Abstract
Among the many molecular imaging techniques, reporter gene imaging has been a dynamic area of research. The HaloTag protein is a modified haloalkane dehalogenase which was designed to covalently bind to synthetic ligands (i.e. the HaloTag ligands [HTL]). Covalent bond formation between the HaloTag protein and the chloroal-kane within the HTL occurs rapidly under physiological conditions, which is highly specific and essentially irreversible. Over the years, HaloTag technology has been investigated for various applications such as in vitro/in vivo imaging, protein purification/trafficking, high-throughput assays, among others. The goal of this study is to explore the use of the HaloTag protein as a novel reporter gene for positron emission tomography (PET) imaging. By attaching a HaloTag -reactive chloroalkane to 1, 4, 7-triazacyclononane-N, N', N“-triacetic acid (NOTA) through hydrophilic linkers, the resulting NOTA-conjugated HTLs were labeled with 64Cu and tested for PET imaging in living mice bearing 4T1-HaloTag-ECS tumors, which stably express the HaloTag protein on the cell surface. Significantly higher uptake of 64Cu-NOTA-HTL-S (which contains a short hydrophilic linker) in the 4T1-HaloTag-ECS than the non-HaloTag-expressing 4T1 tumors was observed, which demonstrated the HaloTag specificity of 64Cu-NOTA-HTL-S and warranted future investigation of the HaloTag protein as a PET reporter gene.
Keywords: Reporter gene, HaloTag, positron emission tomography (PET), cell tracking, cancer, molecular imaging
Introduction
The field of molecular imaging has undergone tremendous expansion over the last decade, partly owing to the increasingly wider availability of scanners dedicated to small animal studies [1, 2]. Among the many molecular imaging techniques, reporter gene imaging has been a dynamic area of research. A wide variety of reporter genes and reporter probes have been developed, and a few of these have entered clinical investigation [3-5]. The most widely-used reporter genes include those for optical (both fluorescence [6] and bioluminescence [7]) and radionuclide-based imaging, namely single-photon emission computed tomography (SPECT) and positron emission tomography (PET) [8, 9]. Many other reporter genes have also been developed for magnetic resonance imaging (MRI) applications [10].
The HaloTag protein is a modified haloalkane dehalogenase which was designed to covalently bind to synthetic ligands (i.e. the HaloTag ligands [HTL]) [11, 12]. Since the bacterial dehalogenase is relatively small (with a molecular weight of 34 kDa) and the enzymatic reaction is foreign to mammalian cells, cross-reactive interference with the endogenous mammalian biochemistry is negligible. A HTL is typically composed of a chloroalkane and a functional tag, such as a fluorescent dye, an affinity handle, a solid surface, among others. Covalent bond formation between the HaloTag protein and the chloroalkane within the HTL occurs rapidly under physiological conditions, which is highly specific and essentially irreversible. Because of its versatility, the HaloTag technology has been investigated for a wide variety of applications such as in vitro optical imaging [13-16], protein purification [17-20], protein trafficking [21], study of protein-protein and protein-DNA interactions [22], in vivo cell labeling [23], in vivo cancer imaging [24], analysis of protein stability [25], high-throughput assays [26], single molecule force spectroscopy [27], ribosome tagging [28, 29], amongothers.
In this study, we explore the use of the HaloTag protein as a novel reporter gene for non-invasive PET imaging. By attaching a HaloTag-reactive chloroalkane to 1, 4, 7-triazacyclononane-N, N', N“-triacetic acid (NOTA) through hydrophilic linkers which contain polyethylene glycol (PEG) chains of different length, the resulting HTLs can labeled with 64Cu (t1/2: 12.7 h) and employed for PET imaging applications in living mice, bearing 4T1 murine breast tumors stably expressing the HaloTag protein on the cell surface. This report serves as a proof-of-principle for future cell tracking of HaloTag-transfected cells with PET, which is highly quantitative and sensitive with superb tissue penetration.
Materials and methods
Reagents
NOTA-p-Bn-SCN was purchased from Macrocyclics (Dallas, TX). Water and all buffers were of Millipore grade and pre-treated with Chelex 100 resin to ensure that the aqueous solution was heavy-metal free. PD-10 desalting columns were purchased from GE Healthcare (Piscataway, NJ). High-performance liquid chromatography (HPLC) solvents (water, acetonitrile, and trifluoroacetic acid [TFA]) were purchased from Thermo Fisher Scientific Inc. 64Cu was produced via a 64Ni(p,n) 64Cu reaction using a cyclotron at the University of Wisconsin - Madison. All other reagents were obtained from Sigma-Aldrich, except when noted in the following text, and used as received.
Syntheses of the HaloTag ligands
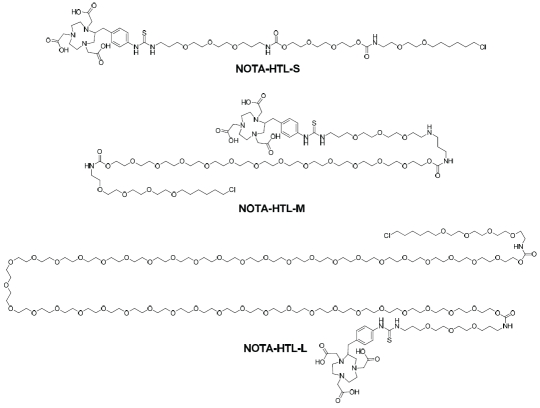
Syntheses of the three NOTA-conjugated HTLs (Figure 1) were straightforward. Since the Halo-Tag-reactive chloroalkane was very hydrophobic, PEG chains with different length (6, ∼18, and >40 ethylene glycol units) were incorporated between the chloroalkane and NOTA. The three compounds were purified by silica gel column chromatography and/or HPLC, and subsequently characterized with 1H-NMR spectroscopy and mass spectrometry. For NOTA-HTL-S (S denotes short, which has 6 ethylene glycol units), the m/z was calculated to be 1096.5 (C48H83CIN7O17S+) and an m/z of 1096.6 was observed in mass spectrometry. For NOTA-HTL-M (M denotes medium, which has 18 ethylene glycol units), the m/z was calculated to be 1668.9 (C74H135CIN7032S+) and an m/z of 1669.0 was observed in mass spectrometry. For NOTA-HTL-L (L denotes long, which has >40 ethylene glycol units), a characteristic bell-shaped mass spectrum was observed in mass spectrometry (since the PEG used was a polymer with a molecular weight of ∼2,000) which matched the calculated m/z.
