Abstract
The mechanisms regulating the emergence of BM prothymocytes remain poorly characterized. Genome-wide transcriptome analyses looking for genes expressed in human prothymocytes led to the identification of AF1q/MLLT11 as a candidate gene conceivably involved in this process. Analysis of AF1q protein subcellular localization and intracellular trafficking showed that despite pronounced karyophily, it was subjected to constitutive nuclear export followed by ubiquitin-mediated degradation in the centrosomal area. Using in vitro assays based on either forced expression or shRNA-mediated silencing of AF1q, we provide evidence that the protein promotes T- over B-cell differentiation in multipotent hematopoietic progenitors. At the molecular level, AF1q confers to multipotent progenitors an increased susceptibility to Delta-like/Notch-mediated signaling. Consistent with these findings, enforced AF1q expression in humanized mice fosters the emergence of BM CD34+CD7+ prothymocytes, enhances subsequent thymus colonization, and accelerates intrathymic T-cell development. In contrast, AF1q silencing provokes a global shift of BM lymphopoiesis toward the B-cell lineage, hinders prothymocyte development, inhibits thymus colonization, and leads to intrathymic accumulation of B cells. Our results indicate that AF1q cooperates with the Notch signaling pathway to foster the emergence of BM prothymocytes and drive subsequent intrathymic specification toward the T-cell lineage.
Introduction
The identity of thymus-colonizing cells has been the subject of prolonged controversy. Despite reports that they corresponded either to multipotent stem/progenitor cells (HPCs) or to early common lymphoid progenitors, current evidence indicates that thymus colonization is ensured by T-lineage–polarized precursors referred to as “prothymocytes.”1 In humans, CD34+CD45RA++CD7+ precursors possess T- over B-cell lymphoid potential, express the thymus-homing receptor CCR9, and undergo selective thymus recruitment in both ex vivo and in vivo colonization assays.2,3 This population emerges in the fetal BM as early as gestational week 8-9, follows bell-curve dynamics during the last 2 trimesters of development, and persists at low levels after birth. The mechanisms regulating the development of BM prothymocytes are still poorly understood. The IL-7 receptor, c-Kit, Wnt/LEF/TCF, and Sonic Hedgehog play a central role in the survival and proliferation of pre-β-selection thymocytes, whereas Notch1 receptor ligation by Delta-like 4 (Dll4) drives their specification toward the T-cell lineage,4,5 but whether and to what extent they also participate in the extrathymic differentiation of prothymocytes remains unclear. Mice carrying targeted deletions of IL-7, Sonic Hedgehog, or Wnt signaling intermediates are characterized by intrathymic accumulation of double-negative (DN)2-3 thymocytes, arguing for a predominant, if not exclusive, post-entry developmental arrest.6,7 Current evidence suggests that Notch signaling plays an important role in the emergence of BM prothymocytes. BM CCR9+VCAM-1− HPCs that correspond to physiologic postnatal thymus colonizers in the mouse up-regulate HES1 and Deltex1 transcript expression and display detectable green fluorescent protein (GFP) mRNA levels in Notch reporter mice.8 Conditional Notch1 inactivation or ectopic expression of Notch inhibitors arrest intrathymic T-cell development before or at the early T-cell lineage progenitor stage and promote accumulation of thymic B cells, whereas constitutive expression of the Notch intracellular domain inhibits B-cell development, leads to extrathymic T-cell differentiation, and drives leukemic transformation.9 The ZBTB7A//LRF proto-oncogene has emerged as an important negative regulator of Notch signaling. Because LRF-deficient progenitors aberrantly express Notch target genes and differentiate into CD4+CD8+ double-positive T cells in the BM, it is assumed that the restriction of Notch signaling imposed by LRF physiologically skews BM lymphoid development toward the B-cell lineage.10 These observations suggest that the outcome of Notch ligation in BM progenitors might not depend solely on the level of Notch ligands or the density of Notch1, but may also rely on fine-tuning of the cell-intrinsic susceptibility to Delta-like/Notch interactions.
ALL1 fused from chromosome 1q (AF1q)/MLLT11 was identified as a mixed lineage leukemia (MLL) fusion partner.11 Complex duplication/translocation events of the AF1q locus have since been reported in hematologic malignancies,12,13 and high AF1q mRNA levels are markers of poor prognosis in leukemia and myelodysplastic syndromes.14 Although AF1q is highly expressed in the thymus, to date, its biologic function remains unknown. In the present study, we show that AF1q promotes early T-cell development in multipotent HPCs through dual restriction of B-cell differentiation and cell-intrinsic sensitization to Delta-like/Notch interactions.
Methods
Plasmid constructs and protein analysis
The full-length AF1q coding sequence was obtained by PCR and cloned in-frame with the HA1 epitope into a pcDNA3-modified vector (HA-AF1q). A2M (L30-32A) and A4M (L24-27-30-32A) mutants were generated by site-directed mutagenesis using the Quick Change Mutagenesis Kit (Stratagene), and cloned into either pcDNA3 or pWPI vectors. HEK-293T cells stably expressing HA-tagged AF1q or A2M were generated by lentiviral transduction. Transient transfections of HeLa and HEK-293T cells were performed using the standard calcium phosphate method. Treatment with leptomycin B (LMB; 2 ng/mL), cycloheximide (CHX, 50 μg/mL, both from Sigma-Aldrich); Lactacystin (10μM; Calbiochem); or TGF-β1 (R&D Systems) was performed for the indicated periods. In vitro ubiquitylation assays and His-tagged protein purification were performed as described previously.15 Cell extracts were prepared in radio-immunoprecipitation assay buffer containing 0.05% SDS (0.05M Tris, pH 8, 0.2M NaCl, 1% NP40, 0.02M EDTA, pH 8, and 0.5% deoxycholate), subjected to SDS-PAGE, transferred by iBlot (Invitrogen) to nitrocellulose membranes, and probed with anti-AF1q (Abnova) or anti–γ-tubulin (Sigma-Aldrich) antibodies. Labeling was developed using horseradish peroxidase–conjugated goat anti–mouse polyclonal antibodies (Pierce). Membranes were processed using the chemiluminescence SuperSignal West Femto Maximum Sensitivity reagent (Thermo Scientific).
Antibodies and immunofluorescence microscopy
Transiently transfected HeLa cells grown on coverslips were washed in PBS and fixed and permeabilized for 10 minutes in PBS, 3% Triton X-100, and 3.7% paraformaldehyde. Nonadherent cells were plated on poly-L-lysine–coated slides, fixed in 2.5% paraformaldehyde, permeabilized in 0.1% Triton X-100, and labeled with the corresponding antibodies. Mouse monoclonal antibodies anti-AF1q and anti–LAP-2 were from Abnova and Transduction Laboratories, respectively. The rabbit anti–γ-tubulin polyclonal antibody was from Abcam; the rat anti-HA tag monoclonal antibody was from Roche. Antibodies were revealed by Alexa Fluor 488– or Alexa Fluor 594–labeled secondary antibodies from Molecular Probes. Slides were counterstained with 4′,6-diamidino-2-phenylindole, dihydrochloride and mounted in Vectashield neutral medium. Fluorescent microscopy was performed either with an Axiovert 2000M microscope or with a confocal LSM510 meta laser microscope (Carl Zeiss Micro-Imaging).
Processing human tissues, cell separation, and in vitro differentiation assays
Umbilical cord blood and thymi were collected according to institutional guidelines and processed as described previously.3 CD34+ HPCs were purified with an EasySep human CD34 Positive Selection Kit (StemCell Technologies; purity > 90%). For FACS sorting, cells were labeled with CD34-PE 5 (PECy5) or CD34− allophycocyanin (APC), CD45RA-PE (BD Pharmingen), and a cocktail of FITC-conjugated antibodies (Lin): CD7 and CD10 (both from Beckman-Coulter Immunotech) and CD19, CD36, and CD41 (all from BD Biosciences). B- and natural killer (NK)–cell differentiation assays have been described elsewhere.16 B and NK cells were monitored by flow cytometry using PE-conjugated CD19 (BioLegend) and CD56 antibodies (BD Biosciences). Assessment of T-cell potential in OP9-Delta-like1 (OP9-DL1) stromal cell cocultures was performed as described previously.3 OP9-DL1 cells were plated in 6-well plates 1 or 2 days before use. Cocultures were conducted in the presence of SCF (10 mg/mL), FMS-related tyrosine kinase 3 ligand (FLT3L; 5 ng/mL), and IL-7 (2 ng/mL). Cells were harvested every 5-7 days for flow cytometric analysis. Labeling was performed with CD7-PE (BD Biosciences), CD1a-APC (BioLegend), CD4-PE (BD Pharmingen), CD8-PECy5 (BD Biosciences), CD3-APC, TCRαβ, and TCRγδ (BD Pharmingen) antibodies.
