Abstract
Dimerization is a critical requirement for the activation of the intracellular kinase domains of receptor tyrosine kinases (RTKs). The single transmembrane (TM) helices of RTKs contribute to dimerization, but the details are not well understood. Work with TM helices in various model systems has revealed a small number of specific dimerization sequence motifs, and it has been suggested that RTK dimerization is modulated by such motifs. Yet questions remain about the universality of these sequence motifs for RTK dimerization, and about how TM domain dimerization in model systems relates to RTK activation in mammalian membranes. To investigate these questions, we designed a 3,888 member combinatorial peptide library based on the TM domain of Neu (ErbB2) as a model RTK. The library contains many closely related, Neu-like sequences, including thousands of sequences with known dimerization motifs. We used an SDS-PAGE-based screen to select peptides that dimerize better than the native Neu sequence, and we assayed the activation of chimeric Neu receptors in mammalian cells with TM sequences selected in the screen. Despite the very high abundance of known dimerization motifs in the library, only a very few dimerizing sequences were identified by SDS-PAGE. About half of those sequences activated the Neu kinase significantly more than the wild-type TM sequence, but none of them activated the kinase less than the wild-type sequence. This work furthers our knowledge about the requirements for membrane protein interactions, and the requirements for RTK activation in cells.
Introduction
Lateral dimerization of transmembrane (TM) α-helical domains plays an important role in receptor tyrosine kinase (RTK)-mediated signal transduction1–3. Since RTK dimers are active while monomers are inactive, the dimerization process controls RTK activity. The TM domains are known to contribute as much as −3 kcal/mol to the dimerization energetics4–7, and defects in dimerization due to single residue mutations in RTK TM domains are known to cause human pathologies, including cancer and dwarfism. TM helix dimerization is believed to be driven by particular sequence motifs8–10. Experiments have successfully identified a few dimerization motifs for TM helix dimers using a variety of model systems, including SDS-PAGE gels, detergent micelles and bicelles, lipid vesicles, bacterial membranes and mammalian membranes (reviewed in8). Yet, despite these successes, we currently do not know the interaction motifs for most RTK TM domains, and more importantly we cannot predict if a particular TM sequence will form dimers in membranes. Even when dimerization has been demonstrated experimentally, a prediction for the TM dimer interaction interface cannot be made with certainty. Finally, we cannot yet design dimerization motifs de novo, underscoring the importance of the work that is yet to be done in this field. The successful de novo design of TM sequences that interact strongly with RTK TM domains would be an important achievement, as it will allow for the development of novel RTK inhibitors that could be used in the clinic.
Information on TM helix dimerization has been derived from synthetic peptides, peptide-protein chimeras and full-length membrane proteins. Numerous methods exist to measure dimerization and such measurements have been made in many different hydrophobic “membrane mimetic” environments11. In detergent micelles, TM helix dimerization has been measured using FRET, analytical ultracentrifugation, and cysteine cross-linking12–14. In lipid bilayers, TM helix dimerization has been characterized using FRET, cysteine cross-linking, and recently, an elegant novel steric trap approach15–17. In bacterial membranes, TM helix dimerization has been studied using genetic reporter assays such as TOXCAT, ToxR and GALLEX18–20. Finally, interactions between TM helices have been probed in mammalian cell membranes using FRET-based assays5,21.
One of the earliest methods used to study TM helix dimerization, SDS-PAGE, is fast, simple, and is used in most laboratories. SDS-PAGE is often the method of choice for initial characterization of TM helix interactions22,23 and has been used to define the most well understood dimerization motif: the GxxxG motif that drives the dimerization of the TM domain of glycophorin A (GpA)24. While often reliable for assaying TM helix dimerization, SDS-PAGE has been shown sometimes to yield misleading results25,26. Thus, the extent of SDS validity is an open question. Here we investigate if SDS-PAGE can be used as a simple high-throughput screening method to identify strongly interacting TM sequences that can activate an RTK better than its own TM domain in mammalian membranes. Furthermore, in this work we address the following questions. (1) How potent and how promiscuous are the known dimerization motifs, as detected by SDS-PAGE? (2) What is the correlation between RTK TM domain dimerization in SDS-PAGE and RTK activation in mammalian membranes? (3) Do small changes in TM domain sequences affect RTK activation? (4) What is the importance of the GxxxG dimerization motif in RTK activation? and (5) How much can RTK activation be varied by changing the TM domain sequence?
The model RTK that we use for this work is rat Neu. Neu (ErbB2 or HER2) belongs to the Epidermal Growth Factor (EGF) receptor family and is unique because it has no ligand. Thus, its activation is regulated exclusively by lateral dimerization. A single residue mutation in the Neu TM domain, V664E, is oncogenic due to increased dimerization and increased activation compared with wild-type27–29. It has been proposed that the effect results from the formation of Glu-mediated hydrogen bonds between the two helices30,31. While this mutation has not been identified in humans thus far, the mutation is activating when engineered into the human sequence.
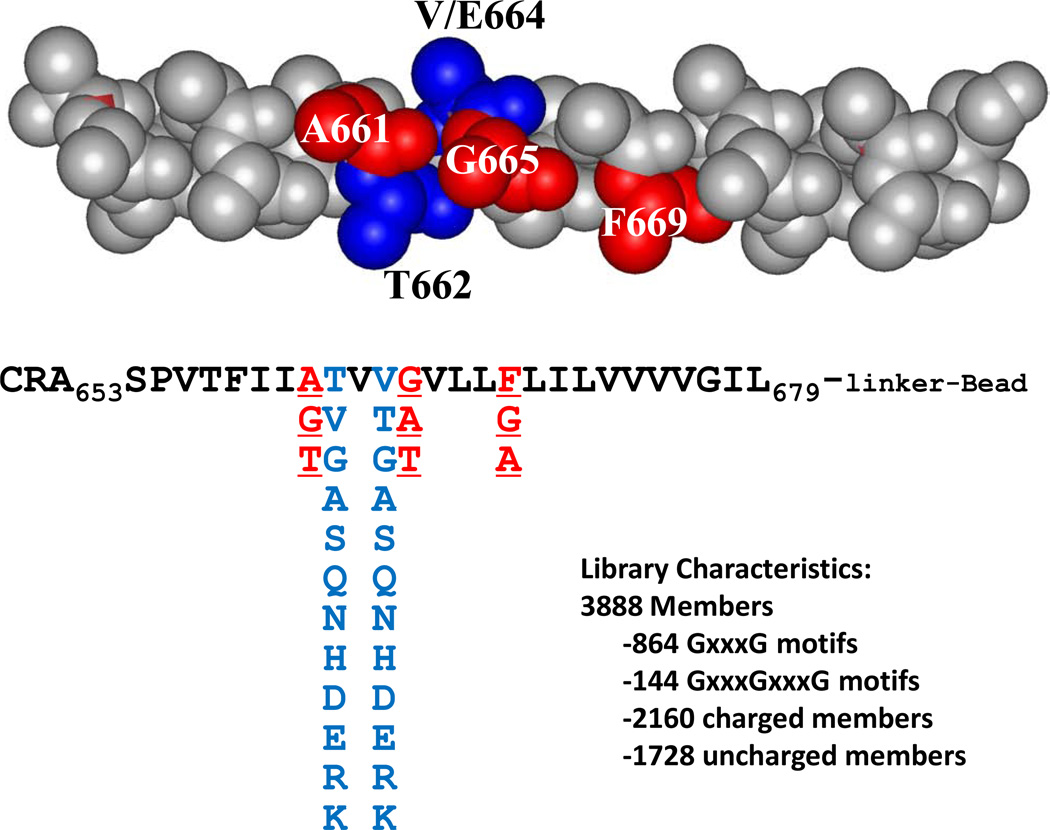
In this work we used the wild-type Neu TM domian, Neu/WT, which is monomeric in SDS-PAGE, and Neu/V664E TM domain, which is dimeric in SDS-PAGE, as the standards to which we compare the dimerization and activation driven by novel Neu TM analog sequences. We designed a peptide library based on the Neu TM domain sequence in which we varied five residues that are likely involved in the interaction interface (Figure 1). The 3,888-member library contained several thousand members with at least one known dimerization motif, including the GxxxG motif identified to be important for GpA dimerization. The library included other motifs of two small residues spaced by three residues, also believed to drive TM helix dimerization and referred to as SmxxxSm motifs (where Sm is a small amino-acid such as Ala, Ser or Thr, in addition to Gly)32. The library also included polar and charged residues, believed to stabilize TM helix dimers via hydrogen bonds or salt-bridges8,33,34.
