Abstract
Background and Aims
Nucleoplasmic Ca2+ regulates cell growth in the liver, but the proteins through which this occurs are unknown.
Methods
We used Rapid Subtraction Hybridization (RaSH) to subtract genes in SKHep1 liver cells expressing the Ca2+ buffer protein parvalbumin (PV) targeted to the nucleus, from genes in cells expressing a mutated form of nuclear-targeted PV which has one of two Ca2+-binding site inactivated. The subtraction permitted selection of genes whose expression was affected by a small alteration in nuclear Ca2+ concentration.
Results
The asparaginyl endopeptidase legumain (LGMN) was identified in this screening. When Ca2+ was buffered in the nucleus of SKHep1 cells, LGMN mRNA was decreased by 97%, in part by a transcriptional mechanism, and decreased expression at the protein level was observed by immunoblot and immunofluorescence. Treatment with Hepatocyte Growth Factor increased LGMN expression. Knockdown of LGMN by siRNA decreased proliferation of SKHep1 cells by ~50% as measured both by BrdU uptake and mitotic index, although an inhibitor of LGMN activity did not affect BrdU incorporation. A significant reduction in the fraction of cells in G2/M phase was seen as well. This was associated with increases in expression of cyclins A and E. Furthermore, LGMN expression was increased in hepatocellular carcinoma cells relative to normal hepatocytes in the same specimens.
Conclusions
These findings suggest a new role for LGMN and provide evidence that nuclear Ca2+ signals regulate cell proliferation in part through modulation of LGMN expression. Increased expression of LGMN may be involved in liver carcinogenesis.
Keywords: Nuclear calcium, hepatocelullar carcinoma, legumain
INTRODUCTION
Ca2+ regulates a wide range of activities in the liver, including bile secretion [32, 35], canalicular contraction [36], metabolism [12], gene transcription [13, 40], apoptosis [30, 31], and growth of liver tumors [42]. The spatial patterns of Ca2+ signals determine the specificity of these signals and which responses are activated [21, 22]. Increases in nucleoplasmic Ca2+ have specific biological effects that differ from the effects of increases in cytosolic Ca2+. These effects include activation of distinct genes and transcription factors [4, 13, 40, 46], activation of intranuclear kinases [8, 9] and regulation of cell proliferation [42]. The nucleus contains the machinery required for local formation of Ca2+ signals, including PIP2, PLC, and InsP3R-gated Ca2+ stores [1, 11, 41], and several mechanisms permit selective activation of Ca2+ signaling pathways within the nucleus. This includes direct coupling between integrin receptors and the nucleus [27] and translocation of receptor tyrosine kinases such as c-met and the insulin receptor to the nucleus [11, 41]. However, the specific targets of Ca2+ signals in the nucleus that are responsible for regulating cell proliferation are not clear.
Legumain (LGMN) is an endopeptidase that was first identified in plants [19] and later in humans and mice [6], and parasites [3] and helminthes [38]. LGMN hydrolyzes peptides and proteins on the carboxyl side of asparaginyl residues. This enzyme is predominantly localized in late endosomes and lysosomes [7] and has been implicated in antigen processing [28, 29], regulation of biosynthesis of lysosomal proteins [45], and extracellular matrix turnover [33]. LGMN also is present in the tumor microenvironment where it is expressed by macrophages and contributes to metastatic behavior by promoting cell migration and tissue invasion [24]. Increased expression of LGMN is associated with poorer differentiation of tumors and a higher degree of necrosis and apoptosis. LGMN co-localizes with integrins at the invading front of tumors and expression of this enzyme is associated with increased invasiveness [10, 26]. Thus, until now the effects of LGMN have been related to its actions as an endopeptidase that modulates the invasive and metastatic potential of cells. Here we identified LGMN as one target of nuclear Ca2+ and characterize the role of this gene in regulating cell proliferation.
MATERIAL AND METHODS
Materials, reagents and cell lines
The SKHep1 liver cell line was from ATCC (Manassas, VA). Cells were grown at 37°C with 5% CO2 in DMEM supplemented with 1% penicillin-streptomycin and 10% fetal bovine serum (Gibco, Grand Island, NY). SuperScript First-Strand Synthesis System for RT-PCR, TRizol reagent and lipofectamine 2000 were from Invitrogen (Eugene, OR). Anti-LGMN polyclonal antibody was from ABCAM (Cambridge, MA). Enhanced chemiluminescence reagent (ECL plus) and horseradish peroxidase (HRP) antibodies were from GE Biosciences (Buckinghamshire, UK). Mouse monoclonal antibodies to α-tubulin, β-actin, anti-gamma tubulin were purchased from Sigma–Aldrich (St. Louis, MO).
PV-DsRed adenovirus constructs and infection conditions
Ca2+ was selectively buffered within the nucleoplasm by expressing parvalbumin (PV) with a nuclear localization sequence fused to DsRed [40]. One of the two Ca2+ binding sites of PV is disrupted in the CD mutant, which was used as well. Recombinant adenoviruses pAd-PV-NLS-DsRed and pAd-PV-NLS-CD-DsRed [42] were used for delivery to SKHep1 cells, which were infected at a multiplicity of infection (MOI) of 100. Cells were used 48 hrs after infection. It has previously been demonstrated that PV-NLS effectively and selectively buffers Ca2+ in the nucleus of transfected cells [40].
Rapid Subtraction Hybridization (RaSH)
RaSH was performed using a modification of the method previously described [17]. Total RNA (25 µg), isolated from cells infected with pAd-PV-NLS-DsRed or pAd-PV-NLS-CD-DsRed (control adenovirus), was used for cDNA synthesis. cDNA then was digested with MboI (Fermentas, Glen Burnie, MD) followed by ligation to adapters XDPN-14 (5’CTGATCACTCGAGA3’) and XDPN-12 (5’GATCTCTCGAGT’3) at 4°C overnight. Mixtures were diluted in 100 µl of 10mM Tris/1mM EDTA, pH 7 and used for PCR amplification with 10 µM XDPN-18 (5’CTGATCA CTCGAGAGATC3’). To obtain TESTER samples, 3µg of the PCR products (DRIVER) were digested with XhoI. The TESTER cDNA (3 µg) was mixed with 10 µg of the DRIVER cDNA in 10 µl of a hybridization solution [0.5 M NaCl / 50nM Tris, pH 7.5 / 0.2% SDS / 40% (vol/vol) formamide] and boiled for 5 min, then incubated at 42°C for 48 hrs. Part of the mixture (3 µl) was ligated with 1 µg of XhoI-digested pZErO-1 (Invitrogen, Carisbad, CA) plasmids and transformed into DH-10B bacteria. The colonies with positive clones were subjected to DNA plasmid extraction and submitted for automated sequencing. Sequences were confirmed by BLASTN.
Real Time PCR
Total RNA was isolated from cells using TRIzol and reverse-transcribed using SuperScript II (Invitrogen). Sense and antisense primer sequences for PCR amplification of β-actin were: 5′-GACGGCCAGGTCATCCTA TTG-3′ and 5′-AGGAAGGCTGGAAAAGAGCC-3′ and 5’-GCATAGGATCCGGCAAAGTC-3’ and 5’-TCCAGTAGATCCA TGGTCAGTGA-3’ for LGMN. DNA templates were amplified by real time PCR using the Sybr Green Method. β-actin was used as an internal control. Experiments were performed in triplicate. After amplification, 10µl of reaction mixture were subjected to electrophoresis on agarose gels and visualized after ethidium bromide staining.
Western blot and CDK 2 kinase assay
Standard methods were used for immunoblots [23]. Cells were lysed at 4°C and fifty micrograms of total cellular protein was separated by SDS-PAGE. Membranes were blocked and then incubated with an affinity-purified polyclonal antibody against human LGMN (1:1000). Membranes were then incubated with peroxidase-conjugated secondary antibody (1:5000). Bands were revealed by enhanced chemiluminescence. For experiments involving HGF stimulation, cells were starved overnight and treated with HGF (100 ng/mL, at 37°C) for the indicated time points prior to protein extraction. CDK2 activity was evaluated by a modified in vitro Histone H1 phosphorylation assay [20].
Immunofluorescence
Confocal immunofluorescence was performed as described [30]. SKHep1 cells were fixed with 4% paraformaldehyde and incubated with anti-LGMN antibody (1:200) for 2 hrs at room temperature followed by incubation with goat anti-rabbit secondary antibody conjugated with Alexa 488 (1:500), for 1 hr. Images were obtained using a Zeiss LSM 510 confocal microscope (Thornwood, NY) and mean fluorescence was quantified with ImageJ software (NIH, Bethesda, MD).
