains implicated in transcriptional regulation by MLL, to be represented in the predicted protein. Even though overall amino acid sequence identity to human MLL is 46.4%, identity and similarity are far greater in the functional domains. Interestingly, domains specifically involved in DNA interactions and epigenetic transcriptional regulation are among the most highly conserved sequences. The CXXC DNA binding region is 98% similar to that in human MLL (2 conservative/ 1 non-conservative substitutions) and contains the same two CGXCXXC motifs and two distal cysteines implicated in properly folding the human MLL CXXC domain for zinc ion binding (Allen, et al 2006). The carboxyl SET domain where histone H3K4 methyltransferase activity resides (Milne, et al 2002) is 94% similar to human. Additionally there is 100% identity at the taspase1 sites required for MLL proteolysis (Hsieh, et al 2003), and high similarity at the SNL sequences, the bromodomain and all four PHDs. Therefore, despite evolutionary distance, we identified high regional amino acid similarity and even identity to human MLL in the zebrafish ortholog. Even though the phylogram tree reflects expected evolutionary divergence of mammals from teleosts, the high conservation of critical functional domains from zebrafish to human, which is much greater than the overall identity, predicts that zebrafish will be useful to model MLL.
Another aspect of the cloning involved characterizing regulatory elements in the 3’UTR. The identification of a AAUAAA polyadenylation signal (PAS) <40 bp upstream of the poly-A tail (Hu, et al 2005), a U-rich element between the PAS and poly-A tail (Hu, et al 2005) and the mRNA instability motifs, AUUUA and UUAUUUAUU (Zubiaga, et al 1995), suggests that the zebrafish mll 3’UTR contains essential regulatory sequences for the mRNA to be fully functional. Although incompletely characterized, the human MLL 3’UTR contains four AUUUA instability motifs and a 3’ PAS (Accession no. AB209508), and other mammalian genes feature these same elements (Hu, et al 2005).
Consistent with the recent genome-wide screen identifying mll amongst maternally supplied SET domain containing transcripts (Sun, et al 2008), our results from temporal RT-PCR, Q-RT-PCR and WISH experiments indicate that mll is maternally supplied. However, our RT-PCR, Q-RT-PCR and WISH experiments took this observation further by also characterizing the developmental timecourse of expression of the zygotic transcript. That mll mRNA is maternally supplied, expressed throughout embryogenesis from when zygotic transcription initiates, and expressed in the adult, indicates that this transcriptional regulator is important during the entire zebrafish lifespan.
The Q-RT-PCR and WISH provide detailed data on the relative abundance of maternal mll mRNA and how zygotic mll expression fluctuates with age and in different tissues; this may provide new leads to developmental roles of Mll in zebrafish and, ultimately, to similar roles of its orthologs in mammals. The whole animal Q-RT-PCR analyses demonstrated that mll expression is highest in early embryogenesis. The abundant mll expression detected in the brain and eye of adults by Q-RT-PCR, and in the head of embryos by WISH, is in keeping with the role of Mll as a maintenance factor for neuronal development in mammals (Yu, et al 1998). By 24 hpf, mll is present both throughout the CNS as well as in haematopoietic tissue in the ICM where primitive erythroid cells are formed (Bertrand, et al 2007, Chen and Zon 2009, Detrich, et al 1995, Gering, et al 1998). By 30–48 hpf, there is a pronounced rostral-caudal gradient. Thus up to 48 hpf, mll transcripts are highly expressed in the rostral CNS and hindbrain, and they are developmentally regulated in the ICM and other tissues.
