Abstract
The c-jun N-terminal kinases (JNKs) are responsive to stress stimuli leading to activation of proapoptotic proteins and transcription. Additionally, JNK mitochondrial localization has been reported. To selectively target mitochondrial JNK signaling, we exploited JNKs interaction with its mitochondrial scaffold, Sab, using small interfering RNAs (siRNAs) and a cell permeable peptide corresponding to the KIM1 domain of Sab. Gene silencing and peptide interference of this interaction disrupted JNK translocation to the mitochondria and reduced phosphorylation of Bcl-2 without significant impact on c-Jun phosphorylation or AP-1 transcription. In contrast, the JNK inhibitory peptide (TI-JIP1) prevented these three functions. Tat-SabKIM1 selectivity was also demonstrated in anisomycin-stressed HeLa cells where Tat-SabKIM1 prevented Bcl-2 phosphorylation, cell death, loss of mitochondrial membrane potential, and superoxide generation, but not c-Jun phosphorylation. Conversely, TI-JIP1 prevented all aforementioned stress-induced events. This probe introduces a means to evaluate JNK-mediated events on the mitochondria without intervening in nuclear functions of JNK.
The c-Jun N-terminal Kinases (JNKs) are serine/threonine protein kinases and members of the mitogen-activated protein kinase (MAPK) superfamily (1). There are three human JNK isoforms. JNK1 and JNK2, are ubiquitously expressed, and JNK3 is expressed in the heart, brain, and testes(1, 2). In response to many stress stimuli, JNK becomes activated via bis-phosphorylation by MAP kinase kinases (MKK4 and MKK7), allowing it to subsequently phosphorylate numerous substrates(1, 3). The most well studied substrates are transcription factors, namely c-jun, that comprise the activator protein-1 (AP-1)(1). Activation of the AP-1 transcription factor initiates proliferation or pro-apoptotic transcription depending on the stimulus(1, 4).
Recently, a new subcellular locale for JNK signaling has emerged. The mitochondria of the cell contain JNK substrates. Mitochondrial JNK (MitoJNK) signaling has been demonstrated in vitro and in vivo using models for DNA damage (5, 6), phorbol ester stress (7), acetaminophen-induced liver injury (8), cardiac oxidative stress (9), anisomycin-induced stress (10), aging (11), and cerebral ischemia (12). Activation of JNK via phosphorylation by upstream MAPK kinases (MAPKKs) (1) causes a small population of JNK to migrate to mitochondria. Recent data from our lab demonstrates that preventing activation of JNKs by treating HeLa cells with N-acetylcysteine (NAC), an antioxidant that prevents JNK activation during stress, inhibits JNK translocation to the mitochondria. Once at the mitochondria catalytically active JNK can dock with a scaffold protein and substrate, Sab(13, 14). The interaction between JNK and Sab occurs through two kinase interaction motifs (KIMs), dubbed KIM1 and KIM2. Evaluation of these two motifs with respect to JNK binding demonstrated that only KIM1 was necessary for JNK binding and JNK-mediated Sab phosphorylation (14). Interestingly, examination of the Sab KIM1 motif as an inhibitor of JNK mediated c-jun phosphorylation clearly demonstrated that the Sab KIM1 peptide was not able to inhibit JNK phosphorylation of c-jun; however, a similar peptide (TI-JIP), from the JNK-interacting protein-1 (JIP1) JNK-binding domain, was able to completely inhibit JNK-mediated c-jun phosphorylation (15).
Once active JNK arrives at the mitochondria, the activated signaling cascade can impact many facets of mitochondrial biology. JNK can use Bcl-2 and other BH3 family proteins as substrates (4, 16, 17). JNK has been demonstrated to specifically phosphorylated Bcl-2 on serine and threonine residues including serine 70, which has been shown to be a necessary modification in apoptosis (4, 6, 17). MitoJNK is able to phosphorylate Bcl-xL during gamma radiation-induced DNA damage in U-937 myeloid lymphoma cells contributing to apoptosis (6). In a myocardial infarction model, MitoJNK was responsible for the release of cytochrome c from the mitochondria (9). MitoJNK also appears to have a role in the regulation of mitochondrial bioenergetics. In acetaminophen-induced liver injury, MitoJNK contributes to a decrease in mitochondrial State III respiration and ATP production (8). Recent studies in anisomycin-stressed primary cortical neurons (10) and aging brain (11) demonstrate that pyruvate dehydrogenase complex (PDHC) subunit E1α is a substrate for mitochondrial JNK (10, 11). In the case of primary cortical neurons, anisomycin stress triggered JNK-dependent phosphorylation of PDHC which decreased the oxidative metabolism of pyruvate (10). This metabolic shift resulted in increased lactate production and decreased ATP production by anisomycin-treated primary cortical neurons.