Transfection of siRNA
Silencer select siRNA sequences specific for LGMN or scrambled control sequences were acquired from Ambion (Austin, TX). Cell cultures were treated with 20 nM of siRNA using lipofectamine 2000 (Invitrogen). Cells were incubated at 37° C in an atmosphere of 5% CO2 for 72 hrs prior to use.
Measurement of BrdU incorporation
Cell proliferation was measured by BrdU incorporation using ELISA (Roche Applied Science). SKHep1 cells were plated in 96-well culture plates, starved for 24 hrs and transfected with siRNA. 72 hrs afterwards, cells were treated for 2 hrs with BrdU labeling solution. Cells were then fixed, and anti-BrdU antibody was added. BrdU incorporation was measured colorimetrically. For experiments employing the legumain inhibitor MV026630, cells were treated for 24 hours with the inhibitor at 25 or 50 µM. Control cells were treated with DMSO alone.
Mitotic index measurements
SKHep1 cells treated with siRNA were labeled for phospho-histone-3 (Upstate Biotechnology, Chicago, IL) by immunofluorescence and then examined by confocal microscopy. Mitotic cells were defined by the presence of DNA condensation and phospho-histone-3 positivity [14]. Mitotic index was calculated as the percentage of mitotic cells per total cell number. At least 20 fields representing a total of >100 cells were visualized for each condition.
Cell cycle analysis
SKHep1 cells transfected with siRNA were trypsinized, washed in phosphate-buffered saline (PBS), fixed 70% Ethanol at 4°C and then washed with PBS. After centrifugation cells were supplemented with RNase (100 µg/ml Sigma) for 5 min and stained with propidium iodide (50 µg/ml). DNA content was determined using a FACSCalibur (BD Biosciences), and the data were analyzed using Flowjo software.
Apoptosis assay
Apoptosis was measured using a caspase-3 activity kit with colorimetric detection (BD Biosciences). Apoptosis was induced using 500 nM staurosporine (Sigma-Aldrich) as a positive control.
Imunofluorescence in human tissue samples
Paraffin-embedded sections of hepatocellular carcinoma (HCC) human liver were pretreated with 10 mmol/L Tris, 1 mmol/L EDTA buffer at 100°C, incubated with anti-LGMN and then labeled with Alexa 488-conjugated secondary antibodies [44]. Nuclei were stained with TO-PRO3. To ensure specificity of staining, images were obtained using machine settings at which no fluorescence was detectable in negative controls labeled with secondary antibodies alone. Specimens were observed by confocal microscopy and quantified with Image J. The diagnosis of hepatocellular carcinoma was made in each case using standard histopathologic criteria [16]. These included the presence of a discreet liver mass composed of a proliferation of hepatocytes exhibiting a variety of disordered growth patterns. Loss of lobular architecture, thickened hepatocellular cell plates, aberrant arteries, abnormal cytologic inclusions, and nuclear pleomorphism were observed to varying degrees in each case. Reticulin stains were used to highlight abnormal cellular arrangements.
Legumain promoter luciferase assay
The plasmids pGL4.10[Luc2] and pRL-CMV, which provide constitutive expression of Renilla luciferase, were obtained from Promega (Madison, WI). For generation of the LGMN reporter vector, a 750 bp sequence upstream of the initiation codon of the LGMN gene was amplified by PCR from SKHep1 genomic DNA with the following primers: 5’-GGACCACCCAGAAACACC-3’ and 5’- CTCGCTTAAGGGCCACTG-3’. The product of this reaction was used as a template in a second round of amplification using primers: 5’- GCGCCTCGAGTTGGCATTCTAAAATAGGGAAGTTAA-3’ and 5’-GCGCAGATCTGCGTGGCATCTGCCAAAA -3’, which introduced a 5' XhoI and 3' BglII restriction site, respectively. The fragment was cloned into the pGL4.10[luc2] to generate the pLGMNprom vector whose identity was confirmed by automated sequencing. Lipofectamine 2000 was used to transfect SKHep1 cells with 10 µg of pLGMNprom alone or in combination with 3 µg of pPV-NLS-DsRed, which expresses a nuclear targeted parvalbumin tagged with DsRed. Luciferase activity was measured with a Dual-Luciferase Reporter Assay (Promega, Madison, WI). Luciferase activity was normalized by Renilla luciferase activity. Each assay was performed in triplicate and the data were compared to empty vector controls.
Statistical analysis
Results are expressed as mean±SEM. Prism software (GraphPad Software, San Diego, CA) was used for data analysis. Statistical significance was tested using Student’s t test or one-way ANOVA followed by Bonferroni post-tests, and p value < 0.05 was taken to indicate statistical significance.
RESULTS
Identification of LGMN as a protein that is sensitive to nuclear Ca2+ by RaSH
To define the spectrum of gene expression changes occurring after buffering nuclear Ca2+, a modified subtraction hybridization technique was used [17]. cDNAs libraries were obtained from SKHep1 cells treated with pAd-PV-NLS-DsRed or pAd-PV-NLS-CD-DsRed. These constructs have been used previously to selectively buffer nuclear Ca2+ in SKHep1 cells [40, 42]. Two cDNA libraries were constructed, one with genes up-regulated and the second one with genes down-regulated when nuclear Ca2+ was buffered (Figure 1). Colonies from the two subtractive libraries were isolated randomly and the PCR-amplified products were sequenced and compared with genes deposited in GenBank. One hundred forty-five insert DNA fragments were amplified by PCR (data not shown). From this pool of positive clones 13 differentially expressed clones were identified (Table 1). Among these 13 clones, expression was down-regulated in three and up-regulated in ten when nuclear Ca2+ was buffered. Three of these genes were chosen for validation by real time PCR: LGMN, transforming growth factor beta regulator 4, and reticulon 4. These three of the thirteen genes were studied further based on literature linking them to cell proliferation and tumor progression. However, additional experiments were only able to demonstrate that expression of LGMN affected cell proliferation, so only that protein was studied in greater detail.
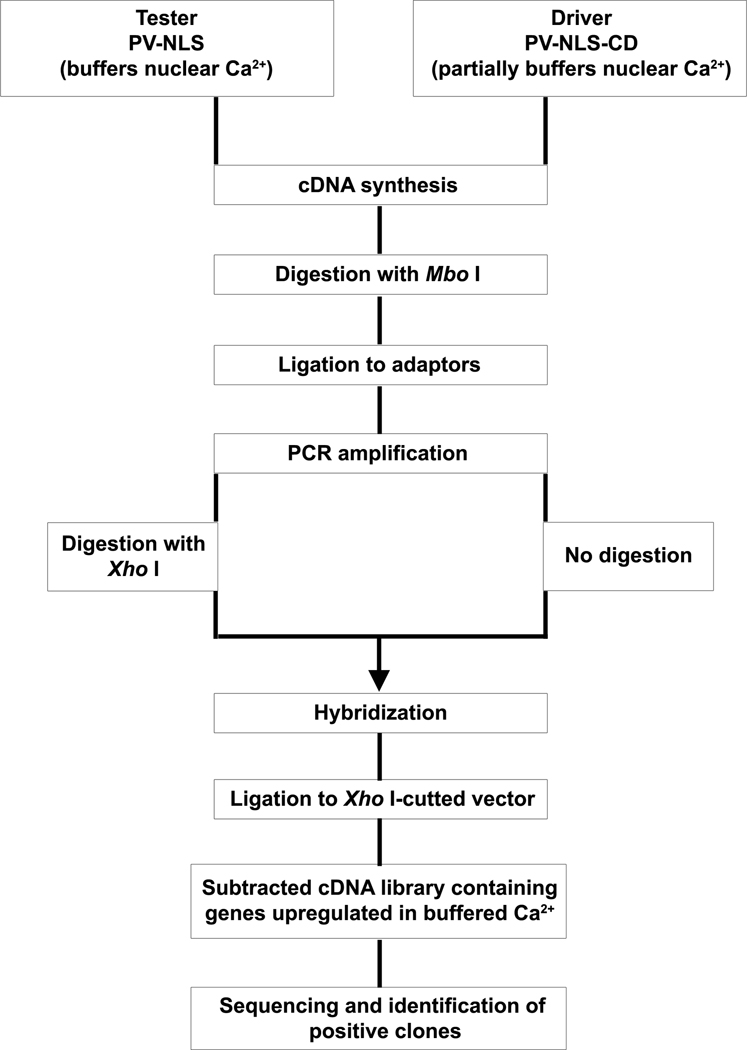
Figure 1. Schematic outline of the RaSH protocol.
Tester and driver libraries were constructed, followed by digestion of only the tester library with XhoI. After hybridization, differentially expressed sequences were cloned into XhoI-digested vectors, resulting in a subtracted cDNA library enriched in genes displaying differential expression. By using the PV-NLS library as the tester and the PV-NLS-CD library as the driver, RaSH was used to produce a subtracted cDNA library enriched in genes up-regulated when nuclear Ca2+ is buffered (adapted from [17]). Down-regulated genes can be isolated using the PV-NLS-CD library as the tester and the PV-NLS library as the driver.
