Abstract
Costello syndrome was first reported based on its characteristic phenotype. Its presentation affects multiple organ systems, including severe failure-to-thrive with macrocephaly, characteristic facial features, hypertrophic cardiomyopathy, papillomata, malignant tumors, and cognitive impairment. Heterozygous germline mutations in the proto-oncogene HRAS have been recognized to cause Costello syndrome, and its inheritance pattern would thus be autosomal dominant. Here we report on the identification of an HRAS mutation c.34G>A, predicting a p.G12S amino acid substitution, in the surviving brother of a previously reported sibling pair, and documentation of the same change in autopsy material from his deceased sister. This represents, to our knowledge, the first molecularly confirmed Costello syndrome in siblings. We did not detect the mutation in a heterozygous state or mosaicism in peripheral white blood cell or cheek swab derived DNA samples from either parent. Using single nucleotide polymorphic markers and allele specific amplification, we clearly identified the mutation in the surviving sibling to be of maternal origin. While we cannot exclude two independently occurring de novo mutations, the complete sharing of polymorphic markers around the mutation site in both siblings supports maternal germ cell mosaicism. Recurrence risk counseling for families with apparently de novo occurring autosomal dominant conditions includes discussion of germ cell mosaicism, and this report underscores the applicability of this concern to Costello syndrome.
Keywords: Costello syndrome, germ cell mosaicism, HRAS, maternal origin, recurrence risk
INTRODUCTION
Costello syndrome encompasses severe failure-to-thrive, cardiac abnormalities including tachyarrhythmia and hypertrophic cardiomyopathy, a predisposition to papillomata and malignant tumors, and neurologic abnormalities including intellectual disability, nystagmus and hypotonia [for review see Gripp and Lin, 2009]. Prior to the identification of its genetic cause, several affected sibling pairs were reported and autosomal recessive inheritance was considered one possible explanation. Costello syndrome is now known to be caused by heterozygous germline mutations in the proto-oncogene HRAS located on chromosome 11p15.5 [Aoki et al., 2005]. Three sibling pairs with a clinical diagnosis of Costello syndrome were previously reported in addition to the family discussed here [Zampino et al., 1993; van den Bosch et al., 2002; Ioan and Fryns, 2002]. Molecular confirmation of the presumed diagnosis is important as demonstrated by sisters with an HRAS and a KRAS mutation [Sovik et al., 2007]. Familial Costello syndrome cases include clinically described twin brothers in whom the documented monozygosity accounts for the familial nature [van den Bosch et al., 2002]. Ioan and Fryns [2002] provided a clinical description of siblings with presumed Costello syndrome and their mother with mild to moderate cognitive impairment, suggesting variable expression of an autosomal dominant condition or somatic mosaicism in the mother. While no molecular data are available in this family [Ioan and Fryns, 2002], it is noteworthy that we reported one female with somatic mosaicism for HRAS p.G12S, who appears to be at risk for offspring with typical Costello syndrome [Gripp et al., 2006]. More recently, we documented transmission from a father with somatic mosaicism for HRAS p.G12S to his son with Costello syndrome [Sol-Church et al., 2009]. These two reports of somatic mosaicism demonstrate a potential mechanism for recurrence of Costello syndrome in siblings [Gripp et al., 2006; Sol-Church et al., 2009].
MATERIALS AND METHODS
In our ongoing clinical and molecular study on Costello syndrome and related disorders, including 86 probands with Costello syndrome and a presumably pathogenic germline HRAS mutation, we are not aware of any recurrence in a sibling of an affected individual in addition to the family reported here.
