Abstract
The GTPase-activating proteins (GAPs) accelerate the hydrolysis of GTP to GDP by small GTPases. The GTPases play diverse roles in many cellular processes, including proliferation, cell motility, endocytosis, nuclear import/export, and nuclear membrane formation. Little is known about GAP-domain proteins in spermatogenesis. We isolated a novel RhoGAP domain-containing tGAP1 protein from male germ cells that exhibits unusual properties. The tGAP1 is expressed at low levels in early spermatogonia. Robust transcription initiates in midpachytene spermatocytes and continues after meiosis. The 175-kDa tGAP1 protein localizes to the cytoplasm of spermatocytes and to the cytoplasm and nucleus in spermatids. The protein contains four GAP domain-related sequences, in contrast to all other GAP proteins that harbor one such domain. No activity toward RhoA, Rac1, or Cdc42 could be detected. Results of transfection studies in various somatic cells indicated that low-level tGAP1 expression significantly slows down the cell cycle. Expression of higher levels of tGAP1 by infection of somatic cells with recombinant adenoviruses demonstrated that tGAP1 efficiently induces apoptosis, which to our knowledge is the first such demonstration for a RhoGAP protein. Based on its subcellular location in spermatids and its activity, tGAP1 may play a role in nuclear import/export.
Keywords: apoptosis, spermatid, spermatogenesis, testis
INTRODUCTION
The development of functional spermatozoa from undifferentiated male germ cells is a coordinated process involving regulated mitotic divisions, meiosis, cell growth, significant differentiation steps in haploid germ cells, significant changes in transcription, and apoptosis. This complicated process takes place in the protected surroundings of seminiferous tubules in the testis, where the only somatic cell, the Sertoli cell, is essential for many of these steps. Failure at any stage of this process will cause the male to become either subfertile or infertile. In addition, cancer may develop, and many different environmental and genetic factors have been implicated.
Normal spermatogenesis depends on a complex network of endocrine, paracrine, and autocrine signaling as well as cell-cell communication. In addition, a balance between proliferation and death of germ cells is established during spermatogenesis [1–3]. It has become clear that spermatogenesis depends on correct temporal and spatial regulation of the cell cycle at mitosis and meiosis [4–6] and involves significant alterations in transcriptional patterns [7–10]. Many testis-specific proteins are synthesized, the contributions of which to sperm-specific structures are now beginning to be understood, and accurate execution of various signaling pathways—including the mitogen-activated protein kinase pathway [9]—is important. In addition, an increasing number of reports have appeared concerning stem cells in testis, their transplantation, and their possible impact on treatment regimens [11]. Some of this work started with observations regarding the effect of vitamin A-deficient diets in male rats, resulting in accumulation of undifferentiated spermatogonia [12], their isolation and characterization [13, 14], and the subsequent transplantation of such cells [11, 12, 15–17].
Currently, little is known about the role of small GTPases and their regulatory partners, guanine nucleotide-exchange factors and GTPase-activating proteins (GAPs), in male germ cells. The GTPases play essential roles in such cellular processes as proliferation, cell motility, endocytosis, nuclear import/export, and nuclear membrane formation. The GTPase superfamily includes members of the Ras, Rho, Rab, Ran, and Arf subfamilies. The Rho subfamily, which includes RhoA, RhoB, RhoC, RhoE, RhoG, Rac1, Rac2, and Cdc42, is involved in various aspects of regulation motility, adhesion, gene expression, cell-cycle progression, and cell division in male germ cells [18–21]. The MgcRacGAP protein [22, 23] belongs to the RhoGAP family and likely is involved in the development and/or function of male germ cells, specifically spermatocytes. It associates with the plasma membrane and binds an anion transporter Tat1 in spermatocytes and spermatids [23, 24]. RhoA, Rac1, and Cdc42, which are substrates for Mgc-RacGAP, are not expressed in male germ cells [24]. Instead, MgcRacGAP binds Rnd2, a Rho-family GTPase, in the Golgi-derived proacrosomal vesicle [24].
We report here the isolation of a novel, testicular GAP domain-containing protein, which we named tGAP1, with the following features: tGAP1 is differentially spliced in spermatogonia, encodes not one but four GAP domains, localizes to the spermatocyte cytoplasm and plasma membrane and to the nucleus in spermatids, and efficiently induces apoptosis in somatic cells.
MATERIALS AND METHODS
RNA Extraction
The RNA was extracted from rat tissues, including testis, liver, lung, kidney, brain, heart, skeletal muscle, intestine, colon, and epididymis, and from NIH3T3 fibroblasts using Trizol reagent (Invitrogen Canada, Inc., Burlington, ON) following the manufacturer’s instructions with minor modifications. Contaminating DNA was removed by incubation of RNA preparations with 10 μg/ml of RNase-free DNase I in conditions suggested by the manufacturer (Pharmacia Diagnostics, Kalamazoo, MI). All investigations were conducted in accordance with the International Guiding Principles for Biomedical Research Involving Animals as promulgated by the Society for the Study of Reproduction and the Canadian Council on Animal Care guidelines.
Reverse Transcription-Polymerase Chain Reaction and Screening cDNA Libraries
The protocol of Liang et al. [25] and Liang and Pardee [26] was applied with minor modifications. One microgram of RNA from the indicated sources was converted to cDNA using primer T12AC, T12CC, or T12GC. One microliter of cDNA was added to standard polymerase chain reaction (PCR) mixtures containing one of the above primers and 5′-GGTACTAAGG-3′ primer and supplemented with [α-35S]dATP. The PCR products were separated by electrophoresis on denaturing urea polyacrylamide gels that, after drying, were exposed to Kodak XAR5 film (Eastman Kodak, Rochester, NY). Gel segments containing differentially expressed cDNAs were excised and incubated in 100 μl of TE (10 mM Tris-HCl [pH 7.4] and 1 mM EDTA), and cDNA was extracted by boiling. The cDNA was reamplified using the same PCR conditions and cloned in the pGEM-T Easy vector (Promega, Nepean, ON, Canada), resulting in clone p5-5.
To isolate cDNAs corresponding to clone p5-5, approximately 2 × 105 recombinants of a cDNA library made from purified rat undifferentiated spermatogonia [27] were screened on duplicate filters using radiolabeled p5-5 as probe, as described previously [28]. Positive plaques were isolated, replated, and rescreened, and three positive clones were sequenced. All recovered cDNA clones were of the same gene, called tGAP1. Distinct tGAP1 cDNA clones (2C, 4A, and 6A) were sequenced. To obtain tGAP1 cDNA from adult male germ cells, reverse transcription (RT)-PCR was carried out using RNA isolated from elutriated spermatids and tGAP1-specific primers. Primers used in the present study were as follows: 1461 (5′-CATACTGACAAAGAGTCTGATGATTTAAAG-3′), CAR1 (5′-CA TAAGGAGCTATGCGGACAG-3′), CAF1 (5′-CCTGAAAATTCACAT TGGAGC-3′), RCF1 (5′-GATGGAACAGACCATTTGGTG-3′), and RC-R1 (5′-CAGTCTTCCTTCAATGTTTTG-3′). Resulting clones were sequenced and compared to clones 2C, 4A, and 6A.
DNA Sequencing and Computer Analysis
Cloned DNA or PCR products were purified using kits from Qiagen (Mississauga, ON) and sequenced using Applied Biosystems (Foster City, CA) automated sequencing units (UC DNA Sequencing Facility). Bioinformatics analysis of sequences was carried out using Omiga version 2 software (Oxford Molecular, Madison, WI) as well as programs available at http://ca.expasy.org, http://www.ensembl.org, and http://www.sanger.ac.uk.
Reverse Transcription-Polymerase Chain Reaction
Single-stranded cDNA was prepared from total RNA of indicated rat tissues using mouse murine leukemia virus reverse transcriptase (Invitrogen Canada) and oligo-dT(12–18) primer (Invitrogen Canada) according to the manufacturer’s instructions. The cDNAs were analyzed for quality and genomic DNA contamination by PCR using β-actin primers (5′-CAA-CACCCCAGCCATGTACG-3′ and 5′-AGGAAGAGGATGCGGCAGTG G-3′) spanning one intron. To analyze expression of tGAP1, these cDNAs were used in PCR reactions using various indicated primer combinations.
Northern Blot Analysis
Northern blot analysis was carried out as described previously [29]. In brief, RNAs extracted from indicated tissues were separated on formaldehyde RNA gels, and RNAs were transferred to Hybond+ filters (Amersham Biosciences, Piscataway, NJ). Filters were probed with radiolabeled PCR products generated using appropriate primers. Prehybridization and hybridization were carried out for 2 h at 65°C and for 14–20 h at 65°C, respectively, after which filters were washed three times (30 min each wash) as described previously [29].
Cell Culture and Transfection
The tGAP1 open reading frame was cloned into the KpnI and NotI sites of vector pCI (Promega). To obtain inducible tGAP1 expression, this insert was also cloned into the pIND vector from the ecdysone-inducible mammalian expression system (Invitrogen Canada). Mouse NIH3T3 cells were grown in Dulbecco modified Eagle medium supplemented with 10% bovine calf serum (Invitrogen Canada). Transient transfections using pCI-tGAP1 vectors were performed in 35-mm tissue-culture dishes using FuGENE6 (Roche Applied Science, Laval, QC, Canada) according to the manufacturer’s instructions using 2 μg of DNA per plate. To generate stable cell lines using the inducible vectors, we carried out cotransfection of NIH3T3 cells, and colony selection was done according to the manufacturer’s instruction (Invitrogen Canada).