The timecourse experiments performed, using more sensitive RT-PCR and Q-RT-PCR and the WISH experiments altogether, indicated that mll transcripts are not only expressed at various times throughout early embryogenesis, but more importantly appear to be expressed during critical timepoints during the primitive and definitive waves through which haematopoiesis progresses in the zebrafish. Although the anatomic sites are different, zebrafish haematopoiesis and blood cell morphology closely parallel those of mammals (Galloway and Zon 2003). Mammalian primitive haematopoiesis occurs in extraembryonic yolk sac blood islands; later in embryogenesis, definitive haematopoiesis in mammals occurs transiently in the yolk sac (Palis 2008) and placenta (Rhodes, et al 2008, Zeigler, et al 2006) and progresses to the AGM, fetal liver (Medvinsky and Dzierzak 1996), and eventually the bone marrow (Morrison, et al 1995). Zebrafish lack extraembryonic yolk sac blood islands, and primitive haematopoiesis giving rise to primitive macrophages and primitive erythroid cells, respectively occurs in the ALM and PLM, which becomes the ICM (Chen and Zon 2009, Detrich, et al 1995, Herbomel, et al 1999, Thompson, et al 1998). Detection of mll transcripts at 12 hpf and 16 hpf is consistent with mll expression during primitive haematopoiesis, and mll transcripts were also present at the start of circulation at 24 hpf. After 24 hpf, haematopoietic ontogeny in zebrafish is believed to progress through transient definitive and definitive waves that respectively generate erythromyeloid progenitor cells in the PBI between 24–36 hpf (Bertrand, et al 2007, Rhodes, et al 2005), and definitive HSCs that migrate from the AGM between 30 hpf and 3 dpf to the CHT (the zebrafish equivalent of mammalian fetal liver), the thymus and, ultimately, to the kidney marrow, which becomes the primary haematopoietic organ in the adult (Chen and Zon 2009, Weinstein, et al 1996). The Q-RT-PCR and WISH timecourse experiments also indicated the presence of mll transcripts during the transient definitive (30 hpf, 32 hpf) and definitive waves of haematopoiesis (48 hpf, 72 hpf, 5 dpf, adult) in the zebrafish.
Two independent experiments established that mll is expressed anatomically in zebrafish haematopoietic tissues: First is that WISH analysis of 24 hpf embryos identified mll transcripts in the ICM, the site of primitive haematopoiesis that gives rise to primitive erythrocytes, indicating mll expression in a haematopoietic tissue of the embryo. Second is that Q-RT-PCR detected enrichment of mll expression in the adult kidney, the definitive haematopoiesis site analogous to the bone marrow in mammals. Given that the first wave of definitive haematopoiesis in zebrafish embryos initiates with committed erythromyeloid progenitors in the posterior blood island (PBI) at ~24 hpf (Bertrand, et al 2007, Chen and Zon 2009) and the ICM also overlaps posteriorly with the anlagen of the PBI, the mll expression in the posterior ICM may also be consistent with the presence of mll transcripts in the nascent PBI, where important cell fate decisions are determined among erythromyeloid progenitors (Rhodes, et al 2005). However, further study would be necessary to show this more definitively. Interestingly, mll expression appears to be absent over the yolk sac, which is not the site of primitive haematopoiesis in zebrafish as it is in mammals, but rather a site of primitive haematopoietic cell migration before the circulation is established (Detrich, et al 1995, Herbomel, et al 1999). Taken together, these results are starting to unravel patterns of mll expression at mammalian-equivalent sites of blood cell production and migration in zebrafish, and they support an important role of Mll in zebrafish haematopoiesis like in mammals, where Mll is essential for primitive yolk sac haematopoiesis (Hess, et al 1997, Yu, et al 1995) and definitive fetal liver haematopoiesis (Yagi, et al 1998), and expressed in haematopoietic cells of adults (Jude, et al 2007).
Thus, the cloning and characterization of the full-length zebrafish mll ORF and flanking UTRs indicates high cross-species conservation of all of the critical functional domains of human MLL, as well as 3’UTR regulatory elements utilized in mammals. The finding by us and others (Sun, et al 2008) that mll transcripts are maternally supplied to zebrafish embryos extends the developmental window when MLL is important to the single cell stage of embryogenesis, providing a foundation to uncover novel, previously unappreciated functions of this oncoprotein in haematopoiesis and development at a stage that, so far, has not been accessed using mice. Enrichment of mll expression in haematopoietic cells of embryos and in the kidney, the site of definitive haematopoiesis in the zebrafish adult (Weinstein, et al 1996), substantiates further utilization of zebrafish for investigating MLL in haematopoiesis and leukaemogenesis. Zebrafish embryos also should enable better definition of how leukaemia is linked to normal MLL functions gone awry in embryogenesis, because MLL leukaemogenesis in infants initiates in utero (Ford, et al 1993). This work lays the groundwork for detailed functional analyses as the next steps in these investigations.