Clinical Reports
The living individual with Costello syndrome (Patient 1; Fig. 1: II-2; CS#258) was enrolled in our ongoing study of Costello syndrome (A. I. duPont Hospital for Children institutional review board #2005-051). The proband was born to a 29-year-old mother and a 25-year-old father, and polyhydramnios was noted when delivery occurred at 36 weeks gestation. Reported previously in Johnson et al. [1998] as Patient 6, his clinical presentation was typical for Costello syndrome, including severe failure-to-thrive and cognitive impairment, orthopedic complications and facial papillomata. Nystagmus and hypotonia were diagnosed in childhood, and loose appearing skin was present. He is now 32 years old (Fig. 2) and shows the coarse facial features and curly hair typical for Costello syndrome. He is unable to live independently due to significant cognitive disability. While he has no known cardiac problems, his only echocardiogram was performed many years ago. Owing to orthopedic problems including severe kyphosis and hip dislocation, he has never been able to walk independently. His height is about 122 cm (−7SD; 50th centile for age 7 years), weight is 21.2 kg (<5th centile; 50th centile for age 6 1/2 years).
Figure 1.

Pedigree and haplotype analysis. Individuals with a Costello syndrome phenotype are identified through filled symbols. Shading of the symbol depicting the abortus (II-2) indicates possible Costello syndrome. The haplotypes at the six polymorphic sites of the HRAS locus were ascertained in II-2 (Patient 1) and the mother by allelic specific amplification. Presence of the HRAS mutation resulting in Costello syndrome is shown as HRAS c.34 Mut. The haplotype of the father is inferred from sequencing and alleles passed on to Patient 1. The genotypes shown in parenthesis for II-6 (Patient 2) were inputted using linkage disequilibrium data and her inferred haplotype is presented in italic.
Figure 2.
Photo of Patient 1 at age 32 years, note coarse facial features with prominent lips and the tight curls.
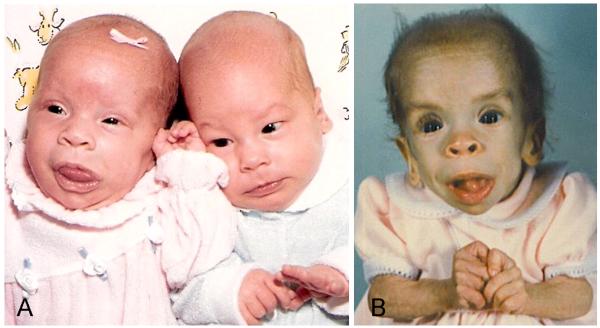
A younger sister (Patient 2; Fig. 1 II-6; CS#307) with a clinical diagnosis of Costello syndrome [Patient 7 in Johnson et al., 1998] was born six years after Patient 1. She was the product of a twin pregnancy (Fig. 3), and her twin sibling was a healthy male. She had a unilateral congenital cataract, severe failure-to thrive and persistent atrial tachycardia [Johnson et al., 1998]. Her facial features included prominent lips and a large tongue as a neonate (Fig. 3). The sparse hair, epicanthal folds and low nasal bridge, prominent lips and lack of subcutaneous fat tissue notable at age 2 years were typical for Costello syndrome (Fig. 3). She died at age 2 years, presumably due to cardiomyopathy and malnutrition, and an autopsy showed dilatation of the right ventricle [Johnson et al., 1998].
Figure 3.

A: Photo of Patient 2 on the left, and her unaffected twin brother on the right, as neonates. Note the patient’s prominent lips and protruding tongue.
B: Patient 2, shortly before her death at age 2 years. Note the cachectic appearance, sparse hair, coarse facial features with prominent mouth and tongue, and the hand position suggestive of ulnar deviation of the fingers.
Their family history was noteworthy for one spontaneous pregnancy loss at 3-4 months gestation (Fig. 1, II-5). The delivering physician suspected this fetus to be affected with Costello syndrome based on its physical appearance, but no further information is available and no additional studies were performed. The mother had eight pregnancies, and six living offspring are not affected by Costello syndrome (Fig. 1).
Molecular Analyses
Peripheral white blood cell and buccal cell genomic DNA was extracted from Patient 1 and his parents using the Qiagen Puregene kit (Gentra Systems, Minneapolis, MN). Patient 2’s DNA was isolated from 23-year-old archived histopathology slides. Briefly, H&E histology slides were sprayed with Frostbite® on both sides and the cover slip removed using a sterile scalpel. The slides were soaked in Histo-Clear™, 2 changes 60 minutes each (with occasional agitation) to remove the mounting medium, and washed sequentially in 100%, 95% and 80% ethanol baths. The slides were rinsed using RNase free water and tissue scraped into lysis buffer using a sterile scalpel. Both liver and kidney genomic DNA was isolated from Patient 2’s archived slides using the Qiagen Puregene kit using the optional proteinase K overnight incubation.
