Abstract
Human germinal center–associated lymphoma (HGAL) and LIM domain only-2 (LMO2) are proteins highly expressed in germinal center (GC) B lymphocytes. HGAL and LMO2 are also expressed in GC-derived lymphomas and distinguish biologically distinct subgroups of diffuse large B-cell lymphomas (DLBCL) associated with improved survival. However, little is known about their regulation. PRDM1/Blimp-1 is a master regulator of terminal B cell differentiation and may also function as a tumor suppressor in the pathogenesis of DLBCL, where it is frequently inactivated by mutations and deletions. We now demonstrate that both HGAL and LMO2 are directly regulated by the transcription repressor PRDM1. In vivo studies demonstrate that PRDM1 directly binds to the recognition sites within the upstream promoters of both HGAL and LMO2. PRDM1 binding suppresses endogenous protein and mRNA levels of HGAL and LMO2. In addition promoter analysis reveals that site specific binding of PRDM1 to the promoters is capable of repressing transcriptional activity. This inhibitory effect of PRDM1 suggests that it has a key role in the loss of HGAL and LMO2 expression upon differentiation of GC B cells to plasma cells and may also contribute to absence of HGAL and LMO2 expression in post-GC lymphoid tumors.
Keywords: Non-Hodgkin’s B Cell Lymphoma, Blimp-1, transcription, HGAL, LMO2
Introduction
Germinal center (GC) reaction is a highly regulated critical step in the generation of humoral immunity. It is characterized by B cell proliferation, immunoglobulin affinity maturation leading to antigen selection, immunoglobulin class-switch, and finally differentiation of B cells into either memory cells or antibody secreting plasma cells [1]. The development of GC B lymphocytes and the subsequent differentiation to memory and plasma cells is tightly regulated and is associated with characteristic changes in gene expression [2, 3]. Despite significant studies on the regulation and function of these gene changes, many important regulatory steps remain to be deciphered.
Gene expression profiling previously identified two genes encoding proteins highly expressed in GC B lymphocytes: human germinal center–associated lymphoma (HGAL), also known as germinal center expressed transcript 2 (GCET2), and LIM domain only-2 (LMO2) [4, 5]. Both genes are induced in GC B cells and silenced during differentiation into plasma cells or memory B cells. HGAL and LMO2 are also expressed in GC-derived lymphomas. Characterization of a large number of diffuse large B-cell lymphomas (DLBCL) has identified HGAL and LMO2 as markers which can be used to distinguish biologically distinct subgroups associated with improved survival [4, 6–10].
The HGAL gene, located on chromosome 3q13, encodes a 178–amino acid (aa) protein with 51% identity and 62% similarity to the murine M17 protein, both exclusively expressed in GC B lymphocytes [6]. Studies in mice revealed that M17 is dispensable for GC formation, immunoglobulin somatic hypermutation, class-switch recombination, and for mounting T cell–dependent antibody responses [11]. However, in contrast to its wild-type littermates, M17-deficient mice exhibited reduced-sized Peyer patches [11]. Recent studies showed that HGAL is involved in motility regulation of GC B-cells and GC-derived malignant lymphoma cells. It inhibits IL-6 and SDF-1–induced migration of malignant lymphoma cells and normal GC B-lymphocytes by interacting with actin and myosin proteins [12, 13] as well as by regulating the RhoA signaling pathway [12, 14]. HGAL-induces activation of RhoA and its downstream effectors by directly binding and activating the RhoA-specific guanine nucleotide exchange factors (RhoGEFs) PDZ-RhoGEF and LARG. This stimulates the GDP-GTP exchange rate of RhoA and results in inhibition of lymphocyte and lymphoma cell motility, induction of transcriptional activation by serum response factor and amplification of RhoA transforming potential [14].
LMO2 gene, located on the short arm of chromosome 11 at band 13 (11p13), is a member of the LIM-only zinc finger protein family and mediates protein-protein interaction in multi-protein transcriptional factor complexes. It was discovered from a recurrent translocation in T-cell acute lymphoblastic leukemia (T-ALL), but its expression is extinguished early in T cell development and is not required for normal development of this lineage [15]. Aberrant expression of LMO2 in immature T cells in the thymus leads to thymocyte self renewal [16], accumulation of early lymphoid precursors and oncogenic transformation leading to childhood T-cell acute lymphoblastic leukemia. LMO2 plays important role in normal endothelial and hematopoietic cells. In the endothelial system it is involved in angiogenesis, playing a critical role in the angiogenetic remodeling of the vasculature [17]. In hematopoietic system, LMO2 expression is restricted to adult hematopoietic stem cells (HSCs) and the erythroid lineage [18] and it is essential for yolk sac erythropoiesis [19]. Chimeric animals produced from homozygous-deficient embryonic stem cells demonstrated abnormal hematopoiesis. We have recently observed specific up-regulation of LMO2 expression in GC B lymphocytes and GC derived DLBCL [2, 5], but its function in these cells is still unknown.
B lymphocyte differentiation into plasma cells is dependent on the transcription factor PR Domain containing 1, with Zinc Finger Domain 1 (PRDM1), also known as B-lymphocyte induced maturation protein-1 (Blimp1). PRDM1 encodes a zinc finger transcriptional repressor described by Turner et al. as an inducer of B cell differentiation [20]. PRDM1 also has a key role in regulating effector function of T cells [21–24] and Natural Killer cells [25, 26]. Stimulation of macrophages and dendritic cells through Toll-like receptors also induces PRDM1 expression suggesting PRDM1 has a role in regulating multiple immune cell types [27, 28]. PRDM1 functions as a transcription repressor by directly binding DNA and acting as a scaffold to recruit multiple co-repressor proteins including the histone H3 methyltransferase, G9a [29], the histone deacetylase HDAC2 [30], the arginine methyltransferase PRMT5 [31] and the histone demethylase LSD1 [32]. In addition, at some gene targets PRDM1 may displace transcriptional activators of the Interferon Regulatory Factor (IRF) family through DNA binding site competition [33].
In B lymphocytes PRDM1 is required for the formation of plasma cells [34]. Conditional knockout of PRDM1 in the B cell compartment leads to an accumulation of activated B cells and a loss of plasma cell differentiation [35, 36]. Conversely, enforced expression of PRDM1 in lymphoma cell lines promotes either partial differentiation or induction of apoptosis [37]. PRDM1 acts as a tumor suppressor in activated B cell (ABC)-like DLBCL, where it is inactivated through multiple mechanisms [38–41]. Gene expression profiling demonstrates that PRDM1 coordinates significant reprogramming of the genes expressed in GC B lymphocytes [42]. A limited number of these reprogrammed genes have been identified as direct targets of PRDM1 repression. These include genes required for maintaining the B cell phenotype and in maintaining cellular proliferation such as CIITA, PAX5, Spi-B, Id3 and c-myc [42–46]. In addition we have recently identified the proliferation genes PCNA and MKI67 as functionally important direct targets of PRDM1 during mantle cell lymphoma therapy [47]. Together these studies have identified PRDM1 as a key regulatory step in the transition from a GC B lymphocyte to a plasma cell; however, many of the functionally important direct targets of PRDM1 in this process remain to be deciphered. This report now establishes HGAL and LMO2 as two GC proteins whose expression is directly regulated by PRDM1.
