Abstract
Peptides within the knottin family have been shown to possess inherent stability, making them attractive scaffolds for the development of therapeutic and diagnostic agents. Given its remarkable stability to proteases, the cyclic peptide kalata B1 was employed as a scaffold to create a large knottin library displayed on the surface of E. coli. A library exceeding 109 variants was constructed by randomizing seven amino acids within a loop of the kalata B1 scaffold and screened using fluorescence-activated cell sorting to identify peptide ligands specific for the active site of human thrombin. Refolded thrombin binders exhibited high nanomolar affinities in solution, slow dissociation rates, and were able to inhibit thrombin’s enzymatic activity. Importantly, 80% of a knottin-based thrombin inhibitor remained intact after a two hour incubation both with trypsin and with chymotrypsin, demonstrating that modifying the kalata B1 sequence did not compromise its stability properties. In addition, the knottin variant mediated 20-fold enhanced affinity for thrombin, when compared to the same seven residue binding epitope constrained by a single disulfide bond. Our results indicate that peptide libraries derived from the kalata B1 scaffold can yield high affinity protein ligands that retain the remarkable protease resistance associated with the parent scaffold. More generally, this strategy may prove useful in the development of stable peptide ligands suitable for in vivo applications.
Peptides are increasingly attractive as therapeutics given their diverse biological functions and potentially high potency and target specificity (1). Yet the development of therapeutic peptides remains challenging given their typically short in vivo half-life that results from rapid renal clearance and from degradation by peptidases in the blood, liver, and kidneys (2). Susceptibility to both exo- and endopeptidases can contribute substantially to the clearance of peptides in circulation, with slow renal filtration increasing exposure to protease degradation (3). Furthermore, peptidase susceptibility may increase the risk of immunogenicity, mediated by antigen-processing proteases (4). Consequently, several strategies to enhance peptide resistance to proteases have been developed including N-terminal acetylation and C-terminal amidation (5), the use of backbone cyclization (6), incorporation of non-canonical or D-amino acids (7–8), and PEGylation (9). Although these approaches have proven useful alone or in combination, there remains a need for general strategies to improve half-life that are compatible with recombinant protein synthesis and preserve the benefits of small size.
An alternative strategy to enhance the intrinsic stability of peptides is the use of naturally occurring disulfide-constrained peptide scaffolds that are resistant to proteolysis. In particular, the knottin family of peptides provides attractive candidate scaffolds for the development of peptidase-resistant peptides because of their compact and stable cystine knot structure formed by three disulfide bonds (10). Two disulfide bonds, along with the backbone, form a ring through which a third disulfide bond threads, forming the cystine knot structure. A variety of knottin family members have been utilized as scaffolds for developing stable peptide ligands through loop grafting of bioactive peptides (11). For example, binding epitopes were successfully grafted into a loop of Ecballium elaterium trypsin inhibitor II (EETI-II). The grafted binder was enriched from an excess of non-binding peptides after surface display on E. coli and screening via fluorescence-activated cell sorting (FACS) (12). Similarly, thrombopoietin receptor binding peptides were inserted into both the Agouti-related protein (AGRP) and the EETI-II peptide scaffolds to yield potent antagonists of platelet production, and when homodimerized became effective agonists (13). The circular peptide kalata B1 was modified by replacing individual loops with a VEGF binding sequence, and many of the scaffold constrained sequences exhibited dramatically improved serum stability compared to the linear epitope (14).
To directly identify knottin variants selective for arbitrary targets, peptide libraries presented within knottin scaffolds have been constructed and displayed on phage or yeast for screening. For example, randomization of a loop in the AGRP scaffold, in conjunction with yeast display library screening using FACS, was used to successfully identify αvβ3 integrin ligands with affinities in the high picomolar to low nanomolar range (15). Kunitz-domain inhibitors are larger than knottins, having approximately 60 amino acids rather than 30 amino acids, and contain three disulfide bonds that are not connected in a knot conformation. Phage display libraries based on two different Kunitz domains—human lipoprotein-associated coagulation inhibitor and bovine pancreatic trypsin inhibitor—have yielded ligands binding to a variety of enzyme targets including neutrophil elastase, chymotrypsin, pancreatic elastase, thrombin, plasmin, and human plasma kallikrein (16–19). A potent plasma kallikrein inhibitor (Ecallantide) with a Ki of 44 pM was developed using this approach and approved for the treatment of hereditary angioedema (20). These examples demonstrate the utility of disulfide-rich peptide libraries.
Despite their stabilizing cystine knot structure, many knottin peptides remain susceptible to degradation by proteases in vivo. Derivatives of AGRP and two squash protease inhibitors were stable to pepsin and elastase, but were extensively degraded by trypsin and chymotrypsin (21). Similarly constrained microproteins, including ω-conotoxin MVIIa and a hybrid of two squash trypsin inhibitors (EETI-II and MCoTI-II), were degraded by trypsin, with the latter also degraded by chymotrypsin (22–23). The cyclotide kalata B1 is a remarkable member of the knottin family because it is completely resistant to trypsin and pepsin degradation for up to 24 hours (24). Furthermore, kalata B1 exhibits exceptional chemical and thermal stability, making it an attractive scaffold for therapeutic design (24). Although native kalata B1 exhibits a circular backbone (25), some acyclic permutants retain the stability of the parent cyclotide (24), exhibit reduced undesired hemolytic activity, fold with the correct disulfide connectivity (26), and are amenable to display technologies. Thus, we constructed a large bacterial display library by randomizing a single loop within kalata B1 and fusing the acyclic scaffold to an engineered bacterial display protein (eCPX) (27–28). The library was screened for binders to human thrombin, a serine protease involved in the blood coagulation cascade. Our results indicate that the kalata B1 scaffold can yield trypsin- and chymotrypsin-resistant thrombin inhibitors and may provide a general methodology to create peptide ligands with increased stability for diagnostic or therapeutic use.
RESULTS AND DISCUSSION
Design and screening of a knottin library
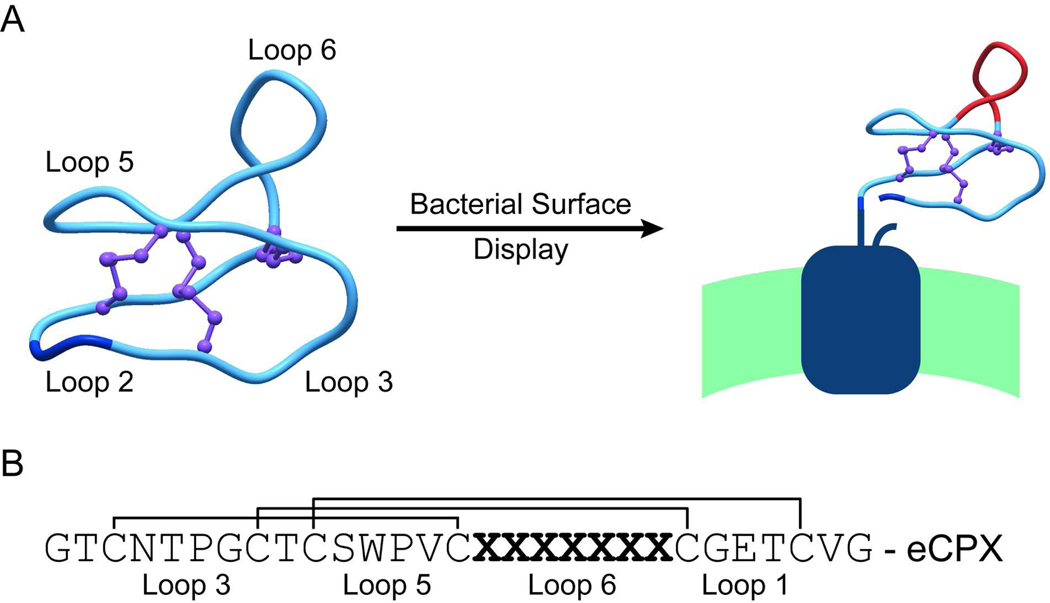
A knottin peptide library was displayed on the cell surface of E. coli by opening the native cyclic structure of kalata B1 between two glycine residues within loop two and fusing the resulting kalata B1 C-terminus to the N-terminus of the eCPX display scaffold (Figure 1) (27). A fully randomized seven amino acid peptide library was created within loop six of kalata B1; we chose this loop because it exhibits a high degree of sequence variability across the cyclotide family and is the longest occurring loop (25). Transformation into E. coli yielded a library of 6×109 independent transformants, providing ~18% coverage of the theoretical library diversity (3.4×1010 sequence variants) based on the use of NNS codons (29).