Adenovirus Constructs
The system for generation of recombinant adenovirus has been described previously [30]. Briefly, the complete tGAP1 coding region (sense orientation; 3879 base-pair [bp] KpnI/NotI restriction fragment), partial tGAP1 antisense orientation (3166-bp XhoI/SalI restriction fragment), and human p53 (1191-bp XbaI/BglII restriction fragment) were subcloned into the pAdTrack-CMV shuttle vector. The shuttle-vector plasmids and a shuttle vector without insert were linearized with PmeI and transformed into Escherichia coli strain BJ5183 by electroporation in a Bio-Rad Gene Pulser electroporator (Bio-Rad, Mississauga, ON, Canada): BJ5183 stably contains the adenoviral backbone vector pAdEasy-1. Bacterial colonies containing recombinants were selected with kanamycin (50 μg/ml) and screened by restriction endonuclease digestions (PacI, EcoRI, and BamHI). The recombinant adenoviral constructs were transformed into XL1Blue bacteria for large-scale amplification. For production of adenoviruses in mammalian cells, recombinant adenoviral constructs were linearized using PacI and transfected into 293T cells using SuperFect (Qiagen). The process of viral production was monitored by visualization of green fluorescent protein (GFP) expression, which is incorporated into the adenoviral genome. Viruses were harvested after 7–10 days. Viruses were purified and concentrated using an Amicon Ultra filter (Millipore Canada Ltd., Nepean, ON) and titrated by serial dilution and infection of cells. Infection rates in indicated cells were monitored using GFP. In all subsequent experiments, infection rates of 90–100% were used for all viruses.
Germ Cell Cytoplasmic and Nuclear Protein Extracts
Elutriated germ cells were prepared as described previously [31] with minor modifications. Briefly, testes were isolated from adult Sprague-Dawley rats. After decapsulation, seminiferous tubules were purified and subsequently treated with collagenase/trypsin to release Sertoli cells and germ cells. Germ cells were fractionated by centrifugal elutriation as described [31]. Fractions were collected containing 90% pure pachytene spermatocytes (fraction 220), 90% pure round spermatids (fraction 90), and 50% pure late spermatids (fraction 50).
For total protein extracts, elutriated cells were extracted in lysis buffer (50 mM Hepes [pH 7.0], 1% [v/v] Nonidet-P40 [Sigma, St. Louis, MO], 1 μg/ml of aprotinin, and 100 μg/ml of PMSF). Protein was quantified by spectrophotometry.
Germ cell nuclear and cytoplasmic protein extracts were prepared as described previously [32] with minor modifications. Briefly, germ cells obtained by elutriation (107 cells) were washed twice with ice-cold PBS and suspended in 400 μl of ice-cold buffer A (10 mM Hepes [pH 8.0], 10 mM KCl, 2 mM MgCl2, 0.1 mM EDTA [pH 8.0], 30 mM sucrose, 1 mM dithiothreitol (DTT), 0.4 mM PMSF, 0.3 μg/ml of leupeptin, 5 μM pepstatin A, and 0.5% Nonidet P-40) and incubated on ice for 1 min. Nuclei were collected by centrifugation at 1500 × g for 10 min at 4°C, and the supernatant (cytoplasmic fraction) was collected, aliquoted, and stored at −80°C. The nuclear pellet was washed twice with ice-cold buffer B (buffer A plus 10 mM CaCl2) and collected by centrifugation at 1500 × g for 10 min at 4°C. The nuclear pellet was resuspended in 100 μl of ice-cold buffer C (50 mM Hepes [pH 7.8], 100 mM KCl, 0.1 mM EDTA [pH 8.0], 15% glycerol, 1 mM DTT, 0.5 mM PMSF, and 5 μM pepstatin A), mixed gently at 4°C for 20 min, and centrifuged for 5 min at 14 000 rpm in a microfuge at 4°C. The supernatant (nuclear extract) was collected, frozen in liquid nitrogen, and stored at −80°C.
Antibody Preparation
The tGAP1 coding sequence in cDNA clone 4A was subcloned into the pMAL-2c vector (New England Biolabs Ltd., Pickering, ON, Canada). MBP-4A fusion protein was purified using an amylose column purification system (New England Biolabs) according to the manufacturer’s instructions. Antibody was raised against the purified protein in New Zealand White rabbits as described previously [28]. Two segments of tGAP (between amino acids 192 and 377 and between amino acids 479 and 662, containing the first and second putative GAP-domains D1 and D2, respectively) were individually cloned in the pMAL-2c vector. The purified fusion proteins MBP-D1 and MBP-D2 were isolated as described above.
Antibodies were affinity-purified and tested for specificity by immunoblotting as described previously [33]. Peptides used for affinity purification were MBP-4A, MBP-D1, and MBP-D2 separately.
Immunohistochemistry
Testes from adult rats were fixed in 4% paraformaldehyde, embedded in paraffin, and analyzed essentially as described previously [34]. Sections were stained using a Ventana Basic DAB Detection and Amplification kit (Ventana Medical System, Inc., Tucson, AZ), representing an indirect biotin-avidin system for detecting rabbit polyclonal primary antibodies. Affinity-purified anti-tGAP1 polyclonal antibody (made using MBP-4A) was used. As control, experiments were carried out using antibodies that had been preincubated with MBP fusion proteins and without primary antibody.
GAP Activity Assays
To analyze potential GTPase activity of tGAP1 (clone 4A) and the putative GAP-domains D1 and D2, in vitro GAP assays were performed. The positive control used in the assays was mouse CdGAP [35]. Briefly, recombinant RhoA, Rac, and Cdc42 (5 μg each) were preloaded with [γ-32P]GTP (10 μCi, 6000 Ci/mmol) in 20 μl of 20 mM Tris-HCl (pH 7.5), 25 mM NaCl, 0.1 mM DTT, and 5 mM EDTA for 10 min at 30°C. After addition of MgCl2 (final concentration, 20 mM), 0.75 μg of preloaded GTPases were diluted with 20 mM Tris-HCl (pH 7.5), 0.1 mM DTT, 1 mM GTP, 0.86 mg/ml of bovine serum albumin, and 0.2 μM of GST-CdGAP (amino acids 3–662); MBP-D1, MBP-D2, or MBP-4A were added to a final concentration of 1.3 μM. Control reactions did not contain GAP protein. The mixture was incubated at room temperature, and 5-μl samples were removed at 0 and 5 min, diluted in 1 ml of ice-cold buffer A (50 mM Tris-HCl [pH 7.5], 50 mM NaCl, and 5 mM MgCl2), and filtered through nitrocellulose filters prewetted with buffer A. Filters were washed with 10 ml of cold buffer A, dried, and counted. The percentage GTP remaining was calculated as GTP bound to the GTPase after 5 min of incubation, with GAP compared to the 0-min time point, which represents 100% GTP remaining bound.
Apoptosis Analysis
Rat IEC18 and monkey COS1 cells were cultured in appropriate medium. Cells were infected with adenoviruses containing no insert or expressing tGAP1 (in the sense or antisense orientation) or p53 at 90% confluence. The efficiency of infection was monitored for GFP using an inverted fluorescence microscope (Leica Microsystems Ltd., Richmond Hill, ON, Canada). After infection, cells were harvested at indicated times and washed with PBS. To determine apoptosis, in indicated cases the cells were treated with annexin V antisera and 7-amino-actinomycin D (7-AAD) using the annexin V-PE Apoptosis Detection kit (BD Biosciences Canada, Mississauga, ON) [36]. Cells were separated by fluorescent-activated cell sorting on a BD FACSVantage SE™ System (Becton Dickinson Canada, Inc., Mississauga, ON), and analysis of flow data was performed using the Cell Quest Pro software of the FacScan unit (Flow Cytometry Core Facility, University of Calgary).
RESULTS
tGAP1 Is a Novel Male Germ Cell Gene
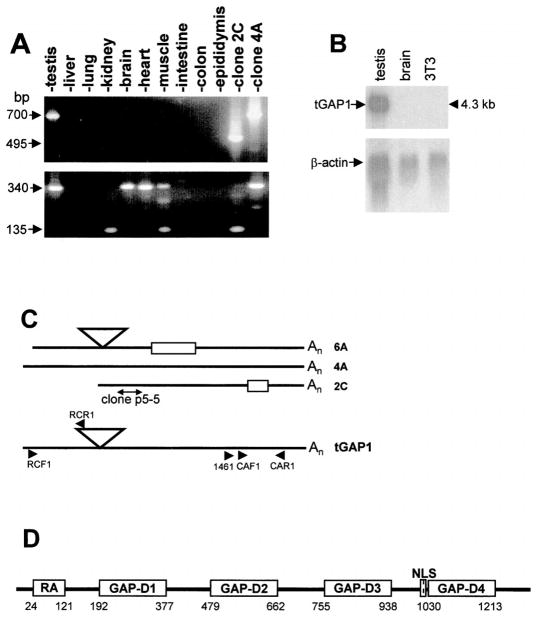
In a comparison of total testis cells and undifferentiated spermatogonia (isolated exactly as described in [13, 14]), we obtained three unique PCR fragments. One, labeled p5-5, represented a novel gene, and the results of its study are presented here. Clone p5-5 was first used to screen an undifferentiated spermatogonial cDNA library. Three cDNA clones (2C, 4A, and 6A) were isolated (GenBank accession no. AY631398, AY631399, and AY631400, respectively) that represent the novel gene, which we named tGAP1. To analyze the tGAP1 mRNA tissue expression pattern, we used primers 1461 and CAR1 (Fig. 1C) in RT-PCR to analyze rat tissues. The results show that by this method, tGAP1 expression is restricted to testis (Fig. 1A, top). Northern blot analysis demonstrated that the size of the adult tGAP1 transcript is approximately 4.3 kilobases (kb) (Fig. 1B). Full-length adult rat testicular tGAP1 cDNA was cloned by RT-PCR (GenBank accession no. AY631396): The tGAP1 open reading frame predicts a 1288-amino acid residue protein. The adult testis tGAP1 cDNA sequence was compared to those of clones 2C, 4A, and 6A, which indicated that it was a composite of 2C, 4A, and 6A (comparison schematically shown in Fig. 1C).