Given that the Sab KIM1 peptide did not impact c-jun phosphorylation(18), we hypothesized that the use of a small peptide resembling the KIM1 motif of Sab can selectively disrupt mitochondrial JNK signaling without impacting JNK-mediated transcriptional events. In this work, we demonstrated that JNK translocated to the outer mitochondrial membrane in anisomycin-treated HeLa cells. Silencing Sab or use of a Sab KIM1 motif peptide prevented JNK translocation to the mitochondria without perturbing nuclear JNK-mediated events. Moreover, disruption of the JNK/Sab interaction prevented adverse mitochondrial phenotypes such as mitochondrial superoxide generation and dissipation of mitochondrial membrane potential during anisomycin stress in cells without disturbing c-jun phosphorylation or AP-1 transcription. These data support that targeting the JNK/Sab interaction is a novel means to investigate MitoJNK signaling.
Results
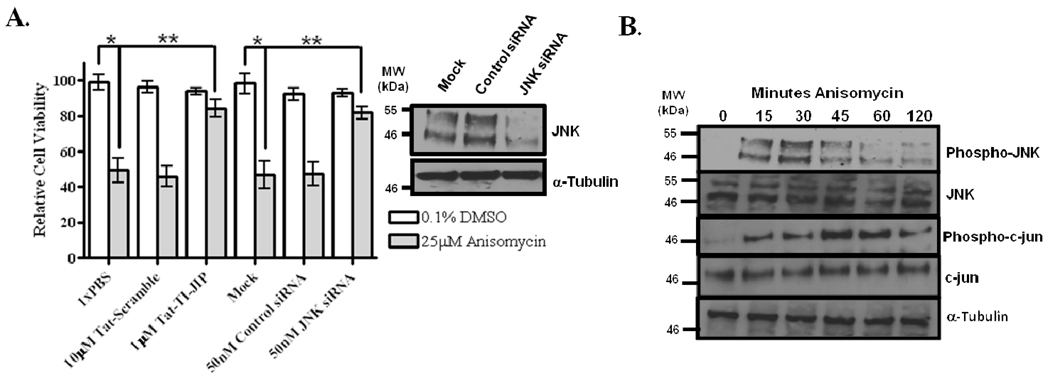
Anisomycin activated JNK and Mediated Cell Death in HeLa Cells
HeLa cells treated with 25µM anisomycin for four hours demonstrated a ~50% decrease in viability when compared to DMSO-treated cells (Figure 1A). Using a small inhibitory, cell permeable peptide of JNK (1µM Tat-TI-JIP), we were able to rescue ~35% of the viability (Figure 1A). Similarly, silencing JNK expression by siRNAs (Figure 1A, Western blot inset) also rescued viability in anisomycin stressed HeLa cells to the same extent as Tat-TI-JIP (Figure 1A). Introduction of 10 µM Tat-Scramble and control siRNA had no protective effect as expected. We further examined JNK activation and signaling during the first two hours of anisomycin-stress using Western blot analysis. Cell lysates were examined 0, 15, 30, 45, 60, and 120 minutes following addition of 25µM anisomycin to the cell culture. Addition of anisomycin increased JNK phosphorylation between 15 and 30 minutes, and then JNK phosphorylation decreased after 30 minutes (Figure 1B). Total JNK abundance remained unchanged during the two hour time course (Figure 1B). Monitoring c-jun phosphorylation on serine 73 during stress revealed that c-jun phosphorylation increased at 15–30 minutes, peaking at 45–60 min, then decreasing following 60 minutes (Figure 1B). c-jun levels remained constant during anisomycin treatment (Figure 1B). Tubulin was used as a loading control (Figure 1B).
Figure 1. Anisomycin induces JNK-mediated cell death in HeLa cells.