Figure 1.
Construction of a library of kalata B1 variants displayed on the cell surface of E. coli. A) The backbone of the cyclic kalata B1 peptide (PDB: 1NB1) was broken in loop two between two glycine residues (shown in dark blue) and fused to the display scaffold eCPX. Loop six was randomized to form the library (shown in red). B) Sequence of the kalata B1 peptide shown with the disulfide bond connectivity, the randomized region, and loop designations.
The kalata B1 library was enriched for α-thrombin binders using three cycles of FACS, with 30 nM biotinylated thrombin in the first two cycles, and a kinetic screen for slow dissociation rates in the third cycle (30). The sequences of individual isolated clones revealed five distinct consensus groups (Table 1). A few of the isolated peptides had eight or nine, rather than seven, amino acids within the randomized region, likely due to PCR errors during library construction or oligonucleotide primer errors. All of the identified groups included a conserved arginine or lysine residue, whereas only two groups contained an acidic residue, aspartic acid. Two of the consensus groups had a glycine pair within the randomized region imparting more flexibility to the loop, and another group exhibited two conserved proline residues, which would add to the rigidity of loop six. Clonal peptide affinities were ranked according to their apparent dissociation rates (Supplementary Figure 1) and clones with the slowest dissociation rate constants from each consensus group were characterized further.
Table 1.
Thrombin-binding kalata B1 variants. The randomized region is bolded with the conserved residues in each consensus group highlighted.
 |
Although kalata B1 has been utilized as a scaffold for grafting (14), the present study, to our knowledge, is the first demonstration that large libraries constructed using the kalata B1 scaffold can rapidly yield high affinity peptide ligands specific for a given protein. When compared to loop grafting, the present method for ligand generation is advantageous because the library is created by loop randomization within the environment of the constrained scaffold, increasing the likelihood that the peptide will retain high affinity in solution and enabling the measurement of displayed peptide function in a structurally relevant context.
Kalata B1 scaffold was important for binding affinity
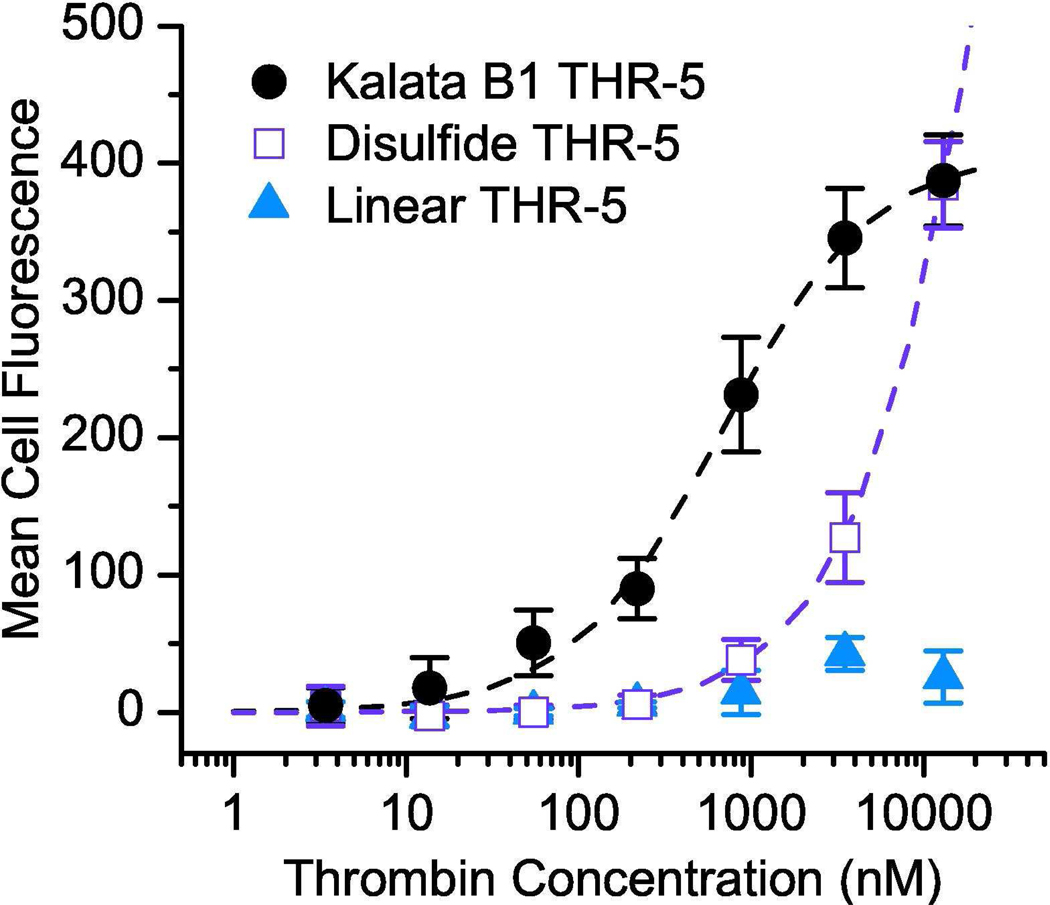
To investigate the effect of the kalata B1 scaffold upon affinity, peptides corresponding to the thrombin-binding region (loop six) of clone THR-5 were constructed as both a single-disulfide constrained peptide and as a linear peptide and displayed on E. coli. Peptide-displaying cells were incubated with a concentration series of α-thrombin and analyzed by flow cytometry in triplicate to measure KDApp values (Figure 2). The kalata B1 scaffold, having three disulfide bonds, exhibited hindered export to the outer membrane when compared to the linear or single-disulfide constrained peptides, suggesting that periplasmic disulfide formation and/or folding may interfere with export (31). For clone THR-5 in the kalata B1 scaffold, the KDApp was calculated to be 670 nM. In contrast, the linear peptide showed negligible binding signal above background at the highest thrombin concentration tested. A single-disulfide constrained loop construct exhibited binding, but the cell fluorescence did not saturate at the highest concentration of thrombin used. However, inspection of the resulting curve suggests that the KDApp of the constrained loop is greater than 15 µM. Similarly, clone THR-24, within the kalata B1 scaffold had a KDApp of 320 nM while the disulfide-constrained and linear peptides exhibited KDApp values greater than 6 µM. Collectively, these results indicate that the kalata B1 scaffold imparts at least a 20-fold increase in affinity to the thrombin-binding epitope relative to a single-disulfide constrained loop. The conformational dependence of the affinity of the binding epitope highlights the importance of screening a knottin library against the desired protein target as opposed to grafting a bioactive loop into the scaffold.
Figure 2.
Measurement of the contribution of the kalata B1 scaffold to the apparent binding affinity for thrombin. The bacterial display clone THR-5 exhibited a KDApp = 670 nM, corresponding to an affinity at least 20-fold greater than that of the same peptide in a single-disulfide constrained loop conformation. The epitope as a linear peptide did not appreciably bind at any of the thrombin concentrations that were assayed.