FIG. 1.
tGAP1 structure and mRNA expression. A) Analysis of tGAP1 mRNA expression. Total RNA was isolated from indicated rat tissues and converted to cDNA. As shown at the top, cDNAs were examined using β-actin gene-specific primers and next used to amplify tGAP1 cDNA using primers 1461 and CAR1. Clones 2C and 4A are alternatively spliced forms of tGAP1 spermatogonial cDNAs used as controls. As shown at the bottom, to uncover low-level expression, reamplification of the first PCR products by seminested PCR was applied using the CAR1 and internal primer CAF1. Note that in the first 30-cycle amplification, only testis-specific expression is seen. After a second round of PCR, tissue specific splicing products are seen in some tissues. B) Northern blot analysis of tGAP mRNA expression. Total RNA extracted from testis, brain, and a fibroblast cell line was examined by Northern blot analysis for expression of tGAP1 (top). The blot was next reprobed using β-actin (bottom). C) Schematic drawing showing an alignment of adult testicular tGAP1 cDNA and three alternatively spliced spermatogonial forms. Clone p5-5 is a differential display RT-PCR (DDRT-PCR) product used to screen a spermatogonia-specific cDNA library. Clones 6A, 4A, and 2C were obtained from this library. Comparison of these clones with tGAP1 cDNA showed 100% similarity, except for the segments that are added or deleted as indicated. The location of primers used in the present study is shown. D) Schematic location of domains on tGAP1 gene. Bioinformatics analysis of the protein sequence predicted for tGAP1 revealed four regions with significant similarity to GAP domains (boxes). Numbers indicate the location of amino acids at the beginning and end of the domains. A region resembling the RalGDS/AF-6 domain (RA) is also shown (see text). Additionally, tGAP1 contains a putative nuclear localization sequence (NLS).
To analyze the possibility that tissues other than testis contain very low levels of tGAP1 transcript that escaped detection in one round of PCR amplifications, a second round of amplification was carried out using nested primers (CAR1 and CAF1). The results (Fig. 1A, bottom) indicate very low-level but detectable tGAP1 mRNA in kidney, brain, heart, and muscle but no expression in liver, lung, intestine, colon, and epididymis. Note that the level of tGAP1 mRNA in brain detected by two rounds of RT-PCR is not detectable by Northern blot analysis. Interestingly, tissue-specific, alternatively spliced tGAP1 transcripts were observed in kidney and muscle in this analysis, resulting in PCR fragments that comigrated with fragments generated using clone 2C (Fig. 1A, bottom).
The tGAP1 sequence analysis had indicated the existence of four repeated regions in the mature testis tGAP1 transcript. Protein analysis showed that these tGAP1 repeats have significant similarity to RhoGAP (GAP) domains (Fig. 1D): The four domains are 30% identical and 49% similar (taking conservative substitutions into account) to human RhoGAP domain. The expectation values for the comparison between the human RhoGAP-domain gnl|CDD|5330 and tGAP1 domains D1, D2, D3, and D4 are 2e-24, 9e-13, 8e-14, and 7e-10, respectively. In addition, the N-terminus of tGAP1 has a region with similarity to a domain termed RA for RalGDS/AF-6 that, in other proteins, interacts with Ras and other small GTPases, and the C-terminus has a sequence similar to a nuclear localization sequence (Fig. 1D). The presence of four putative GAP domains is interesting, because all known RhoGAP and RhoGAP-like proteins—without exception—harbor only one such GAP domain (see Discussion). This analysis shows that tGAP1 might represent a novel member of the family of GAP-domain proteins with an expression that is largely restricted to testis.
tGAP1 Is a Member of a Multigene Family
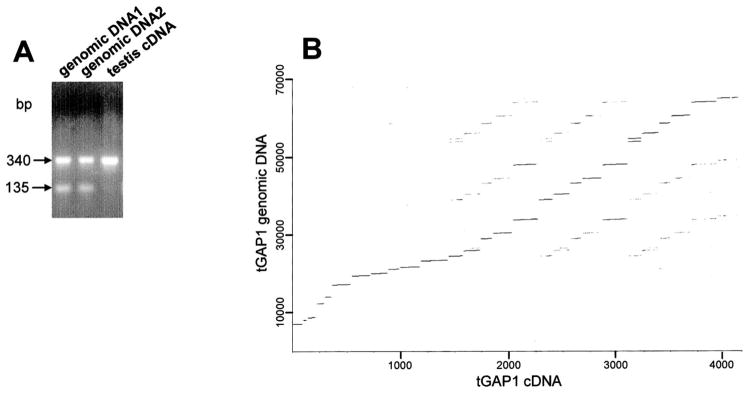
In a PCR analysis of rat genomic DNA using tGAP1 primers CAF1 and CAR1 (Fig. 1C), which should generate one PCR fragment with a size identical to that using adult testis tGAP1 cDNA, we consistently observed two genomic PCR products: One comigrated with the PCR fragment of testis tGAP1 cDNA as expected, but one was smaller (Fig. 2A). Sequence analysis of the larger genomic PCR fragment showed that it was identical to the tGAP1 cDNA sequence in full-length cDNA and that the smaller sequence was similar, but not identical, to that of clone 2C. The synthesis of more than one fragment from genomic DNA suggested the possibility that tGAP1 belongs to a family with at least two members. To investigate this, we performed RT-PCR using adult rat testis RNA and tGAP1 primers RCF1 and RCR1 to obtain the N-terminus of other potential tGAP1-like genes (Fig. 1C). Resulting PCR fragments were cloned into the pCI vector and sequenced. Most clones were identical to tGAP1, but the sequence of one of them, clone F1, was only 88% identical to that of tGAP1. Clone F1 is a partial cDNA sequence, which encodes one putative GAP domain with significant similarity to domain D1 of tGAP1. We named the corresponding gene tGAP2 (Gen-Bank accession no. AY631397).
FIG. 2.
tGAP1 genomic structure. A) Genomic DNA was extracted from two individual rats for PCR. The PCR was carried out using the CAF1 and CAR1 primers and revealed two fragments rather than the expected one fragment. Both PCR fragments were sequenced: The top band is 100% identical to the corresponding tGAP1 cDNA sequence, and the faster-migrating band has a 205-nucleotide deletion. B) A dot-plot comparison between tGAP1 genomic DNA and tGAP1 testicular cDNA to analyze the gene structure. The result shows that tGAP1 consists of 27 exons spread out over 58 kb of genomic DNA.
BLAST analysis of the entire rat genome gave the following results: The tGAP1 gene is located on rat chromosome 2q23 and is 58 kb in size. We compared the nucleotide sequence of this 58-kb genomic fragment with the tGAP1 cDNA sequence in a dot-plot analysis: Regions of homology (representing exons) could be readily observed and indicated that the gene contains 27 exons (Fig. 2B). The second family member, tGAP2, is located on rat chromosome 2q26. The locations of the tGAP1 and tGAP2 genes in the mouse genome are on mouse chromosome 3 region C and on chromosome 3 on the border between regions C and D, respectively.
Developmental Expression of tGAP1 in Rat Testis
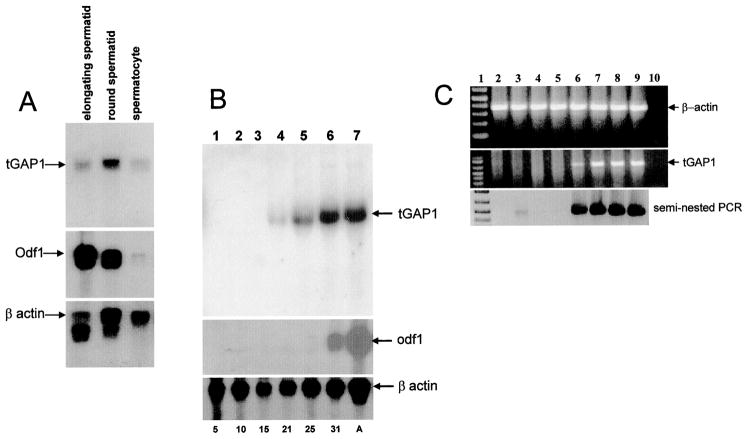
To determine in which male germ cells tGAP1 is transcribed, we carried out the following analyses: First, Northern blot analysis was done using RNA from elutriated germ cell fractions containing pachytene spermatocytes, round spermatids, and elongating spermatids (Fig. 3A). The results show that tGAP1 mRNA is present in spermatocytes, is slightly elevated in round spermatids, and is decreased in elongating spermatids (Fig. 3A). The presence of tGAP1 mRNA in the spermatocyte fractions likely results from transcription of the gene in these cells but also might result from spermatids in the spermatocyte fraction. To distinguish these possibilities, we performed two different assays. In the first, blots were reprobed with the spermatid-specific Odf1 cDNA: The intensity of the Odf1 signal in the spermatocyte fraction is a measure of the level of contamination of spermatocytes with spermatids [31]. The result (Fig. 3A) indicated that contamination of the spermatocyte fraction with spermatids was low. The difference in intensity between signals in the spermatocyte fraction and the spermatid fraction is large for Odf1 but smaller for tGAP1, suggesting that spermatocytes express tGAP1 mRNA. We also performed single-cell RT-PCR on isolated spermatocytes: In all cases, a fragment was amplified, specifically demonstrating that tGAP1 is, indeed, transcribed in pachytene spermatocytes (not shown), which is in agreement with the interpretation from the Northern blot experiment.