(A) Anisomycin stress induces a JNK dependent cell death. Cell viability was monitored following 4 hours of 25µM anisomycin treatment (grey bars) or 0.1% DMSO (white bars). Cells were either pretreated for 30 minutes prior to anisomycin treatment with 10µM Tat-Scramble peptide or 1µM TI-JIP peptide. Additionally, cell viability was also determined by silencing JNK via siRNA-mediated knockdown. Cells were transfected with 50nM siRNA (control or JNK-specific) 66 hours prior to treatment with 25µM anisomycin. Silencing was monitored by Western blot analysis of 30µg cell lysate following 72 hours of silencing (inset). (B) Anisomycin stress activates JNK signaling. JNK signaling was evaluated by Western blot following 0, 5, 10, 15, 30, 60, and 120 minutes of treatment with 25µM anisomycin. Proteins (30µg) were resolved by SDS-PAGE, and antibodies for Phospho-JNK, JNK, Phospho-c-jun, and c-jun were used to detect proteins of interest. α-Tubulin was used as a loading control.
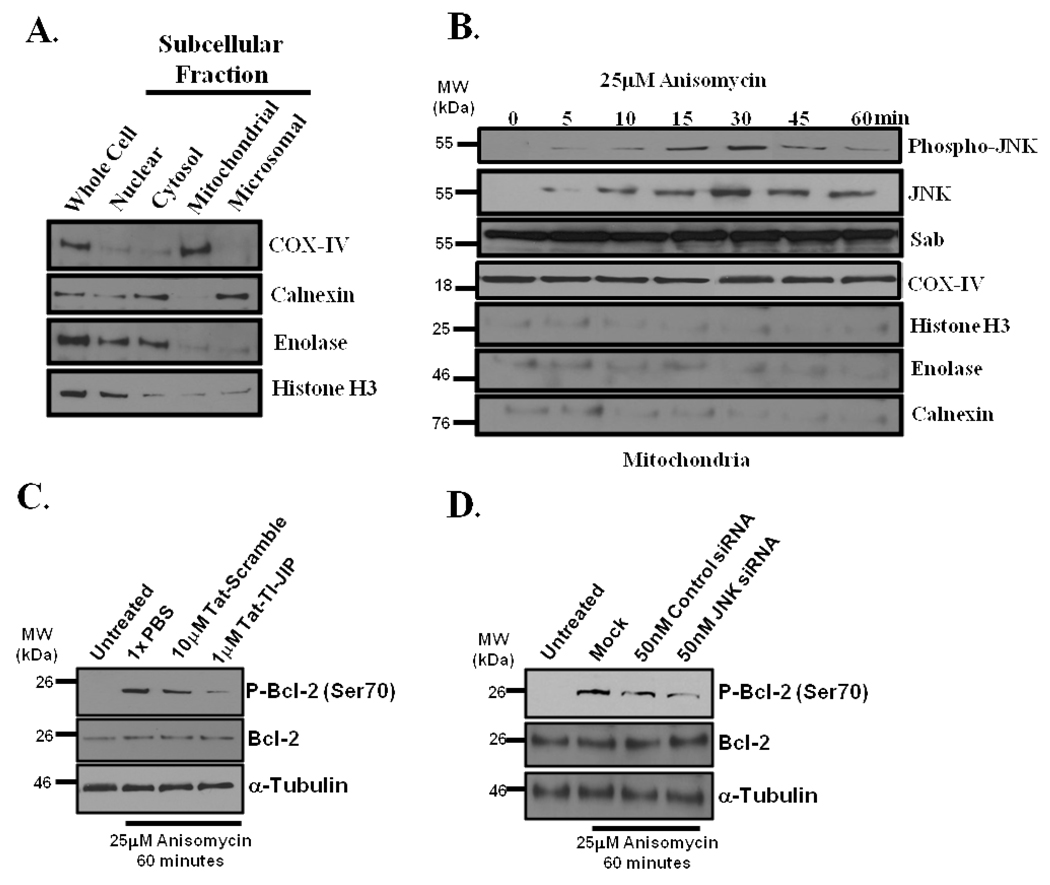
Anisomycin induced JNK translocation to the outer membrane of the mitochondria and JNK mediated Bcl-2 phosphorylation
To evaluate if anisomycin stress provoked JNK translocation to the mitochondria, mitochondria were harvested. In figure 2A, a representative mitochondrial preparation is shown. Western blotting demonstrated the mitochondrial enrichments contained cyclo-oxygenase IV (COX-IV), but very low levels of ER (calnexin), cytosolic (enolase), and nuclear (histone H3) contamination (Figure 2A). Mitochondrial enrichments from HeLa cells stressed with 25µM anisomycin for 0, 15, 30, 45, 60, and 120 minutes were examined for the presence of activated (phosphorylated) JNK. We found detectable levels of phospho-JNK were present on the mitochondria as early as 5 minutes and peaked at 30 minutes following anisomycin treatment (Figure 2B). However, only the 54kDa species was found on the mitochondria; this was confirmed by Western blot analysis for total JNK at the mitochondria (Figure 2B). Sab, the mitochondrial scaffold for JNK, did not have altered abundance on the mitochondria during stress (Figure 2B). Equal mitochondrial loading was assured by a cyclo-oxygenase IV (COX-IV) loading control (Figure 2B). Again, non-mitochondrial contamination was minimal as demonstrated by Western blot analysis of calnexin, enolase, and histone H3. Examination of the proteinase K treated samples and outer mitochondrial membrane enrichments demonstrated JNK was present on the outer mitochondrial membrane (Supplemental Figure S1A and B1B) as described by Hanawa et al. (8).