Acknowledgements
This work was supported by research funding from Eagles Fly for Leukemia to C.A.F., NIH NS050524 to R.B.G., Fondazione Citta’ della Speranza to G.G. and C.A.F., and NIH R01CA153348 to C.A.F. and R.B.G. We thank Amy Kugath for performing zebrafish husbandry and assistance with WISH and Michael Pack for the embryo dissociation protocol for cell sorting.
References
- Allen MD, Grummitt CG, Hilcenko C, Min SY, Tonkin LM, Johnson CM, Freund SM, Bycroft M, Warren AJ. Solution structure of the nonmethyl-CpG-binding CXXC domain of the leukaemia-associated MLL histone methyltransferase. EMBO Journal. 2006;25:4503–4512. doi: 10.1038/sj.emboj.7601340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balgobind BV, Raimondi SC, Harbott J, Zimmermann M, Alonzo TA, Auvrignon A, Beverloo HB, Chang M, Creutzig U, Dworzak MN, Forestier E, Gibson B, Hasle H, Harrison CJ, Heerema NA, Kaspers GJ, Leszl A, Litvinko N, Nigro LL, Morimoto A, Perot C, Pieters R, Reinhardt D, Rubnitz JE, Smith FO, Stary J, Stasevich I, Strehl S, Taga T, Tomizawa D, Webb D, Zemanova Z, Zwaan CM, van den Heuvel-Eibrink MM. Novel prognostic subgroups in childhood 11q23/MLL-rearranged acute myeloid leukemia: results of an international retrospective study. Blood. 2009;114:2489–2496. doi: 10.1182/blood-2009-04-215152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barbazuk WB, Korf I, Kadavi C, Heyen J, Tate S, Wun E, Bedell JA, McPherson JD, Johnson SL. The syntenic relationship of the zebrafish and human genomes. Genome Research. 2000;10:1351–1358. doi: 10.1101/gr.144700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barreau C, Paillard L, Osborne HB. AU-rich elements and associated factors: are there unifying principles? Nucleic Acids Research. 2005;33:7138–7150. doi: 10.1093/nar/gki1012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bertrand JY, Kim AD, Violette EP, Stachura DL, Cisson JL, Traver D. Definitive hematopoiesis initiates through a committed erythromyeloid progenitor in the zebrafish embryo. Development. 2007;134:4147–4156. doi: 10.1242/dev.012385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caldas C, Kim MH, MacGregor A, Cain D, Aparicio S, Wiedemann LM. Isolation and characterization of a pufferfish MLL (mixed lineage leukemia)-like gene (fMll) reveals evolutionary conservation in vertebrate genes related to Drosophila trithorax. Oncogene. 1998;16:3233–3241. doi: 10.1038/sj.onc.1201873. [DOI] [PubMed] [Google Scholar]
- Caslini C, Shilatifard A, Yang L, Hess JL. The amino terminus of the mixed lineage leukemia protein (MLL) promotes cell cycle arrest and monocytic differentiation. Proceedings of the National Academy of Sciences of the United States of America. 2000a;97:2797–2802. doi: 10.1073/pnas.040574897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caslini C, Alarcon AS, Hess JL, Tanaka R, Murti KG, Biondi A. The amino terminus targets the mixed lineage leukemia (MLL) protein to the nucleolus, nuclear matrix and mitotic chromosomal scaffolds. Leukemia. 2000b;14:1898–1908. doi: 10.1038/sj.leu.2401933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chatterjee B, Chin AJ, Valdimarsson G, Finis C, Sonntag JM, Choi BY, Tao L, Balasubramanian K, Bell C, Krufka A, Kozlowski DJ, Johnson RG, Lo CW. Developmental regulation and expression of the zebrafish connexin43 gene. Developmental Dynamics. 2005;233:890–906. doi: 10.1002/dvdy.20426. [DOI] [PubMed] [Google Scholar]