Figure 1. Rational combinatorial library design.
The context of the combinatorial library is the wild-type residues of Neu TM domain. Here the Neu TM domain sequence is represented as an ideal polyalanine α-helix, with the five varied positions shown in color. The potential SmxxxSmxxxSm dimerization motif around residue V664 is shown in red, and the possible residues in the library in those positions are shown below. The residues in positions equivalent to 662 and 664 (in blue) include hydrophobic, polar anionic and cationic residues. The total library contains 3,888 members.
We used SDS-PAGE-based screening to identify sequences within the library that homodimerize more strongly than Neu/WT. A very small number of homodimeric sequences were identified in the SDS-PAGE-based screen, demonstrating that the presence of known dimerization motifs do not guarantee strong interactions. Next, we hypothesized that, if the SDS-PAGE environment is relevant to interactions in mammalian membranes, the selected homodimeric sequences would enhance Neu activation in mammalian membranes. Therefore we measured the activation of chimeric Neu receptors containing these selected TM domain sequences. Some, but not all, of the selected TM domains increased Neu activation in mammalian cell membranes. These results provide answers for the five questions posed above and suggest that SDS-PAGE may be a useful, but not ideal, high-throughput tool for identifying sequences that alter the biological function of RTKs.
Results
Library design
The library used in this work was based on the rat Neu (ErbB2) TM domain sequence. The three dimensional structure for the human ErbB2 TM domain dimer in bicelles was solved using NMR35. The contacts between the two helices occur via the sequence motif Thr652xxxSer656xxxGly660. In the rat sequence, the corresponding motif would be Thr657xxxAla661xxxGly665. While the structure of the rat variant is not available, we hypothesized that a similar combination of SmxxxSm motifs is important for dimerization. The combinatorial peptide library was designed to explore the proposed interaction interface in the Neu wild-type homodimer around the critical Val664 residue which is oncogenic when mutated to glutamate. In the library we fixed all amino acids as the native residues of Neu, including Thr657, except for the five residues around residue Val664, as highlighted in Figure 1. The highlighted residues were varied combinatorially as shown in the figure, and the native residue was always included. Thus the library contained both Neu/WT and Neu/V664E TM domains as members.
Note that a third contiguous SmxxxSm motif is prevented in the native rat sequence by the phenylalanine in position 669. Thus position 669 was also chosen as a combinatorial site. We thus allowed all three of positions 661, 665, and 669 to have Gly or Ala. We also added Thr to positions 661 and 665 and included the native Phe at residue 669 in the library. Thus, most library members have at least two SmxxxSm motifs and some peptides had a contiguous SmxxxSmxxxSmxxxSm motif covering almost four turns of the helix. Many of the small amino acids were glycines. In particular, the 3888-member library also contained over 800 different sequences with at least one GxxxG motif, and 144 sequences with two such motifs arranged contiguously (Gly661xxxGly665xxxGly669).
We also varied the amino acids in positions 662 and 664. These residues are expected to be adjacent to the helical interface and may interact through the side chains reaching across and contacting the neighboring helices36. In the case of residues 662 and 664 we wanted to explore the potential for polar residues to drive homodimerization as the glutamate does in the Val664Glu mutant. Thus in the 662 and 664 positions, the library contained 12 different amino acids, including all of the ionizable amino acids (D,E,H,K and R), several polar amino acids (S,T,N and Q), and several aliphatic amino acids (G,A,V). As a result, about 55% of the library members had at least one ionizable residue in either position 662 or in 664 and almost all library members had one or more polar residues. The library also contained the native valines in positions 662 and 664.
Library screening by SDS-PAGE
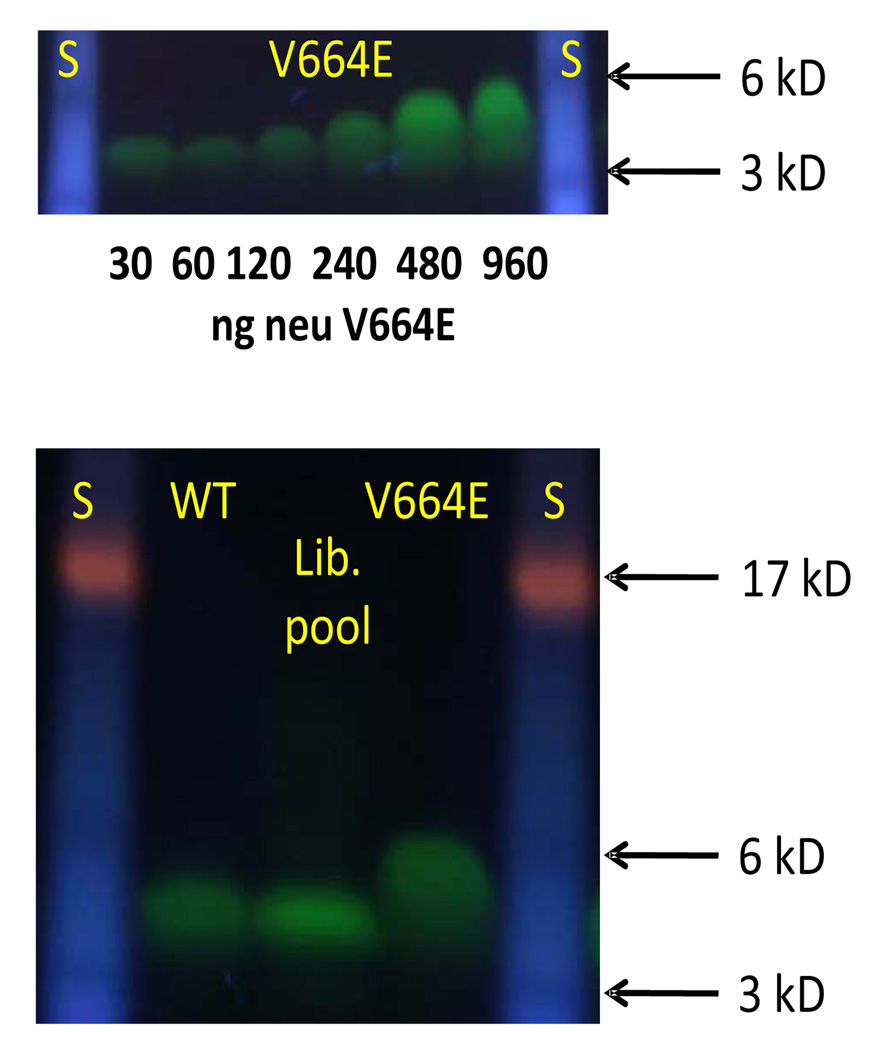
We produced the TM domains of Neu/WT and Neu/V664E using solid phase synthesis with a cysteine at the N-terminus, which was used to label them with Alexafluor488. We first ran the Neu/V664E peptide on an SDS-PAGE gel, while varying the peptide concentration. As we show in Figure 2A, the peptide migrates differently at different concentrations. It exhibits monomer-like mobility at low concentrations (30 ng or 1 µM), migrating at about 3kDa. At about 150 ng (5 µM) the migration of the peptide starts to deviate from pure monomer-like behavior. Dimer-like mobility is observed at high concentrations (≥500 ng/17 µM). This observation is consistent with the law of mass action and suggests that strong dimerization of the mutant takes place in the SDS/polyacrylamide gel environment. In contrast, the wild-type peptide always migrates as a monomer. This is demonstrated in Figure 2B in which we show an SDS-PAGE experiment with 500ng (17 µM) of Neu/WT and Neu/V664E TM domains. Even at this high concentration, the wild-type peptide is monomeric. A pool of ~50 library members were also run on the same gel and had monomer-like mobility.
Figure 2. Behavior of Neu peptides in SDS-PAGE.