Isolated knottins bind thrombin’s active site
To identify the location of the knottin binding site(s) on human thrombin, a competition binding assay was used. Peptide-displaying clones chosen from each consensus group were incubated with biotinylated α-thrombin and a concentration series of a given unlabeled competitor (Supplementary Figure 2). Thus, a decrease in cell fluorescence with increasing concentrations of competitor indicated that the competitor interfered with the binding of α-thrombin to the kalata B1 peptide. First, hirudin, which binds to both the active site and exosite I, inhibited thrombin binding which indicates that the peptides require the active site or exosite I for interaction, rather than exosite II. Second, three direct competitors to the labeled α-thrombin—unlabeled α-thrombin, γ-thrombin (exosite I removed by proteolysis), and thrombin with the active site blocked by the inhibitor PPACK (32)—were assayed for their ability to compete with thrombin. As expected, competition with unlabeled α-thrombin caused a large decrease in cell fluorescence. Increasing concentrations of γ-thrombin caused a similar decrease in fluorescence for all of the peptides assayed, indicating that the peptides do not bind exosite I. Furthermore, when the thrombin active site was pre-blocked by the irreversible inhibitor PPACK, the complex was not able to compete with the labeled α-thrombin. Surprisingly, however, an exosite I-binding DNA aptamer (GGTTGGTGTGGTTGG) (33) competed with the peptides. Conceivably, the DNA aptamer could allosterically alter the active site and block the peptide from interacting with thrombin (34). Alternatively, the competition may be due to steric hindrance or poor specificity since the aptamer is able to bind to two separate thrombin molecules by interacting with exosite I and II simultaneously. Four different clones (THR-5, THR-29, THR-24, and THR-17) exhibited the same behavior in this competition assay. Collectively, these results indicate that the identified peptides bind the active site of thrombin.
Refolded peptides bind with nanomolar affinity and inhibit activity
To characterize their affinity, activity, and protease resistance, multiple thrombin-binding peptides were synthetically prepared and oxidized to form the characteristic cystine knot of kalata B1. The refolded peptides acyclic kalata B1, THR-5, and THR-29 were purified by RP-HPLC (Figure 3). Peptides THR-20 and THR-24, both from the same consensus group, could not be purified in their crude, reduced form due to poor solubility and therefore were not successfully refolded. Kalata B1 is hydrophobic due to large, nonpolar residues in loop five and six that become surface exposed when the cyclic cystine knot is properly formed. Consequently, scaffold engineering may be beneficial to increase the solubility and folding efficiency of acyclic kalata B1 derived ligands.
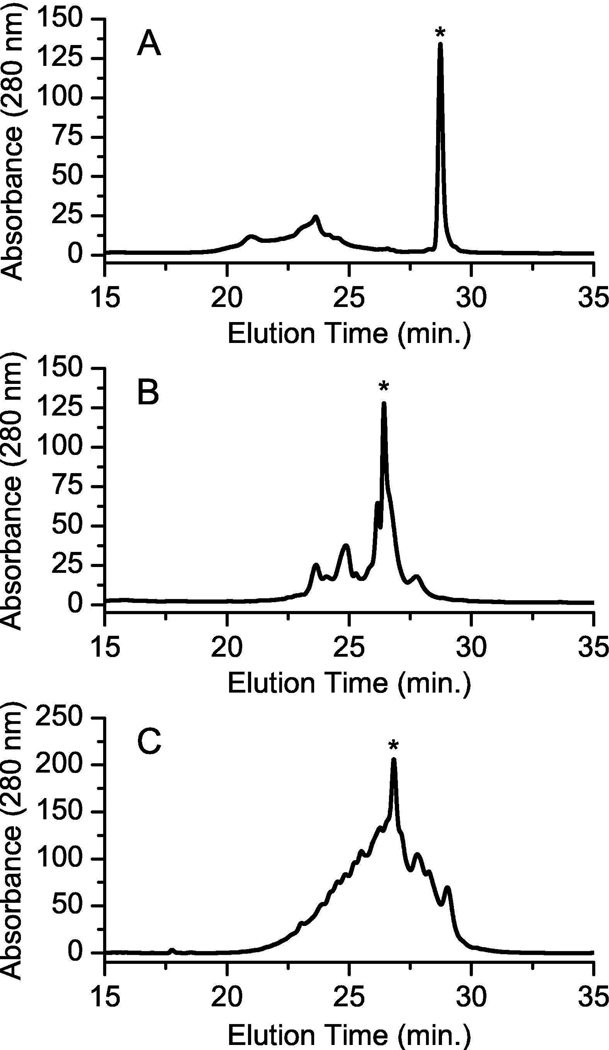
Figure 3.
Purification of the refolded kalata B1 peptides. HPLC absorbance traces for A) kalata B1, B) THR-5, and C) THR-29. The asterisk indicates the refolded product characterized using surface plasmon resonance and trypsin stability assays.
The acyclic kalata B1 refolding mixture exhibited a sharp, late-eluting peak, which has been shown to correspond to the native-like structure for kalata B1 due the surface exposure of hydrophobic residues (26). For the refolded THR-5 mixture, the separation between the misfolded material and the large, late-eluting peak was reduced probably as a result of the randomization of loop six, which disrupted a portion of the surface exposed hydrophobic patch. Purification eliminated the observed shoulder yielding a 90% pure product as determined via LC/MS. The refolded product of THR-29, however, exhibited a broad elution profile suggestive of a more heterogeneous mixture. Peptides corresponding to the indicated elution peaks for the variants (Figure 3) possessed three disulfide bonds as verified via mass spectrometry.
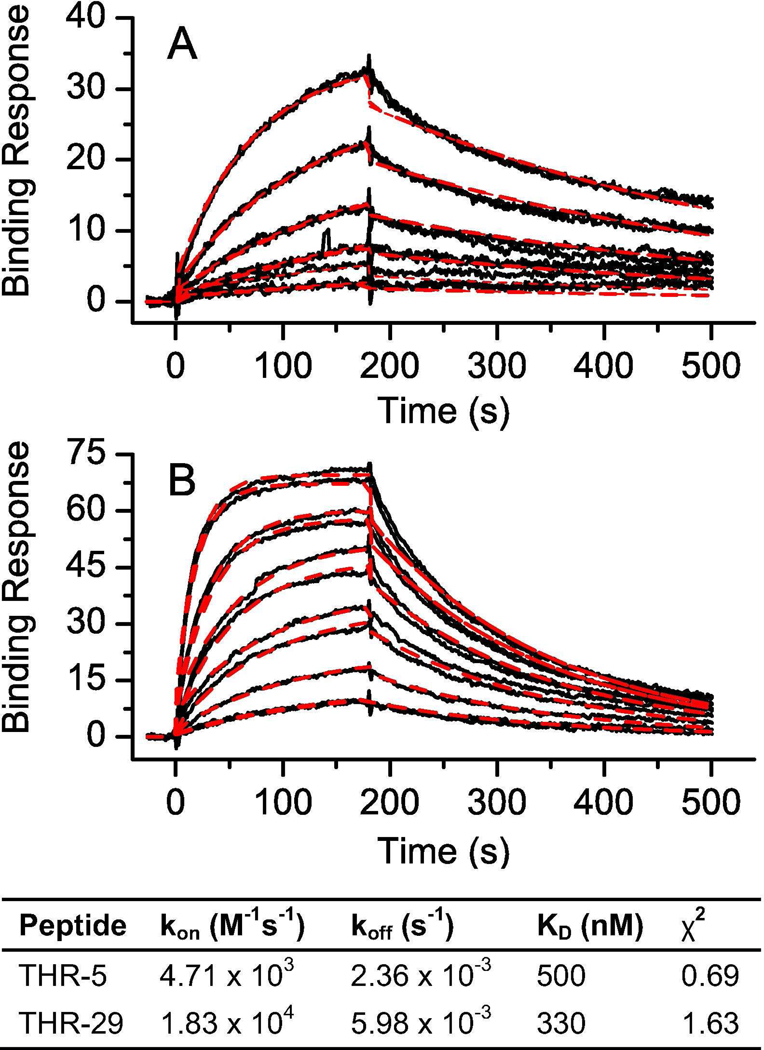
The binding kinetics of the refolded knottins was measured using surface plasmon resonance (Figure 4). Knottin THR-5 exhibited slower association and dissociation rate constants when compared to clone THR-29, and the shape of the kinetic binding response closely resembled that of an antibody. The KD values for both thrombin-binding knottins were between 300 and 500 nM. The native, acyclic kalata B1 peptide produced less than a 5 RU response at the highest peptide concentration tested (10 µM) confirming that the knottin scaffold did not appreciably bind to thrombin.