Table 1.
Sequences identified by RaSH.
| Clone GeneBank | Nomenclature | GeneBank Access | Change* |
|---|---|---|---|
| AT rich interactive domain 1A | ARID 1A | NM_006015.4 | ↑ |
| Legumain | LGMN | NM_005606.5 | ↓ |
| Transforming growth factor beta regulator 4 | TBRG4 | NM_030900.2 | ↓ |
| Reticulon 4 | RTN4 | NM_207521.1 | ↑ |
| Tubulin gamma | TUB1 | NM_001070.3 | ↑ |
| Ras homolog gene family member A | RHOA | NM_001664.2 | ↑ |
| Mps One Binder kinase activator-like 1B | MOB1 | NM_018221.3 | ↑ |
| Similar to ribossomal protein L3 | - | CX816870 | ↑ |
| Splincing factor 3B | - | NM_012426.3 | ↑ |
| Alanyl-tRNA synthetase | AARS | NM_001605.2 | ↑ |
| Serpin peptidase inhibitor member 1 | SERPINE 1 | NM_000602.1 | ↑ |
| Prohibitin 2 | PBHB2 | NM_007531.2 | ↓ |
| Similar to ribossomal protein L27 | - | CX816883 | ↑ |
Change means gene expression level compared to normal control. ↑ means up-regulate; ↓ means down-regulate when nuclear Ca2+ is buffered.
Buffering nuclear Ca2+ inhibits expression of LGMN
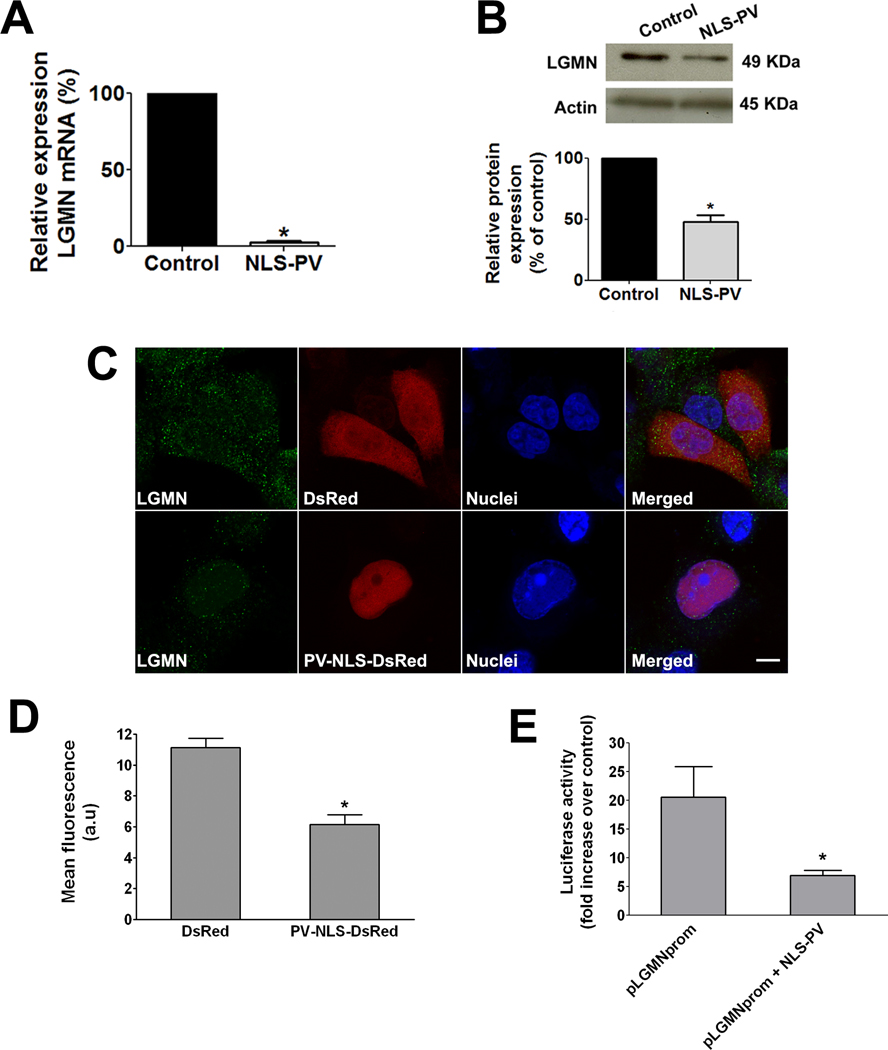
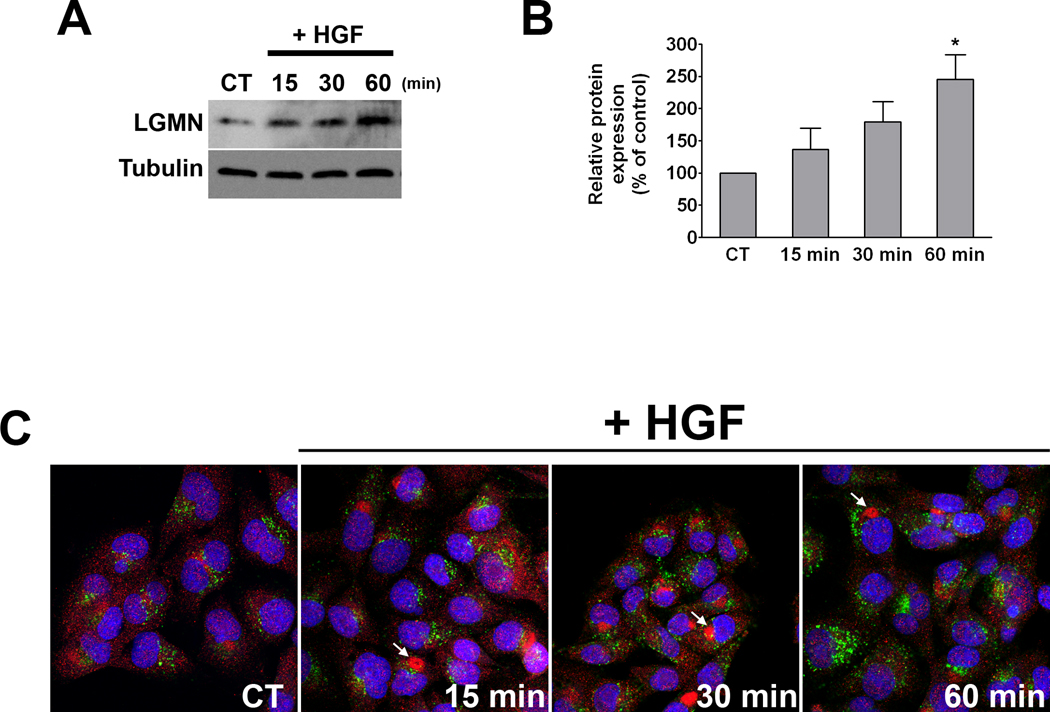
Real time PCR analysis showed that LGMN mRNA decreased by 97±2% (p<0.0001) when nuclear Ca2+ was buffered (Figure 2A). A decrease in LGMN at the protein level was also observed 48 hrs after infection with the adenovirus PV-NLS. Protein expression of LGMN decreased by 52±9% (p<0.01) after nuclear Ca2+ was buffered (Figure 2B). To confirm that expression of this gene was altered in SKHep1 cells expressing PV fused to DsRed within the nucleus, confocal immunofluorescence was performed. Staining for LGMN (Figure 2C) indicated that the expression of this protein was lower in cells expressing DsRed-tagged PV within the nucleus when compared to control cells transfected with DsRed alone (quantified in Figure 2D). Because buffering of nuclear Ca2+ is associated with reduced LGMN mRNA levels, we investigated whether nuclear Ca2+ transcriptionally regulates LGMN expression. A sequence encompassing 750 bp upstream of the LGMN start codon was used as a putative promoter in a dual-reporter luciferase assay. Luciferase activity driven by the LGMN promoter was significantly reduced when nuclear Ca2+ was buffered by expression of parvalbumin in the nucleus (20.57±5.34 pLGMNprom alone as compared to 6.89±0.88 pLGMNprom + PV-NLS; p< 0.05; Figure 2E). These data show that buffering nuclear Ca2+ reduces LGMN expression, in part through transcription regulation. Next we examined whether hepatocyte growth factor (HGF), a potent mitogen for hepatocytes that also selectively increases nuclear Ca2+ in SKHep1 cells [11], modulates LGMN expression. HGF (100 ng/ml) promoted an increase in LGMN expression in a time-dependent manner, reaching its maximum at 60 min (245.7±37.9%, p<0.05; Figures 3A–B). This increase in expression was associated with partial redistribution of LGMN, which changed from a punctate pattern observed in non-stimulated cells to the formation of a large peri-nuclear aggregate in cells treated with HGF (Figure 3C).