Figure 1.
Chemical structures of the three NOTA-conjugated HaloTag ligands.
Stable transfection of 4T1 cells with HaloTag
4T1 murine breast cancer cells were obtained from the American Type Culture Collection (ATCC, Manassas, VA) and cultured in the RPMI 1640 medium (Invitrogen, Carlsbad, CA) with 10% fetal bovine serum, and incubated at 37 °C with 5% CO2. pCI-neo mammalian expression vector (E1841, Promega Corporation) was used for cloning of the HaloTag construct, which was fused to a trans-membrane domain. G418 (V8091, Promega Corporation) was used for selection of the clones. When the cells under selective pressure were expanding at the same rate as non-transfected controls, they were serially diluted. Single colonies were then harvested and confirmed positive by microscopy studies.
Microscopy studies were performed after labeling the transfected 4T1 cells with Alexa Fluor 488-conjugated HaloTag ligand (AF488-HTL, Promega Corporation) followed by TMR-conjugated HaloTag ligand (TMR-HTL, Promega Corporation). AF488-HTL is not cell membrane permeable, therefore it can only label the HaloTag protein expressed on the extracellular surface. On the other hand, TMR-HTL is membrane permeable, which can label the HaloTag protein expressed on the extracellular surface as well as those within the cytoplasm. One positive clone was expanded (termed as “4T1-HaloTag-ECS” where ECS denotes “extracellular surface”) and used for in vitro and in vivo experiments. 4T1 cells stably expressing the HaloTag protein, without fusion to a trans-membrane domain, was also generated using a similar strategy and named as “4T1-HaloTag”. All cells were used for in vitro and in vivo experiments when they reached ∼80% confluence.
In vitro studies of NOTA-HTL-S/M/L
The three NOTA-conjugated HTLs were com-plexed with non-radioactive Cu2+ and tested in the transfected 4T1 cells for their ability to bind to the HaloTag protein in a cellular context, as well as their cell membrane permeability. Stably -transfected 4T1 cells (i.e. 4T1-HaloTag-ECS and 4T1-HaloTag) were each plated in Lab-Tek II chambered coverglass (Nalge Nunc International) and allowed to attach overnight. To assess the ability of each NOTA-conjugated HTL to bind to the HaloTag protein when expressed in mammalian cells, 4T1-HaloTag-ECS cells were first incubated in a 5 μM solution of each NOTA-conjugated HTL in complete media for 15 minutes at 37°C in the presence of 5% CO2. Afterwards, the cells were labeled with 1 μM of AF488-HTL, washed, and imaged. Control 4T1-HaloTag-ECS cells were labeled with 1 μM of AF488-HTLonly.
To assess the cell membrane permeability of NOTA-HTL-S/M/L, 4T1-HaloTag cells were incubated with each ligand and then labeled with 5 μM of TMR-HTL, washed, and imaged. As a control, some 4T1-HaloTag cells were labeled with TMR-HTL only. Microscopy imaging was performed using an Olympus FV500 confocal microscope equipped with a 37°C + 5% CO2 environmental chamber and appropriate filter sets.
Animal model
All animal studies were conducted under a protocol approved by the University of Wisconsin Institutional Animal Care and Use Committee. To generate the tumor model using 4T1 or stably-transfected 4T1 cells, four- to five-week-old female Balb/c mice were purchased from Harlan (Indianapolis, IN) and tumors were established by subcutaneously injecting 2 × 106 cells, suspended in 100 μL of 1:1 mixture of RPMI 1640 and matrigel (BD Biosciences, Franklin lakes, NJ), into the front flank of mice [30, 31]. The tumor sizes were monitored every other day and the animals were subjected to in vivo experiments when the diameter of the tumors reached 6-8 mm (typically ∼10 days after inoculation).
64Cu-labeling of NOTA-HTL-S/M/L
64CuCI2 (111 MBq) was diluted in 300 μL of 0.1 M sodium acetate buffer (pH 6.5) and added to 15 μg of NOTA-HTL-S/M/L. The reaction mixture was incubated for 30 min at 40 °C with constant shaking. 64Cu-NOTA-HTL-L was purified using PD-10 columns with phosphate-buffered saline (PBS) as the mobile phase. 64Cu-NOTA-HTL-S and 64Cu-NOTA-HTL-M was purified by a Dionex Ultimate 3000 HPLC system equipped with a radioactivity and UV detector using a C-18 column. A solvent gradient (A: water with 0.1% TFA; B: acetonitrile with 0.1% TFA) was used, where solvent B was gradually increased from 5% to 75% over a period of 30 min. After collection of the radioactive peak, acetonitrile was removed from the solution with continuous argon flow. The remaining solution was reconstituted into a final concentration of 1 × PBS. The tracers were passed through a 0.2 μm syringe filter before in vivo experiments.
PET imaging and biodistribution studies
Details of the PET scans and biodistribution studies have been reported previously [31]. Briefly, each tumor-bearing mouse was injected with 5-10 MBq of each PET tracer via tail vein and 3 - 15min static PET scans were performed at various time points post-injection (p.i.) in a microPET/microCT Inveon rodent model scanner (Siemens Medical Solutions USA, Inc.). The images were reconstructed using a maximum a posteriori (MAP) algorithm, with no attenuation or scatter correction. Quantitative region-of-interest (ROI) analysis was performed on all PET scans to obtain quantitative data in the unit of percentage of injected dose per gram of tissue (%ID/g). After the last PET scans at 6 h p.i., mice were euthanized and biodistribution studies were carried out to confirm that the quantitative tracer uptake values based on microPET imaging truly represented the actual tracer distribution in the tumor-bearing mice. The radioactivity in the tissue was measured using a gamma-counter (Perkin Elmer) and presented as %ID/g (mean ± SD).
Statistical analysis
Quantitative data were expressed as mean ± SD. Means were compared using Student's t-test. P values < 0.05 were considered statistically significant.
Results
Investigation of NOTA-HTL-S/M/L in 4T1-HaloTag-ECS cells
HaloTag protein expression in the 4T1-HaloTag-ECS cells was confirmed by labeling with AF488-HTL followed by TMR-HTL (Figure 2A). Because the HaloTag protein is membrane bound such that HaloTag faces the outside of the cell when it is in the plasma membrane, AF488-HTL (not cell membrane permeable) binds only to the plasma membrane-bound pool of HaloTag protein in the cells. After AF488-HTL has bound to the HaloTag protein on the cell surface, TMR-HTL was introduced to label the intracellular HaloTag protein. The merged fluorescence image confirmed that the HaloTag protein is expressed in every cell of this stable cell line, both on the cell surface and inside the cytoplasm.