Figure 2. Anisomycin stress triggers JNK translocation to the outer mitochondrial membrane and JNK-mediated phosphorylation of Bcl-2.
(A) Representative mitochondrial enrichment and subcellular fractionation. Mitochondrial purity was determined by Western blotting for ER resident, calnexin; cytosolic enzyme, enolase; and nuclear protein, histone H3. Mitochondria were identified by COX-IV presence. (B) Anisomycin stress-induced JNK translocation to the mitochondria. Mitochondria were prepared from HeLa cells stressed with 25µM anisomycin for 0, 5, 10, 15, 30, 45, and 60 minutes. The mitochondria were monitored for Phospho-JNK, JNK, Sab, and COX-IV by Western analysis, and calnexin, enolase, and histone H3 were monitored as mitochondrial contaminants. COX-IV served as the mitochondria loading control. (C) HeLa cells were pretreated with either 10µM Tat-Scramble or 1µM TI-JIP for 30 minutes prior to incubation with 25µM anisomycin for 60 minutes. Cells were lysed and Western blot analysis was conducted for phosphorylation of Bcl-2 on serine 70, total Bcl-2, and loading control α-tubulin. (D) JNK was silenced with 50nM JNK-specific siRNAs or cells were treated with control siRNAs for 71 hours prior to addition of 25µM anisomycin for 60 minutes. Cells lysates were analyzed for the presence of Phospho-Bcl-2 (Ser70), total Bcl-2, or the loading control, α-tubulin.
To demonstrate that JNK served as an active mitochondrial kinase, we evaluated Bcl-2 phosphorylation in anisomycin-treated HeLa cells, since Bcl-2 phosphorylation on serine 70 has been attributed to JNK during stress(4, 16, 17). HeLa cells were stressed with 25µM anisomycin for 60 minutes in the presence and absence of 10µM Tat-Scramble or 1µM Tat-TI-JIP. Phospho-Bcl-2 levels increased on Ser70 following 60 minutes of anisomycin stress (Figure 2C), and the addition of 10µM Tat-Scramble had minimal impact on Ser70 phosphorylation of Bcl-2; however, 1µM Tat-TI-JIP inhibited most of the Ser70 phosphorylation of Bcl-2 suggesting that JNK-mediated Bcl-2 phosphorylation occurred during anisomycin stress. To confirm that Bcl-2 phosphorylation was in fact JNK-mediated, we silenced JNK expression using siRNAs, and again, anisomycin-induced Bcl-2 phosphorylation on Ser70 was detectable at 60 minutes in mock transfected cells (Figure 2D). Moreover, silencing JNK with 50nM JNK-specific siRNAs reduced the level of Ser70 phosphorylation when compared to anisomycin stressed cells transfected with control siRNAs (Figure 2D).