Results
PRDM1 over-expression suppresses endogenous LMO2 and HGAL expression
Gene expression profiling previously performed by us [2] reveals that expression of HGAL and LMO2 is down-regulated concomitant with induction of PRDM1 during differentiation of human GC lymphocytes to plasma and memory B cells (Figure 1). Consequently, we hypothesized that PRDM1 may regulate the expression of these genes. To address this question, B cell lymphoma cell lines expressing endogenous HGAL and LMO2 proteins were transfected with a PRDM1 expression construct and the changes in mRNA and protein were profiled. Immunoblot analysis of the B cell lymphoma cell line, VAL, revealed that both HGAL and LMO2 protein expression decrease in a dose dependent manner with the increase of PRDM1 (Figure 2A). Similarly the known PRDM1 target, BCL6, also showed a dose dependent suppression by PRDM1. This finding was also observed in the B lymphoma cell line, Raji (Figure 2B). Expression changes at the level of mRNA were analyzed by quantitative reverse transcription PCR (Figure 2C and 2D). Twenty-four hours after PRDM1 transfection, HGAL and LMO2 mRNA levels are suppressed up to 40% in both the VAL and Raji B lymphoma cell lines. The level of suppression is similar to the degree observed with BCL6, a known PRDM1 target. Overall these results show that ectopic expression PRDM1 in germinal cell-derived lymphoma cell lines down-regulates endogenous mRNA and protein expression of both HGAL and LMO2.
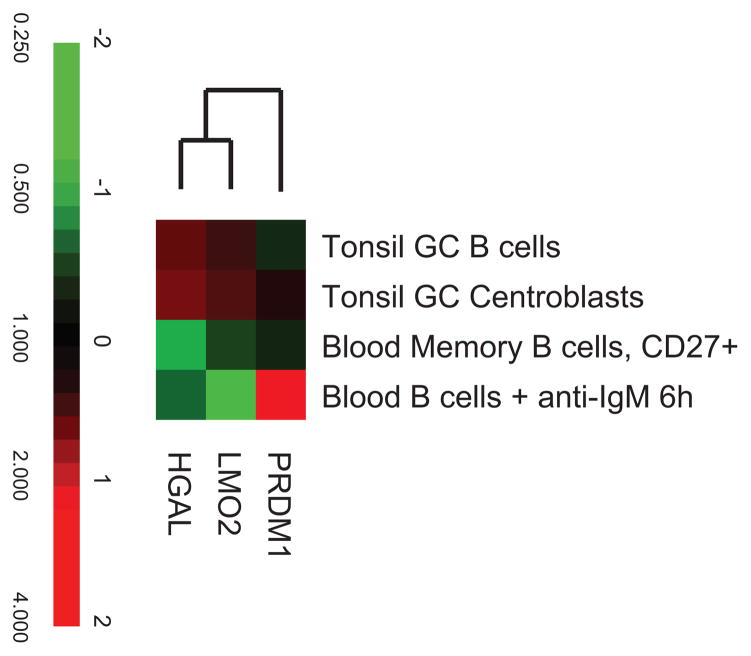
Figure 1. Reciprocal expression of LMO2 and HGAL with PRDM1 in primary B cells.
PRDM1, LMO2 and HGAL mRNA expression was analyzed by cDNA microarrays as was reported previously [2] in GC B-cells and centroblasts, memory B cells and peripheral B-lymphocytes stimulated for 6 hours with anti-IgM. The results show the ratio of hybridization of fluorescent cDNA probes prepared from each experimental mRNA sample to a reference mRNA sample. These ratios are a measure of relative gene expression in each experimental sample and are depicted according to the color scale shown at the bottom.
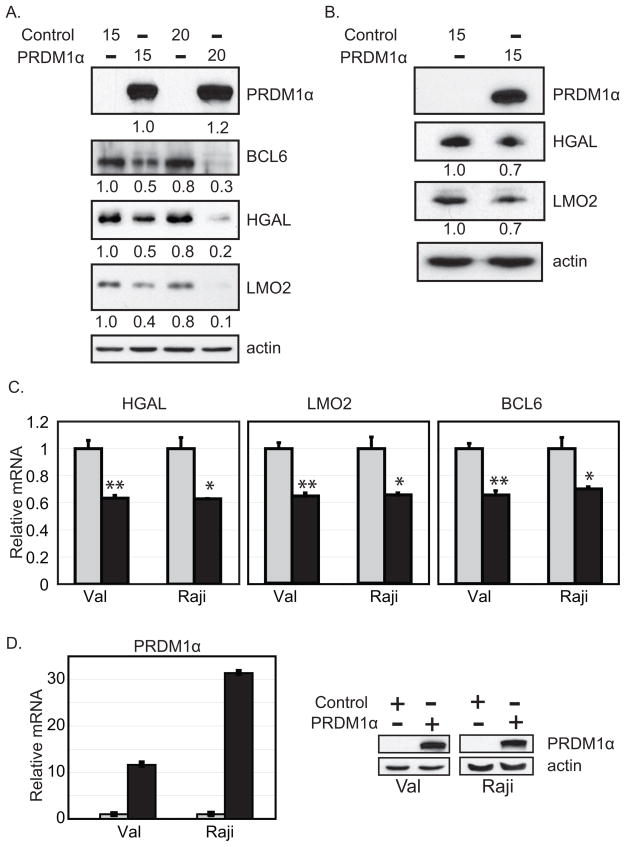
Figure 2. LMO2 and HGAL mRNA and protein levels are decreased by PRDM1. L.
ymphoma cell lines, VAL (panel A) and Raji (panel B) were transfected with 15 or 20 μg of a PRDM1α expression plasmid or empty vector control as indicated at the top of the panel. Forty-eight hours after transfection cellular proteins were resolved by SDS-PAGE and immunoblot analysis performed with the antibodies specific for the proteins indicated on the right side of each panel. The relative intensity of each band was determined by densitometry, normalized to the actin loading control and the value is shown below each lane. PRDM1 expression correlated with a decrease in HGAL and LMO2. BCL6 is a positive control for PRDM1 mediated repression and actin is a loading control. The result is representative of three independent experiments. C) Lymphoma cell lines, VAL and Raji, were transfected with 15 μg of a PRDM1α expression plasmid or empty vector control and the mRNA levels were measured by quantitative RT-PCR. The data shown represents three independent experiments with the error bars representing the SD and p-values indicated by asterisks (**p< 0.003, *p< 0.03). D) Quantitation of PRDM1 mRNA and protein levels in the same experiment shown in panel C (***p< 3×10−7).