FIG. 3.
tGAP1 mRNA expression starts in spermatocytes. A) Male germ cells were elutriated and separated as described in Materials and Methods. Total RNA was extracted from these cells and examined by Northern blot analysis, which was carried out using radiolabeled probes for tGAP1 (top), Odf1 (middle), and β-actin (bottom). The Odf1 is used to verify the purity of germ cells, and the β-actin gene was used as control for RNA integrity and loading. Highest tGAP1 expression was seen in round spermatids. B) To determine the start of tGAP1 mRNA expression in spermatogenesis, RNA was isolated from postnatal rat testes at 5, 10, 15, 21, 25, and 31 days and from adult (A) testis (lanes 1–7, respectively; numbers indicate age) and then examined by Northern blot analysis using as probes tGAP1 (top), Odf1 (middle), and β-actin (bottom). In the rat, spermatogonia are present at Day 6, zygotene cells at Day 15, early pachytene cells at Day 17, midpachytene cells at Day 22, late pachytene cells at Day 26, early round spermatids at Day 30, late round spermatids at Day 35, and elongating spermatids at Day 38 [65]. C) To examine the possibility of low-level tGAP1 mRNA expression during early male germ cell development, RNA was isolated from rat testes and used in RT-PCR with β-actin primers (top) and tGAP primers (1461 and CAR1; middle). The RNAs were isolated from fetal testis (lane 2) and postnatal testes at 5, 10, 15, 21, 25, and 31 days as well as from adults (lanes 3–9, respectively). Lane 1 shows a 100-bp DNA marker. Lane 10 shows a negative control for RT-PCR (no RT reaction but otherwise identical to other PCR conditions). The RT-PCR pattern reflects that of the Northern blot analysis. Reamplification of the first RT-PCR products (bottom) using seminested primers revealed tGAP1 expression in spermatogonia (lane 3), as expected, but not in fetal testis (lane 1).
Second, we performed two developmental expression analyses, one by Northern blot analysis and one by RT-PCR, on RNA isolated from rat testis at different times after birth. Because the first round of spermatogenesis after birth is highly synchronized, the appearance of a tGAP1 transcript at a particular age can be directly correlated with the appearance of differentiating male germ cells at that age. The results of the Northern blot analysis are shown in Figure 3B. These results demonstrate that in this assay, tGAP1 mRNA is first detectable at Day 21 of age, which is consistent with expression in midpachytene spermatocytes (lane 4), whereas—as expected—Odf1 mRNA is first detectable at Day 31, coinciding with the appearance of spermatids (lane 6). Our data indicate that tGAP1 transcription starts in spermatocytes and continues after meiosis. We note a substantial increase in tGAP1 mRNA accumulation concomitant with the appearance of spermatids, suggesting that tGAP1 protein might be more abundant in spermatids than in spermatocytes. Similar RNA preparations were also used in developmental RT-PCR experiments (Fig. 3C). The pattern observed after one round of PCR was in agreement with that of the Northern blot analysis. The tGAP1 mRNA was detectable after a second round of PCR at Day 5 of age, coinciding with the presence of spermatogonia (Fig. 3C, lane 3). No tGAP1 mRNA was detected in fetal tissue even after two rounds of PCR (Fig. 3C, lane 2).
tGAP1 Protein Localizes to the Spermatid Nucleus and Cytoplasm
A rabbit polyclonal antibody was generated against tGAP1 to investigate tGAP1 synthesis in testis. The results are shown in Figure 4A. First, the anti-tGAP1 antibody was shown to specifically immunoprecipitate in vitro-translated, full-length tGAP1 using clone pCI-tGAP1: The apparent size of the tGAP1 protein is estimated to be approximately 175 kDa (lane 1). The anti-tGAP1 antibody also specifically recognized tGAP1 protein synthesized in transiently transfected 293T cells in Western blot assays (lane 2). Second, we used the antibody to analyze the production of tGAP1 during spermatogenesis. Results of Western blot analysis using anti-tGAP1 antibody and extracts from elutriated spermatocytes (lane 6), round spermatids (lane 5), and elongating spermatids (lane 4) indicated that tGAP1 is expressed in these germ cells. Moreover, the intensities of the signals in the Western blots paralleled those in the Northern blot experiment, with peak tGAP1 accumulation being noted in round spermatids. We also observed the presence of two bands, approximately 165 and 175 kDa in size, in germ cells and in total testis, respectively, (lane 3) in the immunoblotting assays lanes 4–6. The ratio between the 175-and 165-kDa tGAP1 proteins changes in the different germ cells during progression through spermatogenesis. We do not, at present, know the origin of the smaller variant.
FIG. 4.
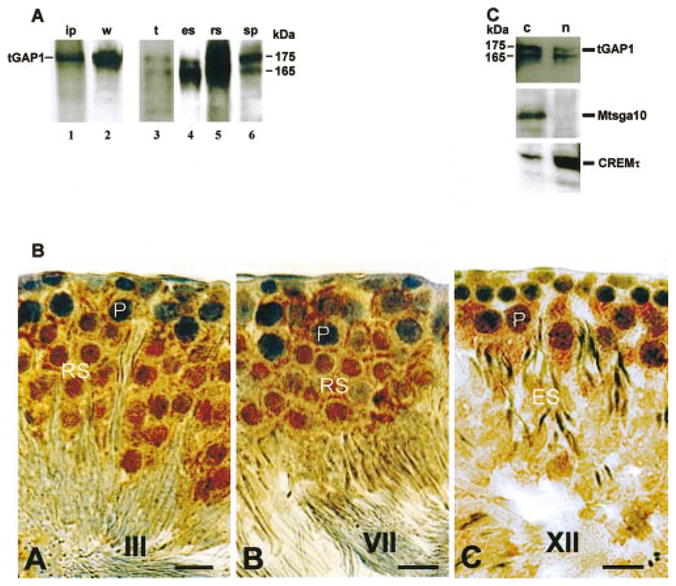
tGAP1 protein accumulates in spermatid nuclei. A) To analyze the antiserum raised against tGAP1, the antiserum was used in immunoprecipitation of in vitro translated tGAP1 (lane 1, ip) and in Western blot analyses of tGAP1 protein expressed in cells transfected with pCI-tGAP1 (lane 2, w), total testis (lane 3, t), and the following elutriated male germ cells: elongating spermatids (lane 4, es), round spermatids (lane 5, rs) and pachytene spermatocytes (lane 6, sp). Note the detection of two tGAP1 proteins in testis and male germ cells. B) Rat testes were prepared for immunohistochemistry as described in Materials and Methods. Immunohistochemistry was carried out using anti-tGAP1 polyclonal antibodies to define the localization of the protein in male germ cells. Staining of germ cells at different stages (I–XIV) of the cycle of the rat seminiferous epithelium is shown. Indicated are stages III, VII, and XII (A–C, respectively), containing spermatocytes (P), round spermatid (RS), and elongated spermatid (ES). Note the cytoplasmic stain in spermatocytes and the nuclear stain in spermatids. Bar = 10 μm. C) Western blot analysis of cytoplasmic (lanes c) and nuclear (lanes n) fractions isolated from testicular germ cells using anti-tGAP1 antiserum. As controls for cross-contamination of cytoplasmic and nuclear fractions, cytoplasmic and nuclear extracts were also analyzed using anti-CREMt antibody, which detects the testicular nuclear CREMτ transcription factor (bottom), and antibodies against Mtsga10 (middle), which is a cytoplasmic protein. Note the presence of tGAP1 in both cytoplasmic and nuclear fractions.
Immunohistochemistry was next carried out using affinity-purified anti-tGAP1 antibody and rat testis sections. We observed tGAP1 staining in the cytoplasm and on cell membranes of pachytene spermatocytes (Fig. 4B). After meiosis, tGAP1 is visible in the cytoplasm but also accumulates in the nucleus of all postmeiotic cells in the seminiferous tubules (Fig. 4B, round spermatids and elongating spermatids). In control experiments, staining was abolished using antibody that was blocked with recombinant tGAP1 protein (not shown).
To obtain further biochemical evidence to support the observation of nuclear tGAP1 protein, we carried out cell fractionation experiments. Nuclear and cytoplasmic extracts were isolated from total testicular germ cells and used for immunoblotting. As control for the quality and degree of cross-contamination of nuclear and cytoplasmic extracts, we used antibodies that recognize the nuclear spermatid transcription factor CREMτ [37] and antibodies that recognize the cytoplasmic protein Mtsga10 [38]. The results using the control antibodies indicated that the cytoplasmic and nuclear extracts are not significantly cross-contaminated: Mtsga10 is predominantly in the cytoplasmic fraction (Fig. 4C, lane c), and CREMτ is predominantly in the nuclear fraction (Fig. 4C, lane n). We next observed that the 175- and 165-kDa tGAP1 proteins are present in germ cell cytoplasm and nucleus (Fig. 4C): The 165-kDa variant appears to be equally distributed, whereas the 175-kDa variant appears to be more cytoplasmic. This result confirms the nuclear location of tGAP1. Together, these data show that tGAP1 variants have multiple subcellular locations depending on the stage of spermatogenesis.
tGAP1 Lacks Detectable RhoA, Cdc42, or Rac GAP Activity
The tGAP1 protein sequence indicated the presence of four putative GAP domains that are 30% identical to human RhoGAP domains. To analyze if tGAP1 has intrinsic GAP activity to known substrates, the first and second GAP domains (D2 is similar to domains D3 and D4) were sub-cloned individually and expressed as MBP-fusion proteins. Next, MBP-D1, MBP-D2, and MBP-4A (containing all four domains) were analyzed by in vitro GAP assays to determine whether they have functional GAP activity toward RhoA, Rac1, and/or Cdc42. The positive control for Cdc42 and Rac was mouse CdGAP. Table 1 shows the results: Percentage GTP remaining represents the ratio of GTP bound to the indicated GTPase after a 5-min incubation with the indicated GAPs compared to GTP bound to the indicated GTPase at the 0-min time point. Clearly, CdGAP is active toward Cdc42 and Rac1, as expected [35]. However, neither tGAP1 nor any of the subcloned putative domains shows activity toward Cdc42, Rac1, or RhoA. This result suggests either that tGAP1 does not have intrinsic GAP activity or, alternatively, that we have not yet found its specific small GTPase substrate. Based on the tGAP1 expression analysis, a small tGAP1 GTPase substrate should be present in the spermatocyte cytoplasm and/or the spermatid nucleus or cytoplasm (see Discussion).