Figure 2. LGMN expression is decreased by buffering nuclear Ca2+.
(A) Real Time quantitative PCR was used to measure the relative expression of LGMN mRNA in the SKHep1 liver cell line. LGMN mRNA was decreased by 97±2% in cells transfected with PV-NLS, relative to nontransfected controls (p< 0.0001). β-actin gene was used to normalize expression in both groups. The data are expressed as mean ± SEM of triplicate measurements and are representative of three separate experiments (p<0.05). (B) Immunoblot of whole-cell protein from SKHep1 cells 48 hr after infection demonstrates that LGMN protein expression is decreased after buffering nuclear Ca2+. Densitometric analysis confirms reduction of LGMN protein expression to 52±9% of controls (p<0.01). Expression of β-actin was used as a loading control. (C) Confocal immunofluorescence confirms decreased LGMN expression (green) in SKHep1 cells transfected with PV-NLS (red). Nuclei are identified by TO-PRO-3 staining (blue). Detection of the DsRed tag on PV-NLS confirms that it is localized to the nucleus. Results are representative of four independent experiments. Scale bar = 10µm. (D) Quantification of LGMN immunofluorescence shows that buffering nuclear Ca2+ reduces LGMN expression by 45% (11.1±0.4 in DsRed group versus 6.1±0.6 a.u. in PV-NLS-DsRed cells; p<0.001). (E) Luciferase assay shows that LGMN promoter activity is inhibited when nuclear Ca2+ is buffered (20.6±5.3 pLGMNprom versus 6.9±0.9 pLGMNprom + PV-NLS; p< 0.05). Data are expressed as mean fold increase ± S.E.M over empty vector in 5 separate experiments.
Figure 3. HGF stimulation increases LGMN expression.
(A) Western blot analysis of total cell lysates prepared from control (non-stimulated) cells and cells stimulated with HGF (100 ng/mL) for the indicated time periods demonstrates a time-dependent increase in LGMN expression. (B) Bar graph shows the densitometric quantification of 4 separate experiments (p<0.05, one-way ANOVA). (C) LGMN localization is altered after HGF treatment. SKHep1 cells were stimulated as above and examined by confocal immunofluorescence. LGMN (red) is present in punctate structures that do not co-localize with the lysosomal marker Lamp-1 (green), in either control or HGF-stimulated cells. Nuclei are labeled by TO-PRO-3 (blue). Upon HGF stimulation, LGMN accumulates near the nucleus (arrows).
LGMN affects cell proliferation
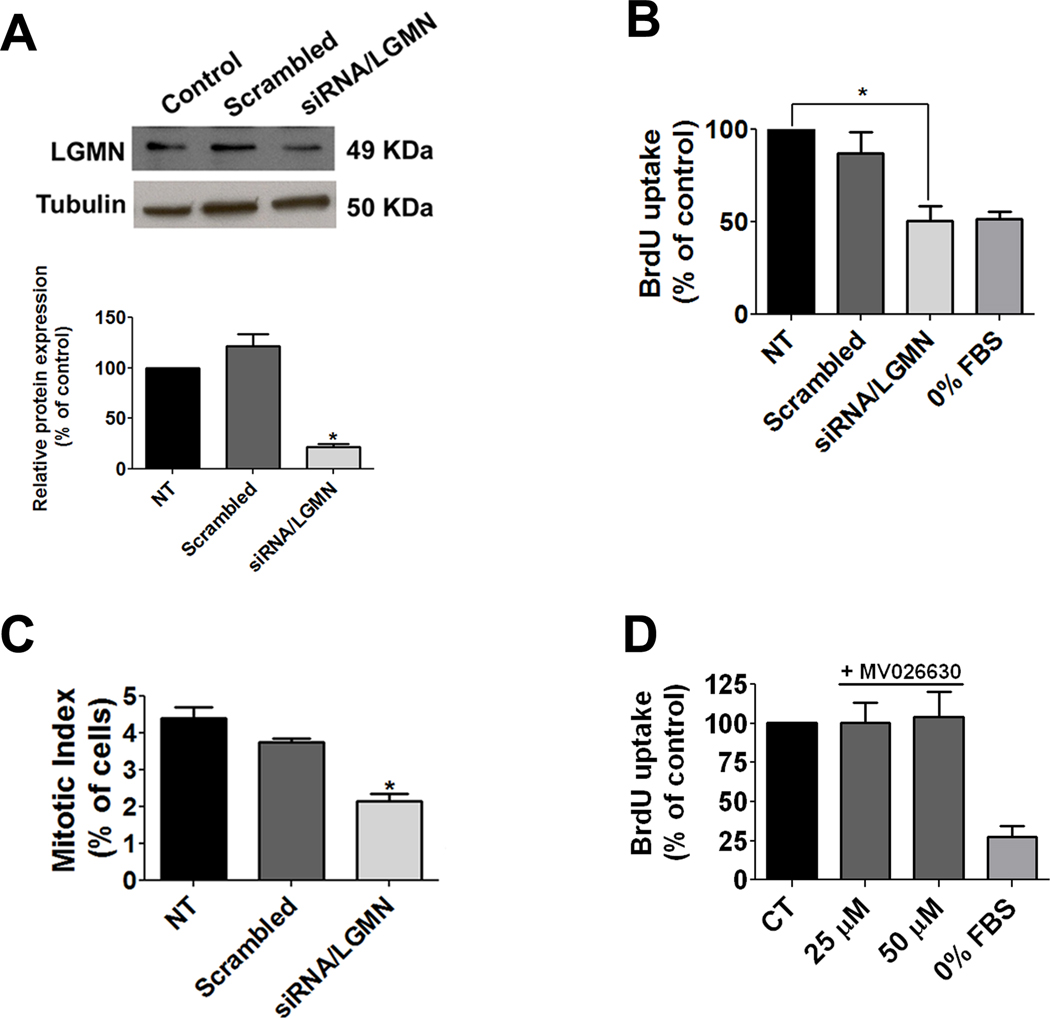
Nuclear Ca2+ regulates proliferation of SKHep1 cells by controlling progression through the cell cycle [42]. Therefore the relationship between LGMN expression and proliferation of these cells was investigated. RNA interference constructs decreased LGMN expression by 78±6% (p<0.0001; Figure 4A). Cell proliferation was assessed by BrdU incorporation in populations of cells in which LGMN was silenced. Knockdown of LGMN decreased BrdU incorporation by 49% (p<0.005; Figure 4B). The fraction of cells in mitosis also was quantified by calculation of the mitotic index. The fraction of cells in mitosis decreased when LGMN was silenced (Figure 4C). Similarly, cell counting showed that proliferation of SKHep1 cells decreased after knockdown of LGMN (data not shown). Because LGMN’s only known biological effect is its endopeptidase activity, BrdU uptake was further investigated in SKHep1 cells in the presence of the chemical inhibitor of LGMN, MV026630 [25]. Inhibition of LGMN enzymatic activity did not alter BrdU incorporation (p>0.05 by one-way ANOVA; Figure 4D), suggesting that LGMN’s proliferative effect is independent of its endopeptidase activity. These results provide evidence that LGMN promotes cell proliferation and suggest that nuclear Ca2+ regulates proliferation of SKHep1 cells in part through modulation of LGMN expression.
Figure 4. Knock down of LGMN inhibits cell proliferation.
(A) Silencing of LGMN. SKHep1 cells were transfected with 20 nM siRNA for LGMN, and incubated for 72 hrs. Immunoblots demonstrate that LGMN but not scrambled siRNA knocks down LGMN expression. α-tubulin serves as a loading control. Densitometry confirms reduction of LGMN to 21.5±3.2% of nontransfected controls (p<0.0001). Results are representative of three separate experiments. (B) Knockdown of LGMN decreases BrdU incorporation in SKHep1 cells to 50.7±8.1% of controls (p<0.0034). (C) Mitotic SKHep1 cells were identified by confocal imaging of phospho histone-3 labeling measured 72 hrs after knockdown of LGMN. The mitotic index is decreased to 2.2±0.7% of control in cells in which LGMN is silenced (p<0.0001). A total of 300 cells were examined in three separate experiments. (D) Inhibition of LGMN activity with MV026630 at either 25 or 50 µM does not alter BrdU uptake in SKHep cells (p>0.05).