PRDM1 binds the HGAL and LMO2 promoters in vivo
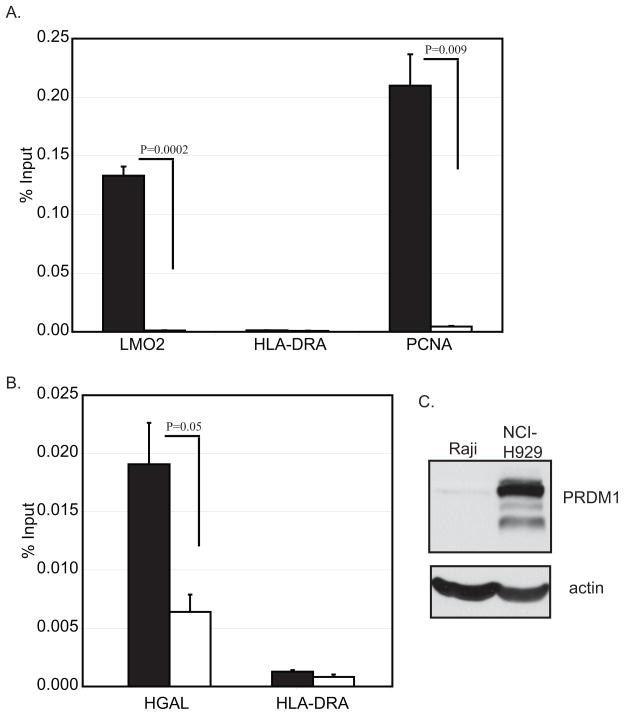
The effect of PRDM1 on HGAL and LMO2 expression could be through direct or indirect transcriptional suppression of these genes. PRDM1 is a direct DNA binding transcription repressor which recognizes the sequence 5′-MAGYGAAAYK-3′ [33]. Bioinformatic search of the HGAL promoter revealed two potential homologies to this sequence located at positions –1608 and –1383 upstream of the transcription initiation site. A similar search of the LMO2 promoter region predicted only one site of high homology at position –1783 relative to the transcription start site. The presence of PRDM1 bound at these promoter regions was determined through chromatin immunoprecipitation from myeloma cell lines which represent differentiated plasma cells and express PRDM1. Robust PRDM1 interaction was observed at the LMO2 promoter in the myeloma cell line NCI-H929 (Figure 3A) and in the myeloma cell line U266 (data not shown). The interaction is highly specific as revealed by the minimal signal obtained with the control antibody. Furthermore the HLA-DRA promoter which we have previously shown not to bind PRDM1 displayed minimal signal intensity [28]. In contrast the PCNA promoter which has been previously demonstrated to bind PRDM1 with very high intensity clearly shows binding similar to that observed with LMO2 [47]. Chromatin immunoprecipitation was also done at the HGAL promoter (Figure 3B). Similar to the LMO2 results, significant PRDM1 binding was clearly detected at the HGAL promoter in the region of the predicted PRDM1 binding sites. The intensity of binding was approximately 7-fold less than observed at LMO2 but remained significantly stronger than either the negative control promoter, HLA-DRA, or the negative control antibody. These findings reveal that endogenous PRDM1 is bound to both LMO2 and HGAL promoters and thus can directly act to suppress these genes.
Figure 3. PRDM1 binds to the LMO2 and HGAL promoters in vivo.
Chromatin immunoprecipitation analysis was performed from the myeloma cell line, NCI-H929, expressing endogenous PRDM1. ChIP analysis was done with both the specific PRDM1 antibody (black bars) and an IgG negative control antibody(white bars). Binding to the promoter regions were assessed by quantitative PCR using primers proximal to the predicted PRDM1 binding sites in the LMO2 promoter (panel A) and the HGAL promoter (panel B). The HLA-DRA promoter was evaluated as a known negative control for PRDM1 binding. The PCNA promoter represents a known positive site of PRDM1 binding. The data are presented as percent input and the error bars represent the SEM of three independent experiments. p-values are indicated. Expression of PRDM1 protein in the NCI-H929 cell line was confirmed by immunoblot analysis (panel C).
PRDM1 directly regulates the HGAL and LMO2 promoters
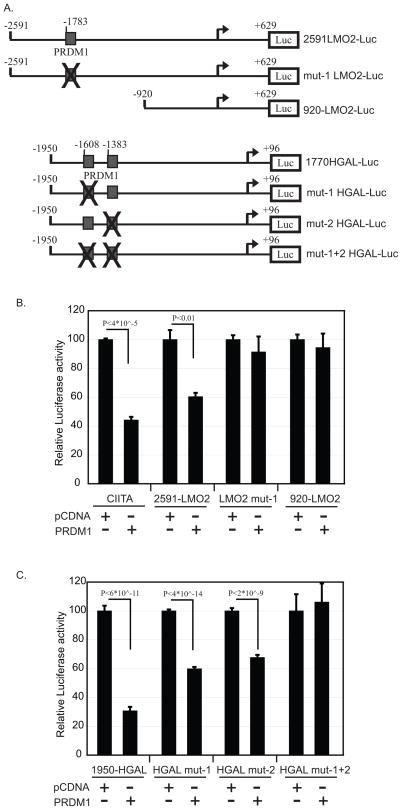
PRDM1 regulates its target genes at the level of transcription. Thus to functionally determine if PRDM1 is a physiological regulator of HGAL and LMO2 transcription we cloned the promoters of both genes. A region of the LMO2 promoter from −2591 to +629 base pairs relative to the transcription initiation site was inserted into a luciferase reporter construct to create 2591LMO2-Luc (Figure 4A). Transfection of this full length, wild type LMO2 promoter into the Raji B lymphoma cell line demonstrated that this region is sufficient to promote robust transcription, similar to the well characterized CIITA promoter [48]. Co-transfection of a PRDM1 expression construct results in an approximate 30% repression of LMO2 promoter activity(Figure 4B). The level of suppression is similar to that observed with the CIITA luciferase construct which we have previously characterized as a direct target of PRDM1 [43]. LMO2 repression by PRDM1 was fully abrogated by either site directed mutagenesis of the predicted PRDM1 binding site (Mutant-LMO2) or 5′ deletion of the PRDM1 binding region (920LMO2). These findings were confirmed in a second B lymphoma cell line, CA46 (data not shown).
Figure 4. PRDM1 directly regulates the LMO2 and HGAL promoters.
A) Schematic of the LMO2 and HGAL luciferase constructs. Grey boxes indicate location of the PRDM1 binding sites and the X indicates the sequence has been mutated to prevent PRDM1 binding. Bent arrow indicates the transcription start site. B) Luciferase analysis of the LMO2 promoter constructs in the Raji B lymphoma cell line. Promoter constructs indicated at the bottom were co-transfected with either a PRDM1 expression plasmid or the control pCDNA plasmid as indicated. The results within each transfection are normalized to the co-transfected pRL-TK signal. The CIITA-luciferase construct is a known PRDM1 target and is shown as a positive control. The results presented represent three independent experiments with the error bars indicating the SD. p-values are shown. C) Luciferase analysis of the HGAL promoter constructs in the Raji B lymphoma cell line. The experiment is as described in panel B and the constructs are shown in panel A. The results presented represent three independent experiments with the error bars indicating the SD. p-values are shown.