Figure 4.
Measurement of thrombin-binding knottin affinity. Response curves were measured using surface plasmon resonance for A) THR-5 and B) THR-29 binding to thrombin immobilized on a CM5 chip. The raw data are shown as a solid black line with the fit (1:1 Langmuir binding model) as a dashed red line. A two-fold dilution series from 78 nM to 2.5 µM was used for each of the peptides with each series run in duplicate. The kinetic values calculated from each of the fits (association rate constant, dissociation rate constant, and KD) are given.
The kalata B1 scaffold conferred antibody-like binding kinetics, characterized by slow association and dissociation rates, to the discovered thrombin-binding epitopes. Interestingly, the therapeutic thrombin inhibitor bivalirudin (Angiomax™) has an association rate constant four orders of magnitude larger than that of THR-5 and THR-29 (4×108 M−1 s−1 versus 103–104 M−1 s−1), balanced by a rapid dissociation rate over two orders of magnitude faster (0.8 s−1 versus 10−3 s−1) (35). Other constrained peptide scaffolds have been shown to exhibit similarly slow dissociation rate constants as those measured here, a feature that may prove useful for in vivo applications. For example, variants of Min-23, a truncated form of EETI-II, bind an anti-Epstein Barr virus antibody with an association rate constant of 3.9−104 M−1 s−1 and a dissociation rate constant of 6.1×10−4 s−1 (36).
To determine whether the soluble kalata B1 variants were effective active site inhibitors, thrombin inhibition by knottin THR-5 was measured using a chromagenic thrombin substrate. Different concentrations of the peptide were assayed with a single concentration of thrombin and substrate (Supplementary Figure 3). An IC50 value of 8.4 ± 0.7 µM was calculated from triplicate experiments. Peptides identified here interact exclusively with the active site and achieve modest inhibitory activity. Many naturally occurring peptides achieve high affinity and inhibitory activity by bivalent binding to both the active site and exosite I of thrombin. For example, derivatives of the approved thrombin inhibitor hirudin have Ki values between 60 and 200 fM (37). In addition, Kunitz-domain inhibitors isolated from ticks also exhibit high thrombin inhibitory activity with Ki values in the low pM to low nM range (38–39).
The affinities of the thrombin-binding knottins could be improved by randomizing an additional loop in the scaffold to generate a peptide inhibitor that mimics the bivalent binding of bivalirudin. Bivalirudin contains an active site binding motif, a short tetra-glycine linker, and an anionic peptide that interacts with exosite I. The active site inhibitor sequence in bivalirudin (D-Phe-Pro-Arg-Pro) does not significantly inhibit thrombin even at concentrations up to 10 µM (40), while THR-5 had an IC50 in the low-µM range. Furthermore, the potency of bivalirudin is reduced by slow hydrolysis within the active site binding ligand (35), whereas the kalata B1 variants are highly resistant to proteolysis. The intact bivalirudin peptide, however, has a Ki of 2 nM (40) illustrating the dramatic effect of bivalent interaction for increasing inhibitory activity. A new library could be constructed based on peptide THR-5 by randomizing loop three in the scaffold (41), and then sorting with more stringent conditions to isolate an inhibitor that interacts with both exosite I and the active site to dramatically improve its affinity.
Variants retain the proteolytic stability of kalata B1
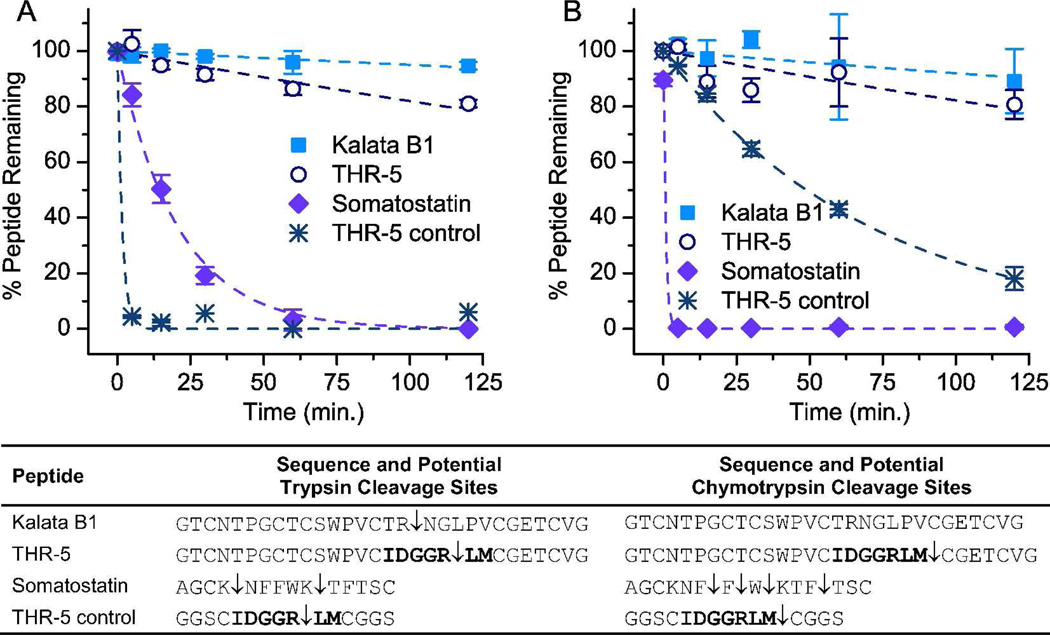
To determine whether the thrombin-binding knottins retained the proteolytic resistance characteristics of the parent kalata B1 peptide, their ability to resist degradation by trypsin (10 µg mL−1) and chymotrypsin (100 µg mL−1) was measured. The trypsin concentration was chosen to enable direct comparisons with previous studies characterizing the trypsin resistance of kalata B1 (24, 42). With both proteases, 80% of knottin THR-5 remained intact following a two hour incubation and exhibited dramatically enhanced resistance compared to the control peptides (Figure 5). The fraction of THR-5 that was degraded by trypsin was hydrolyzed after the arginine residue of the binding epitope in loop six. Chymotrypsin cleaved after the methionine residue also found in the binding loop of THR-5. The acyclic kalata B1 peptide did not show appreciable degradation by trypsin or chymotrypsin over the entire 120 minute incubation as has been observed previously (24).
Figure 5.
Trypsin and chymotrypsin resistance of a thrombin-specific knottin THR-5. The acyclic kalata B1 parent, THR-5, somatostatin, and a cyclic, control peptide corresponding to the binding loop of THR-5 were incubated with A) trypsin and B) chymotrypsin and analyzed by LC/MS. The sequences for each of the peptides are listed along with the potential cleavage sites. Kalata B1 was stable over the two hour incubation period with both proteases, and 80% of THR-5 remained intact. Somatostatin and the THR-5 control were readily degraded by both trypsin and chymotrypsin. The curves were fit to a second-order reaction model and the experiments were performed in duplicate.