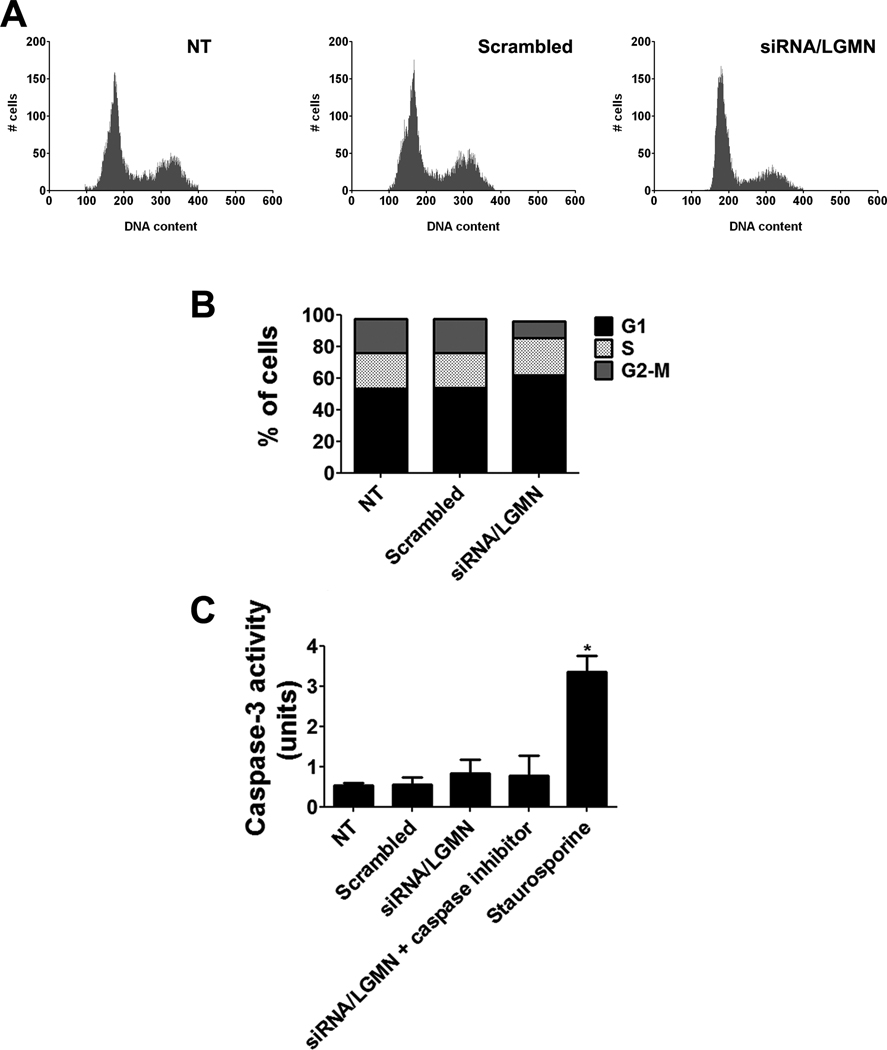
Knockdown of LGMN alters the cell cycle profile rather than apoptosis
In order to understand why reduced expression of LGMN inhibited cell proliferation, we investigated whether the decreased cell proliferation was due to apoptosis or changes in the cell cycle profile. Flow cytometry was used to examine whether reduction in expression of LGMN alters the distribution of cells through the phases of cell cycle. Cells were synchronized in G0 by serum starvation for 24 hrs, and then released into the cell cycle by addition of serum. siRNA was transfected and after 72 hrs the cells were fixed and stained with propidium iodide and submitted to FACS analysis (Figure 5A). In cells in which LGMN was silenced, there was a significant reduction in the fraction of cells in G2/M (25±2% of nontransfected cells, as compared to 7±6% of siRNA-treated cells; p<0.05). Despite the reduction in the fraction of cells in G2/M phase, there was no significant increase in the fractions of cells in G1 or S phase (Figure 5B). No changes were observed in cells transfected with scrambled siRNA. Apoptosis was monitored by caspase-3 activation [2] and no increase was observed in cells treated with LGMN siRNA (Figure 5C). Together, these findings suggest that absence of LGMN causes a decrease in SKHep1 cell proliferation that is not due to an increase in apoptosis.
Figure 5. Cell cycle kinetics after knockdown of LGMN.
(A) Representative FACS cell cycle profiles of non-transfected (NT) and siRNA-transfected SkHep cells 72 hr after treatment with scrambled or LGMN siRNA. (B) In cells in which LGMN was silenced, there was a reduction in the fraction of cells in G2/M phase (7.4±10.9% in LGMN siRNA versus 25.6±3.5% in non-transfected; mean+SD; p<0.05), without a significant increase in the fraction of cells in G1 or S phase. Cell cycle profiles were not changed in cells transfected with scrambled siRNA. Data are mean of 3 independent experiments. (C) Knockdown of LGMN does not induce apoptosis, as measured by caspase-3 activity. Staurosporine (500 nM) was used to induce apoptosis as a positive control for caspase-3, and a caspase-3 inhibitor was used as a negative control. Bar graph shows that caspase-3 activity was not increased in response to knockdown of LGMN (p>0.05, by one-way ANOVA). Results are representative of four independent experiments (*p<0.001).
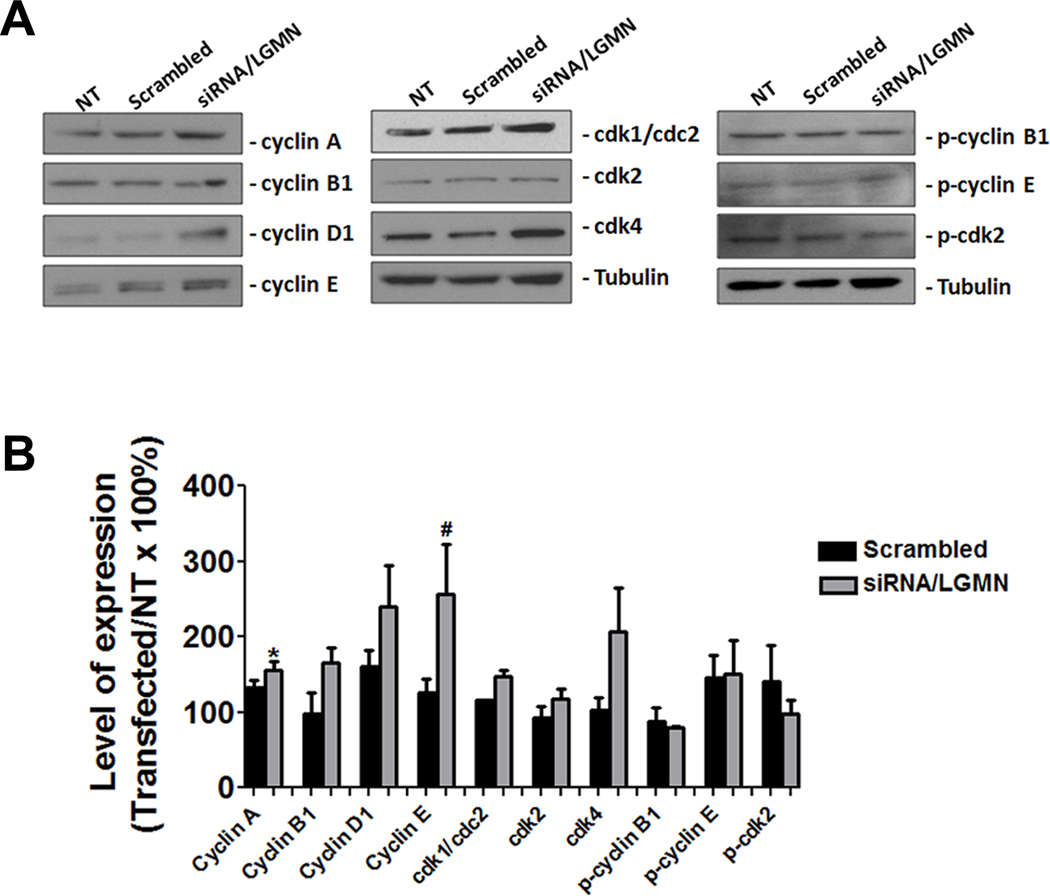
Knockdown of LGMN modulates cell cycle proteins
The changes observed in the cell cycle profile after knock down of LGMN raises the question of whether the decrease in cell proliferation may be due to alterations in expression of checkpoint proteins. To investigate this, we examined expression of these proteins by western blot (Figure 6A). The expression of cyclin D1 and cdk4, which form the complex that regulates the progression through G1 phase, was unchanged, as was cyclin B1, which controls the G2/M transition (Figure 6A). However, the checkpoint proteins of S phase showed increased expression when LGMN was silenced (Figure 6A); both cyclin A and cyclin E expression were significantly increased (p<0.05; Figure 6B). These increases in expression were not followed by an increase in phosphorylation or activity of cdk2 (Supplementary Figure 1), nor were they associated with an increase in PCNA expression. Moreover, there was no change in either phosphorylation of cyclins B1 and E or expression of the CDK inhibitors p21 and p27 (Figure 6A, B and Supplementary Figure 1A, B). Together, these findings suggest that LGMN has only minor effects on the expression profile of checkpoint proteins.