DNA samples from all four family members were amplified using the AmpFlSTR Profiler Plus kit from Applied Biosystems (Foster City, CA), as previously described [Sol-Church et al., 2006] to verify gender and biological relationship. An additional set of 18 chromosome-11 specific short tandem repeats (STRs), included in the ABI Linkage Mapping set v2.0, were used to determine the extent of allele sharing between the two patients. Primers designed to amplify discrete regions of the HRAS locus containing a part of exon 1 and exons 2 to 6, as well as allelic specific primers used in this study are listed in supplemental Table I.
RESULTS
Biologic relationships and gender assignment of the family members were confirmed using the ABI profiling kit. Despite age induced degradation of the DNA samples extracted from Patient 2’s histology slides, 8 of the 10 loci included in the kit amplified. As expected with sample degradation, the largest of the STR markers (D18S51 and D7S820) failed to amplify, but the gender was confirmed by the amelogenin marker (data not shown).
Using primers and conditions included in supplemental Table I, the same de novo germline HRAS mutation c.34G>A, predicting a p.G12S amino acid substitution, was discovered in every DNA sample isolated from Patient 1 and his affected sister, Patient 2 (Fig. 4). This mutation was absent in all parental DNA samples.
Figure 4.
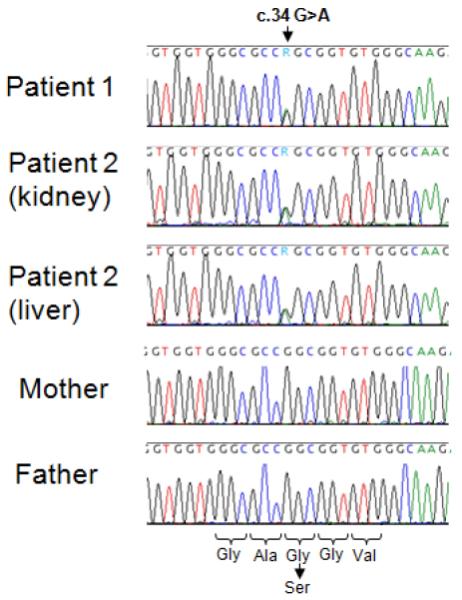
Detection of the germline HRAS mutation in two siblings. The sequence alignments around nucleotide c.34 are shown in the patients and parents. Patient 1’s buccal DNA and Patient 2’s archived liver or kidney DNA samples display the c.34G>A heterozygous mutation, while both parents show wild-type c.34G sequence. Amino acid sequence is depicted along with substitution resulting from the c.34G>A mutation.
In an attempt to identify the parental origin of the mutation in both affected siblings, the family members were genotyped across a ~3.8 kb region of the HRAS locus (Fig. 5). Six known polymorphic sites, including three single nucleotide polymorphisms (SNPs, rs8176330, rs12628 and rs35601764) and three short tandem repeats (STRs, rs112488103, rs113931482 and rs112587690) were used in this study. During our ongoing molecular study of Costello syndrome and survey of the HRAS locus, linkage disequilibrium (LD) was found in 103/106 individuals for rs8176330, rs112488103, and rs35601764. A second LD group was identified in 287/287 genotyped individuals that included markers rs112587690 and rs12628 (data not shown).
Figure 5.
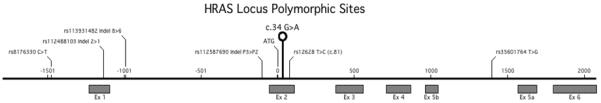
HRAS locus: Schematic representation of the genomic structure of the HRAS gene including the mutation site c.34 G>A and location of six polymorphic sites, as indicated above the sequence by their rs number and alleles. Untranslated exon 1 and coding exons 2 to 6 are represented by boxes. Details of the family members’ genotypes at these sites are shown in Table I and haplotypes in Figure 1.