Figure 2.
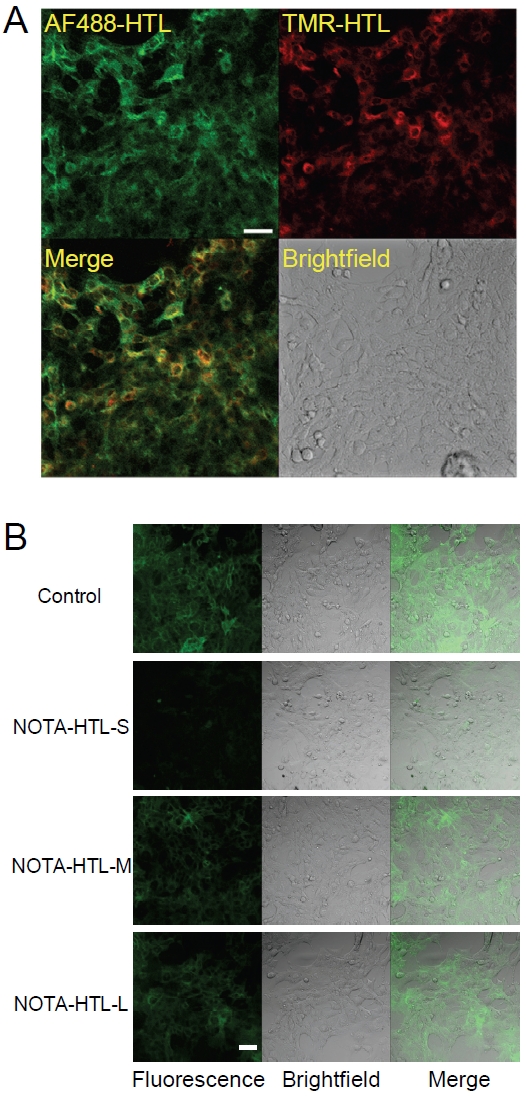
Characterization of 4T1-HaloTag-ECS cells. A. 4T1-HaloTag-ECS cells are labeled with AF488-HTL (not cell membrane permeable) and then with TMR-HTL (cell membrane permeable). Microscopy images confirmed that the cells express the HaloTag protein both on the surface of and inside the cells. B. Binding of NOTA-HTL-S/M/L to the HaloTag protein was assessed by exposing 4T1-HaloTag-ECS cells to each ligand prior to labeling with the cell membrane impermeable AF488-HTL. Confocal images showed significantly less AF488-HTL signal for NOTA-HTL-S, indicative of its efficient binding to HaloTag, but not for NOTA-HTL-M/L Scale bars: 20 μm.
To assess the ability of NOTA-HTL-S/M/L to bind to the HaloTag protein in mammalian cells, 4T1-HaloTag-ECS cells were incubated with each of the NOTA-conjugated HTL (in order to saturate the HaloTag protein on the cell surface) prior to labeling with AF488-HTL. As a control for surface labeling, 4T1-HaloTag-ECS cells were also labeled with AF488-HTL only. It was found that NOTA-HTL-S binds with the highest efficiency to the HaloTag protein when it is expressed on the cell surface (Figure 2B), as indicated by very weak fluorescence signal from AF488-HTL on the 4T1-HaloTag-ECS cell surface (i.e. all the HaloTag protein was pre-bound with NOTA-HTL-S). However, the efficiency of NOTA-HTL-M/L binding to HaloTag protein was significantly lower, since AF488-HTL was still able to bind to the HaloTag protein which resulted in strong fluorescence signal.
Investigation of NOTA-HTL-S/M/L in 4T1-HaloTag cells
Stable expression of the HaloTag protein in 4T1-HaloTag cells was confirmed by microscopy studies after labeling the cells with TMR-HTL (Figure 3A). Uniform fluorescence signal was observed inside all cells, as expected for this stable cell line. To assess the efficiency of NOTA -HTL-S/M/L in entering cells and binding to the intracellular HaloTag protein, 4T1-HaloTag cells were exposed to NOTA-HTL-S/M/L prior to labeling with TMR-HTL. Control 4T1-HaloTag cells were labeled only with TMR-HTL. No significant difference in fluorescence signal was observed between the groups, indicating that none of the NOTA-conjuagted HTLs exhibit significant cell membrane permeability (Figure 3B). Since NOTA -HTL-S/M/L was not able to penetrate the cell membrane and bind to the HaloTag protein expressed inside the cells, the remaining studies were focused on 4T1-HaloTag-ECS cells only.
Figure 3.
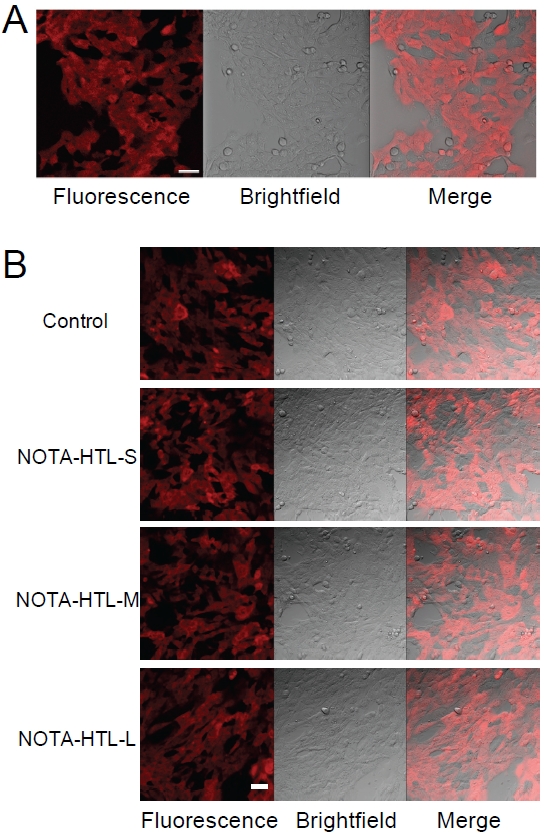
Characterization of 4T1-HaloTag cells. A. The cells are labeled with TMR-HTL and examined under a microscope, which confirmed HaloTag protein expression in the cells. B. Cell membrane permeability of NOTA-HTL-S/M/L was assessed by exposing 4T1-HaloTag cells to each ligand prior to labeling with the cell membrane permeable TMR-HTL. Confocal images showed no significant difference in TMR-HTL labeling between control cells (labeled with TMR-HTL only) and cells previously exposed to each NOTA-conjugated HTL, indicating a lack of cell membrane permeability for NOTA-HTL-S/M/L. Scale bars: 20μm.