(A) The migration of Neu/V664E was examined as a function of peptide concentration from 30 to 960 ng of total peptide, or 1 µM to 32 µM, in each lane in the 10µl of loading solution applied to the gel initially. Molecular weight standards are shown on the left and right. (B) Three samples were examined in this experiment: (i) a pool of library peptides, (ii) purified Neu/WT peptide and (iii) purified Neu/V664E peptide. All peptides were labeled on a C-terminal cysteine with Alexafluor-488. Each lane contains 500 ng of total peptide in 10 µl of loading buffer (17 µM). On the left and right are molecular weight standards with the position of the 3000, 6000 and 17,000 Da standards highlighted.
Thus, to identify peptides in the combinatorial library with strong homo-dimerization propensity, we used the Neu/V664E peptide (with loadings of ≥150 ng/5 µM or higher) as a positive control in an SDS-PAGE “high-throughput” screen. We expected that a strong homodimer will run as a distinct dimer band on the gel, similarly to the mutant Neu/V664E, while a peptide with dimerization tendency between that of the mutant and the wild-type will show a smeared band between the monomer and dimer regions, as the monomer and the dimer are in equilibrium on the timescale of the gel experiment.
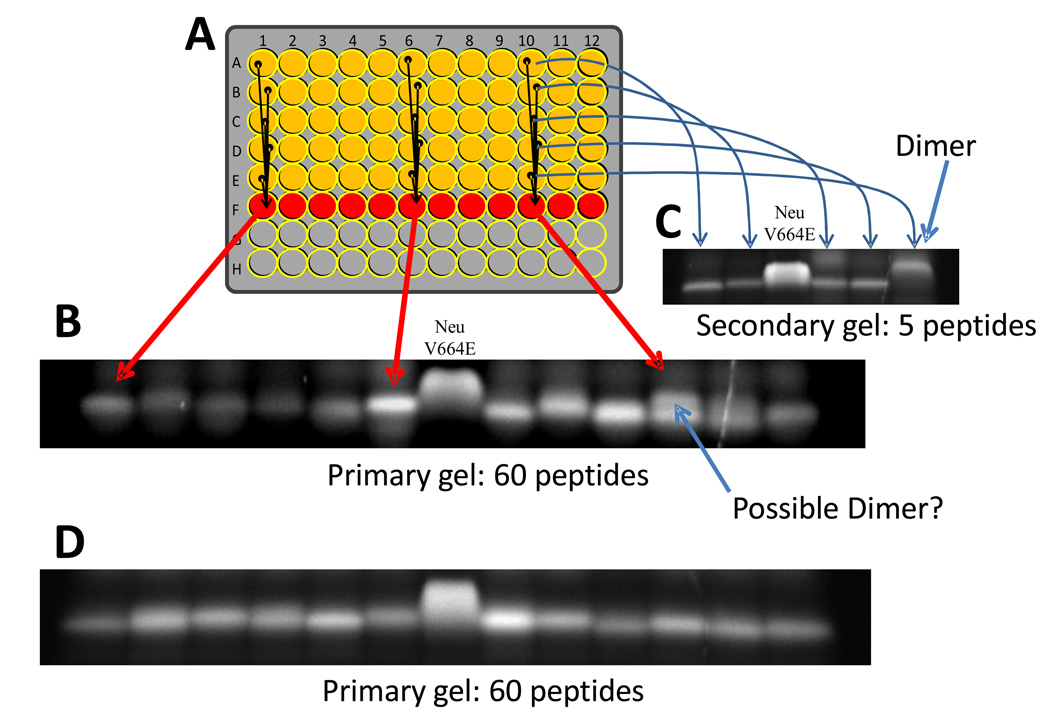
As described in Materials and Methods, in the SDS-PAGE screen each lane contained five randomly selected library members. The majority of members ran as monomers. If a strong homodimer was present in a lane, we saw two bands: a faster migrating band containing four monomeric peptides and a slower migrating dimer band containing one dimeric peptide, with about 20% of the total intensity. Also consistent with our expectations, the presence of a weaker dimer induced band smearing. When a lane with a putative dimer band was identified, we ran a second gel in which each lane contained the remaining half of one of the five peptides that were mixed in the original gel (Figure 3). Again, Neu/V664E TM domain was run as control, to which the migration of the five individual peptides was compared, such that the peptide with homodimerization tendency higher than Neu/WT can be identified. Example images collected during the screen are shown in Figure 3.
Figure 3. SDS-PAGE high throughput screen.
Example gels from the SDS-PAGE based high throughput screen. A: As described in the text, a 12x5 section of a 96-well plate contains one library member per well in SDS gel loading buffer. In the sixth row, a portion of each well above is added to give a mixture of five peptides. B: Sixty peptides are analyzed on a single gel in 12 lanes, with 5 peptides per lane. About 1 µg of Neu/V664E is analyzed in the 8th lane. Most library members migrate as monomers. C: When a putative dimer is identified (e.g. lane 11 in panel B), a second gel is run with the remainder of the five single peptide solutions to identify the dimeric peptide. In this secondary gel, a sample of Neu/V664E is included in lane 3. D: Most gels analyzed contained no dimers, as shown by this typical example.
Using this gel-based high-throughput screen we analyzed 6000 members randomly selected from the library. We thus achieved 80% library coverage overall (calculated using the Poisson equation), i.e. about 80% of the library members were screened at least once (33% exactly once, 25% twice, 13% three times etc) and 20% of the library members were not screened at all. The vast majority of the 6000 random library members screened migrated as homogeneous monomeric bands (see Figure 2B and Figure 3). Dimeric peptides were very rare in the screen, which is surprising when one considers the abundance of GxxxG and SmxxxSm dimerization motifs, as well as the abundance of polar residues in the library (Fig. 1). Overall we observed only 10 putative positive bands in the 100 gels (1200 lanes, 6000 peptides). Seven of these ten peptides showed dimeric bands when the individual peptides were analyzed on a second gel. There were three false positives, most likely heterodimers in the mixtures. The seven putative homodimers were sequenced. The sequences of the positive dimeric peptides, Neu-Pos1 through Neu-Pos6, are given in Table 1 and in Figure 4. Importantly, one sequence (Neu-Pos4: AVVD664AVLLG) was identified twice, independently. Overall, the positives did not have a highly conserved sequence motif. Furthermore, despite the presence of many sequences with polar or charged residues at two different positions in the library (positions 662 and 664), the only polar residue observed in either position in the six positive sequences was the aspartate in position 664 of the Neu-Pos4 peptide. This is the same position where the mutant Neu/V664E sequence has a glutamate. However, three of the four other varied residues in Neu-Pos4 were different from those in Neu/V664E. The other positive sequences were mostly non-polar with Val, Ala, Thr or Gly in positions 662 and 664. The sequences of Neu/WT and Neu/V664E TM domain were not among the selected positives. The non-selection of Neu/WT is not surprising as it is monomeric under the screening conditions. The non-selection of Neu/V664E is probably due to it being missed by chance because of the 80% library coverage in the screened peptides.
Table 1.
Relative activation of Neu chimeras containing the indicated transmembrane domains. Activation was measured in triplicate, as described in the text, and normalized to the activation of the mutant Neu/V664E receptor. Uncertainties are standard errors and N=3 for all measurements.
| Receptor | Sequence | Activation/NeuV664E Mean +/− SE (N=3) |
|---|---|---|
| Neu/V664E | 1 | |
| Neu-Pos4 | 0.860 +/− 0.023 | |
| Neu-Pos2 | 0.779 +/− 0.015 | |
| Neu-Pos1 | 0.681+/− 0.011 | |
| Neu-Ran2 | 0.540 +/− 0.054 | |
| Neu-Ran1 | 0.454 +/− 0.060 | |
| Neu-Pos3 | 0.361 +/− 0.040 | |
| Neu-Pos6 | 0.345 +/− 0.041 | |
| Neu-Pos5 | 0.301 +/− 0.058 | |
| Neu WT | 0.281 +/− 0.051 |
Figure 4. Sequences of Neu-related peptides.
Sequences of Neu/WT and Neu/V664E are shown at the top. Six dimeric sequences (Neu-Pos1 through Neu-Pos6) were identified in the screen. One of the seven sequences (Neu-Pos4) was identified twice independently. At the bottom are two random sequences from two beads out of the thousands of library members that were negative for dimerization in the screen (Neu-Ran1 and Neu-Ran2). In parenthesis is the statistical significance of the difference in activation in mammalian cells, relative to Neu/V664E, compared to the wild-type sequence, relative to Neu/V664E (ns=not significant, p> 0.05).