In contrast, the THR-5 binding loop constrained by a single disulfide bond was completely degraded in less than five minutes by trypsin. In fact, the majority of the degradation occurred within one minute following trypsin addition before the reaction could be fully quenched. Furthermore, an unrelated trypsin-sensitive peptide containing two lysines (somatostatin 14) was rapidly degraded by trypsin. Somatostatin, which is also constrained by a single disulfide bond, was completely cleaved by trypsin at both cleavage sites within one hour. By fitting the peptide degradation curves to a second-order reaction model, THR-5 in the kalata B1 scaffold is degraded by trypsin at least 100-fold slower than the THR-5 epitope constrained by a single disulfide bond.
The two control peptides were also extensively degraded by chymotrypsin. The THR-5 loop control was cleaved following the methionine residue and less than 20% of the intact peptide remained after the two hour incubation. The THR-5 control peptide was degraded eight-fold faster than the kalata B1 variant determined by the ratio of the second-order rate constants. In addition, somatostatin, which contains multiple large, hydrophobic residues that are the preferred substrates for chymotrypsin, was cleaved at two or more sites within five minutes and was extensively degraded at all four cleavage sites within two hours. Thus, these results demonstrate that THR-5 was significantly more stable than the single-disulfide constrained control peptides and its proteolytic resistance approached that of the acyclic, parental kalata B1 sequence.
Trypsin and chymotrypsin were chosen for the stability assay because THR-5 had accessible cleavage sites in the thrombin-binding region (loop six) of the peptide. Because trypsin and chymotrypsin are representative members of large classes of proteases and have high activity combined with broad substrate specificity, they are commonly used to assess the relative proteolytic stability of peptides (21, 24). The stability of THR-5 is notable since other cysteine-constrained peptides are susceptible to trypsin and chymotrypsin degradation. The cystine knot structure of native kalata B1 is believed to be critical for its proteolytic stability. Although the use of NMR is generally required to verify the disulfide connectivity (25), the observations that i) the elution profile for refolded THR-5 contained a large, late-eluting peak indicative of surface exposure of a hydrophobic patch and ii) THR-5 exhibits a trypsin resistance comparable to the acyclic permutant of kalata B1 suggests that THR-5 possesses a native-like cystine knot structure (26). In contrast, THR-29 exhibited multiple elution peaks corresponding to distinct peptide conformers constrained by three disulfide bonds. The prolines present in loop six of THR-29 may have hindered its folding into the native conformation. In addition, backbone N- to C-terminal cyclization could further enhance the stability of THR-5 and other kalata B1 derived peptides by increasing resistance to exopeptidases (43).
Conclusions
In summary, a peptide library derived from the stable kalata B1 scaffold was successfully screened using FACS to yield a diverse set of thrombin binders that could be grouped into multiple distinct families with unique consensus motifs. In addition, bacterial display proved to be a highly effective tool for the generation and quantitative screening of a large library of knottin peptides. While phage display, and to a lesser extent mRNA and DNA display methods, have been applied to identify and/or affinity mature peptides, relatively few reports have described the screening of peptide libraries for novel specificities using yeast or bacterial display methods (44–45). These cell surface display technologies are advantageous because they enable quantitative library screening and clone characterization using flow cytometry. The use of the engineered display scaffold eCPX provided sufficient display levels, although lower than the linear and loop constructs, for screening peptide libraries derived from a complex knottin structure with multiple disulfide bonds. The reduced export of the kalata B1 scaffold did not interfere with the selection of thrombin inhibitors using bacterial display. Although reduced display can decrease the dynamic range of the mean cell fluorescence during screening, low display levels proved useful to avoid avidity effects. Cytometry accurately reported affinities of surface displayed thrombin binders that were within two-fold of the corresponding values measured in solution using surface plasmon resonance. This result contrasts with the poor agreement between cell surface and solution affinities observed for homomultimeric targets, where avidity and rebinding effects can skew apparent affinities (27–28) indicating that the reduced display level may be advantageous. The kalata B1 scaffold also conferred dramatically increased binding affinity and protease resistance when compared to a single-disulfide constrained peptide. The kalata B1 variant THR-5 exhibited more than a 100-fold increased resistance to the major digestive protease trypsin when compared to the epitope constrained by a single disulfide bond. Thus, bacterial displayed knottin libraries coupled with quantitative library screening via FACS provide a highly effective tool for the discovery and engineering of protease resistant peptide ligands.
METHODS
Bacterial strains, plasmids, and growth conditions
All bacterial display experiments utilized the plasmid pBAD33 (Cmr) in E. coli strain MC1061 (46–47). Cells were subcultured 1:50 from an overnight culture into LB medium supplemented with 34 µg mL−1 chloramphenicol and incubated at 37°C for 2 hours with vigorous shaking. To achieve surface display, protein synthesis was induced with 0.04% (w/v) L-arabinose and the cells were incubated at 37°C for an additional 45 minutes.
Construction and screening of a kalata B1 library
A library of kalata B1 variants was constructed by breaking the native cyclic backbone within loop two and fusing the resulting C-terminal glycine to the N-terminus of the eCPX display scaffold (27). The display scaffold eCPX utilizes the native OmpX signal sequence which is cleaved during translocation of the fusion protein to the periplasm. The seven amino acids in loop six of kalata B1 were randomized using NNS degenerate codons to create a library. Assembly PCR was performed using KOD Hot Start DNA polymerase (Novagen) with overlapping primers (Supplementary Table 1, Operon). An additional PCR reaction was used to construct the library with the assembled forward primer, GGCTGAAAATCTTCTCTC as the reverse primer, and pBAD33-eCPX as the template. The PCR product and vector were digested with SfiI (New England Biolabs), ligated, and electroporated into E. coli MC1061, yielding 6×109 transformants.
The knottin library was sorted using one round of magnetic-activated cell sorting (MACS) followed by three cycles of FACS, as described elsewhere (48). Human α-thrombin (Haematologic Technologies or Sigma Aldrich) was first biotinylated using the FluoReporter mini-biotin-XX protein labeling kit (Invitrogen). MACS was performed using 30 nM biotinylated α-thrombin and MyOne streptavidin-coated magnetic beads (Invitrogen) with a bead:cell ratio of 1:100. For the first two rounds of FACS, the cells were incubated with 30 nM biotinylated α-thrombin on ice for 45 minutes and labeled with streptavidin-R-phycoerythrin (SAPE, Invitrogen) at a final concentration of 17 nM. A third cycle of kinetic screening was performed by first incubating the population with α-thrombin (80 nM). The cells were then pelleted by centrifugation and incubated with 50 µM DNA aptamer competitor in PBS (GGTTGGTGTGGTTGG, Operon) at room temperature for one hour (49). The cells were labeled with neutravidin-R-phycoerythrin (NAPE, Invitrogen) on ice prior to sorting. NAPE was used instead of SAPE for this last round of sorting to disfavor the enrichment of streptavidin-binding peptides. Individual clones from both the second and third rounds of selections were isolated by plating, and the composition of the corresponding displayed peptide was determined by plasmid DNA sequencing.
Cell surface characterizations
To verify that the isolated clones did not bind the labeling reagents, they were first assayed for binding to SAPE and NAPE. The dissociation rate for each clone was then measured by labeling cells with biotinylated α-thrombin at a final concentration of 800 nM, washing the cells with PBS, and incubation at room temperature. At specific time intervals, aliquots were removed and labeled with SAPE for cytometric analysis. To avoid rebinding effects, an induction time of 15 minutes was used to reduce the number of peptides on the cell surface.