Genotype data for the family at the HRAS locus (Table I) showed that Patient 1 and his father were homozygous at five sites and heterozygous 8/6 at rs113931482. Interestingly, the mother was heterozygous at all six sites. Due to the level of degradation of Patient 2’s DNA samples, direct genotyping information was obtained for only three of these polymorphic markers. However, based on the LD patterns, Patient 2’s genotype could be inputted at the three missing sites rs8176330, rs112587690 and rs35601764 (Table I), making both siblings identical at all six polymorphic sites. Additional sharing of maternal alleles was observed at 11p using linkage mapping set markers: Of six 11p loci genotyped, both siblings shared four while the other two loci were uninformative (data not shown).
Table I.
Family Genotyped at Six Polymorphic Sites
| Markers | rs8176330 | rs112488103 | rs113931482 | rs112587690 | HRAS c.34 | rs12628 | rs35601764 |
|---|---|---|---|---|---|---|---|
| Patient 1 | C/C | 2/2 | 8/6 | P3/P3 | G/A | T/T | T/T |
| Patient 2* | (C/C) | 2/2 | 8/6 | (P3/P3) | G/A | T/T | (T/T) |
| Mother | C/T | 2/1 | 8/6 | P3/P2 | G/G | T/C | T/G |
| Father | C/C | 2/2 | 8/6 | P3/P3 | G/G | T/T | T/T |
genotypes inputted from linkage disequilibrium are indicated in brackets.
As seen in Table I, the only heterozygous site available to determine mutation parental origin in Patient 1 was the 8/6 repeat region of rs113931482. Figure 6 shows the double sequence profiles typically seen in 8/6 heterozygous individuals carrying one allele with six copies and one allele with 8 copies of the basic CGC repeat, in this case Patient 1 and his mother. In order to determine which of these alleles also carried the mutated c.34A base, allelic specific amplification was used on Patient 1 DNA. During allelic specific amplification (ASA), each individual chromosome is amplified separately and genotypes can resolve into two distinct haplotypes. Figure 6 displays the result of ASA performed with either the mutated (Mut c.34A) or wild type (WT c.34G) allele and clearly indicates that the mutation in Patient 1 originated on the chromosome carrying eight copies of the GCG repeat, while the wild type allele carries six copies. Whether the 8- or 6-copy allele was contributed by the maternal germ cell was determined by ASA using SNP rs12628 allele specific primers (c.81T or c.81C) to generate haplotype information for the Patient’s mother. The sequencing results presented in Figure 6 show that the 81C allele was on the same chromosome as the six GCG repeats, and that the 81T allele which she passed on to Patient 1 (see Table 1) carried eight copies of the GCG repeat. Since Patient 1’s mutation was on the allele carrying the eight repeats, we conclude that his Costello syndrome causing mutation is maternally inherited (Fig. 1). His father contributed the second 81T allele, the wild type HRAS c.34G allele, as well as six copies of the GCG repeat (Fig. 1). The degree of allele sharing between the siblings at the HRAS locus and along 11p suggested that Patient 2’s mutation was also maternally inherited. However, her degraded DNA was not a suitable template for ASA amplification of large amplicons.
Figure 6.