A similar analysis of the HGAL promoter was performed. The region of −1950 to +96 of the HGAL promoter relative to the transcription start site was cloned into a luciferase reporter construct (Figure 4A). Three additional constructs were created in which the predicted PRDM1 binding sites were disrupted by site directed mutagenesis, either individually or simultaneously. Co-transfection of the wild-type HGAL promoter with the PRDM1 expression construct led to an approximate 70% repression of HGAL promoter activity in Raji cells (Figure 4C) and 30% repression in HeLa cells (data not shown). Mutation of either PRDM1 binding site alone partially reduced the degree of repression by PRDM1. Concomitant mutation of both PRDM1 binding sites in the HGAL promoter completely reversed the inhibitory effect of PRDM1 on the HGAL promoter. Together these findings demonstrate that the predicted PRDM1 binding sites in the LMO2 and HGAL promoters are functional sites of PRDM1 mediated repression.
Discussion
This study demonstrates that PRDM1 regulates the GC and GC-DLBCL marker genes, HGAL and LMO2. The repression mediated by PRDM1 is through direct binding to consensus elements present in the upstream region of both promoters. Repression is reflected by decreases in both endogenous mRNA and protein. Furthermore, transcriptional activity from both promoters is specifically inhibited in the presence of PRDM1. These findings identify two novel genes highly and specifically expressed in GC B cells that are down regulated by PRDM1 upon transition from GC B lymphocytes to a differentiated plasma cell. While the full functional spectrum of HGAL and LMO2 in the GC reaction is still unknown, future studies most probably will elucidate the importance of their down-regulation by PRDM1 for success of the terminal differentiation process. Furthermore, since PRDM1 is a tumor suppressor gene, down-regulation of HGAL and LMO2 may also play a role in guarding against malignant transformation.
Previous studies showed that PRDM1 may increase migration of breast cancer cells [49]. Repression of HGAL by PRDM1 identifies the first mechanistic link between B cell migration and PRDM1. HGAL through interaction with RhoA specific guanine nucleotide exchange factors inhibits B cell motility [12, 14]. Thus PRDM1 has the potential to release the inhibition and promote B cell migration. This could be physiologically important in a normal GC to allow the differentiating B cells to egress out of the GC. In DLBCL patients, HGAL expression is associated with improved survival [4, 6, 7]. This suggests that PRDM1 repression of HGAL may be an important regulatory step contributing to the clinical outcome. Despite the growing importance of HGAL little is known about the regulation of its expression. IL-4 stimulation up-regulates HGAL expression and Sp1/Sp3 can activate HGAL transcription but the specific transcription factors that control HGAL expression are poorly understood [6, 50]. Investigations of the activation mechanisms and how PRDM1 counteracts activation will provide important insight into this important regulatory process.
LMO2 is a nuclear protein which can participate in the formation of DNA binding complexes which interact with E-proteins, E12 and E47 [51]. Specific genes regulated by LMO2 in B lymphocytes are not known. However, a clear correlation between LMO2 expression and better overall survival in DLBCL patients has been documented [10]. Interestingly, many of the identified direct targets of PRDM1 including LMO2 are involved in transcriptional activation. This suggests that PRDM1 effects on gene expression reprogramming during B cell differentiation are significantly amplified by a cascade of both direct and indirect gene silencing. Furthermore, LMO2 transcriptional repression by PRDM1 in lymphocytes may have important implications for lymphoma pathogenesis. PRDM1 was demonstrated to function as a tumor suppressor in DLBCL. This effect is mediated by suppression of BCL-6 oncogene and probably additional presently unknown oncogenes. LMO2 is a known T-cell oncogene that also may function as a B–cell oncogene. Studies in B cells aimed to identify genes regulated by LMO2 and examining its oncogeneic role are currently ongoing and will reveal both the role of LMO2 and elucidate the activity of PRDM1 in this lineage.
Several reports have begun to characterize the functional promoter of LMO2 [18, 52–54]. The gene contains three potential promoters which generate transcripts with distinct 5′ untranslated regions but include the complete protein encoding exons 4 to 6 and thus result in identical proteins. The promoter which starts transcription at exon 1, referred to as the distal promoter, conveys tissue specific activity and is the site of activity in hematopoietic cells. In contrast a promoter located upstream of exon 4, referred to as the proximal promoter, is active in a broad range of cell types. The distal promoter is activated by factors of the proline and acidic amino acid-rich protein (PAR) family binding downstream of the transcription initiation site [52]. Recently, an intermediate promoter was shown to be active in T-ALL [54]. This promoter is activated by the ETS factors, ERG1 and FLI1. Long range mapping of histone acetylation and transcription factors recently identified 8 conserved elements across 250 kb of the LMO2 locus potentially involved in the hematopoietic expression of LMO2 [18] and functional domains in T-ALL and B-ALL [54]. Binding of several factors to the hematopoietic enhancers was detected including Gata2, Tal1 and LMO2 itself. Interestingly, a negative regulatory region was reported in the T cell line, Jurkat [53]. The element was resolved to a 205 bp region however the specific element and factor were not able to be identified. This region spans the PRDM1 binding site characterized in this report. This finding is consistent with our data in B lymphocytes and suggests that PRDM1 may have a similar role as a suppressor of LMO2 in acute T cell leukemia cells. The conclusion that LMO2 is directly regulated by PRDM1 is also supported by a recent study utilizing chromatin immunoprecipitation combined with microarray hybridization (ChIP-on-chip) [55]. This report identified LMO2 as a potential gene down-regulated by PRDM1 in the myeloma cell line, U266. Our functional characterization of the LMO2 promoter and specific PRDM1 interaction site in B cells significantly expands this observation.
In conclusion, these findings demonstrate that PRDM1 is a physiological transcriptional repressor of the expression of LMO2 and HGAL genes. This inhibitory effect may mediate the loss of HGAL and LMO2 expression upon differentiation of GC B cells to plasma cells and may contribute, in addition to other presently unknown factors, to the absence of HGAL and LMO2 expression in post-GC lymphoid tumors. It is also possible that the tumor suppressor effect of PRDM1 is at least partially mediated by repression of LMO2 and HGAL genes, which are highly expressed in a subset of DLBCL and potentially have an important role in the pathogenesis of this malignancy. Now that this regulatory pathway has been identified it will be important to define the role of PRDM1 inhibition of HGAL and LMO2 in the pathogenesis and outcome of DLBCL.
Materials & Methods
Cell lines and protein accession numbers
Human non-Hodgkin lymphoma (NHL) cell lines VAL (diffuse large B cell lympnoma), Raji and CA-46 (Burkitt’s lymphoma) were maintained in RPMI medium (Invitrogen, Grand Island, NY) supplemented with 10% fetal bovine serum (HyClone-Themo Scientific, Logan, UT) and 1% penicillin/streptomycin (Invitrogen) and 2mM glutamine(Invitrogen). NCI-H929, a multiple myeloma cell line with a plasma cell phenotype was maintained as above with an additional 0.05 M β-mercaptoetanol (Invitrogen) supplement. Human cervical cancer cell line HeLa was grown in Dulbecco’s Modification of Eagle’s Medium (DMEM) (Invitrogen) similarly supplemented with 10% fetal bovine serum, glutamine and penicillin/streptomycin. All cell lines were cultured at 37°C and 5% CO2. The Uniprot accession numbers associated with the proteins used in this report are: LMO2, P25791; HGAL, Q8N6F7; PRDM1, O75626.