- Chen AT, Zon LI. Zebrafish blood stem cells. Journal of Cellular Biochemistry. 2009;108:35–42. doi: 10.1002/jcb.22251. [DOI] [PubMed] [Google Scholar]
- Christie TL, Mui R, White TW, Valdimarsson G. Molecular cloning, functional analysis, and RNA expression analysis of connexin45.6: a zebrafish cardiovascular connexin. American Journal of Physiology - Heart and Circulatory Physiology. 2004;286:H1623–H1632. doi: 10.1152/ajpheart.00800.2003. [DOI] [PubMed] [Google Scholar]
- Detrich HW, 3rd, Kieran MW, Chan FY, Barone LM, Yee K, Rundstadler JA, Pratt S, Ransom D, Zon LI. Intraembryonic hematopoietic cell migration during vertebrate development. Proceedings of the National Academy of Sciences of the United States of America. 1995;92:10713–10717. doi: 10.1073/pnas.92.23.10713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Djabali M, Selleri L, Parry P, Bower M, Young BD, Evans GA. A Trithorax-like gene is interrupted by chromosome 11q23 translocations in acute leukemias. Nature Genetics. 1992;2:113–118. doi: 10.1038/ng1092-113. [DOI] [PubMed] [Google Scholar]
- Fair K, Anderson M, Bulanova E, Mi H, Tropschug M, Diaz MO. Protein interactions of the MLL PHD fingers modulate MLL target gene regulation in human cells. Molecular Cell Biology. 2001;21:3589–3597. doi: 10.1128/MCB.21.10.3589-3597.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Felix CA, Kim CS, Megonigal MD, Slater DJ, Jones DH, Spinner NB, Stump T, Hosler MR, Nowell PC, Lange BJ, Rappaport EF. Panhandle polymerase chain reaction amplifies MLL genomic translocation breakpoint involving unknown partner gene. Blood. 1997;90:4679–4686. [PubMed] [Google Scholar]
- Ford AM, Ridge SA, Cabrera ME, Mahmoud H, Steel CM, Chan LC, Greaves M. In utero rearrangements in the trithorax-related oncogene in infant leukaemias. Nature. 1993;363:358–360. doi: 10.1038/363358a0. [DOI] [PubMed] [Google Scholar]
- Galloway JL, Zon LI. Ontogeny of hematopoiesis: examining the emergence of hematopoietic cells in the vertebrate embryo. Current Topics in Developmental Biology. 2003;53:139–158. doi: 10.1016/s0070-2153(03)53004-6. [DOI] [PubMed] [Google Scholar]
- Geer LY, Domrachev M, Lipman DJ, Bryant SH. CDART: protein homology by domain architecture. Genome Research. 2002;12:1619–1623. doi: 10.1101/gr.278202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gering M, Rodaway AR, Gottgens B, Patient RK, Green AR. The SCL gene specifies haemangioblast development from early mesoderm. EMBO Journal. 1998;17:4029–4045. doi: 10.1093/emboj/17.14.4029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gu Y, Nakamura T, Alder H, Prasad R, Canaani O, Cimino G, Croce CM, Canaani E. The t(4;11) chromosome translocation of human acute leukemias fuses the ALL-1 gene, related to Drosophila trithorax, to the AF-4 gene. Cell. 1992;71:701–708. doi: 10.1016/0092-8674(92)90603-a. [DOI] [PubMed] [Google Scholar]
- Herbomel P, Thisse B, Thisse C. Ontogeny and behaviour of early macrophages in the zebrafish embryo. Development. 1999;126:3735–3745. doi: 10.1242/dev.126.17.3735. [DOI] [PubMed] [Google Scholar]