Figure 6. Progression of cell cycle after knockdown of LGMN.
(A) Immunoblot of total protein from SKHep1 cells tests expression of various cell cycle regulatory proteins 72 hr after knockdown of LGMN. (B) Densitometric analysis summarizes the results of the western blots. There was a significant increase in expression of cyclin A (*p<0.01) and cyclin E (**p<0.05) in cells transfected with LGMN siRNA relative to non-transfected. Expression of α-tubulin was used as an internal control for protein loading. Data are mean of three independent experiments.
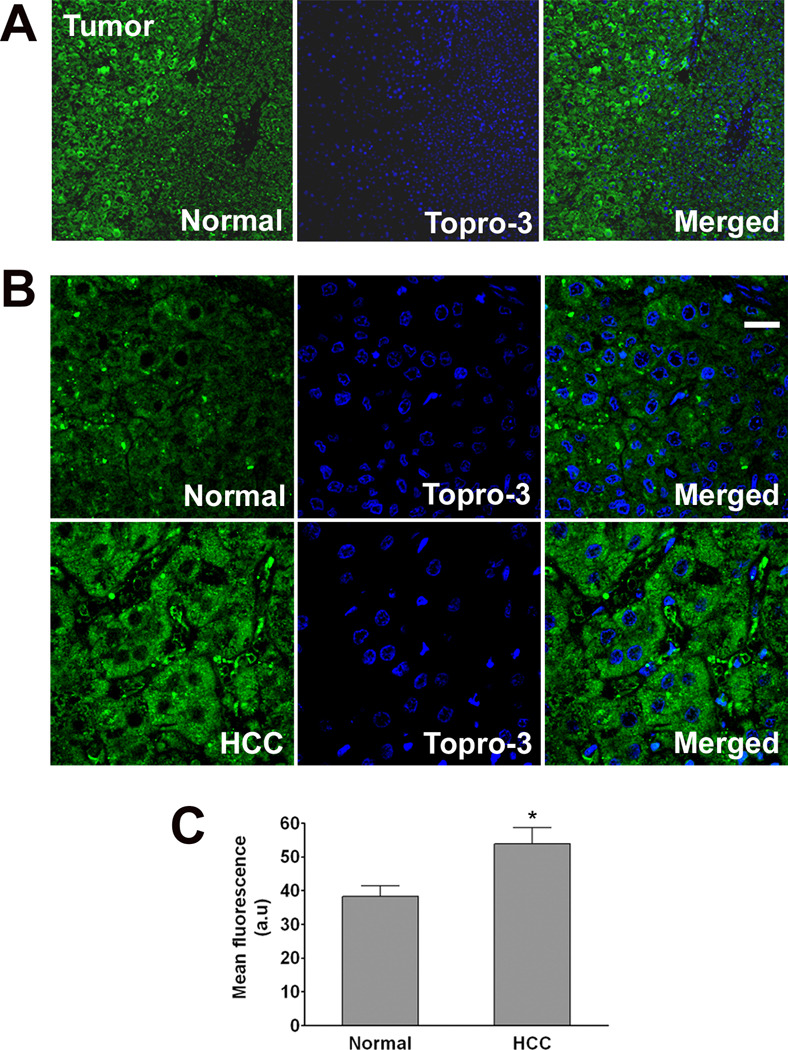
LGMN expression in normal human liver and hepatocellular carcinoma
Among the putative cDNAs identified by RaSH, LGMN is notable because this gene is highly expressed in several types of tumors. Its presence correlates with a poor prognosis and increased tumor invasiveness [24] and it can be used as a prognostic factor in breast cancer [10]. Expression of LGMN also may be involved in early development of colorectal cancer [34]. Therefore, we investigated LGMN expression in hepatocellular carcinoma (HCC). Liver resection specimens from patients diagnosed with HCC were examined by confocal immunofluorescence. Three samples which contain both normal and tumor tissue from each of 5 different patients were examined. LGMN expression was increased in tumor cells (53.9±6.5 a.u.) when compared to normal hepatocytes (38.3±4.3 a.u) in the same tissue samples (p<0.01 by paired t test, Figure 7). These results suggest that LGMN expression is increased in hepatocellular carcinoma, similar to what has been observed in other solids tumors, and is consistent with the idea that the positive effects of LGMN on cell proliferation may promote carcinogenesis.
Figure 7. Expression of LGMN is increased in hepatocellular carcinoma (HCC).
Confocal immunofluorescence images were obtained from paraffin-embedded surgical specimens of tumors from patients with HCC. Immunohistochemical staining was performed to determine expression of LGMN (green) in tumor cells and in nearby normal hepatocytes. To-Pro-3 was used to identify cell nuclei (blue). (A) Low-power (10×) image of carcinoma cells and normal hepatocytes in the same field of view shows that LGMN staining is increased in the HCC. (B) Higher magnification (63×) images confirm increased expression of LGMN and show that it is distributed throughout the cytoplasm in HCC (Scale bar = 30 µm). Findings are representative of what was observed in 3 fields each of specimens from 5 separate patients. (C) Quantification of the average fluorescence in normal and HCC affected areas in the same specimen shows a significant increase in LGMN expression in the carcinoma cells (53.9±4.8 a.u.) as compared to normal hepatocytes (38.3±3.1 a.u.; p<0.01, paired t-test).
DISCUSSION
Intracellular Ca2+ signals regulate cell growth [39], and in hepatocytes, nuclear rather than cytoplasmic Ca2+ is responsible for this regulation [41]. The nucleus contains the machinery needed to generate Ca2+ signals, and can form these signals independent of Ca2+ signals in the cytoplasm [9, 23]. Hepatic mitogens such as insulin [41] and HGF [11] selectively activate this machinery. Buffering nuclear Ca2+ inhibits the growth of liver tumors in particular [42]. Specifically, liver tumors implanted in nude mice grew much more slowly when expressing parvalbumin in their nuclei, but not in their cytosol [42]. The proteins that link nuclear Ca2+ signals to cell proliferation have not been identified, although such Ca2+ signals have effects that would be expected to stimulate cell proliferation. For example, nuclear Ca2+ activates the transcription factors CREB [13] and Elk-1 [40] and stimulates the intranuclear activity of PKC [9] and CaMK-IV [8]. The current work identifies expression of LGMN as a novel target of nuclear Ca2+, and further shows that inhibition of LGMN expression impairs cell proliferation. Moreover, our findings show that the LGMN promoter is sensitive to nuclear Ca2+, and a bioinformatics analysis identified a putative Elk-1 binding site on the promoter (not shown), so it is possible that nuclear Ca2+ regulates LGMN expression through Elk-1.
The findings that LGMN expression is increased in HCC or after treatment of SKHep1 cells with HGF suggest that LGMN may play a role in carcinogenesis in the liver. Although buffering nuclear Ca2+ and knockdown of LGMN each reduce cell proliferation, reduction of LGMN expression does not exactly mimic the effects of PV-NLS. For example, transfection of cells with PV-NLS increases the fraction of cells in G2, and this is associated with a block in early prophase and an increase in the mitotic index [42]. In contrast, LGMN siRNA reduces both the fraction of cells in G2 and the mitotic index. Furthermore, PV-NLS reduces p-cdk1 but does not alter cyclin expression, whereas LGMN siRNA increases expression of cyclins A and E. These results are counterintuitive to the classical model of cell cycle regulation [15] and findings in mouse hepatocytes during liver regeneration [37], both of which predict increased proliferation in cells with higher levels of cyclins due to prolonged CDK2 activation. However, the increased expression of cyclin A and E observed here was not followed by increased CDK2 activity [43]. This unexpected finding is supported by one report demonstrating that cyclin E overexpression promotes cell cycle arrest [18]. Alternatively, the increased expression of cyclin E observed here could be a compensatory up-regulation triggered by the decrease in cell proliferation after knock down of LGMN. Taken together, these results suggest that LGMN has only a minor effect on cell cycle kinetics, and so nuclear Ca2+ may act on cell proliferation only in part by regulating LGMN expression.