Activation of chimeric Neu-receptors with different transmembrane motifs
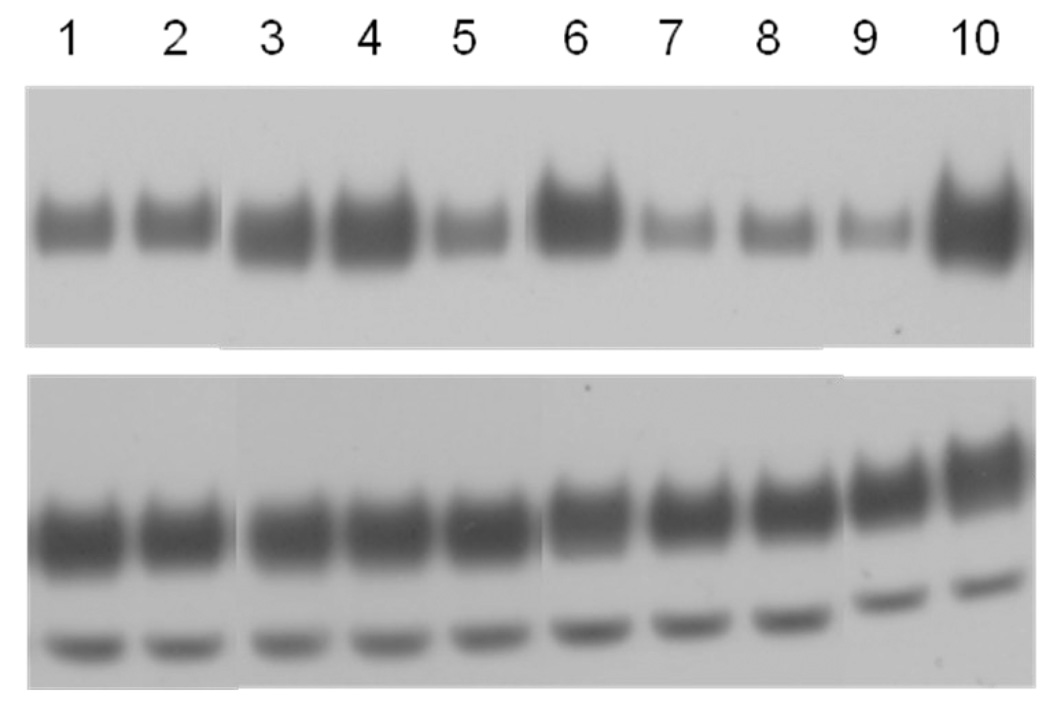
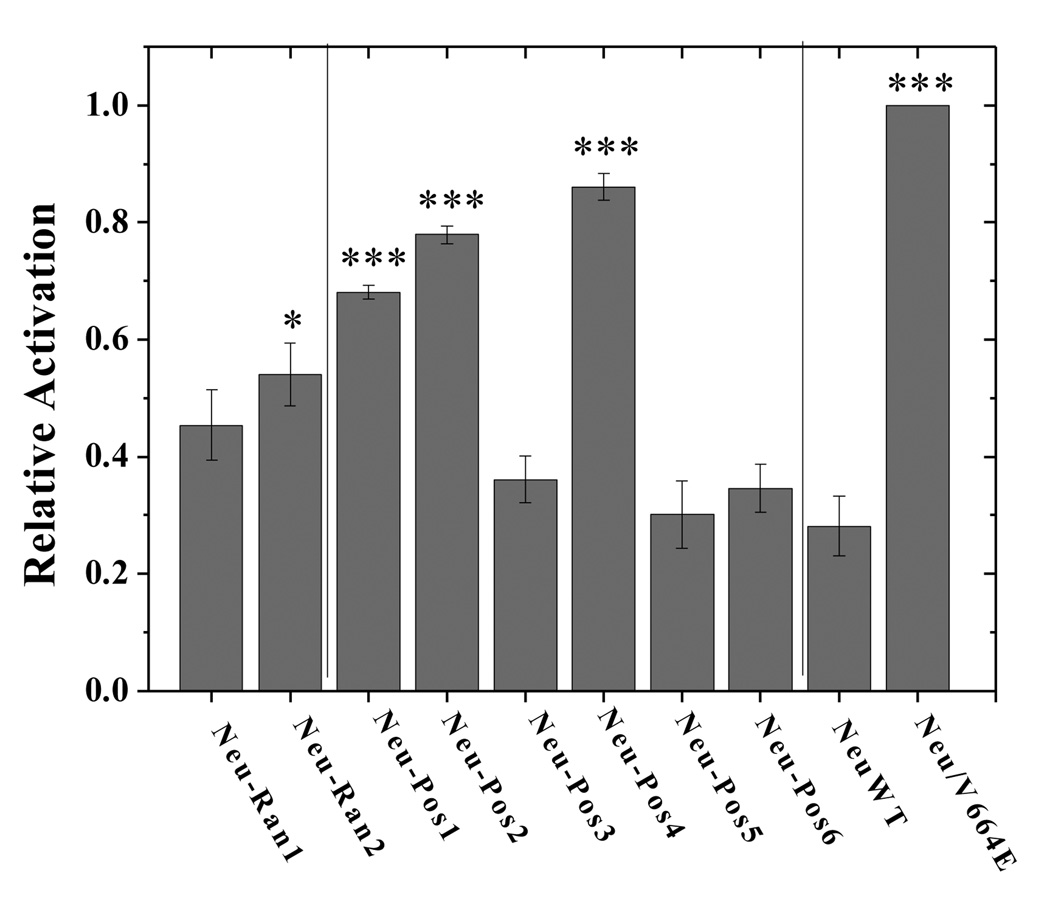
To investigate the Neu activation propensities of the selected transmembrane sequences in mammalian cell membranes, plasmids encoding Neu chimeras with the extracellular and intracellular domains of Neu, and different TM domains identified in the screen and shown in Figure 4 were constructed as described in Materials and Methods. Each of the eight constructed plasmids, together with the plasmids encoding Neu/WT and Neu/V664E were transfected into CHO cells using Fugene HD (Roche Applied Science, IN) following the manufacturer’s protocol. 24 hours after the transfection, the cells were lysed and the lysates were subjected to SDS-PAGE and Western blotting. The membranes were stained with anti-C-Neu antibodies to probe the expression levels of the receptors in the whole cell lysates. The phosphorylation of the receptors was probed using anti-p-ErbB2 antibodies (Cell Signaling Technology, MA) as previously described28. A representative film is shown in Figure 5A. The Western blot films were scanned and processed with ImageQuant TL. To quantify the relative activation levels of the six chimeric receptors, the measured intensities of the anti-p-ErbB2 bands were divided by the intensities of the anti-C-Neu bands. Three sets of independent experiments were performed to determine the averages and the standard deviations. The results are shown in Figure 5B and in Table 1.
Figure 5.
A. Western blot results. Western blots showing staining intensities of anti-Neu antibodies (reporting on Neu expression) and anti-P-ErbB2 antibodies (reporting on Neu activation) for Neu/WT, Neu/V664E and the eight chimeric Neu receptors. 1:Neu-Ran1; 2:Neu-Ran2; 3:Neu-Pos1; 4:Neu-Pos2; 5:Neu-Pos3; 6:Neu-Pos4; 7:Neu-Pos5; 8:Neu-Pos6; 9:Neu/WT; 10:Neu/V664E.
B. Quantification of Western blots. The Western blots, such as the one shown in Figure 5A, were quantified with ImageQuant TL. The activation level of each receptor is obtained from the ratio of the phosphorylated Neu staining intensity to the total Neu staining intensity. The activation levels were normalized to that of Neu/V664E. A one-factor ANOVA test was used to compare the relative activation levels measured for the various Neu chimeras, and the Tukey-Kramer test was used for pairwise comparisons. The chimeric receptors labeled with * have significantly higher activation than Neu/WT (p<0.05) and those labeled *** exhibit highly significant differences (p<0.001).