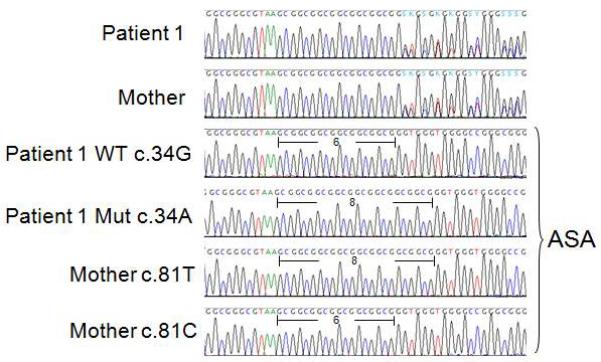
Sequencing of the region around rs113931482. This family is heterozygous 8/6 at this site (Table I) and the double sequencing profiles observed in Patient 1 and his mother are typical of heterozygous individuals carrying alleles with six or eight copies of the CGC trinucleotide repeat. Allelic specific amplification (ASA) was used to discriminate both alleles, and determine their position in relation to either the Patient 1’s c.34 mutation site or his mother’s c.81 SNP. In Patient 1 the WT c.34G appears on the same allele with six copies of the CGC repeat, while the mutated allele c.34A is on the same allele with the eight repeats. The mother’s c.81T allele (which she did not pass to either Patient 1 or 2) is located in cis with six CGC copies. The c.81C allele, which she passed to Patient 1, accompanies eight copies of the CGC unit, revealing that the mutation in Patient 1 originated on the maternally inherited chromosome.
DISCUSSION
The identification of the most common Costello syndrome causing HRAS mutation, c.34G>A, predicting a p.G12S amino acid substitution, in these previously clinically reported siblings [Johnson et al., 1998] documents, to our knowledge, the first molecularly confirmed recurrence in a family. Presence of the identical mutation in both siblings, in the absence of identifiable mosaicism in parental blood and cheek swab derived DNA, is consistent with either (A) two mutations arising independently from each other, (B) presence of the change in one parent’s germ cells, or (C) somatic mosaicism below a level our testing could detect. Though de novo HRAS mutations resulting in Costello syndrome occur predominantly in the paternal germline [Zampino et al., 2007], we previously documented two patients with maternally inherited germline mutations [Sol-Church et al., 2006]. Patient 1 reported here is the third individual with a maternally derived mutation causing Costello syndrome. The unambiguous maternal provenance of the germline mutation in Patient 1 and recurrence of the same mutation in Patient 2 indicates gonadal mosaicism in the mother. Sharing of the maternally inherited allele of the chromosome 11 p-arm and HRAS locus SNPs between siblings suggests both mutations were inherited through the maternal germline. Although previous reports of somatic mosaicism for HRAS mutations [Gripp et al., 2006; Sol-Church et al., 2009; Girisha et al., 2010] would anticipate the recurrence of Costello syndrome in rare families, the family reported here confirms the, albeit low, recurrence risk after the birth of a propositus.
The successful mutation detection in degraded DNA extracted from a 23-year-old sample derived from histopathology slides as the single remaining DNA source for a patient is noteworthy. This proof-of-principle may serve as an example for other cases, when critical information needs to be obtained from long deceased family members. Further, the newly available next-generation sequencing approaches will lead to the identification of novel changes in very rare conditions, and confirmatory studies will need to be performed on unrelated patients with the same rare condition. For exceedingly rare disorders the use of old archival material, possibly autopsy slides as in the case reported here or tissue from a preserved body as reported by Gurrieri et al. [2011], may be the only option.
In conclusion, we report here the first molecular confirmation of siblings with Costello syndrome and the third case of a maternally derived mutation.
Supplementary Material
ACKNOWLEDGMENTS
We thank the patient and his parents for sharing their information. We thank Dana Denning for locating the autopsy slides; Dr. Diana Corao-Uribe for reviewing the autopsy slides, the Nemours Histology core for technical support with slide preparation for DNA extraction, and the Center for Pediatric Research COBRE-funded Molecular Core for sequencing support. Dr. Stevenson is supported by a Doris Duke Charitable Foundation Clinical Scientist Development Award and Dr. Sol-Church by grant 2P20 RR020173-068A1 form the NIH National Center for Research Resources.