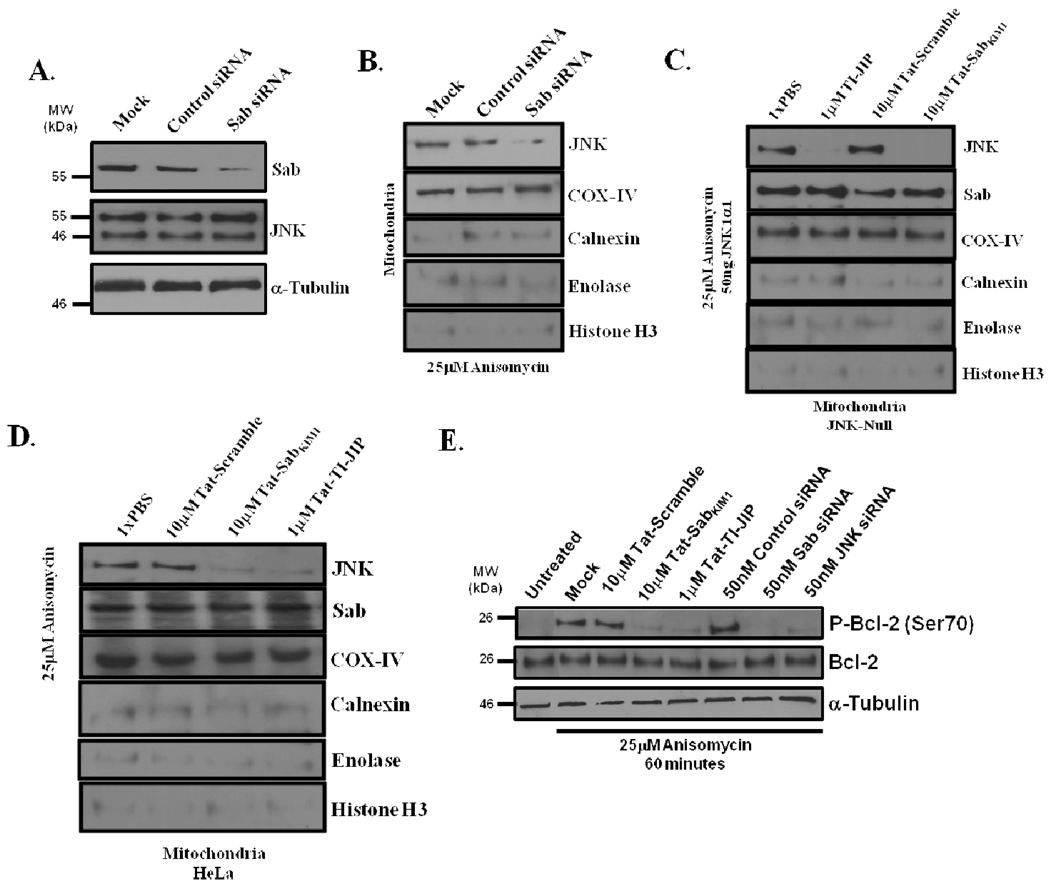
Interfering with the JNK/Sab interaction prevented mitochondrial translocation of JNK and phosphorylation of Bcl-2
JNK and Sab have been shown to interact at the mitochondria (13, 14). To selectively disrupt the interaction between JNK and Sab, we chose to silence Sab expression using siRNA knock-down. Following 72 hours of siRNA transfection, cells were lysed and protein abundance was determined by Western blot analysis. Sab expression was reduced by greater than 70% using Sab-specific siRNAs as compared to control siRNA transfected cells and mock transfected cells (Figure 3A). Moreover, silencing Sab had no impact on JNK expression, and equal loading was validated using tubulin as a control (Figure 3A). We next evaluated by Western analysis if silencing Sab expression could prevent JNK translocation to the mitochondria during anisomycin treatment of cells. After 72 hours of siRNA transfection HeLa cells were treated with 25µM anisomycin. Mock or control siRNA transfected cells had no impact on JNK translocation following 30 minutes of stress (Figure 3B). As expected, silencing Sab prevented JNK translocation to the mitochondria during stress (Figure 3B). COX-IV again was used as a loading control for mitochondria (Figure 3B). Mitochondrial enrichments contained little non-mitochondrial contaminants as determined by Western blot analysis for calnexin, enolase and histone H3 (Figure 3B).
Figure 3. Disruption of the JNK/Sab interaction prevented JNK localization on the mitochondria and JNK-mediated phosphorylation Bcl-2 serine 70.