Since the V664E mutation has been shown to increase Neu activation in mammalian membranes28,37, the activation of Neu/V664E was the standard to which we normalized the measured activation/expression ratio. The wild type Neu activation level was about 30% of the Neu/V664E activation level. The activation of the chimeric Neu receptors varied between 30 and 85% of the Neu/V664E activation level (Table 1). We analyzed the relative activation of the chimeric receptors simultaneously by one-factor ANOVA, using a Tukey-Kramer analysis for pair-wise comparisons. The overall ANOVA p-value is less than 0.0001, indicating a very significant deviation from the null hypothesis that all activation levels are the same. In Table 2 we show the Tukey-Kramer pair-wise statistical significance for the sequences studied. In terms of activation, the sequences cluster into two groups. Three sequences (Neu-Pos4, Neu-Pos2 and Neu-Pos1 in red) are highly activating. Four sequences, (Neu-Pos3, Neu-Ran2, Neu-Pos5, Neu-Pos6 in blue) are not distinguishable from Neu/WT. One sequence (Neu-Ran1 in grey) is intermediate. The two randomly selected sequences are less activating than the three sequences that were identified as positive in the screen, although one of them (Neu-Ran2) is marginally higher in activation than the wild type Neu sequence.
Table 2.
Statistical cross-comparisons of activation. One-factor ANOVA was used to cross-compare all measured activation values (Table 1). The overall p-value was less than 0.0001. P-values for pair-wise comparisons were calculated using the Tukey-Kramer method.
| P4 | P2 | P1 | R2 | R1 | P3 | P6 | P5 | WT | |
|---|---|---|---|---|---|---|---|---|---|
| I | ns | ns | ** | *** | *** | *** | *** | *** | P4 |
| I | ns | * | ** | *** | *** | *** | *** | P2 | |
| I | ns | * | ** | *** | *** | *** | P1 | ||
| I | ns | ns | ns | * | * | R2 | |||
| I | ns | ns | ns | ns | R1 | ||||
| I | ns | ns | ns | P3 | |||||
| I | ns | ns | P6 | ||||||
| I | ns | P5 | |||||||
| I | WT |
NS: P>0.05;
0.01<p<0.05,
0.001<p<0.01;
p<0.001.
I indicates identity, or self-self comparison.
Discussion
Choice of SDS-PAGE as a screening method
Although SDS-PAGE analysis has been used for decades to assess the propensity of transmembrane helices to self associate, questions have been raised about its universality25,26. The best example of a system in which SDS-PAGE seems to perform well is the transmembrane domain of the erythrocyte protein glycophorin A, where dimerization propensities of native and mutant sequences in SDS-PAGE correlate well with the behavior in other systems, including bacterial membranes18,34,38. The widely recognized GxxxG dimerization motif was first identified in this way22 and later was expanded to include similar SmxxxSm dimerization motifs39. Similarly, the self-association of fibroblast growth factor receptor (FGFR) transmembrane domains in SDS gels agrees well with measurements made in synthetic bilayers and in cellular membranes40,41. Fusion peptides, derived from HIV type 1 glycoprotein 41 (gp41), form trimers on SDS PAGE, consistent with the trimeric form of gp41 in membranes42. In other systems, however, the correlation between SDS-PAGE and dimerization in membranes is less clear. For example, we have shown that a single asparagine residue in the middle of a hydrophobic helix can decrease peptide mobility in SDS gels without driving dimerization25.
Both Neu/WT and Neu/V664E TM domains have a propensity for dimerization in cellular membranes, as demonstrated via cross-linking in intact cells37. The cross-linking propensities of the TM domains, and the full-length Neu receptors increase due to the V664E mutation27,28,37. Furthermore, the human wild type Neu (ErbB2) TM sequence self-associates well enough in bicelles for a dimer structure to be determined35. In our SDS-PAGE experiments, only the Neu/V664E peptide is dimeric, while Neu/WT is monomeric (Fig. 2B). The fact that Neu/WT migrates as a monomer can be explained by the fact that SDS is known to destabilize TM helix dimers, i.e., the interactions between TM helices are reduced or even abolished in the SDS environment. Also, compared to other published experiments, our experiments were run at very low peptide concentrations, favoring the monomeric state. Importantly, we showed that the migration of Neu/V664E changes from monomer to dimer with increasing peptide concentration (Figure 2A), demonstrating that we are observing true dimerization rather than anomalously reduced mobility caused by the polar glutamate residue. Because we can distinguish between the biologically-relevant dimerization propensities of Neu/WT and Neu/V664E using SDS-PAGE, this technique was used to screen the whole library shown in Fig. 1 for strong homodimer sequences.
Insights into RTK TM domain dimerization and RTK activation
The experimental design was guided by the five specific questions posed in the Introduction. Here we discuss our findings within the context of these five questions.
How promiscuous are the known dimerization motifs, as detected by SDS-PAGE?
While most of the six selected positive TM sequences have recognizable dimerization motifs, perhaps the most surprising result of the screen was the general lack of dimerization in the library. The 3888-member library contains hundreds of GxxxG dimerization motifs and thousands of SmxxxSm dimerization motifs. Similarly, 55% of the library members have at least one charged amino acid (D,E,K or R) and almost every library member has at least one polar or charged amino acid (S,T,Q,N,H,D,E,K or R). Yet, homodimerization in SDS-PAGE was very rare. More than 99.8% of the peptides we assayed appeared to be monomeric in SDS-PAGE gels, at least at the low concentrations studied. These results suggest that the interactions between TM helices are highly specific, and thus non-promiscuous, in SDS. The fact that the known dimerization motifs did not guarantee interactions suggests that structural context plays a more important role in dimerization than generally accepted.
What is the correlation between RTK TM domain dimerization in SDS-PAGE and RTK activation in mammalian membranes?
The activation of a receptor tyrosine kinase by autophosphorylation requires, minimally, receptor dimerization in the plasma membrane. However, it is also known that the structure of the dimer (i.e. the orientation of the two receptors with respect to one another, the disposition of the TM domain in the membrane, and the conformation of the extracellular domains) also affects the activation propensity1,43. In fact, RTKs are believed to be dimeric but inactive in the absence of ligand and undergo a conformational change upon ligand binding that promotes activation44. With these caveats in mind, we tested the SDS-PAGE-selected dimerization sequences for their ability to drive autophosphorylation (activation) of the kinase domain of Neu. We chose Neu (also known as ErbB2 or HER2) for this study because its activation does not require a ligand45,46. Instead, the only two requirements for Neu activation are dimerization and a phosphorylation-competent dimer structure.
We compared the activation of Neu chimeras with closely related Neu-like TM domains. In addition to Neu/WT, Neu/V664E and the six selected positive sequences, we also tested the activation by two randomly selected negative TM sequences from the library (Neu-Ran1 and Neu-Ran2). Of the six SDS PAGE-selected positive sequences, three activate Neu significantly better than the wild type (see Table 2). The best activation was from Neu-Pos4, which was identified twice independently and has an aspartate in position 664 where the oncogenic mutant Neu/V664E has a glutamate. Neu-Pos1 and Neu-Pos2 are also significantly activating, despite the lack of polar or charged groups other than Thr. Neu-Pos3, 5 and 6 were not more activating than Neu/WT in mammalian membranes despite all three having GxxxG dimerization motifs. Given the differences between an SDS-PAGE environment and a living mammalian cell membrane, and the complexities of RTK activation in living cells, the fact that only half of the positives selected in the gel are also positive in cell membranes is not surprising. The false positives can be explained by factors such as differences in dimerization due to environment, selection of antiparallel dimers in the SDS-PAGE experiment or differences in structure or orientation of the dimer in the cell membranes. Nonetheless, the fact that half of the SDS-PAGE positives are also activating in the cell membrane demonstrates that there is an overlap in the physical chemistry of the interactions in the two very different environments.
What is the importance of the GxxxG dimerization motif in SDS Gel dimerization and in RTK activation?
The best known dimerization motif for TM helices is the GxxxG motif. The GxxxG motif is considered a more stringent determinant for dimerization than the SmxxxSm motif, and has been shown to be essential for glycophorin A TM helix dimerization24. However, the importance of GxxxG in the dimerization of RTKs is uncertain. Many RTK TM domains do not have GxxxG motifs and yet interact well47. It is interesting to note that the three SDS-PAGE positives which do not activate Neu (Neu-Pos3,5 and 6) all contain GxxxG dimerization motifs. In fact, two of them contain GxxxGxxxG tandem motifs. On the other hand, the three positive sequences that do not contain GxxxG motifs all activate Neu better than wild type. Thus, our results show that the GxxxG sequence motif is not an absolute determinant of dimerization in mammalian membranes.