DNA constructs
Expression constructs for PRDM1α have been described previously by Ghosh et.al. [43]. The region consisting of 2591 bp upstream of the human LMO2 transcription initiation site and 629 bp downstream was cloned by PCR into the vector PCR2.1 (Invitrogen) using primers 5′-GGCTCGGCCTAAAACCTTC-3′ and 5′-GAAAGAGAAGCCAGAGTGCC-3′. The initiation site is based on data from the NCBI genome annotation (build 37) and is 208 bp 5′ of the previously reported site [52]. The BamHI-HindIII fragment was then subcloned into the BglII-HindIII sites of PGL3-Basic (Promega, Madison, WI) to create the 2591LMO2-Luc reporter plasmid. The construct 920LMO2-Luc was constructed by subcloning the HincII-HindIII fragment from 2591LMO2-pCDNA2.1 into the SmaI-HindIII sites of pGL3-Basic. The LMO2-mutant reporter plasmid was generated by site-directed mutagenesis (Mutagenex, Inc. Piscataway, NJ) of 2591LMO2-Luc converting the predicted PRDM1 binding site at position –1783 bp from 5′-ACCCTCACTTTCATTTC-3′ to 5′-CCCTCgtcgaCATTTC-3′. All constructs were confirmed by sequencing.
The region consisting of 1950 bp upstream the human HGAL transcription initiation site and 96 bp downstream was amplified from the SUDHL-6 cell line by the Phusion High-Fidelity PCR Master Mix (Finnzymes Oy, Espoo, Finland) using the primers HGAL-FWD 5′-GGAAAGAGCTCGAGTGACCAAACTGGAAACAAC-3′ and HGAL-REV 5′-GGGAAAGCTAGCTTGTGCTCTGACAGGGCAAC-3′. PCR products were digested with SacI and NheI (New England Biolabs, Beverly, MA) and ligated into the pGL3-Basic vector to create the 1950HGAL-Luc construct. Mutagenesis of the predicted PRDM1 binding sites at position –1608 and –1383 of the 1950HGAL-Luc construct was performed using the QuickChange XL Site-Directed Mutagenesis Kit (Stratagene, La Jolla, CA). Primers used for mutagenesis with mutations in lower case are HGAL-mutant#1: 5′-CACAGAAGGTAGGCTTTAAGTCTGGTCgCgTgCtCgTAGTGTAATGCATTTGAGATT GATCCA-3′ and 5′-TGGATCAATCTCAAATGCATTACACTAcGaGcAcGcGACCAGACTTAAAGCCTACC TTCTGTG-3′ and for HGAL-mutant#2: 5′-TATAAAAATTTGTACACACAGTCTTAGAGGACATAcGTgTgtCgTGGCTAAATGCCT AGGAGTGAAATTGC-3′ and 5′-GCAATTTCACTCCTAGGCATTTAGCCAcGacAcACgTATGTCCTCTAAGACTGTGTG TACAAATTTTTATA-3′.
Transfections and luciferase assays
Non-Hogdkin lymphoma cell lines were transfected by electroporation using either a BioRad Gene Pulser II (BioRad Laboratories, Hercules, CA) or an Amaxa Nucleofector II (Lonza, Walkersville, MD). The Gene Pulser conditions used 107 cells electroporated at 200V and 1070μF in 300μl of RPMI supplemented with 10% FBS. The Nucleofector II conditions used 3×106 cells electroporated using program X-001 and solution V for VAL cells and program M-013 and solution V for Raji cells. Transfections for luciferase measurements were performed with 10–15 μg of the luciferase reporter, 1.5 μg of the PRDM1α expression vector or control pCDNA3.1 plasmid and 10 ng of the internal control plasmid pRL-TK (Promega). Cells were cultured for 48 hours after transfection in 10 mL of complete medium and subsequently harvested into 100 μl Passive Lysis Buffer (Promega). HeLa cells were transfected in triplicate using SiPort NeoFX (Ambion, Austin, TX) according to the manufacturer’s instructions. Briefly, 45 ×103 cells per well were seeded with 50 μl of transfection mix (1 μl of SiPort, 75 ng of pRL-TK, 1.25μg of the luciferase reporter plasmid, and 1 μg of the PRDM1α expression plasmid or control pCDNA3.1 per well) in a final volume of 0.5 ml. Cells were cultured for 48 hours after transfection and harvested in Passive Lysis Buffer. All luciferase readings were performed using the 20/20n luminometer (Turner Biosystems, Sunnyvale, CA). Firefly luciferase activity was normalized to Renilla luciferase activity in all experiments.
Chromatin Immunoprecipitation (ChIP)
Chromatin was prepared as previously described [56]. 1.5 × 106 cell equivalents were used in each immunoprecipitation reaction. Primary antibodies were used at 0.5 μg per reaction and incubated over night. The antibodies used were PRDM1 (PRDI-BF1) (Cell Signaling, Danvers, MA) and non specific rabbit IgG (Upstate-Millipore, Billerica, MA). RNA was removed from the immunoprecipitated DNA by treatment with RNase (Ambion) for 30 minutes at 37°C and proteinase K (Roche) for 1 hour at 45°C. Column purification of the immunoprecipitated DNA was done using the PCR purification kit (Qiagen, Valencia, CA). Analysis of the immunoprecipitated DNA was performed by realtime PCR using SyberGreen (Quanta Biosciences, Gaithersburg, MD) in a CFX96 PCR machine (BioRad Laboratories) The specific ChIP primers are LMO2-Fwd: 5′-TGGTGACTGCTGTGGGTAAG-3′, LMO2-Rev: 5′-GCCCACTCACTCTTGCTTTC-3′ and HGAL-Fwd 5′-GGAGTGAAATTGCCAGGTTG -3′ and HGAL-Rev 5′-GAGAAGGGGTCAAGGGAACT -3′. Primers for ChIP analysis of the HLA-DRA promoter are as reported previously [56]. Quality control was carried out for each primer set which included optimization of annealing temperature, melt curve analysis to confirm amplification of a single product and standard curve analysis to confirm PCR efficiency was between 90% and 110%.
Immuno-blotting
Cells were collected 48 hours post-transfection and protein levels were determined by immunoblot. Transfected cells were washed once with ice-cold phosphate-buffered saline (PBS) and homogenized in RIPA buffer (1× PBS, 1% Nonidet P-40, 0.5% sodium deoxycholate, 0.1% sodium dodecyl sulfate (SDS), 10mM phenylmethylsulfonyl fluoride, 1μg/mL aprotinin, and 100mM sodium orthovanadate) on ice for 30 minutes. Cell lysates were centrifuged at 14,000 g for 15 minutes at 4°C to remove insoluble material. Protein concentration of the lysates was determined using a Bradford assay Coomasie Plus (Pierce, Rockford, IL). A total of 40μg of whole-cell lysate per sample was separated on 10% SDS-PAGE, transferred to polyvinylidene difluoride membranes (BioRad Laboratories), and immunoblotted with specific antibodies. LMO2 and HGAL monoclonal antibodies were produced in our laboratory as previously reported [4, 5], PRDM1 antibody was from Cell Signaling Technology, BCL6 antibody was from Santa Cruz Biotechnology Inc (Santa Cruz, CA) and β-Actin antibody from Sigma-Aldrich (St. Louis, MO). Films were scanned and data subjected to densitometric analysis using Scion Image Software (National Institutes of Health [NIH]). Protein levels were normalized to the corresponding loading controls and reported as ratios.