TABLE 1.
Analysis of GAP activity of tGAP1 and tGAP1 domains.
| Small GTPase tested
|
|||
|---|---|---|---|
| RhoA | Cdc42 | Rac1 | |
| No GAP | 60a | 63 | 66 |
| CdGAP | 59 | 3 | 7 |
| D1-domain tGAP1 | 57 | 66 | 54 |
| D2-domain tGAP1 | 56 | 69 | 55 |
| tGAP1b | 62 | 76 | 60 |
Numbers indicate the percentage GTPase-GTP remaining after 5 min of incubation with the indicated GAP-domain proteins in comparison to the amount of GTPase-GTP at time 0.
tGAP1 harbors four (D1–D4) regions with significant similarity to GAP domains.
tGAP1 Affects Mouse Fibroblast Proliferation
To study a possible function of tGAP1, we transfected NIH3T3 mouse fibroblasts, which do not express tGAP1, with pCI-tGAP1 and then selected G418-resistant colonies. However, we were unable to obtain NIH3T3 cells stably expressing tGAP1, indicating that tGAP1 expression might adversely affect the cell cycle or cell growth. We therefore used an inducible mammalian expression system based on ecdysone-directed induction of transcription. Several colonies were obtained containing the inducible tGAP1 expression construct; however, these colonies grew very slowly in comparison to colonies obtained using empty vector. To confirm tGAP1 expression, RNA was extracted for Northern blot analysis from slow-growing, early passage, uninduced colonies. The results showed clearly detectable amounts of tGAP1 mRNA in the absence of ecdysone induction (Fig. 5, lanes 2 and 3), indicating leaky expression from the inducible promoter. We observed that after 8–10 additional passages, these colonies started to grow at the rate of vector-only controls. Northern blot analysis of tGAP1 mRNA in these late-passage, normal-growing cells indicated that expression of tGAP1 was absent both before (lanes 4 and 5) and after ecdysone induction (lanes 6 and 7). Together these results suggest that tGAP1 expression has a negative effect on fibroblasts.
FIG. 5.
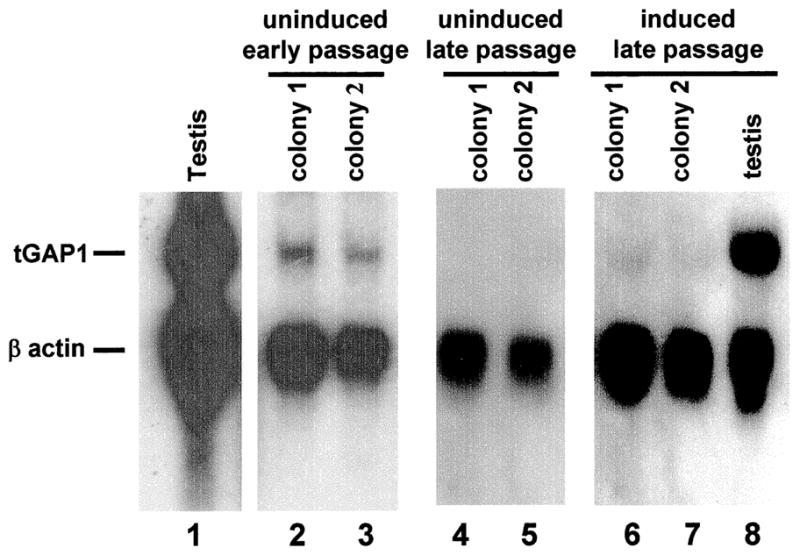
Expression of tGAP1 in transfected fibroblasts. The tGAP1 was cloned in the ecdysone-inducible pIND vector and transfected in NIH3T3 cells that lack endogenous tGAP1 expression (see Fig. 1B) together with the pVgRXR vector. Several stably transfected colonies were obtained, most of which grew slowly, and colonies 1 and 2 were analyzed further. Total RNA was extracted from colonies 1 and 2 before induction (lanes 2 and 3), as well as from testis used as a positive control (lanes 1 and 8) and analyzed for tGAP1 expression. Northern blots were probed simultaneously with radiolabeled tGAP1 and β-actin probes. This analysis showed a leaky expression of tGAP1 mRNA (lanes 2 and 3). After several more passages, colonies started growing at a rate that was normal for fibroblasts. The analysis of tGAP1 mRNA expression by a similar Northern blot assay indicated that tGAP1 expression had essentially disappeared both before and after induction with ecdysone (lanes 4–7).
tGAP1 Induces Apoptosis
Expression of tGAP1 might halt cell division (cell arrest) and/or induce programmed cell death (apoptosis), which would explain our inability to obtain tGAP1-expressing fibroblasts. To investigate the possibility of tGAP1-induced apoptosis, rat IEC18 cells, which do not express endogenous tGAP1 mRNA (see below), were infected with adenoviruses carrying no insert and adenoviruses expressing either tGAP1 or antisense tGAP1, and cells were monitored at different times after infection by fluorescence microscopy (all adenoviruses used express GFP). To confirm tGAP1 expression in infected cells, we first performed Western blot analysis of total cellular proteins extracted 36 h after infection. As shown in Figure 6A, tGAP1 protein expression was evident in cells infected with tGAP1 in the sense orientation (lane 4) but not in cells infected with adenoviruses carrying tGAP1 in the antisense orientation (lane 5). The tGAP1 protein expressed from the adenovirus comigrates with spermatid tGAP1 and tGAP1 expressed in human 293T cells (Fig. 6A, lanes 1 and 2, respectively). Furthermore, we determined that in adenovirus-infected cells, tGAP1 protein accumulates in both cytoplasm and nucleus (Fig. 6B, panel 2), which is in agreement with our observations in testis.
FIG. 6.
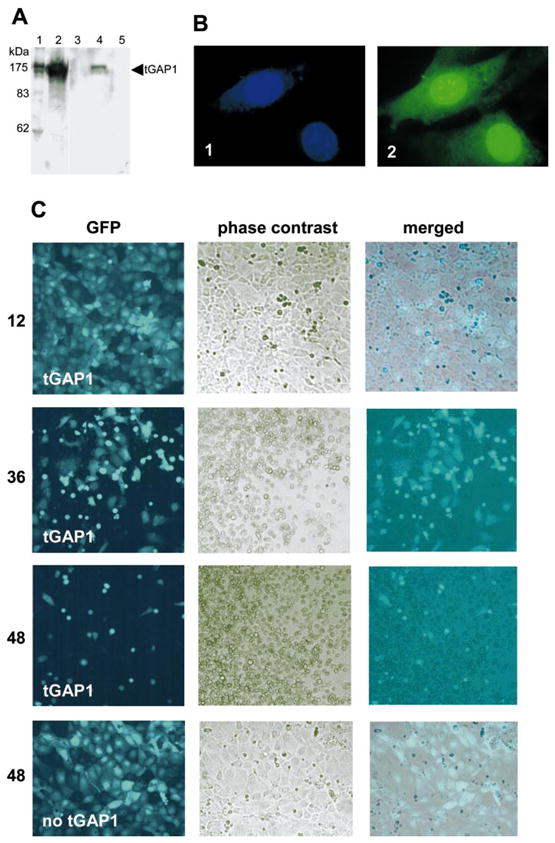
tGAP1 expression causes cell death. A) For efficient transfer of tGAP1 in cells, tGAP1 was cloned in an adenovirus vector that also independently expresses GFP. The expression of tGAP1 protein in rat IEC18 and human 293T epithelial cells infected with tGAP1 adenovirus was examined by Western blot analysis using anti-tGAP1polyclonal antibodies. Lane 1: round spermatid protein extract; lane 2: 293T cells expressing tGAP1; lanes 3–5: rat IEC18 cells infected with adenoviruses lacking an insert, expressing tGAP1, and expressing antisense tGAP1, respectively. B) To analyze the subcellular localization of exogenously expressed tGAP1, NIH3T3 cells were transfected with a GFP-tGAP1 fusion construct. Transfected cells were analyzed by fluorescence microscopy for staining of nuclear DNA with 4′,6′-diami-dino-2-phenylindole (1) and expression of GFP-tGAP1 (2). Note that tGAP1 is present in both nucleus and cytoplasm. Magnification, ×100. C) To analyze if tGAP1 affects cells, rat IEC18 cells were infected with the adenovirus described in A that expresses tGAP1 and GFP. Expression of GFP is indicative of live infected cells. Cell cultures were examined at 12, 36, and 48 h after infection, as indicated, for GFP expression. Both GFP and phase-contrast images of infected cultures at these time points are shown (top three rows of images), as is a merged image. As control, rat IEC18 cells were infected with adenovirus expressing only GFP; these cells showed that adenovirus itself had no effect on the cells after 48 h (bottom). Magnification, ×20.
We next examined a possible cell-death effect of tGAP1 by monitoring GFP expression over time in cells infected with tGAP1-expressing adenoviruses and cells infected with control adenovirus. At 12 h after infection, a large proportion of IEC18 cells is GFP-positive (Fig. 6C, panel 12-tGAP1), indicating efficient infection with tGAP1-expressing adenovirus. At 36 h after infection, most cells changed morphology and became rounded, and some started to detach (Fig. 6C, panel 36-tGAP1). At 48 h after infection, few cells were still alive, as monitored by significantly decreased GFP expression (Fig. 6C, panel 48-tGAP1). In contrast, IEC18 cells infected with control adenovirus were efficiently infected, remained attached, and displayed normal morphology 48 h after infection (Fig. 6C, panel 48-no tGAP1). This suggested that tGAP1 induces cell death or affects the cytoskeleton, resulting in the observed morphological changes.