(A) siRNA-mediated knockdown of Sab expression. Using 50nM siRNA specific for Sab, HeLa cells were transfected and grown for 72 hours. Cell lysates were probed for the relative abundance of Sab, JNK, and α-Tubulin by Western analysis. (B) Silencing Sab prevented JNK translocation to the mitochondria. Cells were treated with 50nM control or Sab-specific siRNA for 66 hours prior to 30 minutes of 25µM anisomycin treatment. Following anisomycin treatment, mitochondrial enrichments were prepared and analyzed by Western blot for the presence of JNK. COX-IV served as a mitochondrial loading control, and calnexin, enolase, and histone H3 were used as controls for subfractional contamination. (C) Use of the Tat-SabKIM1 peptide interfered with JNK mitochondrial translocation in vitro. Mitochondria were extracted from JNK-null fibroblast following 30 minutes of stress with 25µM anisomycin. Next, 50ng of recombinant JNK1α1 was pre-incubated with 10µM Tat-SabKIM1, 10µM Tat-Scramble, or 1µM TI-JIP for 15 minutes. Mitochondria (30µg) were incubated with recombinant JNK1α1 for 30 minutes at 30°C. Mitochondria were recovered by centrifugation and monitored for the presence of JNK and Sab by Western blot. COX-IV was the mitochondrial loading control, while calnexin, enolase, and histone H3 were used to monitor mitochondrial contamination. (D) The Tat-SabKIM1 peptide prevented JNK mitochondrial localization in anisomycin-treated HeLa cells. Cells were pretreated with 10µM Tat-SabKIM1, 10µM Tat-Scramble, or 1µM TI-JIP. Mitochondria were harvested following 30 minutes of 25µM anisomycin exposure. Western blot was used to determine the presence of JNK, Sab, COX-IV (loading control), calnexin (ER contaminant), enolase (cytosolic contaminant), and histone H3 (nuclear contaminant). (E) To disrupt the JNK /Sab interaction, HeLa cells were treated with 10µM Tat-Scramble, 1µM TI-JIP, or 10µM Tat-SabKIM1 peptide for 30 minutes prior to anisomycin stress or gene expression of JNK or Sab was silenced using 50nM of JNK or Sab specific siRNAs prior to anisomycin stress. The cells were then stressed for 60 minutes with 25µM anisomycin. Following stress, cells were lysed, and the relative abundances of Phospho-Bcl-2(Ser70), total Bcl-2, and the loading control, α-tubulin, were evaluated by Western blot.
While siRNAs knockdowns can selectively reduce Sab levels on the mitochondria and prevent JNK mitochondrial localization, siRNA knockdown can vary drastically between cell lines. In addition, we wanted to develop a means to interfere with the JNK/Sab interaction that would easily amenable to potential studies in mammals. Given the in vivo success of the TI-JIP peptide, we decided to design cell permeable peptides of the Sab KIM1 motif (AVVRPGSLDLR) with an HIV-Tat motif attached to enhance cellular penetrance. To extend the half-life in a manner similar to TI-JIP, the Tat-SabKIM1 peptide was designed as the retro-inverso configuration (see sequence in Experimental Procedures). Using a FITC-conjugated version of the peptide, we found that the peptide was cell permeable, and it stained the entirety of the cell as detected by microscopy (Supplemental Figure S2A), and the peptide remained in the cell at concentrations >90% following 24 hours incubation (Supplemental Figure S2B). To demonstrate that the Tat-SabKIM1 peptide could prevent JNK translocation to the mitochondria, we isolated mitochondria from JNK-null fibroblasts following 30 minutes of incubation 25µM anisomycin. The time of stress was required to ‘prime’ the mitochondria for JNK signaling, as unstressed mitochondria did not demonstrate JNK-mediated mitochondrial dysfunction in the presence of JNK1α1 (Supplemental Figure S3A and S3B). We next incubated the mitochondria with PBS, 10µM Tat-SabKIM1 peptide, 10µM Tat-Scrambled peptide, or 1µM TI-JIP peptide, and then incubated with recombinant JNK1α1 for 30 minutes at 37°C. PBS, or Tat-Scramble peptide did not prevent JNK translocation to the mitochondria (Figure 3C); however, either TI-JIP or Tat-SabKIM1 prevented