- Hess JL, Yu BD, Li B, Hanson R, Korsmeyer SJ. Defects in yolk sac hematopoiesis in Mll-null embryos. Blood. 1997;90:1799–1806. [PubMed] [Google Scholar]
- Hilden JM, Dinndorf PA, Meerbaum SO, Sather H, Villaluna D, Heerema NA, McGlennen R, Smith FO, Woods WG, Salzer WL, Johnstone HS, Dreyer Z, Reaman GH. Analysis of prognostic factors of acute lymphoblastic leukemia in infants: report on CCG 1953 from the Children's Oncology Group. Blood. 2006;108:441–451. doi: 10.1182/blood-2005-07-3011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsieh JJ, Cheng EH, Korsmeyer SJ. Taspase1: a threonine aspartase required for cleavage of MLL and proper HOX gene expression. Cell. 2003;115:293–303. doi: 10.1016/s0092-8674(03)00816-x. [DOI] [PubMed] [Google Scholar]
- Hu J, Lutz CS, Wilusz J, Tian B. Bioinformatic identification of candidate cis-regulatory elements involved in human mRNA polyadenylation. RNA. 2005;11:1485–1493. doi: 10.1261/rna.2107305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jude CD, Climer L, Xu D, Artinger E, Fisher JK, Ernst P. Unique and independent roles for MLL in adult hematopoietic stem cells and progenitors. Cell Stem Cell. 2007;1:324–337. doi: 10.1016/j.stem.2007.05.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liedtke M, Cleary ML. Therapeutic targeting of MLL. Blood. 2009;113:6061–6068. doi: 10.1182/blood-2008-12-197061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu H, Cheng EH, Hsieh JJ. Bimodal degradation of MLL by SCFSkp2 and APCCdc20 assures cell cycle execution: a critical regulatory circuit lost in leukemogenic MLL fusions. Genes & Development. 2007;21:2385–2398. doi: 10.1101/gad.1574507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- Medvinsky A, Dzierzak E. Definitive hematopoiesis is autonomously initiated by the AGM region. Cell. 1996;86:897–906. doi: 10.1016/s0092-8674(00)80165-8. [DOI] [PubMed] [Google Scholar]
- Meyer C, Kowarz E, Hofmann J, Renneville A, Zuna J, Trka J, Ben Abdelali R, Macintyre E, De Braekeleer E, De Braekeleer M, Delabesse E, de Oliveira MP, Cave H, Clappier E, van Dongen JJ, Balgobind BV, van den Heuvel-Eibrink MM, Beverloo HB, Panzer-Grumayer R, Teigler-Schlegel A, Harbott J, Kjeldsen E, Schnittger S, Koehl U, Gruhn B, Heidenreich O, Chan LC, Yip SF, Krzywinski M, Eckert C, Moricke A, Schrappe M, Alonso CN, Schafer BW, Krauter J, Lee DA, Zur Stadt U, Te Kronnie G, Sutton R, Izraeli S, Trakhtenbrot L, Lo Nigro L, Tsaur G, Fechina L, Szczepanski T, Strehl S, Ilencikova D, Molkentin M, Burmeister T, Dingermann T, Klingebiel T, Marschalek R. New insights to the MLL recombinome of acute leukemias. Leukemia. 2009;23:1490–1499. doi: 10.1038/leu.2009.33. [DOI] [PubMed] [Google Scholar]
- Milne TA, Briggs SD, Brock HW, Martin ME, Gibbs D, Allis CD, Hess JL. MLL targets SET domain methyltransferase activity to Hox gene promoters. Molecular Cell. 2002;10:1107–1117. doi: 10.1016/s1097-2765(02)00741-4. [DOI] [PubMed] [Google Scholar]
- Morrison SJ, Uchida N, Weissman IL. The biology of hematopoietic stem cells. Annual Review of Cell & Developmental Biology. 1995;11:35–71. doi: 10.1146/annurev.cb.11.110195.000343. [DOI] [PubMed] [Google Scholar]