The thrombin-binding epitope (i.e. randomized loop) of clone THR-5 was constructed as both linear and single-disulfide constrained peptides on the cell surface by recombinantly fusing them to the N-terminus of eCPX. Cells were labeled with 4-fold serial dilutions of biotinylated α-thrombin (3.4 nM to 14 µM) and SAPE and analyzed by flow cytometry. The apparent equilibrium dissociation constant (KDApp) was calculated by fitting the plot of mean cell fluorescence versus thrombin concentration to the equation:
FL is the mean cell fluorescence, FLmax is the maximum observed fluorescence, FLbg is the background cell fluorescence of unlabeled bacteria, and [T] is the thrombin concentration (30).
To determine the thrombin-binding site of each of the consensus groups, peptides were labeled with biotinylated thrombin (875 nM) and a concentration series (5 nM to 20 µM) of unlabeled competitors including α-thrombin, γ-thrombin, PPACK-blocked thrombin (all forms of thrombin from Haematologic Technologies), hirudin (Hyphen Biomed), and a thrombin specific DNA aptamer (Operon) (49).
Refolding of the synthetic kalata B1 peptides
Synthetic, reduced knottin peptides (Biomatik) were purified using RP-HPLC (HP 1100) on a Grace Vydac C18 column. The purified material was then added to the refolding buffer (1:1 0.1 M ammonium bicarbonate:isopropanol, 5 mM reduced glutathione, 0.5 mM oxidized glutathione) until the peptide concentration reached 50 µg mL−1 (50). The refolding reaction was stirred at 4°C for 70 hours and then acidified to pH 3. The peptides were loaded onto a C18 column and purified using a gradient of 5–51% acetonitrile over 36 minutes at a flow rate of 4 mL min−1. The refolded, acyclic kalata B1 mixture showed a sharp elution peak at 28 minutes, while the misfolded and partially folded kalata B1 material eluted earlier as a broad peak. Clone THR-5 also showed a clear elution peak (27 minutes) following the peaks containing misfolded material although the separation between the two species was not as distinct as with kalata B1. The misfolded material contained a mixture of species with both two and three disulfide bonds present. Clone THR-29, however, did not have a clear, late eluting peak. Peptides THR-20 and THR-24 exhibited poor solubility, which prevented successful refolding. All collected fractions were analyzed using mass spectrometry on a Micromass QTOF2 ESI/TOF instrument to determine contents and degree of disulfide connectivity. In addition, the purified peaks were run on an analytical C18 column to determine peptide purity, which was calculated to be over 90% for THR-5 and acyclic kalata B1.
Affinity measurements using surface plasmon resonance
The solution affinities of the refolded peptides were measured using a Biacore 3000 surface plasmon resonance instrument (GE Healthcare). On a CM5 chip, α-thrombin was immobilized through amine coupling. The surface was activated with a 1:1 (v:v) mixture of 0.4 M EDC (N-ethyl-N′-(3-dimethylaminopropyl)carbodiimide) and 0.1 M NHS (N-hydroxysuccinimide). Then 2 µg mL−1 α-thrombin in 10 mM sodium acetate buffer, pH 5, was immobilized to a level of approximately 1600 RU. Unreacted carboxyl groups were deactivated using 1 M ethanolamine, pH 8.5. An additional reference flow cell underwent the same immobilization procedure without the α-thrombin step. A kinetic binding analysis was performed using two-fold serial dilutions of the refolded peptides (78 nM to 2.5 µM) with each of the series run in duplicate. The peptides were injected for 3 minutes at 50 µL min−1 followed by a 6 minute dissociation period with HBS-EP (10 mM HEPES, 150 mM NaCl, 3 mM EDTA, 0.05% (v/v) Tween 20, pH 7.4) as the running buffer. A regeneration step of 10 mM sodium acetate, pH 5, injected for 12 seconds at 100 µL min−1 was performed after each injection of THR-5, while only a single buffer injection was required for the regeneration of THR-29. Response signals were double referenced by subtracting the signals of both a reference flow cell as well as blank buffer injections. BIAevalution software (GE Healthcare) was used to calculate the equilibrium dissociation constant (KD) values by fitting the data to a 1:1 Langmuir binding model.
The response curves did not exactly fit the chosen 1:1 model with the most deviation occurring at high peptide concentrations. To test whether other artifacts may be causing the different shape of the curves, mass transfer limitations and linked reactions were tested using the Biacore wizards. Mass transfer was not a major effect as established by the constant slope of the association curve with increasing flow rates (15, 30, 75 µL min−1). To determine if there were multiple species present in the analyte or if a conformational change occurred during the binding, the peptide was injected for increasing lengths of time and the shape of the dissociation curves were compared. The overlaying dissociation curves resulting from this test indicated that the analyte did not contain more than one binding species.
Inhibition of thrombin by kalata B1 variants
To determine if the thrombin-binding knottins could inhibit the enzymatic activity of thrombin, the peptides were tested with a chromagenic peptide substrate and α-thrombin. A concentration series of refolded THR-5 (470 nM to 18.3 µM) was preincubated with 2 nM α-thrombin (4360 U mg−1) in the following buffer (50 mM Tris-HCl, 150 mM NaCl, 2.5 mM CaCl2, pH 8) for 45 minutes. The assay was initiated by adding 25 µM chromagenic peptide substrate (Pefachrome TH, Pentapharm) and was run at 25°C. The IC50 was calculated by fitting the normalized initial velocities versus inhibitor concentration to the Langmuir isotherm equation (51).
Trypsin and chymotrypsin stability assays
For the trypsin digests, the refolded peptides (final concentration of 0.5 mg mL−1 in HBS-EP buffer) were mixed with 0.1 mg mL−1 trypsin (Sigma Aldrich, 12,500 BAEE U mg−1) to a final peptide:trypsin mass ratio of 50:1 and incubated at 37°C (24). Similarly, chymotrypsin (Thermo Scientific, 56 BTEE U mg−1) was added to the peptides (in HBS buffer with no EDTA or Tween) at a final peptide:chymotrypsin mass ratio of 5:1 and kept at 37°C. Two control peptides, somatostatin 14 (AGCKNFFWKTFTSC, Anaspec) and clone THR-5 as a single-disulfide constrained peptide (GGSCIDGGRLMCGGS, Biomatik), were used as positive controls for the trypsin and chymotrypsin digestion. Following the addition of the protease, aliquots were removed at 0, 5, 15, 30, 60, and 120 minutes and the digestion quenched by the addition of TFA to a final concentration of 0.1% (v/v). Peptides were analyzed by LC/MS (Waters 2695 HPLC system/Micromass QTOF2) with a gradient of 5–46% acetonitrile over 32 minutes and recovery was calculated by integration of the 280 nm signal. To determine relative cleavage rates of the peptides, the stability data (% degradation versus time) were fit to the following second-order rate model:
where kcat/Km is the rate constant, t is time, and [E] is the enzyme concentration which is assumed constant resulting in the kinetics following a pseudo first-order form (52). The ratio of the rate constants for two different peptides gave a relative measure of their stability.
Supplementary Material
ACKNOWLEDGMENTS
We thank J. Pavlovich for his assistance with the LC/MS experiments. This work was supported by NIH NHLBI grant 5 U01 H1080718-04 and U.S. Army Research Office-Institute for Collaborative Biotechnologies grants DAAD19-03-D-0004 and W911NF-09-D-0001.
The abbreviations used are
- FACS
fluorescence-activated cell sorting
- MACS
magnetic-activated cell sorting
- eCPX
enhanced circularly permuted OmpX
- EETI-II
Ecballium elaterium trypsin inhibitor II
- AGRP
Agouti-related protein
- KDApp
apparent equilibrium dissociation constant
- KD
equilibrium dissociation constant
- RP-HPLC
reverse-phase high performance liquid chromatography
Footnotes
Supporting Information Available: This material is available free of charge via the Internet.