Lentiviral transductions
Lentiviral plasmids pWPI and pLVTHM were a gift from Didier Trono (Ecole Polytechnique Fédérale, Lausanne, Switzerland). pLKO.1/shRNAs targeting AF1q were purchased from Open Biosystem and tested as described previously.17 The corresponding AF1q-specific shRNA54 (shAF1q), shRNA58, and scrambled shRNAs (shCtl) were subcloned into pLVTHM. VSV-pseudotyped third-generation lentiviral vectors were produced by transient transfection into 293T cells and purified by ultracentrifugation. Expression titers of GFP vectors were estimated on 293T cells by limiting dilution. CD34+ HPCs were preactivated overnight under serum-free culture conditions (BIT 9500; StemCell Technologies) with SCF (50 ng/mL), FLT3L (50 ng/mL), and thrombopoietin (TPO, 10 ng/mL, all from R&D Systems) before being exposed to the corresponding vectors (107 transducing units).
qRT-PCR and ChIP
Total RNAs were extracted using the RNeasy Mini or Micro Kits (QIAGEN). RNA quality control and quantification were monitored using the RNA 6000 Pico LabChip Kit and the Agilent 2100 Bioanalyzer (Agilent Technologies). RNAs were reverse transcribed using random hexamers with Superscript II reverse transcriptase (Invitrogen Life Technologies). Amplification of mRNAs by quantitative real-time RT-PCR (qRT-PCR) was performed with the corresponding primers (supplemental Table 1, available on the Blood Web site; see the Supplemental Materials link at the top of the online article) and a LightCycler (Roche Diagnostics). PCR products were detected by the SYBR Green dye (Roche). Nested duplex qRT-PCR for the simultaneous amplification of AF1q and HPRT was performed as described previously18 (supplemental Table 1). Gene-expression levels were quantified with LightCycler v.3.5.3 software (Roche Diagnostics). Gene-expression levels were normalized relative to endogenous HPRT as well as to levels detected in Jurkat cells used as an external calibrator. This procedure allows for direct comparison between the diverse biologic samples. ChIP and qRT-PCR analysis was performed as described previously.19 Primers were designed within regulatory (Spen-1, Spen-2, and Spen-3) and conserved intronic regions (Spen-4 and Spen-5) of the Spen locus (supplemental Table 2). Results were normalized with respect to the promoter of the β2-microglobulin gene for histone H3 acetylation or PAX6 gene for histone H3 lysine 9 methylation.
Microarray and statistical analyses
The methodology and results of transcriptome analyses are provided as supplemental Methods. Full microarray data can be downloaded from the GEO database under accession numbers GSE29522 and GSE29523.
Mice
NOD-SCID/γc−/− (NSG) mice (The Jackson Laboratory) were housed in the pathogen-free animal facility of Institut Universitaire d'Hématologie. Use of the mice was approved by the animal care and use committee of the Institut Universitaire d'Hématologie. Six-week-old mice were irradiated with 2.5 Gy 18-24 hours before intravenous injection of CD34+ HPCs (1.5 × 105 cells), exposed to the corresponding lentiviral vectors, and killed 8 weeks after grafting. The BM, spleen, and thymus were harvested and analyzed with a FACSCanto cytometer (Becton Dickinson). Labeling of BM cells was performed with CD45-Alexa-Fluor 700 (BioLegend), CD34-Pacific Blue (BioLegend), CD7-PE (BD Biosciences), CD10-PECy7 (BD Biosciences), and CD19-APC (BioLegend) antibodies. Labeling of thymocytes was performed with CD34-Pacific Blue and CD1a-PE (BioLegend), CD4-APC (BD Biosciences), and CD8-PECy7 (BioLegend).
Results
AF1q nuclear export and proteasome-dependent degradation in the centrosomal area
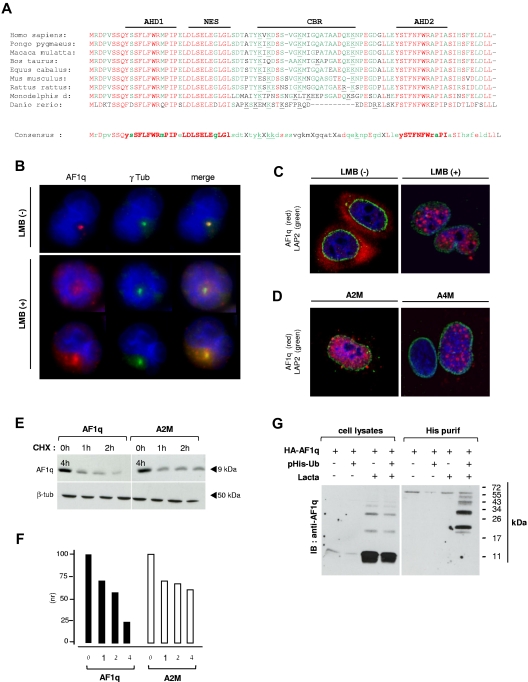
Results of genome-wide transcriptome analyses to screen for genes expressed in umbilical cord blood CD34++CD45RA++CD7+ prothymocytes3 and the fact that the expression of these also increased linearly from CD34++CD44hiCD1a− to CD34+CD1a− and to CD34+CD1a+ thymocytes20 led us to identify AF1q/MLLT11 as a candidate gene possibly involved in regulating the emergence of prothymocytes (supplemental Figure 1 and supplemental Table 3). Bioinformatics analysis and alignment of orthologs showed that AF1q is characterized by the duplication of a conserved canonical domain (referred to as AF1q Homology Domain [AHD]) located at both sides of a central basic region (CBR), and by a prototypic nuclear export signal (NES; Figure 1A). We first examined AF1q subcellular localization in primary thymocytes or leukemia T-cell lines and found that the protein concentrates in the centrosomal area (Figure 1B top panel). Because AF1q displays a predicted NES, we speculated that its centrosomal accumulation depended on prior nuclear export. AF1q no longer accumulated in this area in SupT1 cells treated with LMB, an inhibitor of CRM1-dependent nuclear export. LMB treatment resulted in a heterogeneous staining pattern, with AF1q either reentering the nucleus or remaining confined to the cytoplasm (Figure 1B bottom panel). Consistent with these observations, HA-tagged AF1q localized in the cytoplasm of transiently transfected HeLa cells, whereas it relocated in a speckled pattern in the nucleus after LMB treatment (Figure 1C). Genetic mapping of the NES confirmed these results by showing that mutations of Leu residues 30/32 (A2M) or 27/29/30/32 (A4M) both resulted in constitutive nuclear sequestration of the corresponding variants (Figure 1D).
Figure 1.
Intracellular trafficking and ubiquitin-dependent catabolism of AF1q by the proteasome. (A) Comparison of the amino acid sequence of AF1q orthologs. The AF1q AHD1 and AHD2, NES, and CBR are indicated. ClustalW alignments were performed using the Accelrys Gene v2.5 software. Conserved amino acids are shown in red; consensus residues are in green. Synonymous amino-acid substitutions are indicated in lower-case letters; conserved basic residues of the CBR are underlined. (B) The centrosomal localization of AF1q follows its nuclear export. SupT1 cells were treated with 2 ng/mL of LMB for 2 hours before labeling with AF1q (red) and γ-tubulin (green) antibodies. (C-D) Subcellular localization of wild-type AF1q and of its Δ-NES L30/32A (A2M) or L27/29/30/32A (A4M) mutant derivatives. HeLa cells were transfected with vectors encoding HA-tagged AF1q (C) or the mutant proteins (D). Cells were treated or not with 2 ng/mL of LMB for the last 2 hours and labeled with AF1q (red) and LAP2 (green) antibodies. LAP-2 is a marker of the inner nuclear envelope. Confocal microcopy analysis was performed 48 hours after transfection. (E-F) Constitutive nuclear relocation of AF1q slows down its catabolism. HEK-AF1q (right panel) and HEK-A2M cell lines (left panel) were treated for the indicated periods with CHX and analyzed by immunoblot (E) followed by protein quantification by densitometry (F). Results are normalized relative to protein levels detected in control cells cultured under the same conditions but without CHX. (G) AF1q degradation depends on its prior ubiquitylation. HEK-AF1q cells were transfected or not with a His-tagged ubiquitin (6xHis-Ubiquitin) vector, and treated or not with 10μM Lactacystin (Lacta) for 12 hours before immunoblotting with AF1q antibodies. Left panel shows total cell extracts; right panel, His-purified proteins.