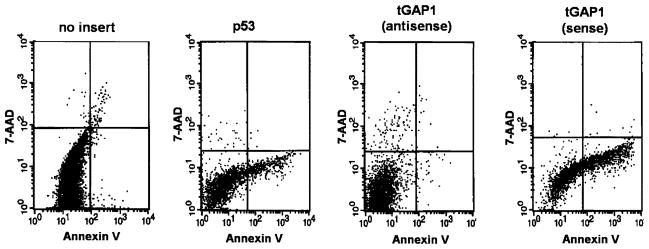
To analyze if tGAP1-induced cell death results from apoptosis, we double-stained infected cells with two markers, annexin V-fluorescein and 7-AAD, to identify early apoptotic intact cells (annexin V-positive/7-AAD-negative cells) and late-apoptotic populations with damaged membranes that permit entry of isothiocyanate-7-AAD (annexin V-positive/7-AAD-positive cells) [36]. Rat IEC18 cells were individually infected for 36 h in the following groups: adenovirus expressing only GFP, adenovirus expressing the p53 gene, adenovirus expressing antisense tGAP1, and adenovirus expressing tGAP1. Infected cells were examined by fluorescent-activated cell sorting. Figure 7 shows the results: The lower-right quadrant reflects early apoptotic cells (annexin V-positive/7-AAD-negative) and clearly shows that tGAP1 induces apoptosis, as exemplified by the presence of abundant annexin V-positive/7-AAD-negative cells. The efficiency of induction of apoptosis by tGAP1 is similar to that of the positive-control p53. As expected, adenovirus without an insert and adenovirus expressing antisense tGAP1 failed to induce apoptosis. Identical results were obtained for COS1 cells infected with these viruses (not shown).
FIG. 7.
tGAP1 induces apoptosis. Rat IEC18 cells were infected with recombinant adenoviruses without an insert (no insert), expressing p53 (p53), tGAP1 in the antisense orientation (tGAP1 antisense) and tGAP1 (tGAP1 sense). Infected cells were treated with annexin V antisera and 7-AAD as described in Materials and Methods. Cells were next separated by flow cytometry. The analysis of flow data were done using the Cell Quest Pro software of the Becton Dickinson FacScan unit and demonstrated that tGAP1 induces apoptosis with an efficiency similar to that of p53, as evidenced by the appearance of cells in the lower hand quadrant (annexin V-positive/7-AAD-negative). Antisense tGAP1 or adenovirus without an insert did not induce apoptosis.
DISCUSSION
The Rho GTPases are a subgroup of the large superfamily of Ras small GTPases that hydrolyze GTP to GDP [39–43]. Currently, more than 18 distinct Rho GTPases have been identified in mammalian cells, but the best-studied family members are RhoA, Rac1, and Cdc42. These proteins have been found to regulate the morphological and migratory properties of mammalian cells, signaling pathways that control gene transcription, cell-cycle entry, and cell survival. The Rho-related enzyme activity is regulated by corresponding GAP proteins that accelerate by orders of magnitude the conversion of GTP to GDP by the small GTPases. These GAP proteins contain a GAP domain with a conserved structure, the so-called arginine finger [44, 45], that inserts into the phosphate-binding pocket of small GTPases and activates GTPase activity [46]. Mutation of the Arg residue reduces enzymatic activity but has no significant effect on the binding of the GAP protein to its substrate [47]. The RhoGAP proteins have, in addition, two or more of a variety of domains with specific function, including SH3 and PH domains [48]. All GAP proteins have one GAP domain, except one putative RIKEN cDNA (GenBank accession no. AK005660) that encodes two putative domains). In contrast, the tGAP1 coding region predicts a protein with four putative GAP domains. Sequence analysis of these four domains indicated that the Arg residue, which is conserved in GAP domains, is not present. It was recently shown that RanGAP, which binds Ran together with a Ran-binding protein, RanBP1, does not employ an arginine finger in this interaction [49]. Attempts to determine GAP activity for tGAP1 have failed for RhoA, Rac, and Cdc42, possibly because tGAP1 domains lack the critical Arg residue. This finding makes sense, because RhoA, Rac1, and Cdc42 are not expressed in male germ cells [24]. This result suggests, therefore, that tGAP1 lacks intrinsic GAP activity or, alternatively, that the tGAP1 substrate remains to be identified.
Analysis of the tGAP1 mRNA expression pattern revealed several interesting features. The predominant site of expression is testis. Expression in other tissues, including brain, heart, and muscle, could only be detected after two successive rounds of PCR. In testis, tGAP1 mRNA synthesis coincides with the appearance of midpachytene spermatocytes, and the presence of readily detectable amounts of tGAP1 mRNA after meiosis could result from stabilization of the transcript or continued transcription. In addition, tGAP1 is expressed at Day 5 of age, coinciding with the presence of spermatogonia. However, whereas meiotic and postmeiotic male germ cells express one tGAP1 mRNA, spermatogonia express several differentially spliced variants. These variants are predicted to encode tGAP1 variant proteins with only three of the four putative GAP domains. In addition, the presence of very small amounts of clone 2C-like mRNA in kidney, full-length mRNA in brain and heart, and both types in muscle suggests the possibility that different tGAP1 proteins have differential activity and/or specificity. Using tGAP1 antibodies, we show that tGAP1 protein expression patterns in testis reflect those of tGAP1 mRNA. However, the protein analysis revealed two noteworthy features: First, the sub-cellular localization of tGAP1 appears to change from predominantly cytoplasmic in spermatocytes to predominantly nuclear in spermatids. Thus, tGAP1 can be added to the increasing number of proteins that have been shown to change location in male germ cells during development [50–52], one of which is Ran [53]. Second, immunoblot results from these germ cells revealed the presence of two tGAP1 protein variants of 175 and 165 kDa. The origin of two proteins instead of one is not yet known, but it might involve posttranslational modifications. Spermatocytes produce more of the 175-kDa protein, whereas elongating spermatids synthesize more of the 165-kDa variant. These two variants are present in cytoplasmic and nuclear fractions. The large size of both suggests an active nuclear import mechanism: Both contain a putative nuclear localization signal just upstream of the last GAP-domain D4. In agreement with our observations in testis, transfected somatic cells and somatic cells infected with adenoviruses expressing tGAP1 also produce both variants (and in a ratio similar to that seen in spermatocytes). This indicates that the mechanism resulting in two products instead of one is not male germ cell-specific.
What could be the function and mechanism of action of the tGAP1 protein during spermatogenesis? Spermatogenesis is the process of germ cell proliferation and differentiation. It has been shown that significant numbers of male germ cells that result from mitosis and meiosis die by an apoptotic process [1, 54, 55]. Germ cell apoptosis may eliminate cells that accumulate mistakes during the many divisions and differentiation steps. The significance of regulating cell population by apoptosis is more apparent when sperm production is halted [56]. In an attempt to generate cell lines that stably express tGAP1, we discovered that tGAP1 prevented colony growth, which is in agreement with other reports concerning the negative effects of GAP domain-containing proteins on cell growth [57–59]. Further experimentation uncovered that tGAP1 efficiently induces apoptosis in somatic cells. The mechanism by which it does this is not known, but our observation represents the first documentation of apoptotic activity for a GAP domain-containing protein. We believe, however, that the possibility that tGAP1 regulates apoptosis in male germ cells is not likely: From the immunocytochemical analysis, it appears that all spermatocytes and all spermatids express tGAP1 in easily detectable quantities, yet apoptosis is a highly selective process. If tGAP1 plays a role in testicular apoptosis, it must itself be under tight control, because its mere expression in somatic cells results in massive apoptosis. It therefore is more likely that the apoptotic pathway induced by tGAP1 in somatic cells is not available in spermatids.
Based on our data, one can, however, envisage different functions for tGAP1 in the regulation of small GTPases. In one model, tGAP1 lacks GTPase stimulatory activity but can still bind GTPases and/or other proteins: It thus would effectively sequester the GTPase (perhaps in the nucleus) and, in that manner, regulate its function. Alternatively, one can propose a model in which tGAP1 regulates a yet-to-be-identified small GTPase, such as Rnd2 or Ran. The testicular Ran GTPase proteins [60] are attractive potential tGAP1-regulated proteins for several reasons: 1) Similar to RanGAPs, all four tGAP1 GAP domains lack the Arg residue; 2) similar to tGAP1, which is present in the spermatid cytoplasm and nucleus, Ran relocates from manchette to nucleus in elongating spermatids [53]; and 3) Ran-binding proteins have been isolated from testis [61, 62]. These two models can also be combined: tGAP1 could sequester Ran in the nucleus, thus impacting import/export in the spermatid nucleus. It is also interesting to note that the protein causing segregation distortion in male Drosophila flies was identified as Sd-RanGAP [63, 64], which encodes a truncated protein that mislocalizes to the nucleus but that retains enzymatic activity. The Sd-RanGAP acts by reducing nuclear RanGTP levels, disrupting the Ran signaling pathway, with significant effects on male fertility.
Our genomic analysis and bioinformatics approach showed that tGAP1 belongs to a multigene family. Both tGAP1 and tGAP2 are significantly related and are clustered in syntenic regions in the rat and mouse genomes. It is not known if the arrangement of tGAP genes is important for their transcriptional regulation, but the discovery of more than one family member suggests a role for these novel multi-GAP domain-containing proteins that is conserved.
Acknowledgments
We thank Ms. Ibtissem Triki for the GAP assays and Dr. F. van Dissel for a gift of the undifferentiated spermatogonial cDNA library.