64Cu-labeling
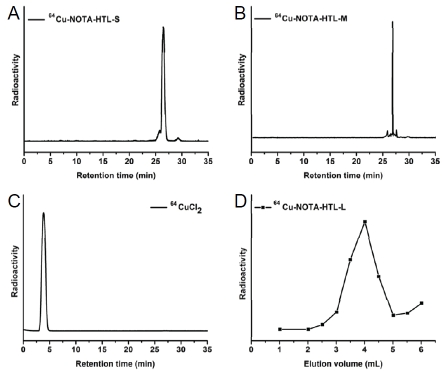
For NOTA-HTL-S/M, 64Cu-labeling including HPLC purification took 100 ± 10 min (n = 6). The crude radio-HPLC profiles of 64Cu-NOTA-HTL-S/M are shown in Figure 4A&B, with a radio-HPLC trace of 64CuCb shown as a reference standard in Figure 4C. 64Cu-NOTA-HTL-S had a sharp peak with a retention time of 26.9 min while 64Cu-NOTA-HTL-M elutes at 26.5 min. For NOTA-HTL-L, 64Cu-labeling including purification using PD-10 columns took 80 ± 10 min (n = 10), with a representative radioactivity elution profile from the column shown in Figure 4D. After purification with HPLC or PD-10 column, the decay-corrected radiochemical yield was >85% for 64Cu-NOTA-HTL-L, -35% for 64Cu-NOTA -HTL-S, and -30% for 64Cu-NOTA-HTL-M, each with a radiochemical purity of > 98%.
Figure 4.
64Cu-labeling of NOTA-HTL-S/M/L. A. A crude radio-HPLC trace of 64CU-NOTA-HTL-S before purification. B. A crude radio-HPLC trace of 64Cu-NOTA-HTL-M before purification. C. A radio-HPLC trace of 64CuCl2 as a reference. D. Size exclusion column chromatography profile of 64Cu-NOTA-HTL-L, which showed no contamination of 64CuCl2 (eluted after 6 mL).
PET imaging
Based on the low molecular weight of 64Cu-NOTA-HTL-S/M/L, which typically undergo fast blood clearance and excretion, the time points of 0.5, 3, and 6 h p.i. were chosen for serial PET scans after intravenous injection of each tracer.
Representative coronal and transaxial slices of mice bearing the 4T1-HaloTag-ECS tumor are shown in Figure 5, and the quantitative data obtained from ROI analysis of the PET images are shown in Figure 6.
Figure 5.
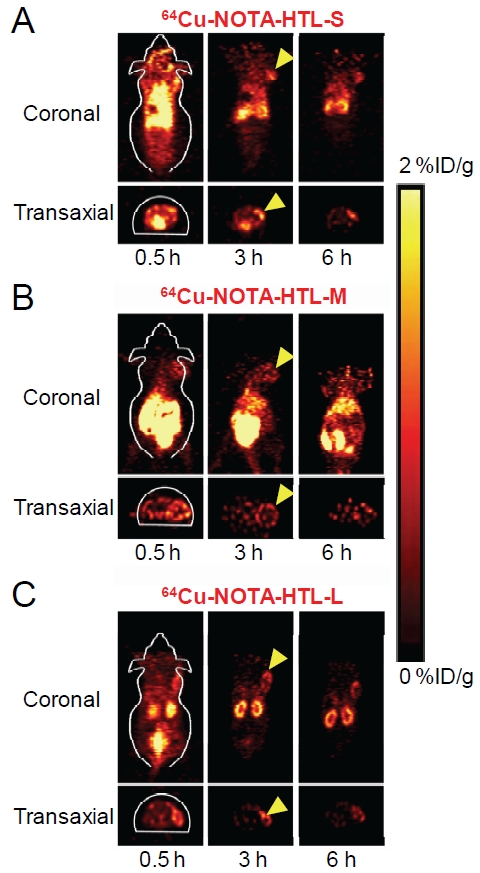
Serial PET images of mice bearing 4T1-HaloTag-ECS tumors at different time points post-injection of 64Cu-NOTA-HTL-S (A), 64Cu-NOTA-HTL-M (B), 64Cu-NOTA-HTL-L (C), respectively. Both coronal and transaxial images are shown. Images are representative of 3 mice per group and arrowheads indicate the tumors.
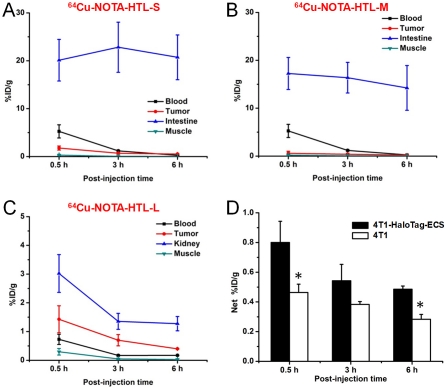
Figure 6.
Time-activity curves of the 4T1-HaloTag-ECS tumor and blood, muscle, intestine/kidney after intravenous injection of 64Cu-NOTA-HTL-S (A), 64Cu-NOTA-HTL-M (B), 64Cu-NOTA-HTL-L (C). D. The “net%ID/g” values, calculated by subtracting the muscle uptake from the tumor uptake, of 64Cu-NOTA-HTL-S in mice bearing 4T1-HaloTag-ECS or 4T1 tumors at different time points post-injection. *: P < 0.05 (n = 3).
Uptake of 64Cu-NOTA-HTL-S and 64Cu-NOTA-HTL-M in the intestines was prominent at all three time points examined, which is typically observed for hydrophobic PET tracers [32, 33]. The intestine uptake of 64Cu-NOTA-HTL-S was 20.1 ± 4.3, 22.8 ± 5.2, and 20.7 ± 4.6 %ID/g at 0.5, 3, and 6 h p.i. respectively (n = 3; Figure 6A). 64Cu-NOTA-HTL-M exhibited similar in vivo kinetics, with intestine uptake of 17.3 ± 3.3, 16.4 ± 3.2, and 14.2 ± 4.7 %ID/g at 0.5, 3, and 6 h p.i. respectively (n = 3; Figure 6B). Both 64Cu -NOTA-HTL-S and 64Cu-NOTA-HTL-M were cleared rapidly from the circulation. The 4T1-HaloTag-ECS tumor uptake of 64Cu-NOTA-HTL-S peaked at 0.5 h p.i. and declined gradually (1.7 ± 0.4, 0.7 ± 0.2, and 0.5 ± 0.1 %ID/g at 0.5, 3, and 6 h p.i. respectively; n = 3; Figure 6A). Compared with 64Cu-NOTA-HTL-S, the 4T1-HaloTag-ECS tumor uptake of 64Cu-NOTA-HTL-M was significantly lower: 0.6 ± 0.4, 0.4 ± 0.2, and 0.2 ± 0.1 %ID/g at 0.5, 3, and 6 h p.i. respectively (n = 3; Figure 6B).