REFERENCES
- Aoki Y, Niihori T, Kawame H, Kurosawa K, Ohashi H, Tanaka Y, Filocamo M, Kato K, Suzuki Y, Kure S, Matsubara Y. Germline mutations in HRAS proto-oncogene cause Costello syndrome. Nat Genet. 2005;37:1038–1040. doi: 10.1038/ng1641. [DOI] [PubMed] [Google Scholar]
- Girisha KM, Lewis LE, Phadke SR, Kutsche K. Costello syndrome with severe cutis laxa and mosaic HRAS G12S mutation. Am J Med Genet Part A. 2010;152A:2861–2864. doi: 10.1002/ajmg.a.33687. [DOI] [PubMed] [Google Scholar]
- Gripp KW, Stabley DL, Nicholson L, Hoffman JD, Sol-Church K. Somatic Mosaicism for an HRAS mutation causes Costello Syndrome. Am J Med Genet Part A. 2006;140A:2163–2169. doi: 10.1002/ajmg.a.31456. [DOI] [PubMed] [Google Scholar]
- Gripp K, Lin A. Costello Syndrome in: GeneReviews at GeneTests: Medical Genetics Information Resource [database online] University of Washington; Seattle: 2009. Copyright. 1997-2009. Available at http://www.genetests.org. [Google Scholar]
- Gurrieri F, Pomponi MG, Pietrobono R, Lucci-Cordisco E, Silvestri E, Storniello G, Neri G. The Simpson–Golabi–Behmel syndrome: A clinical case and a detective story. Am J Med Genet Part A. 2011;155:145–148. doi: 10.1002/ajmg.a.33586. [DOI] [PubMed] [Google Scholar]
- Ioan DM, Fryns JP. Costello syndrome in two siblings and minor manifestations in their mother, further evidence for autosomal dominant inheritance? Genet Couns. 2002;13:353–356. [PubMed] [Google Scholar]
- Johnson JP, Golabi M, Norton ME, Rosenblatt RM, Feldman GM, Yang SP, Hall BD, Fries MH, Carey JC. Costello syndrome: Phenotype, natural history, differential diagnosis, and possible cause. J Pediatr. 1998;133:441–448. doi: 10.1016/s0022-3476(98)70284-7. [DOI] [PubMed] [Google Scholar]
- Sol-Church K, Stabley DL, Nicholson L, Gonzalez IL, Gripp KW. Paternal Bias in Parental Origin of HRAS Mutations in Costello Syndrome. Hum Mutat. 2006;27:736–741. doi: 10.1002/humu.20381. [DOI] [PubMed] [Google Scholar]
- Sol-Church K, Stabley DL, Demmer LA, Agbulos A, Lin AE, Smoot L, Nicholson L, Gripp KW. Male-to-male transmission of Costello syndrome: G12S HRAS germline mutation inherited from a father with somatic mosaicism. Am J Med Genet Part A. 2009;149A:315–321. doi: 10.1002/ajmg.a.32639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sovik O, Schubbert S, Houge G, Steine SJ, Norgard G, Engelsen B, Njolstad PR, Shannon K, Molven A. De novo HRAS and KRAS mutations in two siblings with short stature and neuro-cardio-facio-cutaneous features. J Med Genet. 2007;44:e84. doi: 10.1136/jmg.2007.049361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van den Bosch T, van Schoubroeck D, Fryns JP, Naulaers G, Inion AM, Devriendt K. Prenatal findings in a monozygotic twin pregnancy with Costello syndrome. Prenat Diagn. 2002;22:415–417. doi: 10.1002/pd.333. [DOI] [PubMed] [Google Scholar]
- Zampino G, Mastroiacovo P, Ricci R, Zollino M, Segni G, Martini-Neri ME, Neri G. Costello Syndrome: Further clinical delineation, natural history, genetic definition and nosology. Am J Med Genet. 1993;47:176–183. doi: 10.1002/ajmg.1320470210. [DOI] [PubMed] [Google Scholar]
- Zampino G, Pantaleoni F, Carta C, Cobellis G, Vasta I, Neri C, Pogna EA, DeFeo E, Delogu A, Sarlozy A, Atzeri F, Selicorni A, Rauen KA, Cytrynbaum CS, Weksberg R, Dallapiccola B, Ballabio A, Gelb BD, Neri G, Tartaglia M. Diversity, parental germline origin and phenotypic spectrum of de novo HRAS missense changes in Costello syndrome. Hum Mutat. 2007;28:265–272. doi: 10.1002/humu.20431. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.