Footnotes
Supported by grants from the Canadian Institutes of Health Research (CIHR) to F.A.v.d.H. and from the Canadian Cancer Society to N.L.-V., who is also a recipient of a CIHR new investigator award.
References
- 1.Sinha Hikim AP, Lue Y, Diaz-Romero M, Yen PH, Wang C, Swerdloff RS. Deciphering the pathways of germ cell apoptosis in the testis. J Steroid Biochem Mol Biol. 2003;85:175–182. doi: 10.1016/s0960-0760(03)00193-6. [DOI] [PubMed] [Google Scholar]
- 2.Sinha Hikim AP, Swerdloff RS. Hormonal and genetic control of germ cell apoptosis in the testis. Rev Reprod. 1999;4:38–47. doi: 10.1530/ror.0.0040038. [DOI] [PubMed] [Google Scholar]
- 3.Rodriguez I, Ody C, Araki K, Garcia I, Vassalli P. An early and massive wave of germinal cell apoptosis is required for the development of functional spermatogenesis. EMBO J. 1997;16:2262–2270. doi: 10.1093/emboj/16.9.2262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wolgemuth DJ, Laurion E, Lele KM. Regulation of the mitotic and meiotic cell cycles in the male germ line. Recent Prog Horm Res. 2002;57:75–101. doi: 10.1210/rp.57.1.75. [DOI] [PubMed] [Google Scholar]
- 5.Salazar G, Liu D, Liao C, Batkiewicz L, Arbing R, Chung SS, Lele K, Wolgemuth DJ. Apoptosis in male germ cells in response to cyclin A1-deficiency and cell cycle arrest. Biochem Pharmacol. 2003;66:1571–1579. doi: 10.1016/s0006-2952(03)00513-6. [DOI] [PubMed] [Google Scholar]
- 6.Toppari J, Suominenf JS, Yan W. The role of retinoblastoma protein family in the control of germ cell proliferation, differentiation and survival. APMIS. 2003;111:245–251. doi: 10.1034/j.1600-0463.2003.11101281.x. [DOI] [PubMed] [Google Scholar]
- 7.Eddy EM, O’Brien DA. Gene expression during mammalian meiosis. Curr Top Dev Biol. 1998;37:141–200. [PubMed] [Google Scholar]
- 8.Eddy EM. Male germ cell gene expression. Recent Prog Horm Res. 2002;57:103–128. doi: 10.1210/rp.57.1.103. [DOI] [PubMed] [Google Scholar]
- 9.Sun QY, Breitbart H, Schatten H. Role of the MAPK cascade in mammalian germ cells. Reprod Fertil Dev. 1999;11:443–450. doi: 10.1071/rd00014. [DOI] [PubMed] [Google Scholar]
- 10.Hecht NB. Molecular mechanisms of male germ cell differentiation. Bioessays. 1998;20:555–561. doi: 10.1002/(SICI)1521-1878(199807)20:7<555::AID-BIES6>3.0.CO;2-J. [DOI] [PubMed] [Google Scholar]
- 11.Brinster RL. Germline stem cell transplantation and transgenesis. Science. 2002;296:2174–2176. doi: 10.1126/science.1071607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.De Rooij DG, Grootegoed JA. Spermatogonial stem cells. Curr Opin Cell Biol. 1998;10:694–701. doi: 10.1016/s0955-0674(98)80109-9. [DOI] [PubMed] [Google Scholar]
- 13.van Pelt AM, Van Dissel-Emiliani FM, Gaemers IC, van der Burg MJ, Tanke HJ, De Rooij DG. Characteristics of A spermatogonia and preleptotene spermatocytes in the vitamin A-deficient rat testis. Biol Reprod. 1995;53:570–578. doi: 10.1095/biolreprod53.3.570. [DOI] [PubMed] [Google Scholar]
- 14.van Pelt AM, Morena AR, Van Dissel-Emiliani FM, Boitani C, Gaemers IC, De Rooij DG, Stefanini M. Isolation of the synchronized A spermatogonia from adult vitamin A-deficient rat testes. Biol Reprod. 1996;55:439–444. doi: 10.1095/biolreprod55.2.439. [DOI] [PubMed] [Google Scholar]
- 15.Izadyar F, Creemers LB, Dissel-Emiliani FM, van Pelt AM, De Rooij DG. Spermatogonial stem cell transplantation. Mol Cell Endocrinol. 2000;169:21–26. doi: 10.1016/s0303-7207(00)00346-4. [DOI] [PubMed] [Google Scholar]
- 16.Meachem S, von Schoenfeldt V, Schlatt S. Spermatogonia: stem cells with a great perspective. Reproduction. 2001;121:825–834. doi: 10.1530/rep.0.1210825. [DOI] [PubMed] [Google Scholar]
- 17.Schlatt S. Spermatogonial stem cell preservation and transplantation. Mol Cell Endocrinol. 2002;187:107–111. doi: 10.1016/s0303-7207(01)00706-7. [DOI] [PubMed] [Google Scholar]
- 18.Van Aelst L, D’Souza-Schorey C. Rho GTPases and signaling networks. Genes Dev. 1997;11:2295–2322. doi: 10.1101/gad.11.18.2295. [DOI] [PubMed] [Google Scholar]
- 19.Fujita A, Nakamura K, Kato T, Watanabe N, Ishizaki T, Kimura K, Mizoguchi A, Narumiya S. Ropporin, a sperm-specific binding protein of rhophilin, that is localized in the fibrous sheath of sperm flagella. J Cell Sci. 2000;113:103–112. doi: 10.1242/jcs.113.1.103. [DOI] [PubMed] [Google Scholar]
- 20.Nakamura K, Fujita A, Murata T, Watanabe G, Mori C, Fujita J, Watanabe N, Ishizaki T, Yoshida O. Rhophilin, a small GTPase Rho-binding protein, is abundantly expressed in the mouse testis and localized in the principal piece of the sperm tail. FEBS Lett. 1999;445:9–13. doi: 10.1016/s0014-5793(99)00087-3. [DOI] [PubMed] [Google Scholar]
- 21.Iida H, Kaneko T, Tanaka S, Mori T. Association of the developing acrosome with multiple small Golgi units, the Golgi satellites, in spermatids of the musk shrew, Suncus murinus. J Reprod Fertil. 2000;119:49–58. doi: 10.1530/jrf.0.1190049. [DOI] [PubMed] [Google Scholar]
- 22.Bergeret E, Pignot-Paintrand I, Guichard A, Raymond K, Fauvarque MO, Cazemajor M, Griffin-Shea R. RotundRacGAP functions with Ras during spermatogenesis and retinal differentiation in Drosophila melanogaster. Mol Cell Biol. 2001;21:6280–6291. doi: 10.1128/MCB.21.18.6280-6291.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Toure A, Morin L, Pineau C, Becq F, Dorseuil O, Gacon G. Tat1, a novel sulfate transporter specifically expressed in human male germ cells and potentially linked to RhoGTPase signaling. J Biol Chem. 2001;276:20309–20315. doi: 10.1074/jbc.M011740200. [DOI] [PubMed] [Google Scholar]
- 24.Naud N, Toure A, Liu J, Pineau C, Morin L, Dorseuil O, Escalier D, Chardin P, Gacon G. Rho family GTPase Rnd2 interacts and colocalizes with MgcRacGAP in male germ cells. Biochem J. 2003;372:105–112. doi: 10.1042/BJ20021652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Liang P, Zhu W, Zhang X, Guo Z, O’Connell RP, Averboukh L, Wang F, Pardee AB. Differential display using one-base anchored oligo-dT primers. Nucleic Acids Res. 1994;22:5763–5764. doi: 10.1093/nar/22.25.5763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Liang P, Pardee AB. Differential display of eukaryotic messenger RNA by means of the polymerase chain reaction. Science. 1992;257:967–971. doi: 10.1126/science.1354393. [DOI] [PubMed] [Google Scholar]
- 27.Van den Ham R, Dissel-Emiliani FM, van Peltt AM. Identification of candidate genes involved in gonocyte development. J Androl. 2002;23:410–418. [PubMed] [Google Scholar]
- 28.Shao X, Tarnasky HA, Schalles U, Oko R, van der Hoorn FA. Interactional cloning of the 84-kDa major outer dense fiber protein Odf84. Leucine zippers mediate associations of Odf84 and Odf27. J Biol Chem. 1997;272:6105–6113. doi: 10.1074/jbc.272.10.6105. [DOI] [PubMed] [Google Scholar]
- 29.Modarressi MH, Cameron J, Taylor KE, Wolfe J. Identification and characterization of a novel gene, TSGA10, expressed in testis. Gene. 2001;262:249–255. doi: 10.1016/s0378-1119(00)00519-9. [DOI] [PubMed] [Google Scholar]
- 30.He TC, Zhou S, da Costa LT, Yu J, Kinzler KW, Vogelstein B. A simplified system for generating recombinant adenoviruses. Proc Natl Acad Sci U S A. 1998;95:2509–2514. doi: 10.1073/pnas.95.5.2509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Higgy NA, Zackson SL, van der Hoorn FA. Cell interactions in testis development: overexpression of c-mos in spermatocytes leads to increased germ cell proliferation. Dev Genet. 1995;16:190–200. doi: 10.1002/dvg.1020160211. [DOI] [PubMed] [Google Scholar]
- 32.Oosterhuis JH, van der Hoorn FA. Testis-specific TTF-D binds to single-stranded DNA in the c-mos and Odf1 promoters and activates Odf1. J Biol Chem. 1999;274:11708–11712. doi: 10.1074/jbc.274.17.11708. [DOI] [PubMed] [Google Scholar]
- 33.Oko R, Maravei D. Protein composition of the perinuclear theca of bull spermatozoa. Biol Reprod. 1994;50:1000–1014. doi: 10.1095/biolreprod50.5.1000. [DOI] [PubMed] [Google Scholar]