Because the centrosomal area is a major site of protein catabolism, we assessed the effect of Lactacystin, a pharmacologic inhibitor of the proteasome, on AF1q degradation. Direct side-by-side comparison of their degradation kinetics showed that the nuclear-sequestered A2M derivative persisted for ≥ 4 hours in cells compared with ≤ 2 hours for wild-type AF1q (Figure 1E). Densitometry analysis showed that whereas decay of both proteins was similar during the first hours, the A2M mutant was more stable thereafter (Figure 1F), arguing for the existence of 2 distinct protein pools with different turnovers. To examine whether AF1q centrosomal catabolism depended on prior ubiquitylation, HEK-293T cells stably expressing wild-type AF1q (HEK-AF1q cells) were treated for 12 hours with Lactacystin before probing cell extracts with anti-AF1q antibodies. This yielded a characteristic ladder of high-molecular-weight AF1q species compatible with polyubiquitylation (Figure 1G left panel). In addition, HEK-AF1q cells were transfected with a His-tagged ubiquitin expression vector and treated for 12 hours with Lactacystin before isolation of ubiquitylated proteins: His-purified proteins strongly reacted with anti-AF1q antibodies (Figure 1G right panel), indicating that AF1q degradation depends on prior ubiquitylation. These data suggest that AF1q constitutive CRM1-dependent export from the nucleus is followed by ubiquitin-mediated degradation by the proteasome.
AF1q antagonizes early B-cell development
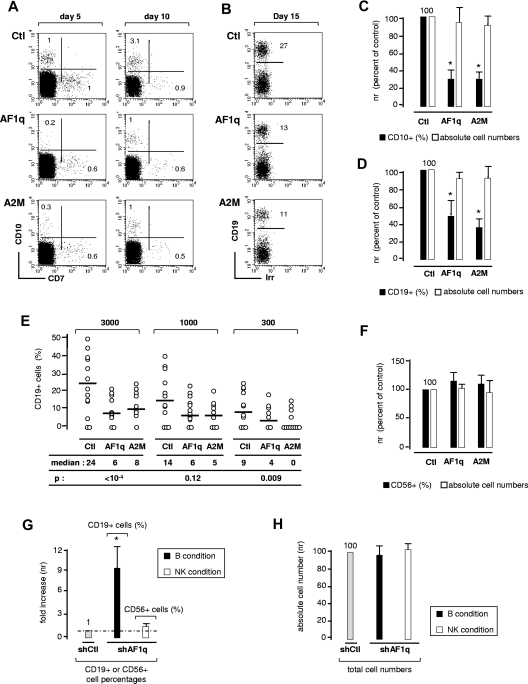
Because AF1q is considered an oncogenic protein, we first investigated the effect of the wild-type protein and of nuclear-sequestered mutant A2M on CD34+ HPC survival and proliferation. CD34+ cells were transduced with the corresponding lentiviral vectors, sorted based on GFP expression, and cultured for 5-12 days with different cytokine combinations. Irrespective of the conditions, neither protein affected cell survival or proliferation, nor did they interfere with the differentiation of granulocytes (CD15+CXCR1+), macrophages (CD1a+CD14+), or dendritic cells (CD1a+CD14−; data not shown). The effect of AF1q on the emergence of lymphoid precursors was then assessed. FACS analysis of AF1q- or A2M-transduced CD34+Lin− HPCs cultured for 5-7 days under serum-free conditions showed that, whereas neither affected total cell yields, both elicited a 3- to 5-fold decrease in CD7−CD10+ pre-pro-B-cell percentages (Figure 2A and 2C), but did not promote the emergence of CD7+CD10− T-cell precursors.
Figure 2.
Effect of AF1q on B-cell differentiation. (A,C) AF1q antagonizes the emergence of CD7−CD10+ pre-pro-B cells in serum-free cultures. CD34+Lin− HPCs were transduced with AF1q, A2M, or the empty vector (Ctl), sorted based on GFP expression, and cultured for 5-11 days with SCF, FLT3L, TPO, and IL-7 under serum-free conditions. (A) Flow cytometry dot-plots of GFP+-gated cells. Numbers in quadrants indicate the percentage of each population. Data are from 1 of 4 independent experiments. (C) Relative quantification of the effect exerted by AF1q or A2M on CD7−CD10+ pre-pro-B-cell differentiation or overall cell proliferation. To limit experimental variability due to donor effect, results are normalized relative to CD10+ cell percentages and absolute cell numbers in cultures of empty vector–transduced cells (Ctl). Data are means ± SD percentages of 4 independent experiments. Statistically significant differences (P < .05, Student t test) in percentages or absolute numbers between AF1q/A2M and the control conditions are marked by asterisks. (B,D) AF1q antagonizes B-cell development in cocultures onto MS5 cells. (B) CD34+ HPCs transduced as above with AF1q, A2M, or the empty vector were seeded onto MS5 cells and cultured for 2 weeks in the absence of exogenous cytokines before absolute cell numbers and CD19+GFP+ B-cell percentages were determined. Data are representative of 1 of 4 independent experiments. (D) Relative quantification of the effect exerted by AF1q and A2M on B-cell development (CD19+ B-cell percentages) or global cell expansion (total cell numbers); results are normalized relative to percentages of CD19+ cells and absolute cell numbers in cultures of empty vector–transduced cells (Ctl). Irr indicates irrelevant antibody. Data are means ± SD percentages of 4 independent experiments. Statistically significant differences are indicated. (E) Limiting-dilution analysis. CD34+ HPCs were transduced and cultured as above. Positive wells were scored on day 14 and analyzed by FACS as indicated above. Empty circles represent CD19+ B-cell percentages. FACS analysis was restricted to cell-containing wells. Medians and P values are indicated. Statistical significance was determined by the Wilcoxon test. (F) AF1q does not affect NK-cell development. CD34+ HPCs transduced with AF1q, A2M, or the empty vector and sorted as above were seeded onto MS5 cells and cultured for 3 weeks with SCF, FLT3L, IL-2, IL-7, and IL-15 before absolute cell numbers and CD56+GFP+ NK-cell percentages were determined. Results are normalized relative to CD56+ cell percentages and absolute cell numbers in cultures of empty vector–transduced cells (Ctl). Means ± SD percentages of 4 independent experiments are shown. (G-H) AF1q deficiency enhances B-cell differentiation. CD34+ HPCs transduced with lentiviral vectors driving the expression of shRNA54 (shAF1q) or scrambled shRNAs (shCtl) were seeded onto MS5 cells and cultured under conditions that promote B-cell (black bars) or NK-cell (empty bars) development. CD19+ and CD56+ cell percentages (G) and absolute numbers (H) were determined by FACS. Results are normalized relative to the control condition (shCtl). Means ± SD of 4 independent experiments are shown. Statistically significant differences are indicated.
To investigate the effect of AF1q on B-cell development, AF1q- or A2M-transduced cells were cultured for 2 weeks on MS5 stromal cells in the absence of cytokines, a condition that constrains myeloid cell outgrowth but also results in reduced cell expansion (5- to 15-fold).21 Although total cell yields did not change, CD19+ B-cell percentages decreased by 2.5- and 3.4-fold in AF1q and A2M-transduced cell cultures, respectively (Figure 2B and 2D). Whether AF1q or A2M affects B-cell progenitor frequency was assessed in limiting-dilution experiments. Analysis based on Poisson distribution showed that, despite an overall similar albeit limited cloning efficiency (1:100-1:200), B-cell progenitor frequencies were lower for A2M- (1:800-1:1200) than for wild-type AF1q- or empty vector–transduced cells (1:300-1:500), indicating that the nuclear-sequestered mutant was more efficient in this respect. In addition, those clones that derived from AF1q- or A2M-transduced cells had lower CD19+ cell percentages than the clones in control wells (Figure 2E), indicating that both proteins affect the capacity of clonable cells to differentiate along the B-cell pathway. CD34+CD10+CD19− pre-pro-B cells and CD34+CD10+CD19+ pro-B cells were then transduced with A2M or the empty vector and cultured for 2 weeks onto MS5 cells. Whereas A2M antagonized acquisition of CD24 and CD19 by the pre-pro-B cells, it had only a minor effect on pro-B cells, confirming that AF1q predominantly affects early the stages of B-cell development (supplemental Figure 2A). We next examined the effect of AF1q on NK-cell differentiation. AF1q-, A2M-, or empty vector–transduced cells were cultured under appropriate conditions, and CD56+ NK-cell percentages were assessed after 3 weeks. This showed that neither protein affected NK-cell development (Figure 2F and data not shown). In agreement with these findings, FACS analysis of mixed B/NK clones derived from A2M- or empty vector–transduced single cells cultured under conditions that promoted B- and NK-cell development confirmed the selective effect of AF1q on B-cell differentiation. Whereas A2M still did not affect CD56+ cell percentages, it decreased by 4-fold the percentage of CD19+ B cells per clone (median: 10% vs 47%; supplemental Figure 2B).