Do small changes in TM domain sequences affect RTK activation?
We know that the single amino acid change, Val664Glu, causes a significant increase in dimerization and activation of Neu27,28,37. But is this particular mutant a special case, or can other small sequence changes similarly affect RTK activation? Because only five residues are varied in the library, the chimeric Neu proteins tested here allow us to address the question. The selected sequences differ from wild-type by five or fewer residues, as shown in Table 1. Yet, despite the small differences in sequence, the biological effects of the SDS-PAGE selected positive sequences cover the whole known range of activation levels, from normal (WT) to oncogenic (V664E). Thus, small changes in sequence can have biologically significant effects on activation that are similar to the effect of the V664E mutation. These findings demonstrate the importance of TM helix dimerization in RTK activation, and suggest that RTKs are poised to respond to subtle changes with biologically relevant responses.
How much can RTK activation be varied by changing the TM domain sequence?
Interestingly, none of the positive sequences activated the Neu tyrosine kinase more than Neu/V664E and none of the random sequences activated less than Neu/WT. While the small number of sequences tested here does not allow us to draw a definitive conclusion, these results suggest that there are natural limits to RTK activation levels, at least for Neu (ErbB2). Most likely, these limits are set by the structure of the phosphorylation-competent dimer43.
Utility of SDS-PAGE in identifying TM sequences that modulate RTK activation
The work described in this paper evaluated the utility of SDS-PAGE as a screening method for the high-throughput identification of novel TM sequences which can over-activate Neu. The results demonstrated that SDS-PAGE could be a useful high throughput screening tool for identification of TM sequences that dimerize. In the context of the Neu library used here, SDS-PAGE was surprisingly stringent, with very few members showing evidence of strong dimerization. Of the six positive sequences identified based on dimerization in SDS-PAGE, three activated the Neu tyrosine kinase in mammalian cell membranes. Given the relative ease and low cost of SDS-PAGE-based screening, a 50% success rate is more than adequate for identifying novel homo-dimerizing sequences that activate RTKs. The method could potentially be used to identify heterodimerizing sequences, which could be used as RTK inhibitors.
It is possible that the interactions in SDS-PAGE are only a small subset of the interactions that occur between the Neu-like sequences in mammalian membranes. Thus, the SDS-PAGE screen may be selecting for only this limited subset of interactions. In the future, we will test this idea by screening the same library for homodimers in other detergents, in bicelles, and in lipid bilayers.
Materials and Methods
Library Synthesis
The combinatorial peptide library was synthesized as previously described48,49. Briefly, peptides were synthesized on 300 µm polystyrene/PEG Tentagel synthesis beads, starting with a photolabile linker. At the combinatorial sites, the resin was separated into different vessels for coupling of different amino acids, followed by remixing. Beta-branched amino acids (V, I and T) and residues following β-branched amino acids were double coupled. A ninhydrin color test was done after each coupling to verify completion and double couplings were done when needed. In this type of “split and recombine” library, each bead has about 1 nmol of a single peptide sequence. The quality of the synthesis was verified with mass spectrometry, Edman degradation and HPLC which showed single-sequence, full-length peptides. After the addition of an N-terminal cysteine residue, the peptide side chains were deprotected with reagent B, followed by extensive washing in dimethylformamide and dichloromethane. Labeling of the terminal cysteine was done with Alexafluor488-maleimide in dimethylformamide, followed by extensive washing of the beads in phenol, dimethylformamide and dichloromethane. After the final washing with dichloromethane, the beads were spread in a glass Petri dish, dried overnight and then cleaved with 365 nm UV light for 5 hours. Each bead released about 200 pmol of dye-labeled peptide after cleavage.
High-throughput screening by SDS PAGE
SDS-PAGE screening was done with 60 sequences on each gel. Sixty single beads containing peptide library members with dry-cleaved photolabile linker were placed in 96 well plates in 5 rows and 12 columns. Hexafluoroisopropanol (HFIP, 100 µl) was added to each well and the plate was placed on a heating block at 80°C in a fume hood until the HFIP had evaporated. This procedure extracts about 200 pmoles of peptide from each bead. At this point 10 µl of SDS gel loading buffer was added to each of the 60 wells and they were incubated for 1 hour to dissolve the peptide. Next, half the volume, or 5 µl, from each well in a column was added to the sixth well in the same column, giving an equimolar mixture of 5 peptides in 25 µl. Care was taken not to transfer the beads into the sixth well. Finally half of that combined solution, or 12.5 µl, of each of the twelve sixth row wells was loaded onto an Invitrogen NuPAGE™ 12% Bis-Tris SDS PAGE gel (15 well, 1 mm thick) giving 12 lanes in the gel with about 50 pmoles (150 ng) of each of five peptides in each lane (60 peptides per gel). In the seventh lane of each gel we loaded 500 pmol (1500 ng) of Neu/V664E as a dimer standard. The gels were developed with 1X NuPAGE™ MES-SDS running buffer at 200V for 30 minutes. Since the majority of library members are monomeric and migrate identically to Neu/WT, the library peptides served as an internal monomer control. After electrophoresis, we placed the gel in a container of running buffer and photographed it immediately under UV light to identify possible peptide dimers by Alexafluor fluorescence. All test lanes contained a distinct monomer band at a position near that of Neu/WT. Most lanes also contained a faint higher band slightly above the position of Neu/V664E which we showed to be a reproducible, non-peptide artifact that was extracted from the synthesis beads. Putative dimer bands, which were rare, migrated within the limits set by Neu/WT and Neu/V664E.
If a lane containing a putative dimer was identified, a second gel was used to examine the five peptides from that lane individually, using the remaining volume of loading buffer for each of the individual peptides. Comparison to Neu/V664E was made to identify sequences with a dimerization propensity. If one of the five individual peptides migrated closer to the dimer position than the others, or showed a smeared band between monomer and dimer, we considered that peptide to be positive.
Plasmids construction of Neu-chimeric proteins with different TM domain motifs
The Neu/WT and Neu/V664E plasmids were a generous gift from D. J. Donoghue at UCSD. To replace the TM domain of Neu by the selected TM peptide motifs, two restriction enzyme sites were created in the plasmid of pcDNA3.1-Neu using multi-site mutagenesis kit (Stratagene). First, a NheI site in the multi-cloning site region of the pcDNA3.1(+) plasmid was removed. The primers for removing the NheI site were: forward. 5'- GGA GAC CCA AGC TGG CCA GCG TTT AAA CTT AAG C -3', reverse. 5'- GCT TAA GTT TAA ACG CTG GCC AGC TTG GGT CTC C -3'. The primers for generating a NheI site before the transmembrane domain were: forward, 5'- CCA GCA GAG CAG AGA GCT AGC CCG GTG ACA TTC ATC -3', reverse, 5'- GAT GAA TGT CAC CGG GCT AGC TCT CTG CTC TGC TGG -3'. The primers for generating a EcoRI site after the transmembrane domain were: forward, 5'- GTG GTG GTC GTT GGA ATT CTA ATC AAA CGA AGG AGA CAG -3'. Reverse, 5'- CTG TCT CCT TCG TTT GAT TAG AAT TCC AAC GAC CAC CAC -3'. After the two restriction enzyme sites were generated, the plasmid was double digested. The DNA oligomers encoding the selected TM peptides, except Neu-Pos4, were synthesized by Invitrogen and then double digested by NheI and EcoRI. They were then ligated to the digested Neu in a pcDNA3.1 (+) vector. After the plasmid encoding Neu-Pos2 was constructed, primers were designed to mutate it to Neu-Pos4. The final plasmids were amplified and the successful substitution of the TM domain with the sequences in Table 1 was confirmed by sequencing provided by Genewiz (Germantown, MD).
Cell Culture
Chinese Hamster Ovary (CHO) cells were cultured in DMEM (Invitrogen, CA) supplemented with 10% Fetal Bovine Serum (FBS, Thermo Scientific) and Non-essential amino acids (Gibco, CA). Cells were maintained in an incubator with 5% CO2 at 37°C.