With significantly lower hydrophobicity than 64Cu -NOTA-HTL-S/M, the intestine uptake of 64Cu-NOTA-HTL-L was much lower since it undergoes renal clearance instead of hepatobiliary clearance (Figure 5). 64Cu-NOTA-HTL-L was cleared rapidly from the tumor-bearing mice and kidney uptake of the tracer was 3.0 ± 0.7, 1.4 ± 0.3, and 1.3 ± 0.2 %ID/g at 0.5, 3, and 6 h p.i. respectively (n = 3; Figure 6C). The 4T1-HaloTag-ECS tumor uptake of 64Cu-NOTA-HTL-L is comparable to that of 64Cu-NOTA-HTL-S: 1.4 ± 0.5, 0.7 ± 0.2, and 0.4 ± 0.1 %ID/g at 0.5, 3, and 6 h p.i. respectively (n = 3; Figure 6C).
Since Cu-NOTA-HTL-L does not bind to the HaloTag protein expressed on the surface of 4T1-HaloTag-ECS cells (due to the interference by the long PEG chain), as evidenced by microscopy studies (Figure 2B), the tumor uptake of 64Cu-NOTA-HTL-L is likely due to passive uptake caused by the enhanced permeability and retention (EPR) effect of this tracer, which is not applicable to 64Cu-NOTA-HTL-S because of its low molecular weight. For 64Cu-NOTA-HTL-M, the tumor uptake is significantly lower than both 64Cu-NOTA-HTL-S and 64Cu-NOTA-HTL-L, because not only does Cu-NOTA-HTL-M not bind to the HaloTag protein expressed on the cell surface (Figure 2B), it also does not have large enough molecular weight to take advantage of the EPR effect for enhanced tumor uptake.
To confirm that the 4T1-HaloTag-ECS tumor uptake of 64Cu-NOTA-HTL-S was HaloTag specific, this tracer was also injected into three mice bearing normal 4T1 tumors without HaloTag expression. To eliminate as many variables as possible thereby ensuring a fair and direct comparison, the uptake in the adjacent muscle was subtracted from the tumor uptake and the “net %ID/g” values are presented in Figure 6D. The net %ID/g values of 64Cu-NOTA-HTL-S in the 4T1 -HaloTag-ECS tumors were 0.8 ± 0.1, 0.5 ± 0.1, and 0.5 ± 0.1 %ID/g at 0.5, 3, and 6 h p.i. respectively, while the net %ID/g values in the in normal 4T1 tumors were 0.5 ± 0.1, 0.4 ± 0.1, and 0.3 ± 0.1%ID/g at 0.5, 3, and 6 h p.i. respectively (n = 3). At 0.5 h and 6 h p.i., the difference in net %ID/g values in the two tumor types reached statistical significance, which was not the case at 3 h p.i. due to the large variance between mice in the 4T1-HaloTag-ECS group.
Biodistribution studies
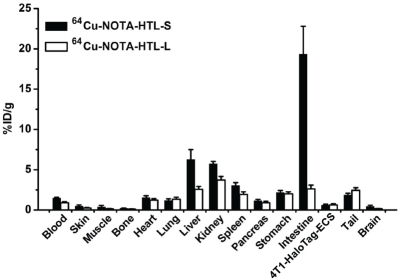
After the last PET scans at 6 h p.i., the mice were euthanized. Tumors and major tissues were collected for biodistribution studies to further validate the in vivo PET data. Besides the intestines, the kidneys and the liver also had significant tracer uptake at 6 h p.i. for 64Cu-NOTA-HTL-S (Figure 7). Uptake in all tissues was low for 64Cu-NOTA-HTL-L (< 5 %ID/g) due to fast clearance of the tracer. Overall, the biodistribution data was in good agreement with the quantitative data obtained from ROI analysis of the non-invasive PET scans.
Figure 7.
Biodistribution of 64Cu-NOTA-HTL-S and 64Cu-NOTA-HTL-L at 6 h post-injection in mice bearing the 4T1-HaloTag-ECS tumors (n = 3).
Discussion
In this proof-of-principle study, we report the first use of the HaloTag protein as a reporter gene for PET imaging. 4T1 murine breast cancer cells were stably transfected to express the HaloTag protein either on the extracellular surface (i.e. 4T1-HaloTag-ECS) or inside the cells (i.e. 4T1-HaloTag). Based on the in vitro studies, only NOTA-HTL-S could bind to the HaloTag protein on the extracellular surface efficiently while none of the three NOTA-conjugated HTLs was able to penetrate the cell membrane. Therefore, in vivo studies were primarily focused on 64Cu-NOTA-HTL-S in the 4T1-HaloTag-ECS tumor model. Significantly higher tracer uptake in the 4T1-HaloTag-ECS than the 4T1 tumor was observed, which demonstrated the HaloTag specificity of 64Cu-NOTA-HTL-S and warranted future investigation of the HaloTag protein as a PET reporter gene.
In vivo stability of 64Cu-chelates has been a hotly debated topic over the last decade. Many chelators have been designed and investigated for 64Cu-labeling [34]. Recently, an elegant study compared the effect of the bifunctional chelator on the biodistribution of a 64Cu-labeled antibody [35]. It was found that differences in the thermodynamic stability of these 64Cu-chelator complexes were not associated with significant differences in tumor uptake of the tracer. However, there were significant differences in tracer concentration in other tissues, in particular those involved in tracer clearance (e.g. liver and spleen). It is now generally agreed that NOTA is one of the best chelators for 64Cu-labeling, which is also used here. In pilot studies, we also labeled DOTA-HTLs (where DOTA denotes 1, 4, 7, 10-tetraazacyclododecane-1, 4, 7, 10-tetraacetic acid) with 64Cu and tested them in vivo. No significant difference was observed between the NOTA- and DOTA-based tracers. Comparing to antibodies which have relatively long circulation half-lives (hours to days) [36-38], the short circulation half-lives of these 64Cu-labeled HTLs (< 0.5 h) make the difference between NOTA- and DOTA-based tracers insignificant.