Whether endogenous AF1q influences B- or NK-cell differentiation was finally examined by transducing CD34+ HPCs with a lentiviral vector producing shRNA targeting AF1q (shAF1q) (supplemental Figure 3A-B). AF1q silencing did not lead to alteration in overall cell growth or survival, but increased up to 9-fold the generation of CD19+ B cells and only marginally affected CD56+ NK-cell yields in bulk cultures (Figure 2G-H). To assess the stage at which AF1q inhibits B-cell differentiation, we compared the effect of AF1q silencing on the capacity of sorted CD34+CD45RA−Lin− (referred to as CD45RA−Lin− thereafter) and CD34+CD45RA+Lin− HPCs to generate B or NK cells. The CD45RA−Lin− population is enriched in multipotent HPCs, whereas the CD45RA+Lin− cells comprise mixed lympho-granulo-macrophagic precursors.22,23 We found that AF1q silencing increased by 3- to 5-fold the CD19+ B-cell percentages in cultures initiated with early CD45RA−Lin− progenitors (supplemental Figure 3C), while not affecting the capacity of CD45RA+Lin− cells to generate B or NK cells. Although the hypothesis that AF1q interferes with the growth or survival of a minor cell subset cannot be absolutely ruled out, these data indicate that AF1q antagonizes entry of multipotent HPCs into the B-cell pathway.
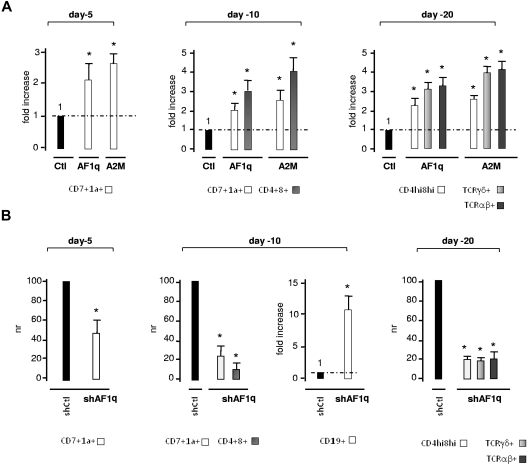
AF1q promotes T-cell development from multipotent HPCs
We next investigated whether AF1q synergizes with Delta-like/Notch-induced signaling to promote CD45RA−Lin− HPC entry into the T-cell pathway. Culturing CD45RA−Lin− HPCs transduced with AF1q- or A2M-lentiviral vectors onto OP9-DL1 cells24 showed that AF1q species augmented the culture day 5 percentages of CD7+CD1a− pro-T cells by 2.1- and 2.5-fold, respectively (Figure 3A and supplemental Figure 4A). Such was also the case subsequently for CD7+CD1a+ precursors that reached 30% of AF1q- or A2M-transduced cells by day 10 compared with only 10%-15% in the control condition. Consistent with these findings, post-β-selection double-positive CD4+CD8+ T cells were detected on day 10 in cultures of AF1q- or A2M-transduced cells, earlier than in the control condition. The stimulatory effect of AF1q and A2M on T-cell development persisted until culture day 20, leading to an up to 4-fold increase of TCRαβ+ or TCRγδ+ cell percentages. In agreement with these observations, we also found that AF1q- and A2M-transduced cells displayed increased capacity to reconstitute murine thymus lobes in fetal thymic organ cultures3 (data not shown). To determine whether AF1q also acts in a nonredundant manner during early T-cell development, CD45RA−Lin− HPCs were transduced with the shAF1q lentiviral vector before coculture on OP9-DL1 cells (Figure 3B and supplemental Figure 4B). By day 5, AF1q silencing led to a 2-fold reduction in CD7+CD1a− cell percentages as well as to delayed maturation of T-cell precursors, only a minority of which had already acquired membrane CD1a by day 10. Interestingly, a small number of CD19+ B cells (3%-5%) were detected at that time in the cultures. By day 20, AF1q silencing had led to a 5- to 7-fold decrease in the percentages of double-positive CD4hiCD8hi and of mature TCRαβ+ or TCRγδ+ cells. Neither forced AF1q/A2M expression nor shRNA-mediated AF1q silencing affected the overall cell growth in OP9-DL1 cocultures. We concluded that AF1q synergizes with Delta-like/Notch interactions to promote T-cell development from multipotent HPCs.
Figure 3.
Effect of AF1q on the kinetics of T-cell development in OP9-DL1 cell cocultures. (A) Forced expression of AF1q promotes T-cell development. CD34+CD45RA−Lin− HPCs transduced with AF1q, A2M, or the empty vector (Ctl) were sorted based on GFP expression and cultured onto OP9-DL1 cells with SCF, FLT3L, and IL-7. GFP+ cells were FACS analyzed at the indicated time points for determining the percentages of CD7+CD1a−, CD7+CD1a+, CD4+CD8+, CD4+CD8hi, TCRαβ+, and TCRγδ+ cells. Results are normalized relative to percentages in cultures of empty vector–transduced cells (Ctl), and are presented as -fold increases. Means ± SD percentages of 4 independent experiments are shown. Statistically significant differences (P < .05; Student t test) between AF1q/A2M and the control conditions are marked by asterisks. Note that AF1q/A2M did not significantly affect overall cell yields (data not shown). The corresponding dot-plot analyses are shown in supplemental Figure 5A. (B) AF1q deficiency impairs T-cell development. CD34+CD45RA−Lin− HPCs transduced with vectors driving the expression of AF1q-specific shRNA54 (shAF1q) or scrambled shRNAs (shCtl) were seeded onto OP9-DL1 cells, cultured, and processed as described. Results are normalized relative to percentages in cultures of cells transduced with scrambled shRNAs (shCtl), and are presented as normalized ratios (NR) or -fold increase. Means ± SD percentages of 4 independent experiments are shown. Statistically significant differences are indicated. Dot-plot analyses are presented in supplemental Figure 5B. Note also that AF1q deficiency does not affect cell proliferation (data not shown).
The AF1q locus is not a downstream target of Notch in multipotent HPCs
We then examined the transcriptional regulation of AF1q in CD45RA−Lin− HPCs. Limited cell numbers were cultured for 48 hours as triplicates in 96-well plates without serum but with SCF, FLT3L, and TPO. Nested qRT-PCR showed that none of the cytokines affected AF1q transcript levels (data not shown). Because SCF and FLT3L allowed for better cell viability, this condition was used as an internal calibrator thereafter. CD45RA−Lin− HPCs were then treated with IL-7, Wnt3a, or Sonic Hedgehog or cultured onto Notch ligand Delta1ext-IgG before qRT-PCR analysis. None of these affected AF1q transcript levels (supplemental Figure 5A). To screen for other potential activators of AF1q transcription, the HPCs were treated with graded doses of GM-CSF, TNF-α, TGF-β1, retinoic acid, IL-3, or IL-15 alone or in combination. Only TGF-β1 elicited a dose-dependent increase of AF1q transcript levels, and BMP4, another TGFβ super-family member, exerted a comparable activity. Because TGF-β1 regulates the stability of various nuclear proteins,25 we examined whether it affected AF1q protein catabolism (supplemental Figure 5B). Dose-response and time-course experiments showed that adding TGF-β1 to CHX-treated HEK-AF1q cells delayed AF1q degradation. Neither TGF-β1 nor Delta1ext-Ig antagonized AF1q nuclear export (data not shown).