RNA isolation, reverse transcription, and Real-time polymerase chain reaction
Total cellular RNA was isolated from transfected cells using the Trizol reagent (Invitrogen) according to the manufacturer’s instructions. RNA (2 μg) was reverse transcribed using the High-Capacity cDNA Archive kit (Applied Biosystems, Foster City, CA) according to the manufacturer’s protocol while incubated at 25°C for 10 minutes and 37°C for 120 minutes. Real-time polymerase chain reaction measurements were performed using the ABI PRISMs 7900HT Sequence Detection System Instrument and software (Applied Biosystems, Carlsbad, CA), as previously reported [9]. Commercially available Assays-on-Demand were used for measurement of expression of LMO2, HGAL, BCL6 and PRDM1 and were normalized to the 18S endogenous control.
Statistical methods
Quantitative RT-PCR, quantitative PCR, and promoter-luciferase assays were evaluated by two-tailed paired t tests. p-values are presented in each figure.
Acknowledgments
We wish to thank the staff of the Molecular Genomics Core and Flow Cytometry Core at the H. Lee Moffitt Cancer Center.
KLW is supported by National Institutes of Health (NIH) grants CA080990 and ISL is supported by NIH CA109335 and NIH CA122105, the Dwoskin Familyand Fidelity Foundations.
References
- 1.MacLennan IC. Germinal centers. Annu Rev Immunol. 1994;12:117–139. doi: 10.1146/annurev.iy.12.040194.001001. [DOI] [PubMed] [Google Scholar]
- 2.Alizadeh AA, Eisen MB, Davis RE, Ma C, Lossos IS, Rosenwald A, Boldrick JC, Sabet H, Tran T, Yu X, et al. Distinct types of diffuse large B-cell lymphoma identified by gene expression profiling. Nature. 2000;403:503–511. doi: 10.1038/35000501. [DOI] [PubMed] [Google Scholar]
- 3.Klein U, Tu Y, Stolovitzky GA, Keller JL, Haddad J, Jr, Miljkovic V, Cattoretti G, Califano A, Dalla-Favera R. Transcriptional analysis of the B cell germinal center reaction. Proc Natl Acad Sci U S A. 2003;100:2639–2644. doi: 10.1073/pnas.0437996100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Natkunam Y, Lossos IS, Taidi B, Zhao S, Lu X, Ding F, Hammer AS, Marafioti T, Byrne GE, Jr, Levy S, et al. Expression of the human germinal center-associated lymphoma (HGAL) protein, a new marker of germinal center B-cell derivation. Blood. 2005;105:3979–3986. doi: 10.1182/blood-2004-08-3112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Natkunam Y, Zhao S, Mason DY, Chen J, Taidi B, Jones M, Hammer AS, Hamilton Dutoit S, Lossos IS, Levy R. The oncoprotein LMO2 is expressed in normal germinal-center B cells and in human B-cell lymphomas. Blood. 2007;109:1636–1642. doi: 10.1182/blood-2006-08-039024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lossos IS, Alizadeh AA, Rajapaksa R, Tibshirani R, Levy R. HGAL is a novel interleukin-4-inducible gene that strongly predicts survival in diffuse large B-cell lymphoma. Blood. 2003;101:433–440. doi: 10.1182/blood-2002-06-1931. [DOI] [PubMed] [Google Scholar]
- 7.Azambuja D, Lossos IS, Biasoli I, Morais JC, Britto L, Scheliga A, Pulcheri W, Natkunam Y, Spector N. Human germinal center-associated lymphoma protein expression is associated with improved failure-free survival in Brazilian patients with classical Hodgkin lymphoma. Leuk Lymphoma. 2009;50:1830–1836. doi: 10.3109/10428190903242628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Durnick DK, Law ME, Maurer MJ, Natkunam Y, Levy R, Lossos IS, Kurtin PJ, McPhail ED. Expression of LMO2 is associated with t(14;18)/IGH-BCL2 fusion but not BCL6 translocations in diffuse large B-cell lymphoma. Am J Clin Pathol. 2010;134:278–281. doi: 10.1309/AJCPATUP1D0HGCUG. [DOI] [PubMed] [Google Scholar]
- 9.Lossos IS, Czerwinski DK, Alizadeh AA, Wechser MA, Tibshirani R, Botstein D, Levy R. Prediction of survival in diffuse large-B-cell lymphoma based on the expression of six genes. N Engl J Med. 2004;350:1828–1837. doi: 10.1056/NEJMoa032520. [DOI] [PubMed] [Google Scholar]
- 10.Natkunam Y, Farinha P, Hsi ED, Hans CP, Tibshirani R, Sehn LH, Connors JM, Gratzinger D, Rosado M, Zhao S, et al. LMO2 protein expression predicts survival in patients with diffuse large B-cell lymphoma treated with anthracycline-based chemotherapy with and without rituximab. J Clin Oncol. 2008;26:447–454. doi: 10.1200/JCO.2007.13.0690. [DOI] [PubMed] [Google Scholar]
- 11.Christoph T, Rickert R, Rajewsky K. M17: a novel gene expressed in germinal centers. Int Immunol. 1994;6:1203–1211. doi: 10.1093/intimm/6.8.1203. [DOI] [PubMed] [Google Scholar]
- 12.Lu X, Chen J, Malumbres R, Cubedo Gil E, Helfman DM, Lossos IS. HGAL, a lymphoma prognostic biomarker, interacts with the cytoskeleton and mediates the effects of IL-6 on cell migration. Blood. 2007;110:4268–4277. doi: 10.1182/blood-2007-04-087775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lu X, Kazmierczak K, Jiang X, Jones M, Watt J, Helfman DM, Moore JR, Szczesna-Cordary D, Lossos IS. Germinal center-specific protein human germinal center associated lymphoma directly interacts with both myosin and actin and increases the binding of myosin to actin. FEBS J. 2011 doi: 10.1111/j.1742-4658.2011.08109.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Jiang X, Lu X, McNamara G, Liu X, Cubedo E, Sarosiek KA, Sanchez-Garcia I, Helfman DM, Lossos IS. HGAL, a germinal center specific protein, decreases lymphoma cell motility by modulation of the RhoA signaling pathway. Blood. 2010;116:5217–5227. doi: 10.1182/blood-2010-04-281568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.McCormack MP, Forster A, Drynan L, Pannell R, Rabbitts TH. The LMO2 T-cell oncogene is activated via chromosomal translocations or retroviral insertion during gene therapy but has no mandatory role in normal T-cell development. Mol Cell Biol. 2003;23:9003–9013. doi: 10.1128/MCB.23.24.9003-9013.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.McCormack MP, Young LF, Vasudevan S, de Graaf CA, Codrington R, Rabbitts TH, Jane SM, Curtis DJ. The Lmo2 oncogene initiates leukemia in mice by inducing thymocyte self-renewal. Science. 2010;327:879–883. doi: 10.1126/science.1182378. [DOI] [PubMed] [Google Scholar]