Reporter gene imaging in vivo is technically challenging [3]. In this study, the 64Cu-labeled HTLs need to circulate in the blood in mice, ex-travasate, bind to the HaloTag protein expressed on the surface of 4T1-HaloTag-ECS cells, and undergo covalent linkage with the HaloTag protein to prevent rapid washout from the tumor thereby achieving sufficient tumor contrast. Given the short circulation half-lives of these HTLs, all these processes must happen in a short time period while the unbound tracers quickly clear from the hepatobiliary or renal pathway. Low level of absolute tumor uptake is common for reporter gene-based imaging, most of which are less than a few %ID/g.
A number of strategies can be adopted to improve the absolute tumor uptake and tumor contrast of radiolabeled HTLs in future studies. First, the expression level of the HaloTag protein was only moderate in the 4T1-HaloTag-ECS cells. Since this is a proof-of-principle study, the major goal was to demonstrate that HaloTag can be used as a reporter gene. With this goal already achieved here, it is expected that further increasing the HaloTag protein expression level can lead to higher tumor uptake in subsequent studies. Second, development of future generation HTLs may give better in vivo performance than the ones investigated here. Since the key component of a HTL involved in HaloTag binding is the chloroalkane tail, HTLs can tolerate a wide variety of modifications to optimize its hydrophobicity/hydrophilicity, which can dramatically affect the clearance route and phar-macokinetics. Third, multimerization may also be used for improved tumor uptake and retention, which has been widely adopted for peptide-and small molecule-based imaging agents [39, 40]. Attaching multiple HTLs to various nanopar-ticles with linkers of suitable length should be explored in the future. However, one has to bear in mind that since the HaloTag protein is expressed on the tumor cells but not on the tumor vessels, the nanoparticles used have to be small enough with efficient extravasation to achieve optimal tumor uptake. Further development and optimization of the HaloTag-based reporter gene system is currently underway.
Combination of different imaging modalities can provide complementary information and allow scientists to interrogate various biological events from different perspectives. Multimodality reporter genes have been well documented in the literature. Typically, these multimodality reporter genes rely on the fusion of different reporter proteins that can each be used for imaging with one technique [41-46]. However, fusion of multiple reporter proteins is technically challenging. In addition, the overall larger size may also affect the expression and/or function of each reporter protein. HaloTag has the potential to be a multimodality reporter gene with one single protein. A previous report has already used the HaloTag protein for in vivo optical imaging [24], whereas here we demonstrate its application for PET imaging. The same HTL may potentially be labeled with a variety of image tags for other imaging modalities, such as 99mTc/123I for SPECT and Gd3+/iron oxide nanoparticles for MRI, respectively. Differently labeled HTLs can be used for imaging HaloTag-expressing cells with different techniques, which will greatly facilitate the potential widespread use of HaloTag-based reporter gene imaging. One caveat is that the sensitivity of MRI is very low, therefore certain signal amplification strategies will be needed for MRI-based imaging of HaloTag protein expression. We envision that PET/fluorescence imaging is the most powerful combination, since PET has superb sensitivity, excellent quantification capability, unlimited tissue penetration, and is clinically relevant [47], while optical imaging is inexpensive, sensitive, and can provide a convenient means for ex vivo validation of the PET data through microscopy studies.
One major potential application of HaloTag-based reporter gene imaging is to track cells in vivo, independent of the cell types (e.g. immune cells, stem cells, cancer cells, induced pluripotent stem cells, etc.). Although such cell tracking is still at an early stage far from routine clinical applications [48, 49], pilot studies where PET imaging was used for tracking genetically engineered cells in cancer patients have already been reported [4, 5], which warrants optimism that reporter gene-based imaging will gradually gain popularity in future clinical studies.
In conclusion, herein we report the proof-of-principle that the HaloTag protein can be used as a reporter gene for PET imaging in vivo, in a murine tumor model stably transfected with the protein on the cell surface. Further optimization of the HaloTag-based reporter gene system for non-invasive multimodality molecular imaging is currently underway.
Acknowledgments
This work is supported, in part, by the University of Wisconsin Carbone Cancer Center, NCRR 1UL1RR025011, a DOD BCRP Postdoctoral Fellowship, a DOD PCRP IDEA Award, the NIH through the UW Radiological Sciences Training Program 5 T32 CA009206-32, and Promega Corporation.