AF1q increases the susceptibility of multipotent HPCs to Delta-like/Notch–induced signals
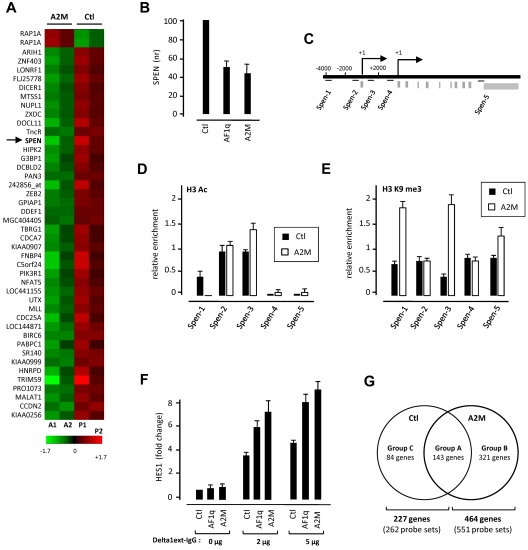
We next compared the gene-expression profiles of CD45RA−Lin− and CD45RA+Lin− HPCs transduced with nuclear-sequestered A2M or the empty vector. Statistical analysis of the microarray data confirmed the selective targeting by AF1q of the more immature cell population: A2M modulated expression of 44 probe sets (43 annotated genes) in CD45RA−Lin− HPCs relative to only 1, ELK4, in the CD45RA+Lin− cells (Figure 4A and data not shown). Of the 43 genes for which expression changed in CD45RA−Lin− cells, 42 were down-regulated (−2.9 ≤ FC ≤ −2.02) and only 1, RAP1a, was up-regulated (supplemental Table 4). SPEN, which encodes a corepressor that competes with the Notch intracellular domain for binding to RBPJ,26 was in the top 2 genes that A2M down-regulated. qRT-PCR confirmed that SPEN transcript levels decreased ≥ 2-fold in AF1q- or A2M-transduced CD45RA−Lin− HPCs (Figure 4B). Analysis of CD45RA−Lin− HPCs by ChIP showed an A2M-induced repressive trimethylation of histone H3 at lysine 9 on the Spen locus but failed to detect association with SPEN regulatory regions (Figure 4C-E and data not shown). AF1q did not interfere with SPEN expression levels in CD45RA+Lin− HPCs or T-cell leukemia cell lines, arguing for the selectivity of its effect (data not shown). Down-modulation of SPEN in pre-DN1 thymocytes was in step with AF1q up-regulation, thus strengthening the physiologic pertinence of our data (supplemental Figure 6).
Figure 4.
AF1q potentiates Notch signaling. (A) Gene-expression profiling of CD34+CD45RA−Lin− HPCs transduced with nuclear-sequestered AF1q (A2M). CD34+CD45+Lin− HPCs were FACS sorted, exposed to A2M (A1-A2) or control (P1-P2) vectors, sorted based on GFP expression, and subjected to global gene-expression analysis 72 hours later. Results are from 2 independent experiments (Exp1: A1/P1; Exp2: A2/P2). Statistical analysis, performed with the R package “locfdr” (lfdr < 2%), showed that 44 probe sets were differentially expressed between A2M (A1-A2)- and empty vector (P1-P2)–transduced CD34+CD45RA−Lin− HPCs. Results are presented as a heat map of the average expression levels (up: red; down: green). (See also supplemental Table 2.) (B) AF1q down-modulates SPEN transcript levels in CD34+CD45RA−Lin− HPCs. qRT-PCR analysis of CD34+CD45RA−Lin− cells transduced with AF1q, A2M, or the empty vector (Ctl). Expression levels (nr) are normalized relative to those in cells transduced with the empty vector. Means ± SD percentages of 3 independent experiments are shown. (C-E) AF1q induces histone H3 modifications at the Spen locus. (C) Diagram illustrating the genomic context of Spen promoter loci. Regions amplified by site-specific qPCR are indicated by bars. (D-E) CD34+CD45RA−Lin− cells were transduced as above with A2M or the empty vector and cultured for 72 hours before ChIP analysis via anti-H3ac (D) and anti-H3K9me3 (E) antibodies. Results are means of 2 ChIP experiments analyzed in triplicate. (F-G) Forced AF1q expression broadens the repertoire of Notch-responsive genes in CD34+CD45RA−Lin− HPCs. (F) CD34+CD45RA−Lin− HPCs were transduced with AF1q, A2M, or the empty vector (Ctl), sorted based on GFP expression, and cultured for 3 more days onto graded doses of plastic-immobilized Delta1ext-IgG before qRT-PCR analysis. Expression levels are normalized relative to those in cells transduced with the empty vector. Means ± SD of 3 independent samples are shown. (G) Venn diagram detailing shared and distinct gene expression between Notch-stimulated A2M- and empty vector–transduced cells. CD34+CD45RA−Lin− HPCs were transduced with A2M or the empty vector (Ctl), and cultured as above before being subjected to global gene-expression analysis. Statistical analysis performed with the R package “locfdr” (lfdr < 5%) showed that, after Notch activation, 551 probe sets were up-regulated in A2M-transduced cells, relative to only 262 for their homologs transduced with the empty vector (see also supplemental Table 5A-D).
To evaluate the relationships between AF1q and the Notch signaling pathway, AF1q- or A2M-transduced CD45RA−Lin− HPCs were cultured for 72 hours on 2 or 5 μg of plastic-immobilized Delta1ext-IgG27 before qRT-PCR analysis. Both AF1q and A2M increased Notch-mediated expression of the Notch-target gene HES1 (Figure 4F). In contrast, neither Notch signaling nor A2M or AF1q affected LRF, Notch1, or Notch2 expression levels in CD45RA−Lin− cells (data not shown). To investigate whether AF1q synergizes with Notch signaling to promote the polarization of CD45RA−Lin− HPCs toward the T-cell lineage, A2M-transduced and control cells were cultured as above on graded doses of plastic-immobilized Delta1ext-IgG before gene-expression profiling. Analysis of the microarray data showed that Notch signaling up-regulated the expression of 551 probe sets (464 annotated genes) in A2M-transduced cells relative to only 262 (227 annotated genes) in controls (Figure 4G). Notch-responsive genes segregated into 3 groups, referred to as A, B, and C. Group A (172 probe sets, 143 annotated genes, supplemental Table 5A) was composed of genes for which expression increased to overall similar extents in A2M-transduced and control cells, and contained several T-cell genes. Group B, constituted of genes overexpressed only in A2M-transduced cells, was the most represented because it comprised 379 of the 641 probes sets up-regulated by Notch (321 annotated genes; supplemental Table 5B) and included most T-lineage–affiliated transcription factors (ETS1, GATA3, TOX, MYB, RUNX3, IKZF2, and SOX4), Notch targets, DTX4, and Nrarp, as well as diverse Wnt signaling pathway intermediates (SFRP5, FZD2, FRZD3, and FZD7). For the majority of group B genes, we also observed higher expression in Notch-activated control cells, although differences were not statistical significant in this case. Group C, encompassing genes overexpressed in control cells (90 probe sets; 84 annotated genes), was more heterogeneous and segregated into 2 subgroups: C1 comprising genes whose expression also increased, albeit to a lesser extent, in A2M-transduced cells, and C2 comprising the majority of genes that remained at baseline in A2M-transduced cells (supplemental Table 5C-D). These data indicate that AF1q increases the susceptibility of multipotent HPCs to Delta-like/Notch interactions, possibly through down-modulation of SPEN. This may also account for accelerated T-cell differentiation in OP9-DL1 cultures.
AF1q promotes the emergence of BM prothymocytes in humanized mice
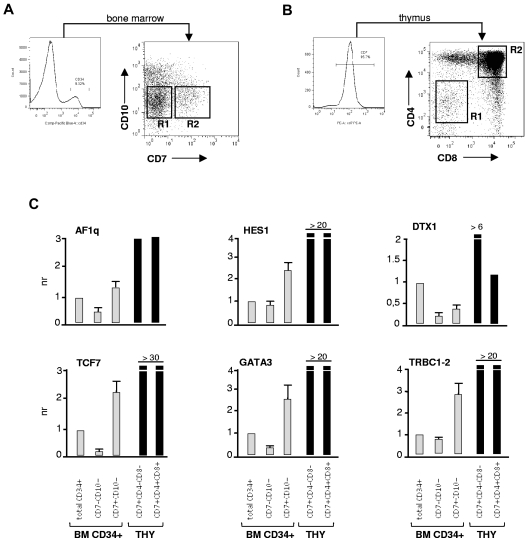
NSG mice were used to examine the effects of AF1q on T-cell lineage commitment and differentiation in vivo.28,29 We first investigated whether NSG mice engrafted with CD34+ HPCs supported the emergence of BM prothymocytes. Analysis of recipient mice 8 weeks after grafting showed that this was the case because we detected the presence of a population of BM CD34+CD7+ HPCs (0.8%-5% of human cells), all of which coexpressed CD45RA and CD2 (data not shown). CD34+CD7+ cells were also detected at low levels in the spleen (0.01%-0.1% of human cells), indicating that they circulated in the blood, and they accounted for the majority of early thymic immigrants (data not shown). To confirm that BM CD34+CD7+ HPCs corresponded to prothymocytes, this population was sorted and analyzed by qRT-PCR (Figure 5A-B). Relative to the more immature CD34+CD7−CD10− cells, CD34+CD7+ HPCs expressed 2- to 8-fold greater AF1q, HES1, TCF7, GATA3, and TRBC1-2 transcript levels (Figure 5C), confirming that they were primed for the T-lineage differentiation program and displayed ongoing Notch signaling. The 2 populations did not differ with regard to SPEN expression (data not shown), indicating that SPEN down-modulation follows entry into the thymus.