REFERENCES
- 1.Pichereau C, Allary C. Therapeutic peptides under the spotlight. Eur. Biopharm. Rev. 2005:88–91. [Google Scholar]
- 2.Sato AK, Viswanathan M, Kent RB, Wood CR. Therapeutic peptides: technological advances driving peptides into development. Curr. Opin. Biotechnol. 2006;17:638–642. doi: 10.1016/j.copbio.2006.10.002. [DOI] [PubMed] [Google Scholar]
- 3.Werle M, Bernkop-Schnürch A. Strategies to improve plasma half life time of peptide and protein drugs. Amino Acids. 2006;30:351–367. doi: 10.1007/s00726-005-0289-3. [DOI] [PubMed] [Google Scholar]
- 4.McGregor DP. Discovering and improving novel peptide therapeutics. Curr. Opin. Pharm. 2008;8:616–619. doi: 10.1016/j.coph.2008.06.002. [DOI] [PubMed] [Google Scholar]
- 5.Stromstedt AA, Pasupuleti M, Schmidtchen A, Malmsten M. Evaluation of strategies for improving proteolytic resistance of antimicrobial peptides by using variants of EFK17, an internal segment of LL-37. Antimicrob. Agents Chemother. 2009;53:593–602. doi: 10.1128/AAC.00477-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Clark RJ, Fischer H, Dempster L, Daly NL, Rosengren KJ, Nevin ST, Meunier FA, Adams DJ, Craik DJ. Engineering stable peptide toxins by means of backbone cyclization: Stabilization of the -conotoxin MII. Proc. Natl. Acad. Sci. USA. 2005;102:13767–13772. doi: 10.1073/pnas.0504613102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Harris AG. Somatostatin and somatostatin analogues: pharmacokinetics and pharmacodynamic effects. Gut. 1994;35:S1–S4. doi: 10.1136/gut.35.3_suppl.s1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Tang Y, Tirrell DA. Biosynthesis of a highly stable coiled-coil protein containing hexafluoroleucine in an engineered bacterial host. J. Am. Chem. Soc. 2001;123:11089–11090. doi: 10.1021/ja016652k. [DOI] [PubMed] [Google Scholar]
- 9.Ramon J, Saez V, Baez R, Aldana R, Hardy E. PEGylated Interferon-2b: A Branched 40K Polyethylene Glycol Derivative. Pharm. Res. 2005;22:1375–1387. doi: 10.1007/s11095-005-5278-4. [DOI] [PubMed] [Google Scholar]
- 10.Nygren P, Skerra A. Binding proteins from alternative scaffolds. J. Immunol. Methods. 2004;290:3–28. doi: 10.1016/j.jim.2004.04.006. [DOI] [PubMed] [Google Scholar]
- 11.Chiche L, Heitz A, Gelly JC, Gracy J, Chau PTT, Ha PT, Hernandez JF, Le-Nguyen D. Squash inhibitors: from structural motifs to macrocyclic knottins. Curr. Protein Peptide Sci. 2004;5:341–349. doi: 10.2174/1389203043379477. [DOI] [PubMed] [Google Scholar]
- 12.Christmann A, Walter K, Wentzel A, Kratzner R, Kolmar H. The cystine knot of a squash-type protease inhibitor as a structural scaffold for Escherichia coli cell surface display of conformationally constrained peptides. Protein Eng. Des. Sel. 1999;12:797–806. doi: 10.1093/protein/12.9.797. [DOI] [PubMed] [Google Scholar]
- 13.Krause S, Schmoldt HU, Wentzel A, Ballmaier M, Friedrich K, Kolmar H. Grafting of thrombopoietin-mimetic peptides into cystine knot miniproteins yields high-affinity thrombopoietin antagonists and agonists. FEBS J. 2006;274:86–95. doi: 10.1111/j.1742-4658.2006.05567.x. [DOI] [PubMed] [Google Scholar]
- 14.Gunasekera S, Foley FM, Clark RJ, Sando L, Fabri LJ, Craik DJ, Daly NL. Engineering Stabilized Vascular Endothelial Growth Factor-A Antagonists: Synthesis, Structural Characterization, and Bioactivity of Grafted Analogues of Cyclotides. J. Med. Chem. 2008;51:7697–7704. doi: 10.1021/jm800704e. [DOI] [PubMed] [Google Scholar]
- 15.Silverman AP, Levin AM, Lahti JL, Cochran JR. Engineered Cystine-Knot Peptides that Bind alpha(v)beta(3) Integrin with Antibody-Like Affinities. J. Mol. Biol. 2009;385:1064–1075. doi: 10.1016/j.jmb.2008.11.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Markland W, Ley AC, Ladner RC. Iterative optimization of high-affinity protease inhibitors using phage display. 2. Plasma kallikrein and thrombin. Biochemistry. 1996;35:8058–8067. doi: 10.1021/bi952629y. [DOI] [PubMed] [Google Scholar]
- 17.Markland W, Ley AC, Lee SW, Ladner RC. Iterative optimization of high-affinity proteases inhibitors using phage display. 1. Plasmin. Biochemistry. 1996;35:8045–8057. doi: 10.1021/bi9526286. [DOI] [PubMed] [Google Scholar]
- 18.Kiczak L, Kasztura M, Koscielska-Kasprzak K, Dadlez M, Otlewski J. Selection of potent chymotrypsin and elastase inhibitors from M13 phage library of basic pancreatic trypsin inhibitor (BPTI) Biochim. Biophys. Acta. 2001;1550:153–163. doi: 10.1016/s0167-4838(01)00282-5. [DOI] [PubMed] [Google Scholar]
- 19.Roberts BL, Markland W, Ley AC, Kent RB, White DW, Guterman SK, Ladner RC. Directed evolution of a protein: selection of potent neutrophil elastase inhibitors displayed on M13 fusion phage. Proc. Natl. Acad. Sci. USA. 1992;89:2429–2433. doi: 10.1073/pnas.89.6.2429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lehmann A. Ecallantide (DX-88), a plasma kallikrein inhibitor for the treatment of hereditary angioedema and the prevention of blood loss in on-pump cardiothoracic surgery. Expert Opin Biol Ther. 2008;8:1187–1199. doi: 10.1517/14712598.8.8.1187. [DOI] [PubMed] [Google Scholar]
- 21.Werle M, Schmitz T, Huang HL, Wentzel A, Kolmar H, Bernkop-Schnürch A. The potential of cystine-knot microproteins as novel pharmacophoric scaffolds in oral peptide drug delivery. J. Drug Targeting. 2006;14:137–146. doi: 10.1080/10611860600648254. [DOI] [PubMed] [Google Scholar]
- 22.Zheng K, Lubman DM, Rossi DT, Nordblom GD, Barksdale CM. Elucidation of peptide metabolism by on-line immunoaffinity liquid chromatography mass spectrometry. Rapid Commun. Mass Spectrom. 2000;14:261–269. doi: 10.1002/(SICI)1097-0231(20000229)14:4<261::AID-RCM868>3.0.CO;2-X. [DOI] [PubMed] [Google Scholar]
- 23.Werle M, Kafedjiiski K, Kolmar H, Bernkop-Schnürch A. Evaluation and improvement of the properties of the novel cystine-knot microprotein McoEeTI for oral administration. Int. J. Pharm. 2007;332:72–79. doi: 10.1016/j.ijpharm.2006.09.028. [DOI] [PubMed] [Google Scholar]
- 24.Colgrave ML, Craik DJ. Thermal, chemical, and enzymatic stability of the cyclotide kalata B1: the importance of the cyclic cystine knot. Biochemistry. 2004;43:5965–5975. doi: 10.1021/bi049711q. [DOI] [PubMed] [Google Scholar]
- 25.Rosengren KJ, Daly NL, Plan MR, Waine C, Craik DJ. Twists, Knots, and Rings in Proteins STRUCTURAL DEFINITION OF THE CYCLOTIDE FRAMEWORK. J. Biol. Chem. 2003;278:8606–8616. doi: 10.1074/jbc.M211147200. [DOI] [PubMed] [Google Scholar]