LGMN is an asparaginyl endopeptidase that hydrolyzes peptides and proteins on the carboxyl side of asparagine residues [7]. Studies in LGMN knockout mice have determined that LGMN is expressed predominantly in late endosomes and lysosomes, where it is involved in processing of cathepsins and lysosomal degradation [45]. LGMN is highly expressed in carcinomas of the breast, colon, and prostate, and in several central nervous system neoplasms, compared with the corresponding normal tissues, in which there is little or no LGMN expression [24]. These findings in other types of tumors are similar to what we report here in HCC. LGMN knockout mice have hepatomegaly, which is attributed to extramedullary hematopoiesis rather than hepatocyte proliferation because liver histology in these mice is normal [5]. However, the incidence of spontaneous HCC has not been studied in these animals, nor has LGMN expression been studied in xenograft models of liver tumors. LGMN is also involved in extracellular matrix remodeling, including fibronectin degradation, which is enhanced by LGMN overexpression [33]. LGMN-containing vesicles are localized to the invading edge of tumor cells, and tumor cells that overexpress LGMN have increased invasive and migratory activity [24]. Collectively, these findings had been interpreted to suggest that LGMN promotes neoplasia by creating a tumor micro-environment that facilitates metastatic behavior. However, the current work suggests that LGMN may have direct effects on cell proliferation as well. Moreover, LGMN co-localizes with integrins [24], which also can directly modulate cell growth. Further work will be needed to understand how LGMN enhances cell proliferation in the liver.
Supplementary Material
(A) Representative western blots of p21, p27 and PCNA expression after 72 hours of knock down of LGMN. Tubulin was used as a loading control. LGMN western blot demonstrates the efficiency of the knockdown. (B) Densitometric analysis of 3 independent experiments shows that expression of p21, p27 and PCNA was similar in control and LGMN siRNA-treated cells. LGMN expression was reduced by approximately 92% in siRNA-treated cells as compared to control (*p<0.0001). (C) CDK2 activity is similar in control and siRNA-treated cells. Top: Representative western blot of phosphorylated Histone H1 after incubation with immunoprecipitated CDK2 from SKHep1 cells under control conditions and after knockdown of LGMN. IgG indicates samples in which normal mouse IgG was used instead of anti-CDK2 antibody. Bottom: Representative CDK2 western blot shows that similar amounts of CDK2 were immunoprecipitated.
ACKNOWLEDGEMENTS
This work was supported by grants from HHMI (MF Leite), CNPq (CAD Jardim and MF Leite), CAPES (VA Andrade), and FAPEMIG (MF Leite), and NIH grants DK45710, DK57751, DK61747, and DK34989 (MH Nathanson). Confocal imaging was supported by CEMEL (UFMG). The authors thank Ewa Menet (Cell Sorter Facility, Yale School of Medicine) for assistance with FACS experiments.
List of abbreviations
- RaSH
Rapid Subtraction Hybridization
- PV
Parvalbumin
- LGMN
Legumain
- BrdU, HCC
hepatocellular carcinoma
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERENCES
- 1.Berridge MJ, Bootman MD, Roderick HL. Calcium signalling: dynamics, homeostasis and remodelling. Nat Rev Mol Cell Biol. 2003;4(7):517–529. doi: 10.1038/nrm1155. [DOI] [PubMed] [Google Scholar]
- 2.Boatright KM, Salvesen GS. Mechanisms of caspase activation. Curr Opin Cell Biol. 2003;15(6):725–731. doi: 10.1016/j.ceb.2003.10.009. [DOI] [PubMed] [Google Scholar]
- 3.Caffrey CR, Mathieu MA, Gaffney AM, Salter JP, Sajid M, Lucas KD, et al. Identification of a cDNA encoding an active asparaginyl endopeptidase of Schistosoma mansoni and its expression in Pichia pastoris. FEBS Lett. 2000;466(2–3):244–248. doi: 10.1016/s0014-5793(99)01798-6. [DOI] [PubMed] [Google Scholar]
- 4.Carrion AM, Link WA, Ledo F, Mellstrom B, Naranjo JR. DREAM is a Ca2+− regulated transcriptional repressor. Nature. 1999;398(6722):80–84. doi: 10.1038/18044. [DOI] [PubMed] [Google Scholar]
- 5.Chan CB, Abe M, Hashimoto N, Hao C, Williams IR, Liu X, et al. Mice lacking asparaginyl endopeptidase develop disorders resembling hemophagocytic syndrome. Proc Natl Acad Sci U S A. 2009;106(2):468–473. doi: 10.1073/pnas.0809824105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chen JM, Dando PM, Rawlings ND, Brown MA, Young NE, Stevens RA, et al. Cloning, isolation, and characterization of mammalian legumain, an asparaginyl endopeptidase. J Biol Chem. 1997;272(12):8090–8098. doi: 10.1074/jbc.272.12.8090. [DOI] [PubMed] [Google Scholar]
- 7.Chen JM, Dando PM, Stevens RA, Fortunato M, Barrett AJ. Cloning and expression of mouse legumain, a lysosomal endopeptidase. Biochem J. 1998;335(Pt 1):111–117. doi: 10.1042/bj3350111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Deisseroth K, Heist EK, Tsien RW. Translocation of calmodulin to the nucleus supports CREB phosphorylation in hippocampal neurons. Nature. 1998;392(6672):198–202. doi: 10.1038/32448. [DOI] [PubMed] [Google Scholar]
- 9.Echevarria W, Leite MF, Guerra MT, Zipfel WR, Nathanson MH. Regulation of calcium signals in the nucleus by a nucleoplasmic reticulum. Nat Cell Biol. 2003;5(5):440–446. doi: 10.1038/ncb980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gawenda J, Traub F, Luck HJ, Kreipe H, von Wasielewski R. Legumain expression as a prognostic factor in breast cancer patients. Breast Cancer Res Treat. 2007;102(1):1–6. doi: 10.1007/s10549-006-9311-z. [DOI] [PubMed] [Google Scholar]
- 11.Gomes DA, Rodrigues MA, Leite MF, Gomez MV, Varnai P, Balla T, et al. c-Met must translocate to the nucleus to initiate calcium signals. J Biol Chem. 2008;283(7):4344–4351. doi: 10.1074/jbc.M706550200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hajn¢czky G, Robb-Gaspers LD, Seitz MB, Thomas AP. Decoding of cytosolic calcium oscillations in the mitochondria. Cell. 1995;82:415–424. doi: 10.1016/0092-8674(95)90430-1. [DOI] [PubMed] [Google Scholar]
- 13.Hardingham GE, Chawla S, Johnson CM, Bading H. Distinct functions of nuclear and cytoplasmic calcium in the control of gene expression. Nature. 1997;385:260–265. doi: 10.1038/385260a0. [DOI] [PubMed] [Google Scholar]
- 14.Hendzel MJ, Wei Y, Mancini MA, Van Hooser A, Ranalli T, Brinkley BR, et al. Mitosis-specific phosphorylation of histone H3 initiates primarily within pericentromeric heterochromatin during G2 and spreads in an ordered fashion coincident with mitotic chromosome condensation. Chromosoma. 1997;106(6):348–360. doi: 10.1007/s004120050256. [DOI] [PubMed] [Google Scholar]
- 15.Hochegger H, Takeda S, Hunt T. Cyclin-dependent kinases and cell-cycle transitions: does one fit all? Nat Rev Mol Cell Biol. 2008;9(11):910–916. doi: 10.1038/nrm2510. [DOI] [PubMed] [Google Scholar]
- 16.Ishak KG, Goodman ZD, Stocker JT. Atlas of tumor pathology. Washington: Armed Forces Institute of Pathology; 2001. Tumors of the liver and intrahepatic bile ducts. [Google Scholar]
- 17.Jiang H, Kang DC, Alexandre D, Fisher PB. RaSH, a rapid subtraction hybridization approach for identifying and cloning differentially expressed genes. Proc Natl Acad Sci U S A. 2000;97(23):12684–12689. doi: 10.1073/pnas.220431297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Keck JM, Summers MK, Tedesco D, Ekholm-Reed S, Chuang LC, Jackson PK, et al. Cyclin E overexpression impairs progression through mitosis by inhibiting APC(Cdh1) J Cell Biol. 2007;178(3):371–385. doi: 10.1083/jcb.200703202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kembhavi AA, Buttle DJ, Knight CG, Barrett AJ. The two cysteine endopeptidases of legume seeds: purification and characterization by use of specific fluorometric assays. Arch Biochem Biophys. 1993;303(2):208–213. doi: 10.1006/abbi.1993.1274. [DOI] [PubMed] [Google Scholar]