Figure 5.
Phenotypic characterization of early lymphoid progenitors in humanized mice. (A-B) Gating procedures. Irradiated NSG mice were injected intravenously with CD34+ HPCs (1.5 × 105 cells), and the BM and thymus were harvested 8 weeks later. Single-cell suspensions obtained from 3 mice were pooled and labeled with the corresponding antibodies. Gates are set on human CD45+ cells. (A) CD34++ BM cells were sorted based on CD7 and CD10 expression. (B) Thymocytes were sorted based on CD7, CD4, and CD8 expression. The corresponding cell populations were then analyzed by qRT-PCR. (C) qRT-PCR analysis of sorted CD34+CD7−CD10− and CD34+CD7+CD10− BM HPCs, and of CD7+CD4−CD8− and CD7+CD4+CD8+ thymocytes of recipient mice 8 weeks after grafting (see also supplemental Figure 5B). Expression levels are normalized relative to those detected in total nonfractionated BM CD34+ HPCs. For BM cells, results are means ± SD of 2 independent experiments. For thymocytes, results are from one experiment.
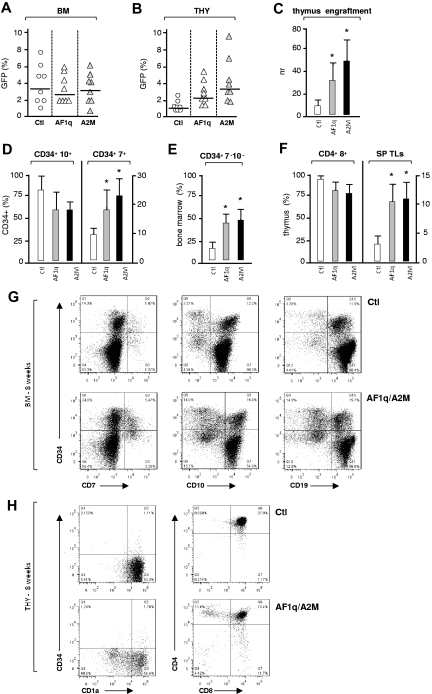
To investigate whether AF1q-enforced expression affected the emergence of prothymocytes, NSG mice were grafted with CD34+ HPCs exposed to lentiviral vectors driving expression of AF1q or A2M or to the empty vector and analyzed 8 weeks later. Comparable levels of BM GFP+CD45+ cells were found in all 3 conditions (median, 2.6%-3.5%), indicating that neither AF1q nor A2M affected engraftment (Figure 6A). In mice reconstituted with empty vector–transduced cells, GFP+ cell percentages among human CD45+ cells were 5- to 10-fold lower in the thymus than in the BM (Figure 6B; median, 0.8%), indicating that these cells competed poorly for thymus colonization. AF1q- and A2M-enforced expression compensated for this defect and led to 3 to 4.5-fold greater levels of GFP+CD45+ thymocytes, respectively (median, 2.2% and 3.6%, respectively; Figure 6B-C). A2M again generated a more pronounced phenotype than wild-type AF1q, but the difference was not statistically significant. Phenotypic characterization of GFP+CD45+ BM cells showed that AF1q/A2M–enforced expression resulted in a 2- to 4-fold increase in the percentages of CD34+CD7+ prothymocytes (Figure 6D,G). Whereas a limited decrease in the levels of CD34+CD10+ B-cell precursors was observed (median, 65% vs 77%), forced expression of AF1q/A2M antagonized B-cell development at later stages. Median percentages of CD34−CD10+19+ B cells decreased from 82% to 54% in NSG mice reconstituted with AF1q/A2M–transduced cells (P = .04, Student t test; Figure 6D,G). AF1q/A2M–enforced expression also elicited a 2.5-fold increase in the level of multipotent CD34+CD7−CD10− HPCs (Figure 6E and G). Flow cytometric analysis of the CD45+GFP+ thymocytes showed increased levels of single-positive mature T cells in AF1q/A2M mice (Figure 6F and H), suggesting accelerated T-cell development.
Figure 6.
Effect of forced AF1q expression on T-cell development in humanized mice. (A-B) CD34+ HPCs (1.5 × 105 cells) exposed to the AF1q, A2M, or empty vectors were injected intravenously into irradiated NSG mice, and the BM, spleen, and thymus were harvested 8 weeks later. Single-cell suspensions were analyzed by multicolor flow cytometry to determine the extent of CD45+GFP+ chimerism and for phenotyping the cells. Percentages of GFP+ cells among CD45+ human cells present in the BM (A) or the thymus (B) of individual recipient mice are shown (transduction efficiency: 14% ± 2%; 8 mice per condition; 2 experiments). Median percentages are indicated by a horizontal bar. (C) Effect of AF1q or A2M on thymus colonization. The efficiency of thymus engraftment was calculated as the ratio of GFP+CD45+ cell percentages detected in the thymus and BM of individual mice. Statistically significant differences (P < .05; Student t test) between AF1q/A2M and control mice are marked by asterisks. (D-F) Percentages of CD7+ and CD10+ lymphoid precursors (D) or of multipotent CD7−CD10− HPCs (E) among CD34+GFP+CD45+ BM cells. (F) percentages of CD4+CD8+ and CD4+/−CD8+/− single-positive T cells among thymic GFP+CD45+ cells. Means ± SD percentages are shown. Statistically significant differences are marked as above. (G-H) Flow cytometric analysis of CD45+GFP+ cells in the BM (H) and thymi (I) of individual mice at week 8 after grafting. Numbers in quadrants indicate the percentages of each population.
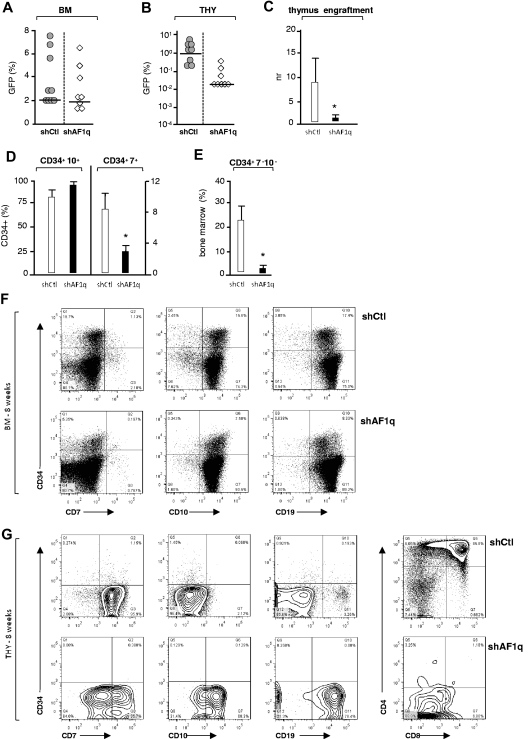
Analysis of mice grafted with CD34+ HPCs exposed to vectors driving the expression of shAF1q or scrambled shRNA showed that, although AF1q silencing did not affect BM engraftment at week 8 (median, 1.8% vs 2.1%), it did inhibit thymus colonization (Figure 7A-C). Relative to the BM, CD45+GFP+ cell percentages decreased to barely detectable levels in the thymi of mice reconstituted with AF1q-deficient cells (median, 0.05% vs 1%), indicating that AF1q-deficient progenitors have a decreased capacity to enter and settle in the thymus. Analysis of the impact of AF1q silencing on BM hematopoiesis found a 2.5-fold reduction in the percentages of CD34+ progenitors, the CD34 expression of which was also decreased (Figure 7F). Consistent with the above reported defect in thymopoiesis, AF1q silencing caused a 3- to 5-fold reduction in the percentages of BM CD34+CD7+ prothymocytes, as well as a corresponding increase of CD34+/−CD10+19+ cells, which represented the majority of BM cells (median, 98% vs 89%; Figure 7D,F). Interestingly, AF1q silencing also led to almost complete disappearance of the most immature CD34+CD7−CD10− HPCs, the percentages of which decreased by 8- to 10-fold (Figure 7E-F). Analysis of the few CD45+GFP+ cells isolated from the thymi of AF1q-deficient mice showed that T-cell development was arrested at the CD7loCD4loCD8− DN stage and disclosed a corresponding accumulation of CD19+ B cells that could reach up to 80% of donor-derived AF1q-deficient thymic cells (Figure 7G). These data indicate that, in the absence of AF1q, T-cell instructive signals are diminished, thus favoring B-cell development in the BM and thymus.