- 17.Yamada Y, Pannell R, Forster A, Rabbitts TH. The oncogenic LIM-only transcription factor Lmo2 regulates angiogenesis but not vasculogenesis in mice. Proc Natl Acad Sci U S A. 2000;97:320–324. doi: 10.1073/pnas.97.1.320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Landry JR, Bonadies N, Kinston S, Knezevic K, Wilson NK, Oram SH, Janes M, Piltz S, Hammett M, Carter J, et al. Expression of the leukemia oncogene Lmo2 is controlled by an array of tissue-specific elements dispersed over 100 kb and bound by Tal1/Lmo2, Ets, and Gata factors. Blood. 2009;113:5783–5792. doi: 10.1182/blood-2008-11-187757. [DOI] [PubMed] [Google Scholar]
- 19.Warren AJ, Colledge WH, Carlton MB, Evans MJ, Smith AJ, Rabbitts TH. The oncogenic cysteine-rich LIM domain protein rbtn2 is essential for erythroid development. Cell. 1994;78:45–57. doi: 10.1016/0092-8674(94)90571-1. [DOI] [PubMed] [Google Scholar]
- 20.Turner CA, Jr, Mack DH, Davis MM. Blimp-1, a novel zinc finger-containing protein that can drive the maturation of B lymphocytes into immunoglobulin-secreting cells. Cell. 1994;77:297–306. doi: 10.1016/0092-8674(94)90321-2. [DOI] [PubMed] [Google Scholar]
- 21.Gong D, Malek TR. Cytokine-dependent Blimp-1 expression in activated T cells inhibits IL-2 production. J Immunol. 2007;178:242–252. doi: 10.4049/jimmunol.178.1.242. [DOI] [PubMed] [Google Scholar]
- 22.Kallies A, Hawkins ED, Belz GT, Metcalf D, Hommel M, Corcoran LM, Hodgkin PD, Nutt SL. Transcriptional repressor Blimp-1 is essential for T cell homeostasis and self-tolerance. Nat Immunol. 2006;7:466–474. doi: 10.1038/ni1321. [DOI] [PubMed] [Google Scholar]
- 23.Martins GA, Cimmino L, Shapiro-Shelef M, Szabolcs M, Herron A, Magnusdottir E, Calame K. Transcriptional repressor Blimp-1 regulates T cell homeostasis and function. Nat Immunol. 2006;7:457–465. doi: 10.1038/ni1320. [DOI] [PubMed] [Google Scholar]
- 24.Rutishauser RL, Martins GA, Kalachikov S, Chandele A, Parish IA, Meffre E, Jacob J, Calame K, Kaech SM. Transcriptional repressor Blimp-1 promotes CD8(+) T cell terminal differentiation and represses the acquisition of central memory T cell properties. Immunity. 2009;31:296–308. doi: 10.1016/j.immuni.2009.05.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kallies A, Carotta S, Huntington ND, Bernard NJ, Tarlinton DM, Smyth MJ, Nutt SL. A role for Blimp1 in the transcriptional network controlling natural killer cell maturation. Blood. 2011;117:1869–1879. doi: 10.1182/blood-2010-08-303123. [DOI] [PubMed] [Google Scholar]
- 26.Smith MA, Maurin M, Cho HI, Becknell B, Freud AG, Yu J, Wei S, Djeu J, Celis E, Caligiuri MA, et al. PRDM1/Blimp-1 controls effector cytokine production in human NK cells. J Immunol. 2010;185:6058–6067. doi: 10.4049/jimmunol.1001682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Chan YH, Chiang MF, Tsai YC, Su ST, Chen MH, Hou MS, Lin KI. Absence of the transcriptional repressor Blimp-1 in hematopoietic lineages reveals its role in dendritic cell homeostatic development and function. J Immunol. 2009;183:7039–7046. doi: 10.4049/jimmunol.0901543. [DOI] [PubMed] [Google Scholar]
- 28.Smith MA, Wright G, Wu J, Tailor P, Ozato K, Chen X, Wei S, Piskurich JF, Ting JP, Wright KL. Positive regulatory domain 1 (PRDM1) and IRF8/Pu.1 counter regulate MHC Class II transactivator (CIITA) expression during dendritic cell maturation. J Biol Chem. 2011;286:7893–7904. doi: 10.1074/jbc.M110.165431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Gyory I, Wu J, Fejer G, Seto E, Wright KL. PRDI-BF1 recruits the histone H3 methyltransferase G9a in transcriptional silencing. Nat Immunol. 2004;5:299–308. doi: 10.1038/ni1046. [DOI] [PubMed] [Google Scholar]
- 30.Yu J, Angelin-Duclos C, Greenwood J, Liao J, Calame K. Transcriptional repression by blimp-1 (PRDI-BF1) involves recruitment of histone deacetylase. Mol Cell Biol. 2000;20:2592–2603. doi: 10.1128/mcb.20.7.2592-2603.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ancelin K, Lange UC, Hajkova P, Schneider R, Bannister AJ, Kouzarides T, Surani MA. Blimp1 associates with Prmt5 and directs histone arginine methylation in mouse germ cells. Nat Cell Biol. 2006;8:623–630. doi: 10.1038/ncb1413. [DOI] [PubMed] [Google Scholar]
- 32.Su ST, Ying HY, Chiu YK, Lin FR, Chen MY, Lin KI. Involvement of histone demethylase LSD1 in Blimp-1-mediated gene repression during plasma cell differentiation. Mol Cell Biol. 2009;29:1421–1431. doi: 10.1128/MCB.01158-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kuo TC, Calame KL. B lymphocyte-induced maturation protein (Blimp)-1, IFN regulatory factor (IRF)-1, and IRF-2 can bind to the same regulatory sites. J Immunol. 2004;173:5556–5563. doi: 10.4049/jimmunol.173.9.5556. [DOI] [PubMed] [Google Scholar]
- 34.Shapiro-Shelef M, Lin KI, McHeyzer-Williams LJ, Liao J, McHeyzer-Williams MG, Calame K. Blimp-1 is required for the formation of immunoglobulin secreting plasma cells and pre-plasma memory B cells. Immunity. 2003;19:607–620. doi: 10.1016/s1074-7613(03)00267-x. [DOI] [PubMed] [Google Scholar]