- 20.Krasinska L, Besnard E, Cot E, Dohet C, Mechali M, Lemaitre JM, et al. Cdk1 and Cdk2 activity levels determine the efficiency of replication origin firing in Xenopus. EMBO J. 2008;27(5):758–769. doi: 10.1038/emboj.2008.16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Leite MF. Ca2+ Signaling in the Liver. In: Arias IM, editor. The Liver Biology and Pathobiology. 5th ed. Wiley; 2009. pp. 485–510. [Google Scholar]
- 22.Leite MF, Hirata K, Pusl T, Burgstahler AD, Okazaki K, Ortega JM, et al. Molecular Basis for Pacemaker Cells in Epithelia. JBiolChem. 2002;277:16313–16323. doi: 10.1074/jbc.M109207200. [DOI] [PubMed] [Google Scholar]
- 23.Leite MF, Thrower EC, Echevarria W, Koulen P, Hirata K, Bennett AM, et al. Nuclear and cytosolic calcium are regulated independently. Proc Natl Acad Sci U S A. 2003;100(5):2975–2980. doi: 10.1073/pnas.0536590100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Liu C, Sun C, Huang H, Janda K, Edgington T. Overexpression of legumain in tumors is significant for invasion/metastasis and a candidate enzymatic target for prodrug therapy. Cancer Res. 2003;63(11):2957–2964. [PubMed] [Google Scholar]
- 25.Loak K, Li DN, Manoury B, Billson J, Morton F, Hewitt E, et al. Novel cell-permeable acyloxymethylketone inhibitors of asparaginyl endopeptidase. Biol Chem. 2003;384(8):1239–1246. doi: 10.1515/BC.2003.136. [DOI] [PubMed] [Google Scholar]
- 26.Luo Y, Zhou H, Krueger J, Kaplan C, Lee SH, Dolman C, et al. Targeting tumor-associated macrophages as a novel strategy against breast cancer. J Clin Invest. 2006;116(8):2132–2141. doi: 10.1172/JCI27648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Maniotis AJ, Chen CS, Ingber DE. Demonstration of mechanical connections between integrins, cytoskeletal filaments, and nucleoplasm that stabilize nuclear structure. Proc Natl Acad Sci U S A. 1997;94(3):849–854. doi: 10.1073/pnas.94.3.849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Manoury B, Hewitt EW, Morrice N, Dando PM, Barrett AJ, Watts C. An asparaginyl endopeptidase processes a microbial antigen for class II MHC presentation. Nature. 1998;396(6712):695–699. doi: 10.1038/25379. [DOI] [PubMed] [Google Scholar]
- 29.Manoury B, Mazzeo D, Li DN, Billson J, Loak K, Benaroch P, et al. Asparagine endopeptidase can initiate the removal of the MHC class II invariant chain chaperone. Immunity. 2003;18(4):489–498. doi: 10.1016/s1074-7613(03)00085-2. [DOI] [PubMed] [Google Scholar]
- 30.Mendes CC, Gomes DA, Thompson M, Souto NC, Goes TS, Goes AM, et al. The type III inositol 1,4,5-trisphosphate receptor preferentially transmits apoptotic Ca2+ signals into mitochondria. J Biol Chem. 2005;280(49):40892–40900. doi: 10.1074/jbc.M506623200. [DOI] [PubMed] [Google Scholar]
- 31.Minagawa N, Kruglov EA, Dranoff JA, Robert ME, Gores GJ, Nathanson MH. The anti-apoptotic protein Mcl-1 inhibits mitochondrial Ca2+ signals. J Biol Chem. 2005;280(39):33637–33644. doi: 10.1074/jbc.M503210200. [DOI] [PubMed] [Google Scholar]
- 32.Minagawa N, Nagata J, Shibao K, Masyuk AI, Gomes DA, Rodrigues MA, et al. Cyclic AMP regulates bicarbonate secretion in cholangiocytes through release of ATP into bile. Gastroenterology. 2007;133(5):1592–1602. doi: 10.1053/j.gastro.2007.08.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Morita Y, Araki H, Sugimoto T, Takeuchi K, Yamane T, Maeda T, et al. Legumain/asparaginyl endopeptidase controls extracellular matrix remodeling through the degradation of fibronectin in mouse renal proximal tubular cells. FEBS Lett. 2007;581(7):1417–1424. doi: 10.1016/j.febslet.2007.02.064. [DOI] [PubMed] [Google Scholar]
- 34.Murthy RV, Arbman G, Gao J, Roodman GD, Sun XF. Legumain expression in relation to clinicopathologic and biological variables in colorectal cancer. Clin Cancer Res. 2005;11(6):2293–2299. doi: 10.1158/1078-0432.CCR-04-1642. [DOI] [PubMed] [Google Scholar]
- 35.Nathanson MH, Gautam A, Bruck R, Isales CM, Boyer JL. Effects of Ca2+ agonists on cytosolic Ca2+ in isolated hepatocytes and on bile secretion in the isolated perfused rat liver. Hepatology. 1992;15:107–116. doi: 10.1002/hep.1840150119. [DOI] [PubMed] [Google Scholar]
- 36.Nathanson MH, Gautam A, Ng OC, Bruck R, Boyer JL. Hormonal regulation of paracellular permeability in isolated rat hepatocyte couplets. Am J Physiol. 1992;262(6 Pt 1):G1079–G1086. doi: 10.1152/ajpgi.1992.262.6.G1079. [DOI] [PubMed] [Google Scholar]
- 37.Nevzorova YA, Tschaharganeh D, Gassler N, Geng Y, Weiskirchen R, Sicinski P, et al. Aberrant cell cycle progression and endoreplication in regenerating livers of mice that lack a single E-type cyclin. Gastroenterology. 2009;137(2):691–703. doi: 10.1053/j.gastro.2009.05.003. 703 e691–696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Oliver EM, Skuce PJ, McNair CM, Knox DP. Identification and characterization of an asparaginyl proteinase (legumain) from the parasitic nematode, Haemonchus contortus. Parasitology. 2006;133(Pt 2):237–244. doi: 10.1017/S0031182006000229. [DOI] [PubMed] [Google Scholar]
- 39.Patel R, Holt M, Philipova R, Moss S, Schulman H, Hidaka H, et al. Calcium/calmodulin-dependent phosphorylation and activation of human Cdc25-C at the G2/M phase transition in HeLa cells. J Biol Chem. 1999;274(12):7958–7968. doi: 10.1074/jbc.274.12.7958. [DOI] [PubMed] [Google Scholar]
- 40.Pusl T, Wu JJ, Zimmerman TL, Zhang L, Ehrlich BE, Berchtold MW, et al. Epidermal growth factor-mediated activation of the ETS domain transcription factor Elk-1 requires nuclear calcium. J Biol Chem. 2002;277(30):27517–27527. doi: 10.1074/jbc.M203002200. [DOI] [PubMed] [Google Scholar]
- 41.Rodrigues MA, Gomes DA, Andrade VA, Leite MF, Nathanson MH. Insulin induces calcium signals in the nucleus of rat hepatocytes. Hepatology. 2008 doi: 10.1002/hep.22424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Rodrigues MA, Gomes DA, Leite MF, Grant W, Zhang L, Lam W, et al. Nucleoplasmic calcium is required for cell proliferation. J Biol Chem. 2007;282(23):17061–17068. doi: 10.1074/jbc.M700490200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Sherr CJ, Roberts JM. CDK inhibitors: positive and negative regulators of G1-phase progression. Genes Dev. 1999;13(12):1501–1512. doi: 10.1101/gad.13.12.1501. [DOI] [PubMed] [Google Scholar]
- 44.Shibao K, Hirata K, Robert ME, Nathanson MH. Loss of inositol 1,4,5-trisphosphate receptors from bile duct epithelia is a common event in cholestasis. Gastroenterology. 2003;125(4):1175–1187. doi: 10.1016/s0016-5085(03)01201-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Shirahama-Noda K, Yamamoto A, Sugihara K, Hashimoto N, Asano M, Nishimura M, et al. Biosynthetic processing of cathepsins and lysosomal degradation are abolished in asparaginyl endopeptidase-deficient mice. J Biol Chem. 2003;278(35):33194–33199. doi: 10.1074/jbc.M302742200. [DOI] [PubMed] [Google Scholar]
- 46.Thompson M, Andrade VA, Andrade SJ, Pusl T, Ortega JM, Goes AM, et al. Inhibition of the TEF/TEAD transcription factor activity by nuclear calcium and distinct kinase pathways. Biochem Biophys Res Commun. 2003;301(2):267–274. doi: 10.1016/s0006-291x(02)03024-3. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(A) Representative western blots of p21, p27 and PCNA expression after 72 hours of knock down of LGMN. Tubulin was used as a loading control. LGMN western blot demonstrates the efficiency of the knockdown. (B) Densitometric analysis of 3 independent experiments shows that expression of p21, p27 and PCNA was similar in control and LGMN siRNA-treated cells. LGMN expression was reduced by approximately 92% in siRNA-treated cells as compared to control (*p<0.0001). (C) CDK2 activity is similar in control and siRNA-treated cells. Top: Representative western blot of phosphorylated Histone H1 after incubation with immunoprecipitated CDK2 from SKHep1 cells under control conditions and after knockdown of LGMN. IgG indicates samples in which normal mouse IgG was used instead of anti-CDK2 antibody. Bottom: Representative CDK2 western blot shows that similar amounts of CDK2 were immunoprecipitated.