Figure 7.
Effect of AF1q deficiency on T-cell development in humanized mice. (A-B) NSG mice were injected with CD34+ HPCs (1.5 × 105 cells) exposed to shAF1q or scrambled shRNA vectors and analyzed as described in Figure 6. The percentages of GFP+ cells among CD45+ human cells present in the BM (A) or thymi (B) of individual mice are shown (transduction efficiency: 12% ± 3%; 8 mice per condition, 2 experiments). Median percentages are indicated by a horizontal bar. (C) Effect of AF1q deficiency on thymus colonization. The efficiency of thymus engraftment was calculated and expressed as above. Statistically significant differences are marked by asterisks. (D-E) Percentages of CD7+ and CD10+ lymphoid precursors (D) or of multipotent CD7−CD10− HPCs (E) among CD34+GFP+CD45+ BM cells. Statistically significant differences are marked by asterisks. (F-G) Flow cytometric analysis of CD45+GFP+ cells in the BM (F) and thymi (G) of individual mice at week 8 after grafting. Numbers in quadrants indicate the percentages of each population.
Discussion
In the present study, we identify AF1q as a novel upstream regulator of lymphoid development in humans. Analysis of AF1q intracellular trafficking found that it is constitutively exported from the nucleus and then subjected to ubiquitin-mediated degradation by the proteasome in the centrosomal area. Because this suggested that AF1q biologic function might depend on its subcellular localization, we compared the effects of the wild-type protein and also its nuclear-sequestered mutant A2M on hematopoiesis. Neither affected in vitro HPC growth and survival nor led to myelodysplasia or leukemia transformation in vivo in humanized mice, indicating that AF1q does not possess intrinsic transforming capacity. Neither AF1q nor A2M influenced erythroid-, GM-, or NK-cell development in vitro, but both displayed a modulatory effect on T-cell compared with B-cell lymphoid potential of multipotent CD34+ HPCs. Whereas forced AF1q or A2M expression antagonized B-cell differentiation, both stimulated T-cell development. In OP9-DL1 cocultures, AF1q and A2M enhanced the Notch-dependent generation of CD7+CD1a−/+ T-cell precursors and accelerated subsequent T-cell development. Evidence of a functional link between AF1q and the Notch pathway is substantiated by the finding that AF1q/A2M conferred an increased susceptibility to Notch ligation to multipotent HPCs, which was characterized by more efficient induction of the Notch target gene HES1, as well as of several T lineage–affiliated genes.
Consistent with our in vitro data, AF1q/A2M promoted the differentiation of BM prothymocytes in NSG mice, enhanced thymus colonization, and accelerated subsequent intrathymic development, whereas AF1q silencing generated an opposite phenotype. A more limited effect of AF1q on early B-cell development was observed in reconstitution experiments, probably because of the well-documented B-lineage developmental bias of human hematopoiesis in NSG mice.28,30 In vivo analysis of the impact of AF1q silencing on lymphoid development strengthened the relationship between AF1q and Notch signaling. As reported for mice deficient in Notch1 signaling,31–34 AF1q silencing arrested intrathymic T-cell development at the CD4−CD8− DN stage and led to the accumulation of intrathymic B cells. Although this may increase the susceptibility of CD34+ HPCs to Delta-like4/Notch1 interactions, we do not favor the hypothesis that down-modulation of SPEN is pivotal for the impact of AF1q on lymphoid development in the BM. Unlike forced expression of AF1q or of Notch intracellular domain, SPEN deficiency does not affect B-cell development, but generates instead a thymic phenotype characterized by both enhanced generation of early thymic progenitors and subsequent intrathymic block at the DN2 stage.35 The molecular mechanisms by which AF1q exerts its repressive effect on gene expression require further investigation. Even when tethered to a synthetic promoter, AF1q and A2M do not affect the transcriptional activity of a Luciferase reporter gene (A.P., unpublished data), indicating that AHDs do not correspond to docking sites for coactivators or corepressors. We nonetheless found that AF1q induces H3K9me3-repressive histone marks at the SPEN locus, suggesting that it may be part of a repressive chromatin complex.
Our data support a model in which TGF/BMP–induced up-regulation of AF1q in CD34+CD7−CD10− HPCs promotes Notch-dependent emergence of BM prothymocytes (supplemental Figure 7). Later, possibly through down-modulation of SPEN, the increase in AF1q expression in early T-cell-lineage progenitors cooperates with Notch signaling to drive intrathymic commitment toward the T-cell lineage.
Supplementary Material
Acknowledgments
The authors thank Niclas Setterblad, Micaël Yagello, Michel Schmitt, and Christelle Doliger (Service Commun d'Imagerie Cellulaire et Moléculaire, Institut Universitaire d'Hématologie, Paris, France) for their assistance; Simona Denti, Armelle Regnault, and Valerie Lallemand (Institut Universitaire d'Hématologie, Paris) for discussion and help in experimental procedures; Sebastien Jauliac and Françoise Pflumio for critical comments on the manuscript; Juan Carlos Zuniga-Pflucker (University of Toronto, Sunnybrook Research Institute, Toronto, ON) for the OP9-DL1 cells; Didier Trono (Ecole Polytechnique Fédérale, Lausanne, Switzerland) for the third-generation lentiviral vectors; and Jerôme Larghero (Banque de Sang Placentaire, Hôpital Saint Louis, Paris) and the Hôpital Pierre Rouques “Les Bluets” for providing umbilical cord blood samples.
This work was supported by Inserm, the Association pour la Recherche sur le Cancer, the Comité de Paris de la Ligue Nationale Contre le Cancer, and the National Institutes of Health.
Footnotes
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Authorship
Contribution: A.P. performed the research and wrote the manuscript; M.D. analyzed the data; N.M., M.R.-S., E.N., H.B.-M., K.K., and V.P. performed the research; M.P. and N.L. performed the mouse studies; I.D.B. contributed vital new reagents and helped to analyze the data; J.G.G. wrote the manuscript; M.G. and F.S. designed the research and wrote the manuscript; and B.C. designed and directed the research and wrote the manuscript.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Pr Bruno Canque, MD, PhD, Inserm U944 and Université de Paris 7/CNRS UMR 7212, Institut Universitaire d'Hématologie, Centre Hayem, 1 avenue Claude Vellefaux 75475, Paris cedex 10, France; e-mail: bruno.canque@gmail.com.