Western Blots
Transfection of the plasmids in CHO cells was performed using Fugene HD (Roche Applied Science, IN) following the manufacturer’s protocol. After culturing the cells in the incubator for 24 hours following transfection, cells were lysed with lysis buffer (25 mM Tris-HCl, 0.5% TritonX-100, 20 mM NaCl, 2 mM EDTA, 2 mM NaVO4 and protease inhibitor (Roche Applied Science, IN)). After centrifugation at 15,000g for 15 minutes at 4°C, the supernatant was collected and the pellet was discarded. Protein concentration was determined using the BCA™ assay kit (Pierce, IL). The lysate was loaded into 3–8% NuPAGE® Novex® Tris-Acetate mini gels (Invitrogen, CA). Proteins were separated by electrophoresis, and transferred onto a nitrocellulose membrane. The membrane was blocked with milk for one hour at room temperature, and then stained with either anti-C-Neu antibodies (Santa Cruz Technology, CA) to assay expression or anti-p-ErbB2 antibodies (Cell Signaling Technology, MA) to assay phosphorylation. The secondary antibodies were anti-rabbit HRP conjugated antibody (Promega, WI). ECL™ detection reagent (GE Healthcare life sciences, UK) was used to reveal the bands on the films. The bands were quantified using ImageQuant TL (GE Healthcare life sciences, UK).
Quantification of Western blotting
The western blot films were scanned and processed with ImageQuant TL as described elsewhere28,50,51. At least three sets of independent experiment were performed in order to determine the average and the standard deviations. A one-factor ANOVA was used to compare the relative activations values measured for the various Neu chimeras, and the Tukey-Kramer test was used for pairwise comparisons.
Research highlights.
SDS-PAGE can be used as a high-throughput screen for TM helix dimerization
Interactions between TM helices are highly specific in SDS PAGE
Homodimer sequences selected in SDS PAGE can activate RTKs in cells
Small changes in RTK TM sequence can have significant biological effects
Acknowledgement
Supported by GM 60000 and GM68619.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Reference List
- 1.Li E, Hristova K. Role of receptor tyrosine kinase transmembrane domains in cell signaling and human pathologies. Biochemistry. 2006;45:6241–6251. doi: 10.1021/bi060609y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Li E, Hristova K. Receptor Tyrosine Kinase transmembrane domains: function, dimer structure, and dimerization energetics. Cell Adhesion and Migration. 2010;4:249–254. doi: 10.4161/cam.4.2.10725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hubert P, Sawma P, Duneau JP, Khao J, Henin J, Bagnard D, Sturgis J. Single-spanning transmembrane domains in cell growth and cell-cell interactions: More than meets the eye? Cell Adhesion and Migration. 2010;v:313–324. doi: 10.4161/cam.4.2.12430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Li E, You M, Hristova K. SDS-PAGE and FRET suggest weak interactions between FGFR3 TM domains in the absence of extracellular domains and ligands. Biochemistry. 2005;44:352–360. doi: 10.1021/bi048480k. [DOI] [PubMed] [Google Scholar]
- 5.Chen L, Placone J, Novicky L, Hristova K. The extracellular domain of fibroblast growth factor receptor 3 inhibits ligand-independent dimerization. Science Signaling. 2010;3:ra86. doi: 10.1126/scisignal.2001195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chen L, Merzlyakov M, Cohen T, Shai Y, Hristova K. Energetics of ErbB1 transmembrane domain dimerization in lipid bilayers. Biophys. J. 2009;96:4622–4630. doi: 10.1016/j.bpj.2009.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Artemenko EO, Egorova NS, Arseniev AS, Feofanov AV. Transmembrane domain of EphA1 receptor forms dimers in membrane-like environment. Biochim. Biophys. Acta. 2008;1778:2361–2367. doi: 10.1016/j.bbamem.2008.06.003. [DOI] [PubMed] [Google Scholar]
- 8.MacKenzie KR. Folding and stability of alpha-helical integral membrane proteins. Chem. Rev. 2006;106:1931–1977. doi: 10.1021/cr0404388. [DOI] [PubMed] [Google Scholar]
- 9.Sternberg MJE, Gullick WJ. A Sequence Motif in the Transmembrane Region of Growth Factor Receptors with Tyrosine Kinase Activity Mediates Dimerization. Protein Eng. 1990;3:245–248. doi: 10.1093/protein/3.4.245. [DOI] [PubMed] [Google Scholar]
- 10.Mendrola JM, Berger MB, King MC, Lemmon MA. The single transmembrane domains of ErbB receptors self-associate in cell membranes. J. Biol. Chem. 2002;277:4704–4712. doi: 10.1074/jbc.M108681200. [DOI] [PubMed] [Google Scholar]
- 11.Hong H, Joh NH, Bowie JU, Tamm LK. Methods for Measuring the Thermodynamic Stability of Membrane Proteins. Methods in Enzymology: Biothermodynamics,Vol 455, Part A. 2009;455:213–236. doi: 10.1016/S0076-6879(08)04208-0. [DOI] [PubMed] [Google Scholar]
- 12.Fleming KG, Ackerman AL, Engelman DM. The effect of point mutations on the free energy of transmembrane α-helix dimerization. J. Mol. Biol. 1997;272:266–275. doi: 10.1006/jmbi.1997.1236. [DOI] [PubMed] [Google Scholar]
- 13.Fisher LE, Engelman DM, Sturgis JN. Detergents modulate dimerization, but not helicity, of the glycophorin A transmembrane domain. J. Mol. Biol. 1999;293:639–651. doi: 10.1006/jmbi.1999.3126. [DOI] [PubMed] [Google Scholar]
- 14.Cristian L, Lear JD, DeGrado WF. Determination of membrane protein stability via thermodynamic coupling of folding to thiol-disulfide interchange. Protein Sci. 2003;12:1732–1740. doi: 10.1110/ps.0378603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.You M, Li E, Wimley WC, Hristova K. FRET in liposomes: measurements of TM helix dimerization in the native bilayer environment. Analytical Biochemistry. 2005;340:154–164. doi: 10.1016/j.ab.2005.01.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Cristian L, Lear JD, DeGrado WF. Use of thiol-disulfide equilibria to measure the energetics of assembly of transmembrane helices in phospholipid bilayers. Proceedings of the National Academy of Sciences of the United States of America. 2003;100:14772–14777. doi: 10.1073/pnas.2536751100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hong H, Blois TM, Cao Z, Bowie JU. Method to measure strong protein-protein interactions in lipid bilayers using a steric trap. Proceedings of the National Academy of Sciences of the United States of America. 2010;107:19802–19807. doi: 10.1073/pnas.1010348107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Russ WP, Engelman DM. TOXCAT: A measure of transmembrane helix association in a biological membrane. Proc. Natl. Acad. Sci. USA. 1999;96:863–868. doi: 10.1073/pnas.96.3.863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Langosch D, Brosig B, Kolmar H, Fritz HJ. Dimerisation of the glycophorin a transmembrane segment in membranes probed with the ToxR transcription activator. J. Mol. Biol. 1996;263:525–530. doi: 10.1006/jmbi.1996.0595. [DOI] [PubMed] [Google Scholar]
- 20.Schneider D, Engelman DM. GALLEX, a measurement of heterologous association of transmembrane helices in a biological membrane. J. Biol. Chem. 2003;278:3105–3111. doi: 10.1074/jbc.M206287200. [DOI] [PubMed] [Google Scholar]
- 21.Chen L, Novicky L, Merzlyakov M, Hristov T, Hristova K. Measuring the Energetics of Membrane Protein Dimerization in Mammalian Membranes. J. Am. Chem. Soc. 2010;132:3628–3635. doi: 10.1021/ja910692u. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lemmon MA, Flanagan JM, Hunt JF, Adair BD, Bormann BJ, Dempsey CE, Engelman DM. Glycophorin-A dimerization is driven by specific interactions between transmembrane α-helices. J. Biol. Chem. 1992;267:7683–7689. [PubMed] [Google Scholar]