References
- 1.Massoud TF, Gambhir SS. Molecular imaging in living subjects: seeing fundamental biological processes in a new light. Genes Dev. 2003;17:545–580. doi: 10.1101/gad.1047403. [DOI] [PubMed] [Google Scholar]
- 2.Weissleder R, Pittet MJ. Imaging in the era of molecular oncology. Nature. 2008;452:580–589. doi: 10.1038/nature06917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kang JH, Chung JK. Molecular-genetic imaging based on reporter gene expression. J Nucl Med. 2008;49(Suppl 2):164S–179S. doi: 10.2967/jnumed.107.045955. [DOI] [PubMed] [Google Scholar]
- 4.Jacobs A, Voges J, Reszka R, Lercher M, Gossmann A, Kracht L, Kaestle C, Wagner R, Wienhard K, Heiss WD. Positron-emission tomography of vector-mediated gene expression in gene therapy for gliomas. Lancet. 2001;358:727–729. doi: 10.1016/s0140-6736(01)05904-9. [DOI] [PubMed] [Google Scholar]
- 5.Yaghoubi SS, Jensen MC, Satyamurthy N, Budhiraja S, Paik D, Czernin J, Gambhir SS. Noninvasive detection of therapeutic cytolytic T cells with 1SF-FHBG PET in a patient with glioma. NatClin Pract Oncol. 2009;6:53–58. doi: 10.1038/ncponc1278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Giepmans BN, Adams SR, Ellisman MH, Tsien RY. The fluorescent toolbox for assessing protein location and function. Science. 2006;312:217–224. doi: 10.1126/science.1124618. [DOI] [PubMed] [Google Scholar]
- 7.Contag CH, Bachmann MH. Advances in in vivo bioluminescence imaging of gene expression. Annu Rev Biomed Eng. 2002;4:235–260. doi: 10.1146/annurev.bioeng.4.111901.093336. [DOI] [PubMed] [Google Scholar]
- 8.Herschman HR. Noninvasive imaging of reporter gene expression in living subjects. Adv Cancer Res. 2004;92:29–80. doi: 10.1016/S0065-230X(04)92003-9. [DOI] [PubMed] [Google Scholar]
- 9.Min JJ, Gambhir SS. Molecular imaging of PET reporter gene expression. Handb Exp Pharmacol. 2008:277–303. doi: 10.1007/978-3-540-77496-9_12. [DOI] [PubMed] [Google Scholar]
- 10.Gilad AA, Ziv K, McMahon MT, van Zijl PC, Neeman M, Bulte JW. MRI reporter genes. J Nucl Med. 2008;49:1905–1908. doi: 10.2967/jnumed.108.053520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Los GV, Wood K. The HaloTag: a novel technology for cell imaging and protein analysis. Methods Mol Biol. 2007;356:195–208. doi: 10.1385/1-59745-217-3:195. [DOI] [PubMed] [Google Scholar]
- 12.Los GV, Encell LP, McDougall MG, Hartzell DD, Karassina N, Zimprich C, Wood MG, Learish R, Ohana RF, Urh M, Simpson D, Mendez J, Zimmerman K, Otto P, Vidugiris G, Zhu J, Darzins A, Klaubert DH, Bulleit RF, Wood KV. HaloTag: a novel protein labeling technology for cell imaging and protein analysis. ACS Chem Biol. 2008;3:373–382. doi: 10.1021/cb800025k. [DOI] [PubMed] [Google Scholar]
- 13.Zhang Y, So MK, Loening AM, Yao H, Gambhir SS, Rao J. HaloTag protein-mediated site-specific conjugation of bioluminescent proteins to quantum dots. Angew Chem Int Ed Engl. 2006;45:4936–4940. doi: 10.1002/anie.200601197. [DOI] [PubMed] [Google Scholar]
- 14.Lang C, Schulze J, Mendel RR, Hansch R. HaloTag: a new versatile reporter gene system in plant cells. J Exp Bot. 2006;57:2985–2992. doi: 10.1093/jxb/erl065. [DOI] [PubMed] [Google Scholar]
- 15.So MK, Yao H, Rao J. HaloTag protein-mediated specific labeling of living cells with quantum dots. Biochem Biophys Res Commun. 2008;374:419–423. doi: 10.1016/j.bbrc.2008.07.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Takemoto K, Matsuda T, McDougall M, Klaubert DH, Hasegawa A, Los GV, Wood KV, Miyawaki A, Nagai T. Chromophore-assisted light inactivation of HaloTag fusion proteins labeled with eosin in living cells. ACS Chem Biol. 2011;6:401–406. doi: 10.1021/cb100431e. [DOI] [PubMed] [Google Scholar]
- 17.Hata T, Nakayama M. Rapid single-tube method for small-scale affinity purification of polyclonal antibodies using HaloTag Technology. J Biochem Biophys Methods. 2007;70:679–682. doi: 10.1016/j.jbbm.2007.01.014. [DOI] [PubMed] [Google Scholar]
- 18.Motejadded H, Kranz B, Berensmeier S, Franzreb M, Altenbuchner J. Expression, one-step purification, and immobilization of HaloTag(TM) fusion proteins on chloroalkane-functionalized magnetic beads. Appl Biochem Biotechnol. 2010;162:2098–2110. doi: 10.1007/s12010-010-8985-1. [DOI] [PubMed] [Google Scholar]
- 19.Chumanov RS, Kuhn PA, Xu W, Burgess RR. Expression and purification of full-length mouse CARM1 from transiently transfected HEK293T cells using HaloTag technology. Protein Expr Purif. 2011;76:145–153. doi: 10.1016/j.pep.2010.11.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ohana RF, Hurst R, Vidugiriene J, Slater MR, Wood KV, Urh M. HaloTag-based purification of functional human kinases from mammalian cells. Protein Expr Purif. 2011;76:154–164. doi: 10.1016/j.pep.2010.11.014. [DOI] [PubMed] [Google Scholar]
- 21.Svendsen S, Zimprich C, McDougall MG, Klaubert DH, Los GV. Spatial separation and bidirectional trafficking of proteins using a multi-functional reporter. BMC Cell Biol. 2008;9:17. doi: 10.1186/1471-2121-9-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Urh M, Hartzell D, Mendez J, Klaubert DH, Wood K. Methods for detection of protein-protein and protein-DNA interactions using HaloTag. Methods Mol Biol. 2008;421:191–209. doi: 10.1007/978-1-59745-582-4_13. [DOI] [PubMed] [Google Scholar]
- 23.Schroder J, Benink H, Dyba M, Los GV. In vivo labeling method using a genetic construct for nanoscale resolution microscopy. Biophys J. 2009;96:L01–03. doi: 10.1016/j.bpj.2008.09.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kosaka N, Ogawa M, Choyke PL, Karassina N, Corona C, McDougall M, Lynch DT, Hoyt CC, Levenson RM, Los GV, Kobayashi H. In vivo stable tumor-specific painting in various colors using dehalogenase-based protein-tag fluorescent ligands. Bioconjug Chem. 2009;20:1367–1374. doi: 10.1021/bc9001344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Yamaguchi K, Inoue S, Ohara O, Nagase T. Pulsechase experiment for the analysis of protein stability in cultured mammalian cells by covalent fluorescent labeling of fusion proteins. Methods Mol Biol. 2009;577:121–131. doi: 10.1007/978-1-60761-232-2_10. [DOI] [PubMed] [Google Scholar]