- 34.Shao X, Tarnasky HA, Lee JP, Oko R, van der Hoorn FA. Spag4, a novel sperm protein, binds outer dense-fiber protein Odf1 and localizes to microtubules of manchette and axoneme. Dev Biol. 1999;211:109–123. doi: 10.1006/dbio.1999.9297. [DOI] [PubMed] [Google Scholar]
- 35.Lamarche-Vane N, Hall A. CdGAP, a novel proline-rich GTPase-activating protein for Cdc42 and Rac. J Biol Chem. 1998;273:29172–29177. doi: 10.1074/jbc.273.44.29172. [DOI] [PubMed] [Google Scholar]
- 36.Herault O, Colombat P, Domenech J, Degenne M, Bremond JL, Sensebe L, Bernard MC, Binet C. A rapid single-laser flow cytometric method for discrimination of early apoptotic cells in a heterogeneous cell population. Br J Haematol. 1999;104:530–537. doi: 10.1046/j.1365-2141.1999.01203.x. [DOI] [PubMed] [Google Scholar]
- 37.Delmas V, van der Hoorn F, Mellstrom B, Jegou B, Sassone-Corsi P. Induction of CREM activator proteins in spermatids: downstream targets and implications for haploid germ cell differentiation. Mol Endocrinol. 1993;7:1502–1514. doi: 10.1210/mend.7.11.8114765. [DOI] [PubMed] [Google Scholar]
- 38.Modarressi MH, Behnam B, Cheng M, Taylor KE, Wolfe J, van der Hoorn FA. Tsga10 encodes a 65-kDa protein that is processed to the 27-kDa fibrous sheath protein. Biol Reprod. 2004;70:608–615. doi: 10.1095/biolreprod.103.021170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Marshall C. How do small GTPase signal transduction pathways regulate cell cycle entry? Curr Opin Cell Biol. 1999;11:732–736. doi: 10.1016/s0955-0674(99)00044-7. [DOI] [PubMed] [Google Scholar]
- 40.Quimby BB, Dasso M. The small GTPase Ran: interpreting the signs. Curr Opin Cell Biol. 2003;15:338–344. doi: 10.1016/s0955-0674(03)00046-2. [DOI] [PubMed] [Google Scholar]
- 41.Wittmann T, Waterman-Storer CM. Cell motility: can Rho GTPases and microtubules point the way? J Cell Sci. 2001;114:3795–3803. doi: 10.1242/jcs.114.21.3795. [DOI] [PubMed] [Google Scholar]
- 42.Moon SY, Zheng Y. Rho GTPase-activating proteins in cell regulation. Trends Cell Biol. 2003;13:13–22. doi: 10.1016/s0962-8924(02)00004-1. [DOI] [PubMed] [Google Scholar]
- 43.Peck J, Douglas G, Wu CH, Burbelo PD. Human RhoGAP domain-containing proteins: structure, function and evolutionary relationships. FEBS Lett. 2002;528:27–34. doi: 10.1016/s0014-5793(02)03331-8. [DOI] [PubMed] [Google Scholar]
- 44.Fidyk NJ, Cerione RA. Understanding the catalytic mechanism of GTPase-activating proteins: demonstration of the importance of switch domain stabilization in the stimulation of GTP hydrolysis. Biochemistry. 2002;41:15644–15653. doi: 10.1021/bi026413p. [DOI] [PubMed] [Google Scholar]
- 45.Via A, Ferre F, Brannetti B, Valencia A, Helmer-Citterich M. Three-dimensional view of the surface motif associated with the P-loop structure: cis and trans cases of convergent evolution. J Mol Biol. 2000;303:455–465. doi: 10.1006/jmbi.2000.4151. [DOI] [PubMed] [Google Scholar]
- 46.Resat H, Straatsma TP, Dixon DA, Miller JH. The arginine finger of RasGAP helps Gln-61 align the nucleophilic water in GAP-stimulated hydrolysis of GTP. Proc Natl Acad Sci U S A. 2001;98:6033–6038. doi: 10.1073/pnas.091506998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Haberland J, Gerke V. Conserved charged residues in the leucine-rich repeat domain of the Ran GTPase activating protein are required for Ran binding and GTPase activation. Biochem J. 1999;343:653–662. [PMC free article] [PubMed] [Google Scholar]
- 48.Jenna S, Lamarche-Vane N. The RhoGAP superfamily of proteins. In: Symons Marc., editor. Rho GTPases. New York: Kluwer Academic Press/Plenum Press; 2003. pp. 68–87. [Google Scholar]
- 49.Seewald MJ, Korner C, Wittinghofer A, Vetter IR. RanGAP mediates GTP hydrolysis without an arginine finger. Nature. 2002;415:662–666. doi: 10.1038/415662a. [DOI] [PubMed] [Google Scholar]
- 50.Inselman A, Eaker S, Handel MA. Temporal expression of cell cycle-related proteins during spermatogenesis: establishing a timeline for onset of the meiotic divisions. Cytogenet Genome Res. 2003;103:277–284. doi: 10.1159/000076813. [DOI] [PubMed] [Google Scholar]
- 51.Sutou S, Miwa K, Matsuura T, Kawasaki Y, Ohinata Y, Mitsui Y. Native tesmin is a 60-kDa protein that undergoes dynamic changes in its localization during spermatogenesis in mice. Biol Reprod. 2003;68:1861–1869. doi: 10.1095/biolreprod.102.005603. [DOI] [PubMed] [Google Scholar]
- 52.Sudhakar L, Rao MR. Stage-dependent changes in localization of a germ cell-specific lamin during mammalian spermatogenesis. J Biol Chem. 1990;265:22526–22532. [PubMed] [Google Scholar]
- 53.Kierszenbaum AL, Gil M, Rivkin E, Tres LL. Ran, a GTP-binding protein involved in nucleocytoplasmic transport and microtubule nucleation, relocates from the manchette to the centrosome region during rat spermiogenesis. Mol Reprod Dev. 2002;63:131–140. doi: 10.1002/mrd.10164. [DOI] [PubMed] [Google Scholar]
- 54.Print CG, Loveland KL. Germ cell suicide: new insights into apoptosis during spermatogenesis. Bioessays. 2000;22:423–430. doi: 10.1002/(SICI)1521-1878(200005)22:5<423::AID-BIES4>3.0.CO;2-0. [DOI] [PubMed] [Google Scholar]
- 55.Allan DJ, Harmon BV, Roberts SA. Spermatogonial apoptosis has three morphologically recognizable phases and shows no circadian rhythm during normal spermatogenesis in the rat. Cell Prolif. 1992;25:241–250. doi: 10.1111/j.1365-2184.1992.tb01399.x. [DOI] [PubMed] [Google Scholar]
- 56.Martincic DS, Klun IV, Zorn B, Vrtovec HM. Germ cell apoptosis in the human testis. Pflugers Arch. 2001;442:R159–R160. doi: 10.1007/s004240100007. [DOI] [PubMed] [Google Scholar]
- 57.Wang Z, Tseng CP, Pong RC, Chen H, McConnell JD, Navone N, Hsieh JT. The mechanism of growth-inhibitory effect of DOC-2/DAB2 in prostate cancer. Characterization of a novel GTPase-activating protein associated with N-terminal domain of DOC-2/DAB2. J Biol Chem. 2002;277:12622–12631. doi: 10.1074/jbc.M110568200. [DOI] [PubMed] [Google Scholar]
- 58.Wolf RM, Draghi N, Liang X, Dai C, Uhrbom L, Eklof C, Westermark B, Holland EC, Resh MD. p190RhoGAP can act to inhibit PDGF-induced gliomas in mice: a putative tumor suppressor encoded on human chromosome 19q13.3. Genes Dev. 2003;17:476–487. doi: 10.1101/gad.1040003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Menna PL, Skilton G, Leskow FC, Alonso DF, Gomez DE, Kazanietz MG. Inhibition of aggressiveness of metastatic mouse mammary carcinoma cells by the beta2-chimaerin GAP domain. Cancer Res. 2003;63:2284–2291. [PubMed] [Google Scholar]
- 60.Lopez-Casas PP, Lopez-Fernandez LA, Parraga M, Krimer DB, del Mazo J. Developmental regulation of expression of Ran/M1 and Ran/M2 isoforms of Ran-GTPase in mouse testis. Int J Dev Biol. 2003;47:307–310. [PubMed] [Google Scholar]
- 61.Anway MD, Li Y, Ravindranath N, Dym M, Griswold MD. Expression of testicular germ cell genes identified by differential display analysis. J Androl. 2003;24:173–184. doi: 10.1002/j.1939-4640.2003.tb02660.x. [DOI] [PubMed] [Google Scholar]
- 62.Wang LF, Zhu HD, Miao SY, Cao DF, Wu YW, Zong SD, Koide SS. Molecular cloning and characterization of a novel testis-specific nucleoporin-related gene. Arch Androl. 1999;42:71–84. doi: 10.1080/014850199262904. [DOI] [PubMed] [Google Scholar]
- 63.Kusano A, Staber C, Ganetzky B. Nuclear mislocalization of enzymatically active RanGAP causes segregation distortion in Drosophila. Dev Cell. 2001;1:351–361. doi: 10.1016/s1534-5807(01)00042-9. [DOI] [PubMed] [Google Scholar]
- 64.Kusano A, Staber C, Chan HY, Ganetzky B. Closing the (Ran)GAP on segregation distortion in Drosophila. Bioessays. 2003;25:108–115. doi: 10.1002/bies.10222. [DOI] [PubMed] [Google Scholar]
- 65.Petrie RG, Jr, Morales CR. Receptor-mediated endocytosis of testicular transferrin by germinal cells of the rat testis. Cell Tissue Res. 1992;267:45–55. doi: 10.1007/BF00318690. [DOI] [PubMed] [Google Scholar]