- Murayama E, Kissa K, Zapata A, Mordelet E, Briolat V, Lin HF, Handin RI, Herbomel P. Tracing hematopoietic precursor migration to successive hematopoietic organs during zebrafish development. Immunity. 2006;25:963–975. doi: 10.1016/j.immuni.2006.10.015. [DOI] [PubMed] [Google Scholar]
- Palis J. Ontogeny of erythropoiesis. Current Opinion in Hematology. 2008;15:155–161. doi: 10.1097/MOH.0b013e3282f97ae1. [DOI] [PubMed] [Google Scholar]
- Panzer JA, Gibbs SM, Dosch R, Wagner D, Mullins MC, Granato M, Balice-Gordon RJ. Neuromuscular synaptogenesis in wild-type and mutant zebrafish. Developmental biology. 2005;285:340–357. doi: 10.1016/j.ydbio.2005.06.027. [DOI] [PubMed] [Google Scholar]
- Payne E, Look T. Zebrafish modelling of leukaemias. British Journal of Haematology. 2009;146:247–256. doi: 10.1111/j.1365-2141.2009.07705.x. [DOI] [PubMed] [Google Scholar]
- Pieters R, Schrappe M, De Lorenzo P, Hann I, De Rossi G, Felice M, Hovi L, LeBlanc T, Szczepanski T, Ferster A, Janka G, Rubnitz J, Silverman L, Stary J, Campbell M, Li CK, Mann G, Suppiah R, Biondi A, Vora A, Valsecchi MG. A treatment protocol for infants younger than 1 year with acute lymphoblastic leukaemia (Interfant-99): an observational study and a multicentre randomised trial. Lancet. 2007;370:240–250. doi: 10.1016/S0140-6736(07)61126-X. [DOI] [PubMed] [Google Scholar]
- Rhodes J, Hagen A, Hsu K, Deng M, Liu TX, Look AT, Kanki JP. Interplay of pu.1 and gata1 determines myelo-erythroid progenitor cell fate in zebrafish. Developmental Cell. 2005;8:97–108. doi: 10.1016/j.devcel.2004.11.014. [DOI] [PubMed] [Google Scholar]
- Rhodes KE, Gekas C, Wang Y, Lux CT, Francis CS, Chan DN, Conway S, Orkin SH, Yoder MC, Mikkola HK. The emergence of hematopoietic stem cells is initiated in the placental vasculature in the absence of circulation. Cell Stem Cell. 2008;2:252–263. doi: 10.1016/j.stem.2008.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rowley JD, Olney HJ. International workshop on the relationship of prior therapy to balanced chromosome aberrations in therapy-related myelodysplastic syndromes and acute leukemia: overview report. Genes Chromosomes Cancer. 2002;33:331–345. doi: 10.1002/gcc.10040. [DOI] [PubMed] [Google Scholar]
- Song HD, Sun XJ, Deng M, Zhang GW, Zhou Y, Wu XY, Sheng Y, Chen Y, Ruan Z, Jiang CL, Fan HY, Zon LI, Kanki JP, Liu TX, Look AT, Chen Z. Hematopoietic gene expression profile in zebrafish kidney marrow. Proceedings of the National Academy of Sciences of the United States of America. 2004;101:16240–16245. doi: 10.1073/pnas.0407241101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun XJ, Xu PF, Zhou T, Hu M, Fu CT, Zhang Y, Jin Y, Chen Y, Chen SJ, Huang QH, Liu TX, Chen Z. Genome-wide survey and developmental expression mapping of zebrafish SET domain-containing genes. PLoS ONE. 2008;3:e1499. doi: 10.1371/journal.pone.0001499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takeda S, Chen DY, Westergard TD, Fisher JK, Rubens JA, Sasagawa S, Kan JT, Korsmeyer SJ, Cheng EH, Hsieh JJ. Proteolysis of MLL family proteins is essential for taspase1-orchestrated cell cycle progression. Genes & Development. 2006;20:2397–2409. doi: 10.1101/gad.1449406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamura K, Dudley J, Nei M, Kumar S. MEGA4: Molecular Evolutionary Genetics Analysis (MEGA) software version 4.0. Molecular Biology and Evolution. 2007;24:1596–1599. doi: 10.1093/molbev/msm092. [DOI] [PubMed] [Google Scholar]