References
- 1.Rodewald HR, Kretzschmar K, Takeda S, Hohl C, Dessing M. Identification of pro-thymocytes in murine fetal blood: T lineage commitment can precede thymus colonization. EMBO J. 1994;13(18):4229–4240. doi: 10.1002/j.1460-2075.1994.tb06743.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Awong G, Herer E, Surh CD, Dick JE, La Motte-Mohs RN, Zuniga-Pflucker JC. Characterization in vitro and engraftment potential in vivo of human progenitor T cells generated from hematopoietic stem cells. Blood. 2009;114(5):972–982. doi: 10.1182/blood-2008-10-187013. [DOI] [PubMed] [Google Scholar]
- 3.Haddad R, Guimiot F, Six E, et al. Dynamics of thymus-colonizing cells during human development. Immunity. 2006;24(2):217–230. doi: 10.1016/j.immuni.2006.01.008. [DOI] [PubMed] [Google Scholar]
- 4.Besseyrias V, Fiorini E, Strobl LJ, et al. Hierarchy of Notch-Delta interactions promoting T cell lineage commitment and maturation. J Exp Med. 2007;204(2):331–343. doi: 10.1084/jem.20061442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Koch U, Fiorini E, Benedito R, et al. Delta-like 4 is the essential, nonredundant ligand for Notch1 during thymic T cell lineage commitment. J Exp Med. 2008;205(11):2515–2523. doi: 10.1084/jem.20080829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Crompton T, Outram SV, Hager-Theodorides AL. Sonic hedgehog signalling in T-cell development and activation. Nat Rev Immunol. 2007;7(9):726–735. doi: 10.1038/nri2151. [DOI] [PubMed] [Google Scholar]
- 7.Rothenberg EV, Taghon T. Molecular genetics of T cell development. Annu Rev Immunol. 2005;23:601–649. doi: 10.1146/annurev.immunol.23.021704.115737. [DOI] [PubMed] [Google Scholar]
- 8.Lai AY, Kondo M. Identification of a bone marrow precursor of the earliest thymocytes in adult mouse. Proc Natl Acad Sci U S A. 2007;104(15):6311–6316. doi: 10.1073/pnas.0609608104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Maillard I, Fang T, Pear WS. Regulation of lymphoid development, differentiation, and function by the Notch pathway. Annu Rev Immunol. 2005;23:945–974. doi: 10.1146/annurev.immunol.23.021704.115747. [DOI] [PubMed] [Google Scholar]
- 10.Maeda T, Merghoub T, Hobbs RM, et al. Regulation of B versus T lymphoid lineage fate decision by the proto-oncogene LRF. Science. 2007;316(5826):860–866. doi: 10.1126/science.1140881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Tse W, Zhu W, Chen HS, Cohen A. A novel gene, AF1q, fused to MLL in t(1;11) (q21;q23), is specifically expressed in leukemic and immature hematopoietic cells. Blood. 1995;85(3):650–656. [PubMed] [Google Scholar]
- 12.Lestou VS, Ludkovski O, Connors JM, Gascoyne RD, Lam WL, Horsman DE. Characterization of the recurrent translocation t(1;1)(p36.3; q21.1-2) in nonHodgkin lymphoma by multicolor banding and fluorescence in situ hybridization analysis. Genes Chromosomes Cancer. 2003;36(4):375–381. doi: 10.1002/gcc.10181. [DOI] [PubMed] [Google Scholar]
- 13.Le Baccon P, Leroux D, Dascalescu C, et al. Novel evidence of a role for chromosome 1 pericentric heterochromatin in the pathogenesis of B-cell lymphoma and multiple myeloma. Genes Chromosomes Cancer. 2001;32(3):250–264. doi: 10.1002/gcc.1189. [DOI] [PubMed] [Google Scholar]
- 14.Tse W, Meshinchi S, Alonzo TA, et al. Elevated expression of the AF1q gene, an MLL fusion partner, is an independent adverse prognostic factor in pediatric acute myeloid leukemia. Blood. 2004;104(10):3058–3063. doi: 10.1182/blood-2003-12-4347. [DOI] [PubMed] [Google Scholar]
- 15.Lallemand-Breitenbach V, Jeanne M, Benhenda S, et al. Arsenic degrades PML or PML-RARalpha through a SUMO-triggered RNF4/ubiquitin-mediated pathway. Nat Cell Biol. 2008;10(5):547–555. doi: 10.1038/ncb1717. [DOI] [PubMed] [Google Scholar]
- 16.Haddad R, Guardiola P, Izac B, et al. Molecular characterization of early human T/NK and B-lymphoid progenitor cells in umbilical cord blood. Blood. 2004;104(13):3918–3926. doi: 10.1182/blood-2004-05-1845. [DOI] [PubMed] [Google Scholar]
- 17.Moffat J, Grueneberg DA, Yang X, et al. A lentiviral RNAi library for human and mouse genes applied to an arrayed viral high-content screen. Cell. 2006;124(6):1283–1298. doi: 10.1016/j.cell.2006.01.040. [DOI] [PubMed] [Google Scholar]
- 18.Peixoto A, Monteiro M, Rocha B, Veiga-Fernandes H. Quantification of multiple gene expression in individual cells. Genome Res. 2004;14(10A):1938–1947. doi: 10.1101/gr.2890204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Maës J, Maleszewska M, Guillemin C, et al. Lymphoid-affiliated genes are associated with active histone modifications in human hematopoietic stem cells. Blood. 2008;112(7):2722–2729. doi: 10.1182/blood-2008-02-140806. [DOI] [PubMed] [Google Scholar]
- 20.Soulier J, Clappier E, Cayuela JM, et al. HOXA genes are included in genetic and biologic networks defining human acute T-cell leukemia (T-ALL). Blood. 2005;106(1):274–286. doi: 10.1182/blood-2004-10-3900. [DOI] [PubMed] [Google Scholar]
- 21.Berardi AC, Meffre E, Pflumio F, et al. Individual CD34+CD38lowCD19-CD10- progenitor cells from human cord blood generate B lymphocytes and granulocytes. Blood. 1997;89(10):3554–3564. [PubMed] [Google Scholar]
- 22.Canque B, Camus S, Dalloul A, et al. Characterization of dendritic cell differentiation pathways from cord blood CD34(+)CD7(+)CD45RA(+) hematopoietic progenitor cells. Blood. 2000;96(12):3748–3756. [PubMed] [Google Scholar]
- 23.Fritsch G, Buchinger P, Printz D, et al. Rapid discrimination of early CD34+ myeloid progenitors using CD45-RA analysis. Blood. 1993;81(9):2301–2309. [PubMed] [Google Scholar]
- 24.Awong G, La Motte-Mohs RN, Zuniga-Pflucker JC. In vitro human T cell development directed by notch-ligand interactions. Methods Mol Biol. 2008;430:135–142. doi: 10.1007/978-1-59745-182-6_9. [DOI] [PubMed] [Google Scholar]
- 25.McMahon S, Charbonneau M, Grandmont S, Richard DE, Dubois CM. Transforming growth factor beta1 induces hypoxia-inducible factor-1 stabilization through selective inhibition of PHD2 expression. J Biol Chem. 2006;281(34):24171–24181. doi: 10.1074/jbc.M604507200. [DOI] [PubMed] [Google Scholar]
- 26.Tanigaki K, Honjo T. Regulation of lymphocyte development by Notch signaling. Nat Immunol. 2007;8(5):451–456. doi: 10.1038/ni1453. [DOI] [PubMed] [Google Scholar]
- 27.Varnum-Finney B, Wu L, Yu M, et al. Immobilization of Notch ligand, Delta-1, is required for induction of notch signaling. J Cell Sci. 2000;113(pt 23):4313–4318. doi: 10.1242/jcs.113.23.4313. [DOI] [PubMed] [Google Scholar]
- 28.Ishikawa F, Yasukawa M, Lyons B, et al. Development of functional human blood and immune systems in NOD/SCID/IL2 receptor {gamma} chain(null) mice. Blood. 2005;106(5):1565–1573. doi: 10.1182/blood-2005-02-0516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ito M, Hiramatsu H, Kobayashi K, et al. NOD/SCID/gamma (c) (null) mouse: an excellent recipient mouse model for engraftment of human cells. Blood. 2002;100(9):3175–3182. doi: 10.1182/blood-2001-12-0207. [DOI] [PubMed] [Google Scholar]
- 30.Shultz LD, Lyons BL, Burzenski LM, et al. Human lymphoid and myeloid cell development in NOD/LtSz-scid IL2R gamma null mice engrafted with mobilized human hemopoietic stem cells. J Immunol. 2005;174(10):6477–6489. doi: 10.4049/jimmunol.174.10.6477. [DOI] [PubMed] [Google Scholar]
- 31.Feyerabend TB, Terszowski G, Tietz A, et al. Deletion of Notch1 converts pro-T cells to dendritic cells and promotes thymic B cells by cell-extrinsic and cell-intrinsic mechanisms. Immunity. 2009;30(1):67–79. doi: 10.1016/j.immuni.2008.10.016. [DOI] [PubMed] [Google Scholar]
- 32.Izon DJ, Aster JC, He Y, et al. Deltex1 redirects lymphoid progenitors to the B cell lineage by antagonizing Notch1. Immunity. 2002;16(2):231–243. doi: 10.1016/s1074-7613(02)00271-6. [DOI] [PubMed] [Google Scholar]
- 33.Maillard I, Weng AP, Carpenter AC, et al. Mastermind critically regulates Notch-mediated lymphoid cell fate decisions. Blood. 2004;104(6):1696–1702. doi: 10.1182/blood-2004-02-0514. [DOI] [PubMed] [Google Scholar]
- 34.Radtke F, Wilson A, Stark G, et al. Deficient T cell fate specification in mice with an induced inactivation of Notch1. Immunity. 1999;10(5):547–558. doi: 10.1016/s1074-7613(00)80054-0. [DOI] [PubMed] [Google Scholar]
- 35.Tsuji M, Shinkura R, Kuroda K, Yabe D, Honjo T. Msx2-interacting nuclear target protein (Mint) deficiency reveals negative regulation of early thymocyte differentiation by Notch/RBP-J signaling. Proc Natl Acad Sci U S A. 2007;104(5):1610–1615. doi: 10.1073/pnas.0610520104. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.