- 26.Hartzell DD, Trinklein ND, Mendez J, Murphy N, Aldred SF, Wood K, Urh M. A functional analysis of the CREB signaling pathway using HaloCHIP-chip and high throughput reporter assays. BMC Genomics. 2009;10:497. doi: 10.1186/1471-2164-10-497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Taniguchi Y, Kawakami M. Application of HaloTag protein to covalent immobilization of recombinant proteins for single molecule force spectroscopy. Langmuir. 2010;26:10433–10436. doi: 10.1021/la101658a. [DOI] [PubMed] [Google Scholar]
- 28.Gallo S, Beugnet A, Biffo S. Tagging of functional ribosomes in living cells by HaloTag(R) technology. In Vitro Cell Dev Biol Anim. 2011;47:132–138. doi: 10.1007/s11626-010-9370-7. [DOI] [PubMed] [Google Scholar]
- 29.Zhou ZP, Shimizu Y, Tadakuma H, Taguchi H, Ito K, Ueda T. Single molecule imaging of the trans-translation entry process via anchoring of the tagged ribosome. J Biochem. 2011;149:609–618. doi: 10.1093/jb/mvr010. [DOI] [PubMed] [Google Scholar]
- 30.Wang H, Cai W, Chen K, Li ZB, Kashefi A, He L, Chen X. A new PET tracer specific for vascular endothelial growth factor receptor 2. Eur J Nucl Med Mol Imaging. 2007;34:2001–2010. doi: 10.1007/s00259-007-0524-0. [DOI] [PubMed] [Google Scholar]
- 31.Hong H, Yang Y, Zhang Y, Engle JW, Barnhart TE, Nickles RJ, Leigh BR, Cai W. Positron emission tomography imaging of CD105 expression during tumor angiogenesis. Eur J Nucl Med Mol Imaging. 2011;38:1335–1343. doi: 10.1007/s00259-011-1765-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Cai W, Zhang X, Wu Y, Chen X. A thiol-reactive 18F-labeling agent, N-[2-(4-18F-fluorobenzamido)ethyl]maleimide (1SF-FBEM), and the synthesis of RGD peptide-based tracer for PET imaging of avb3 integrin expression. J Nucl Med. 2006;47:1172–1180. [PMC free article] [PubMed] [Google Scholar]
- 33.Zhang X, Cai W, Cao F, Schreibmann E, Wu Y, Wu JC, Xing L, Chen X. 18-labeled bombesin analogs for targeting GRP receptor-expressing prostate cancer. J Nucl Med. 2006;47:492–501. [PubMed] [Google Scholar]
- 34.Wadas TJ, Wong EH, Weisman GR, Anderson CJ. Coordinating radiometals of copper, gallium, indium, yttrium, and zirconium for PET and SPECT imaging of disease. Chem Rev. 110:2858–2902. doi: 10.1021/cr900325h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Dearling JL, Voss SD, Dunning P, Snay E, Fahey F, Smith SV, Huston JS, Meares CF, Treves ST, Packard AB. Imaging cancer using PET-the effect of the bifunctional chelator on the biodis-tribution of a 64Cu-labeled antibody. Nucl Med Biol. 2011;38:29–38. doi: 10.1016/j.nucmedbio.2010.07.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Cai W, Chen K, He L, Cao Q, Koong A, Chen X. Quantitative PET of EGFR expression in xeno-graft-bearing mice using 64Cu-labeled cetuxi-mab, a chimeric anti-EGFR monoclonal antibody. Eur J Nucl Med Mol Imaging. 2007;34:850–858. doi: 10.1007/s00259-006-0361-6. [DOI] [PubMed] [Google Scholar]
- 37.Cai W, Ebrahimnejad A, Chen K, Cao Q, Li ZB, Tice DA, Chen X. Quantitative radioim-munoPET imaging of EphA2 in tumour-bearing mice. Eur J Nucl Med Mol Imaging. 2007;34:2024–2036. doi: 10.1007/s00259-007-0503-5. [DOI] [PubMed] [Google Scholar]
- 38.Cai W, Wu Y, Chen K, Cao Q, Tice DA, Chen X. In vitro and in vivo characterization of 64Cu-labeled Abegrin™, a humanized monoclonal antibody against integrin avb3. Cancer Res. 2006;66:9673–9681. doi: 10.1158/0008-5472.CAN-06-1480. [DOI] [PubMed] [Google Scholar]
- 39.Mammen M, Chio S, Whitesides GM. Polyvalent interactions in biological systems: implications for design and use of multivalent ligands and inhibitors. Angew Chem Int Ed Engl. 1998;37:2755–2794. doi: 10.1002/(SICI)1521-3773(19981102)37:20<2754::AID-ANIE2754>3.0.CO;2-3. [DOI] [PubMed] [Google Scholar]
- 40.Cai W, Niu G, Chen X. Imaging of integrins as biomarkers for tumor angiogenesis. Curr Pharm Des. 2008;14:2943–2973. doi: 10.2174/138161208786404308. [DOI] [PubMed] [Google Scholar]
- 41.Ray P, De A, Min JJ, Tsien RY, Gambhir SS. Imaging tri-fusion multimodality reporter gene expression in living subjects. Cancer Res. 2004;64:1323–1330. doi: 10.1158/0008-5472.can-03-1816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ray P, Tsien R, Gambhir SS. Construction and validation of improved triple fusion reporter gene vectors for molecular imaging of living subjects. Cancer Res. 2007;67:3085–3093. doi: 10.1158/0008-5472.CAN-06-2402. [DOI] [PubMed] [Google Scholar]
- 43.Ray P, Wu AM, Gambhir SS. Optical biolumi-nescence and positron emission tomography imaging of a novel fusion reporter gene in tumor xenografts of living mice. Cancer Res. 2003;63:1160–1165. [PubMed] [Google Scholar]
- 44.Kesarwala AH, Prior JL, Sun J, Harpstrite SE, Sharma V, Piwnica-Worms D. Second-generation triple reporter for bioluminescence, micro-positron emission tomography, and fluorescence imaging. Mol Imaging. 2006;5:465–474. [PubMed] [Google Scholar]
- 45.Luker GD, Luker KE, Sharma V, Pica CM, Dahlheimer JL, Ocheskey JA, Fahrner TJ, Milbrandt J, Piwnica-Worms D. In vitro and in vivo characterization of a dual-function green fluorescent protein-HSV1-thymidine kinase reporter gene driven by the human elongation factor 1 alpha promoter. Mol Imaging. 2002;1:65–73. doi: 10.1162/15353500200201118. [DOI] [PubMed] [Google Scholar]
- 46.Hong H, Yang Y, Cai W. Imaging gene expression in live cells and tissues. Cold Spring Harb Protoc. 2011 doi: 10.1101/pdb.top103. 2011: pdb top103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Phelps ME. PET: the merging of biology and imaging into molecular imaging. J Nucl Med. 2000;41:661–681. [PubMed] [Google Scholar]
- 48.Hong H, Yang Y, Zhang Y, Cai W. Non-invasive imaging of human embryonic stem cells. Curr Pharm Biotechnol. 2010;11:685–692. doi: 10.2174/138920110792246500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Hong H, Yang Y, Zhang Y, Cai W. Non-invasive cell tracking in cancer and cancer therapy. Curr Top Med Chem. 2010;10:1237–1248. doi: 10.2174/156802610791384234. [DOI] [PMC free article] [PubMed] [Google Scholar]