- 23.Lemmon MA, Flanagan JM, Treutlein HR, Zhang J, Engelman DM. Sequence specificity in the dimerization of transmembrane alpha-helices. Biochemistry. 1992;31:12719–12725. doi: 10.1021/bi00166a002. [DOI] [PubMed] [Google Scholar]
- 24.MacKenzie KR, Prestegard JH, Engelman DM. A transmembrane helix dimer: Structure and implications. Science. 1997;276:131–133. doi: 10.1126/science.276.5309.131. [DOI] [PubMed] [Google Scholar]
- 25.Walkenhorst WF, Merzlyakov M, Hristova K, Wimley WC. Polar residues in transmembrane helices can decrease electrophoretic mobility in polyacrylamide gels without causing helix dimerization. Biochim. Biophys. Acta. 2009;1788:1321–1331. doi: 10.1016/j.bbamem.2009.02.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Rath A, Glibowicka M, Nadeau VG, Chen G, Deber CM. Detergent binding explains anomalous SDS-PAGE migration of membrane proteins. Proceedings of the National Academy of Sciences of the United States of America. 2009;106:1760–1765. doi: 10.1073/pnas.0813167106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Weiner DB, Liu J, Cohen JA, Williams WV, Greene MI. A Point Mutation in the Neu Oncogene Mimics Ligand Induction of Receptor Aggregation. Nature. 1989;339:230–231. doi: 10.1038/339230a0. [DOI] [PubMed] [Google Scholar]
- 28.He L, Hristova K. Pathogenic activation of receptor tyrosine kinases in mammalian membranes. J. Mol. Biol. 2008;384:1130–1142. doi: 10.1016/j.jmb.2008.10.036. [DOI] [PubMed] [Google Scholar]
- 29.Schechter AL, Stern DF, Vaidyanathan L, Decker SJ, Drebin JA, Greene MI, Weinberg RA. The Neu Oncogene - An Erb-B-Related Gene Encoding A 185,000-Mr Tumor-Antigen. Nature. 1984;312:513–516. doi: 10.1038/312513a0. [DOI] [PubMed] [Google Scholar]
- 30.Smith SO, Smith C, Shekar S, Peersen O, Ziliox M, Aimoto S. Transmembrane interactions in the activation of the Neu receptor tyrosine kinase. Biochemistry. 2002;41:9321–9332. doi: 10.1021/bi012117l. [DOI] [PubMed] [Google Scholar]
- 31.Smith SO, Smith CS, Bormann BJ. Strong hydrogen bonding interactions involving a buried glutamic acid in the transmembrane sequence of the neu/erbB-2 receptor. Nature Struct. Biol. 1996;3:252–258. doi: 10.1038/nsb0396-252. [DOI] [PubMed] [Google Scholar]
- 32.Senes A, Gerstein M, Engelman DM. Statistical analysis of amino acid patterns in transmembrane helices: The GxxxG motif occurs frequently and in association with -branched residues at neighboring positions. J. Mol. Biol. 2000;296:921–936. doi: 10.1006/jmbi.1999.3488. [DOI] [PubMed] [Google Scholar]
- 33.Dawson JP, Weinger JS, Engelman DM. Motifs of serine and threonine can drive association of transmembrane helices. J. Mol. Biol. 2002;316:799–805. doi: 10.1006/jmbi.2001.5353. [DOI] [PubMed] [Google Scholar]
- 34.Sulistijo ES, Jaszewski TM, MacKenzie KR. Sequence-specific dimerization of the transmembrane domain of the "BH3-only" protein BNIP3 in membranes and detergent. J. Biol. Chem. 2003;278:51950–51956. doi: 10.1074/jbc.M308429200. [DOI] [PubMed] [Google Scholar]
- 35.Bocharov EV, Mineev KS, Volynsky PE, Ermolyuk YS, Tkach EN, Sobol AG, Chupin VV, Kirpichnikov MP, Efremov RG, Arseniev AS. Spatial structure of the dimeric transmembrane domain of the growth factor receptor ErbB2 presumably corresponding to the receptor active state. J. Biol. Chem. 2008;283:6950–6956. doi: 10.1074/jbc.M709202200. [DOI] [PubMed] [Google Scholar]
- 36.Russ WP, Engelman DM. The GxxxG motif: A framework for transmembrane helix-helix association. J. Mol. Biol. 2000;296:911–919. doi: 10.1006/jmbi.1999.3489. [DOI] [PubMed] [Google Scholar]
- 37.He LJ, Shobnam N, Hristova K. Specific inhibition of a pathogenic receptor tyrosine kinase by its transmembrane domain. Biochimica et Biophysica Acta-Biomembranes. 2011;1808:253–259. doi: 10.1016/j.bbamem.2010.08.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Duong MT, Jaszewski TM, Fleming KG, MacKenzie KR. Changes in apparent free energy of helix-helix dimerization in a biological membrane due to point mutations. J. Mol. Biol. 2007;371:422–434. doi: 10.1016/j.jmb.2007.05.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Curran AR, Engelman DM. Sequence motifs, polar interactions and conformational changes in helical membrane proteins. Cur. Opinion Struc. Biol. 2003;13:412–417. doi: 10.1016/s0959-440x(03)00102-7. [DOI] [PubMed] [Google Scholar]
- 40.Li E, You M, Hristova K. FGFR3 dimer stabilization due to a single amino acid pathogenic mutation. J. Mol. Biol. 2006;356:600–612. doi: 10.1016/j.jmb.2005.11.077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.You M, Li E, Hristova K. The achondroplasia mutation does not alter the dimerization energetics of FGFR3 transmembrane domain. Biochemistry. 2006;45:5551–5556. doi: 10.1021/bi060113g. [DOI] [PubMed] [Google Scholar]
- 42.Kliger Y, Aharoni A, Rapaport D, Jones P, Blumenthal R, Shai Y. Fusion peptides derived from the HIV type 1 glycoprotein 41 associate within phospholipid membranes and inhibit cell-cell fusion - Structure-function study. J. Biol. Chem. 1997;272:13496–13505. doi: 10.1074/jbc.272.21.13496. [DOI] [PubMed] [Google Scholar]
- 43.Bell CA, Tynan JA, Hart KC, Meyer AN, Robertson SC, Donoghue DJ. Rotational coupling of the transmembrane and kinase domains of the Neu receptor tyrosine kinase. Mol. Biol. Cell. 2000;11:3589–3599. doi: 10.1091/mbc.11.10.3589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Moriki T, Maruyama H, Maruyama IN. Activation of preformed EGF receptor dimers by ligand-induced rotation of the transmembrane domain. J. Mol. Biol. 2001;311:1011–1026. doi: 10.1006/jmbi.2001.4923. [DOI] [PubMed] [Google Scholar]
- 45.Cho HS, Mason K, Ramyar KX, Stanley AM, Gabelli SB, Denney DW, Leahy DJ. Structure of the extracellular region of HER2 alone and in complex with the Herceptin Fab. Nature. 2003;421:756–760. doi: 10.1038/nature01392. [DOI] [PubMed] [Google Scholar]
- 46.Burgess AW, Cho HS, Eigenbrot C, Ferguson KM, Garrett TPJ, Leahy DJ, Lemmon MA, Sliwkowski MX, Ward CW, Yokoyama S. An open-and-shut case? Recent insights into the activation of EGF/ErbB receptors. Molecular Cell. 2003;12:541–552. doi: 10.1016/s1097-2765(03)00350-2. [DOI] [PubMed] [Google Scholar]
- 47.Finger C, Escher C, Schneider D. The Single Transmembrane Domains of Human Receptor Tyrosine Kinases Encode Self-Interactions. Science Signaling. 2009;2 doi: 10.1126/scisignal.2000547. [DOI] [PubMed] [Google Scholar]
- 48.Rausch JM, Marks JR, Wimley WC. Rational combinatorial design of pore-forming beta-sheet peptides. Proceedings of the National Academy of Sciences of the United States of America. 2005;102:10511–10515. doi: 10.1073/pnas.0502013102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Rathinakumar R, Wimley WC. Biomolecular engineering by combinatorial design and high-throughput screening: Small, soluble peptides that permeabilize membranes. J. Am. Chem. Soc. 2008;130:9849–9858. doi: 10.1021/ja8017863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.He L, Horton WA, Hristova K. The physical basis behind achondroplasia, the most common form of human dwarfism. J. Biol. Chem. 2010;285:30103–30114. doi: 10.1074/jbc.M109.094086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Chen F, Degnin C, Laederich MB, Horton AW, Hristova K. The A391E mutation enhances FGFR3 activation in the absence of ligand. Biochimica et Biophysica Acta-Biomembranes. 2011;1808:2045–2050. doi: 10.1016/j.bbamem.2011.04.007. [DOI] [PMC free article] [PubMed] [Google Scholar]