- 35.Shapiro-Shelef M, Lin KI, Savitsky D, Liao J, Calame K. Blimp-1 is required for maintenance of long-lived plasma cells in the bone marrow. J Exp Med. 2005;202:1471–1476. doi: 10.1084/jem.20051611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Savitsky D, Calame K. B-1 B lymphocytes require Blimp-1 for immunoglobulin secretion. J Exp Med. 2006;203:2305–2314. doi: 10.1084/jem.20060411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Messika EJ, Lu PS, Sung Y-J, Yao T, Chi J-T, Chien Y-h, Davis MM. Differential Effect of B Lymphocyte-induced Maturation Protein (Blimp-1) Expression on Cell Fate during B Cell Development. J Exp Med. 1998;188:515–525. doi: 10.1084/jem.188.3.515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Pasqualucci L, Compagno M, Houldsworth J, Monti S, Grunn A, Nandula SV, Aster JC, Murty VV, Shipp MA, Dalla-Favera R. Inactivation of the PRDM1/BLIMP1 gene in diffuse large B cell lymphoma. J Exp Med. 2006;203:311–317. doi: 10.1084/jem.20052204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Tam W, Gomez M, Chadburn A, Lee JW, Chan WC, Knowles DM. Mutational analysis of PRDM1 indicates a tumor-suppressor role in diffuse large B-cell lymphomas. Blood. 2006;107:4090–4100. doi: 10.1182/blood-2005-09-3778. [DOI] [PubMed] [Google Scholar]
- 40.Garcia JF, Roncador G, Sanz AI, Maestre L, Lucas E, Montes-Moreno S, Fernandez Victoria R, Martinez-Torrecuadrara JL, Marafioti T, Mason DY, et al. PRDM1/BLIMP-1 expression in multiple B and T-cell lymphoma. Haematologica. 2006;91:467–474. [PubMed] [Google Scholar]
- 41.Mandelbaum J, Bhagat G, Tang H, Mo T, Brahmachary M, Shen Q, Chadburn A, Rajewsky K, Tarakhovsky A, Pasqualucci L, et al. BLIMP1 is a tumor suppressor gene frequently disrupted in activated B cell-like diffuse large B cell lymphoma. Cancer Cell. 2010;18:568–579. doi: 10.1016/j.ccr.2010.10.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Shaffer AL, Lin KI, Kuo TC, Yu X, Hurt EM, Rosenwald A, Giltnane JM, Yang L, Zhao H, Calame K, et al. Blimp-1 orchestrates plasma cell differentiation by extinguishing the mature B cell gene expression program. Immunity. 2002;17:51–62. doi: 10.1016/s1074-7613(02)00335-7. [DOI] [PubMed] [Google Scholar]
- 43.Ghosh N, Gyory I, Wright G, Wood J, Wright KL. Positive regulatory domain I binding factor 1 silences class II transactivator expression in multiple myeloma cells. J Biol Chem. 2001;276:15264–15268. doi: 10.1074/jbc.M100862200. [DOI] [PubMed] [Google Scholar]
- 44.Lin KI, Angelin-Duclos C, Kuo TC, Calame K. Blimp-1-dependent repression of Pax-5 is required for differentiation of B cells to immunoglobulin M-secreting plasma cells. Mol Cell Biol. 2002;22:4771–4780. doi: 10.1128/MCB.22.13.4771-4780.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Lin Y, Wong K, Calame K. Repression of c-myc transcription by Blimp-1, an inducer of terminal B cell differentiation. Science. 1997;276:596–599. doi: 10.1126/science.276.5312.596. [DOI] [PubMed] [Google Scholar]
- 46.Piskurich JF, Lin KI, Lin Y, Wang Y, Ting JP, Calame K. BLIMP-I mediates extinction of major histocompatibility class II transactivator expression in plasma cells. Nat Immunol. 2000;1:526–532. doi: 10.1038/82788. [DOI] [PubMed] [Google Scholar]
- 47.Desai S, Maurin M, Smith MA, Bolick SC, Dessureault S, Tao J, Sotomayor E, Wright KL. PRDM1 is required for mantle cell lymphoma response to bortezomib. Mol Cancer Res. 2010;8:907–918. doi: 10.1158/1541-7786.MCR-10-0131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Ghosh N, Piskurich JF, Wright G, Hassani K, Ting JP, Wright KL. A novel element and a TEF-2-like element activate the major histocompatibility complex class II transactivator in B-lymphocytes. J Biol Chem. 1999;274:32342–32350. doi: 10.1074/jbc.274.45.32342. [DOI] [PubMed] [Google Scholar]
- 49.Wang X, Belguise K, O’Neill CF, Sanchez-Morgan N, Romagnoli M, Eddy SF, Mineva ND, Yu Z, Min C, Trinkaus-Randall V, et al. RelB NF-kappaB represses estrogen receptor alpha expression via induction of the zinc finger protein Blimp1. Mol Cell Biol. 2009;29:3832–3844. doi: 10.1128/MCB.00032-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Steinke JW, Hodsdon W, Parenti S, Ostraat R, Lutz R, Borish L, Hagman J. Identification of an Sp factor-dependent promoter in GCET, a gene expressed at high levels in germinal center B cells. Mol Immunol. 2004;41:1145–1153. doi: 10.1016/j.molimm.2004.06.031. [DOI] [PubMed] [Google Scholar]
- 51.Wadman IA, Osada H, Grutz GG, Agulnick AD, Westphal H, Forster A, Rabbitts TH. The LIM-only protein Lmo2 is a bridging molecule assembling an erythroid, DNA-binding complex which includes the TAL1, E47, GATA-1 and Ldb1/NLI proteins. EMBO J. 1997;16:3145–3157. doi: 10.1093/emboj/16.11.3145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Crable SC, Anderson KP. A PAR domain transcription factor is involved in the expression from a hematopoietic-specific promoter for the human LMO2 gene. Blood. 2003;101:4757–4764. doi: 10.1182/blood-2002-09-2702. [DOI] [PubMed] [Google Scholar]
- 53.Hammond SM, Crable SC, Anderson KP. Negative regulatory elements are present in the human LMO2 oncogene and may contribute to its expression in leukemia. Leuk Res. 2005;29:89–97. doi: 10.1016/j.leukres.2004.05.013. [DOI] [PubMed] [Google Scholar]
- 54.Oram SH, Thoms JA, Pridans C, Janes ME, Kinston SJ, Anand S, Landry JR, Lock RB, Jayaraman PS, Huntly BJ, et al. A previously unrecognized promoter of LMO2 forms part of a transcriptional regulatory circuit mediating LMO2 expression in a subset of T-acute lymphoblastic leukaemia patients. Oncogene. 2010;29:5796–5808. doi: 10.1038/onc.2010.320. [DOI] [PubMed] [Google Scholar]
- 55.Doody GM, Care MA, Burgoyne NJ, Bradford JR, Bota M, Bonifer C, Westhead DR, Tooze RM. An extended set of PRDM1/BLIMP1 target genes links binding motif type to dynamic repression. Nucleic Acids Res. 2010;38:5336–5350. doi: 10.1093/nar/gkq268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Desai S, Bolick SC, Maurin M, Wright KL. PU.1 regulates positive regulatory domain I-binding factor 1/Blimp-1 transcription in lymphoma cells. J Immunol. 2009;183:5778–5787. doi: 10.4049/jimmunol.0901120. [DOI] [PMC free article] [PubMed] [Google Scholar]