- 26.Daly NL, Craik DJ. Acyclic Permutants of Naturally Occurring Cyclic Proteins. J. Biol. Chem. 2000;275:19068–19075. doi: 10.1074/jbc.M000450200. [DOI] [PubMed] [Google Scholar]
- 27.Rice JJ, Daugherty PS. Directed evolution of a biterminal bacterial display scaffold enhances the display of diverse peptides. Protein Eng. Des. Sel. 2008;21:435–442. doi: 10.1093/protein/gzn020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kenrick SA, Daugherty PS. Bacterial display enables efficient and quantitative peptide affinity maturation. Protein Eng. Des. Sel. 2010;23:9–17. doi: 10.1093/protein/gzp065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Patrick WM, Firth AE, Blackburn JM. User-friendly algorithms for estimating completeness and diversity in randomized protein-encoding libraries. Protein Eng. Des. Sel. 2003;16:451–457. doi: 10.1093/protein/gzg057. [DOI] [PubMed] [Google Scholar]
- 30.Boder ET, Wittrup KD. Optimal screening of surface-displayed polypeptide libraries. Biotechnol. Prog. 1998;14:55–62. doi: 10.1021/bp970144q. [DOI] [PubMed] [Google Scholar]
- 31.Georgiou G, Segatori L. Preparative expression of secreted proteins in bacteria: status report and future prospects. Curr. Opin. Biotechnol. 2005;16:538–545. doi: 10.1016/j.copbio.2005.07.008. [DOI] [PubMed] [Google Scholar]
- 32.Bode W, Turk D, Karshikov A. The refined 1.9-angstrom X-ray crystal-structure of D-Phe-Pro-Arg chloromethylketone-inhibited human alpha-thrombin. Protein Sci. 1992;1:426–471. doi: 10.1002/pro.5560010402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Paborsky LR, McCurdy SN, Griffin LC, Toole JJ, Leung LL. The single-stranded DNA aptamer-binding site of human thrombin. J. Biol. Chem. 1993;268:20808–20811. [PubMed] [Google Scholar]
- 34.Petrera NS, Stafford AR, Leslie BA, Kretz CA, Fredenburgh JC, Weitz JI. Long range communication between exosites 1 and 2 modulates thrombin function. J. Biol. Chem. 2009;284:25620–25629. doi: 10.1074/jbc.M109.000042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Parry MAA, Maraganore JM, Stone SR. Kinetic Mechanism for the Interaction of Hirulog with Thrombin. Biochemistry. 1994;33:14807–14814. doi: 10.1021/bi00253a020. [DOI] [PubMed] [Google Scholar]
- 36.Souriau C, Chiche L, Irving R, Hudson P. New Binding Specificities Derived from Min-23, a Small Cystine-Stabilized Peptidic Scaffold. Biochemistry. 2005;44:7143–7155. doi: 10.1021/bi0481592. [DOI] [PubMed] [Google Scholar]
- 37.Warkentin TE. Bivalent direct thrombin inhibitors: hirudin and bivalirudin. Best Practice & Research Clinical Haematology. 2004;17:105–125. doi: 10.1016/j.beha.2004.02.002. [DOI] [PubMed] [Google Scholar]
- 38.Maritz-Olivier C, Stutzer C, Jongejan F, Neitz AWH, Gaspar ARM. Tick anti-hemostatics: targets for future vaccines and therapeutics. Trends Parasitol. 2007;23:397–407. doi: 10.1016/j.pt.2007.07.005. [DOI] [PubMed] [Google Scholar]
- 39.Macedo-Ribeiro S, Almeida C, Calisto BM, Friedrich T, Mentele R, Sturzebecher J, Fuentes-Prior P, Pereira PJB. Isolation, Cloning and Structural Characterisation of Boophilin, a Multifunctional Kunitz-Type Proteinase Inhibitor from the Cattle Tick. PLoS One. 2008;3 doi: 10.1371/journal.pone.0001624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Maraganore JM, Bourdon P, Jablonski J, Ramachandran KL, Fenton JW. Design and characterization of hirulogs: a novel class of bivalent peptide inhibitors of thrombin. Biochemistry. 1990;29:7095–7101. doi: 10.1021/bi00482a021. [DOI] [PubMed] [Google Scholar]
- 41.Clark RJ, Daly NL, Craik DJ. Structural plasticity of the cyclic-cystine-knot framework: implications for biological activity and drug design. Biochem. J. 2006;394:85–93. doi: 10.1042/BJ20051691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Simonsen SM, Sando L, Rosengren KJ, Wang CK, Colgrave ML, Daly NL, Craik DJ. Alanine scanning mutagenesis of the prototypic cyclotide reveals a cluster of residues essential for bioactivity. J. Biol. Chem. 2008;283:9805–9813. doi: 10.1074/jbc.M709303200. [DOI] [PubMed] [Google Scholar]
- 43.Iwai H, Plückthun A. Circular [beta]-lactamase: stability enhancement by cyclizing the backbone. FEBS Lett. 1999;459:166–172. doi: 10.1016/s0014-5793(99)01220-x. [DOI] [PubMed] [Google Scholar]
- 44.Daugherty PS. Protein engineering with bacterial display. Curr. Opin. Struct. Biol. 2007;17:474–480. doi: 10.1016/j.sbi.2007.07.004. [DOI] [PubMed] [Google Scholar]
- 45.Gai SA, Wittrup KD. Yeast surface display for protein engineering and characterization. Curr. Opin. Struct. Biol. 2007;17:467–473. doi: 10.1016/j.sbi.2007.08.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Guzman LM, Belin D, Carson MJ, Beckwith J. Tight regulation, modulation, and high-level expression by vectors containing the arabinose PBAD promoter. J. Bacteriol. 1995;177:4121–4130. doi: 10.1128/jb.177.14.4121-4130.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Casadaban MJ, Cohen SN. Analysis of gene control signals by DNA fusion and cloning in Escherichia coli. J. Mol. Biol. 1980;138:179–207. doi: 10.1016/0022-2836(80)90283-1. [DOI] [PubMed] [Google Scholar]
- 48.Kenrick S, Rice J, Daugherty P. Flow Cytometric Sorting of Bacterial Surface-Displayed Libraries. Current Protocols in Cytometry. 2007;42:4.6.1–4.6.27. doi: 10.1002/0471142956.cy0406s42. [DOI] [PubMed] [Google Scholar]
- 49.Bock LC, Griffin LC, Latham JA, Vermaas EH, Toole JJ. Selection of single-stranded DNA molecules that bind and inhibit human thrombin. Nature. 1992;355:564–566. doi: 10.1038/355564a0. [DOI] [PubMed] [Google Scholar]
- 50.Daly NL, Love S, Alewood PF, Craik DJ. Chemical synthesis and folding pathways of large cyclic polypeptides: studies of the cystine knot polypeptide kalata B1. Biochemistry. 1999;38:10606–10614. doi: 10.1021/bi990605b. [DOI] [PubMed] [Google Scholar]
- 51.Copeland RA. Enzymes: a practical introduction to structure, mechanism, and data analysis. 2nd ed. Wiley-Vch; 2000. [Google Scholar]
- 52.Boulware KT, Daugherty PS. Protease specificity determination by using cellular libraries of peptide substrates (CLiPS) PNAS. 2006;103:7583–7588. doi: 10.1073/pnas.0511108103. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.