- Tatusov RL, Koonin EV, Lipman DJ. A genomic perspective on protein families. Science. 1997;278:631–637. doi: 10.1126/science.278.5338.631. [DOI] [PubMed] [Google Scholar]
- Thompson MA, Ransom DG, Pratt SJ, MacLennan H, Kieran MW, Detrich HW, 3rd, Vail B, Huber TL, Paw B, Brownlie AJ, Oates AC, Fritz A, Gates MA, Amores A, Bahary N, Talbot WS, Her H, Beier DR, Postlethwait JH, Zon LI. The cloche and spadetail genes differentially affect hematopoiesis and vasculogenesis. Developmental biology. 1998;197:248–269. doi: 10.1006/dbio.1998.8887. [DOI] [PubMed] [Google Scholar]
- Tkachuk DC, Kohler S, Cleary ML. Involvement of a homolog of Drosophila Trithorax by 11q23 chromosomal translocations in acute leukemias. Cell. 1992;71:691–700. doi: 10.1016/0092-8674(92)90602-9. [DOI] [PubMed] [Google Scholar]
- Weinstein BM, Schier AF, Abdelilah S, Malicki J, Solnica-Krezel L, Stemple DL, Stainier DY, Zwartkruis F, Driever W, Fishman MC. Hematopoietic mutations in the zebrafish. Development. 1996;123:303–309. doi: 10.1242/dev.123.1.303. [DOI] [PubMed] [Google Scholar]
- Xia ZB, Anderson M, Diaz MO, Zeleznik-Le NJ. MLL repression domain interacts with histone deacetylases, the polycomb group proteins HPC2 and BMI-1, and the corepressor C-terminal-binding protein. Proceedings of the National Academy of Sciences of the United States of America. 2003;100:8342–8347. doi: 10.1073/pnas.1436338100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yagi H, Deguchi K, Aono A, Tani Y, Kishimoto T, Komori T. Growth disturbance in fetal liver hematopoiesis of Mll-mutant mice. Blood. 1998;92:108–117. [PubMed] [Google Scholar]
- Yokoyama A, Kitabayashi I, Ayton PM, Cleary ML, Ohki M. Leukemia proto-oncoprotein MLL is proteolytically processed into 2 fragments with opposite transcriptional properties. Blood. 2002;100:3710–3718. doi: 10.1182/blood-2002-04-1015. [DOI] [PubMed] [Google Scholar]
- Yu BD, Hess JL, Horning SE, Brown GA, Korsmeyer SJ. Altered Hox expression and segmental identity in Mll-mutant mice. Nature. 1995;378:505–508. doi: 10.1038/378505a0. [DOI] [PubMed] [Google Scholar]
- Yu BD, Hanson RD, Hess JL, Horning SE, Korsmeyer SJ. MLL, a mammalian trithorax-group gene, functions as a transcriptional maintenance factor in morphogenesis. Proceedings of the National Academy of Sciences of the United States of America. 1998;95:10632–10636. doi: 10.1073/pnas.95.18.10632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeigler BM, Sugiyama D, Chen M, Guo Y, Downs KM, Speck NA. The allantois and chorion, when isolated before circulation or chorio-allantoic fusion, have hematopoietic potential. Development. 2006;133:4183–4192. doi: 10.1242/dev.02596. [DOI] [PubMed] [Google Scholar]
- Zubiaga AM, Belasco JG, Greenberg ME. The nonamer UUAUUUAUU is the key AU-rich sequence motif that mediates mRNA degradation. Molecular Cell Biology. 1995;15:2219–2230. doi: 10.1128/mcb.15.4.2219. [DOI] [PMC free article] [PubMed] [Google Scholar]