JNK translocation to the mitochondria (Figure 3C). Also, the use of TI-JIP or Tat-SabKIM1 did not alter the levels of Sab on the mitochondria when compared to the other treatments (Figure 3C). COX-IV served as the mitochondrial loading control in Figure 3C. Additionally, calnexin, enolase, and histone H3 contamination was minimal (Figure 3C). Moreover, TI-JIP and Tat-SabKIM1 were sufficient to prevent JNK1α1 phosphorylation of isolated mitochondria from anisomycin-stressed JNK-null MEFs (Supplemental Figure S4). To confirm this observation in anisomycin-stressed HeLa cells again, cells were preincubated with PBS, 10µM Tat-Scrambled peptide, 1µM Tat-TI-JIP peptide, or 10µM Tat-SabKIM1 peptide, and then stressed with 25µM anisomycin for 30 minutes. Mitochondria were harvested from the cells, and JNK localization was determined by Western blot analysis. As in the experiment utilizing JNK-null cells and recombinant JNK1α1, incubating the HeLa cells with 1µM Tat-TI-JIP or 10µM Tat-SabKIM1 prevented endogenous JNK translocation to the mitochondria without impacting Sab expression (Figure 3D). As expected, PBS or Tat-Scramble did not inhibit JNK migration to the mitochondria (Figure 3D). Equivalent mitochondrial loading was confirmed by COX-IV loading control (Figure 3D) and non-mitochondrial contamination was monitored by Western blot.
To elucidate if JNK translocation was required for Bcl-2 phosphorylation during anisomycin stress, we monitored Bcl-2 Ser70 phosphorylation in the presence and absence of mitochondrial JNK signaling. First, we employed the Tat-SabKIM1 peptide to block JNK mitochondrial migration during anisomycin stress. Anisomycin-induced increases in Bcl-2 Ser70 phosphorylation were not impacted by pretreatment with 10µM Tat-Scramble (Figure 3E). Pretreatment of cells with 10µM Tat-SabKIM1 peptide reduced Bcl-2 Ser70 phosphorylation to a level very similar to pretreatment with 1µM TI-JIP (Figure 3E). To specifically determine that the JNK/Sab interaction was required for Bcl-2 phosphorylation, we used siRNAs to knockdown Sab expression prior to anisomycin stress. Compared to mock transfected cells or cells transfected with control siRNAs, cells silencing Sab expression displayed lower Bcl-2 phosphorylation on Ser70 (Figure 3E); similarly, cells silencing JNK had a decrease in Bcl-2 phosphorylation on Ser70 (Figure 3E).
JIP and Sab peptides have different binding affinities and inhibition profiles with respect to JNK
Our group has previously demonstrated that the JIP peptide is a potent inhibitor of JNK1α1 and JNK3α1 catalytic activity(19, 20). Given that the cell permeable versions of JIP and Sab peptides had similar impact on JNK translocation to the mitochondria, albeit at 10-fold higher concentrations for Sab, we evaluated the binding affinity between JNK and the two peptides. JNK3α1 had a 25-fold greater affinity for the JIP peptide compared to the Sab peptide as measured in a fluorescence polarization assay (Table 1). Additionally, the JIP peptide inhibited JNK3α1 phosphorylation of Sab protein at a 12-fold lower concentration than the Sab peptide did (Table 1). Similarly, the JIP peptide potently inhibited JNK3α1 phosphorylation of c-jun and ATF2, while the Sab peptide had no impact on JNK3α1 phosphorylation of these two substrates (Table 1). The scrambled peptide displayed no binding or inhibition with respect to JNK3α1 (Table 1).
Table 1.
Binding and Inhibitory Data for JNK-interacting Peptides for JNK3α1.
| Peptide | Kd* | IC50 -vs-Sab |
IC50 -vs-c-jun |
IC50 -vs-ATF2 |
|---|---|---|---|---|
| TI-JIP | 0.45±0.06µM | 18±9nM | 38±12nM | 27±15nM |
| SabKIM1 | 11±2.4µM | 218±85nM | n.i.*** | n.i.*** |
| Scramble | n.b.** | n.i.*** | n.i.*** | n.i.*** |
Kd was determined using the fluorescent polarization assay described in the methods.
No binding was observed for peptide up to 100µM.
The peptide demonstrated less than 10% inhibition at a concentration of 10µM. IC50s were determined using a Kinase-Glo assay and the data were fit to a 4-parameter nonlinear regression model.
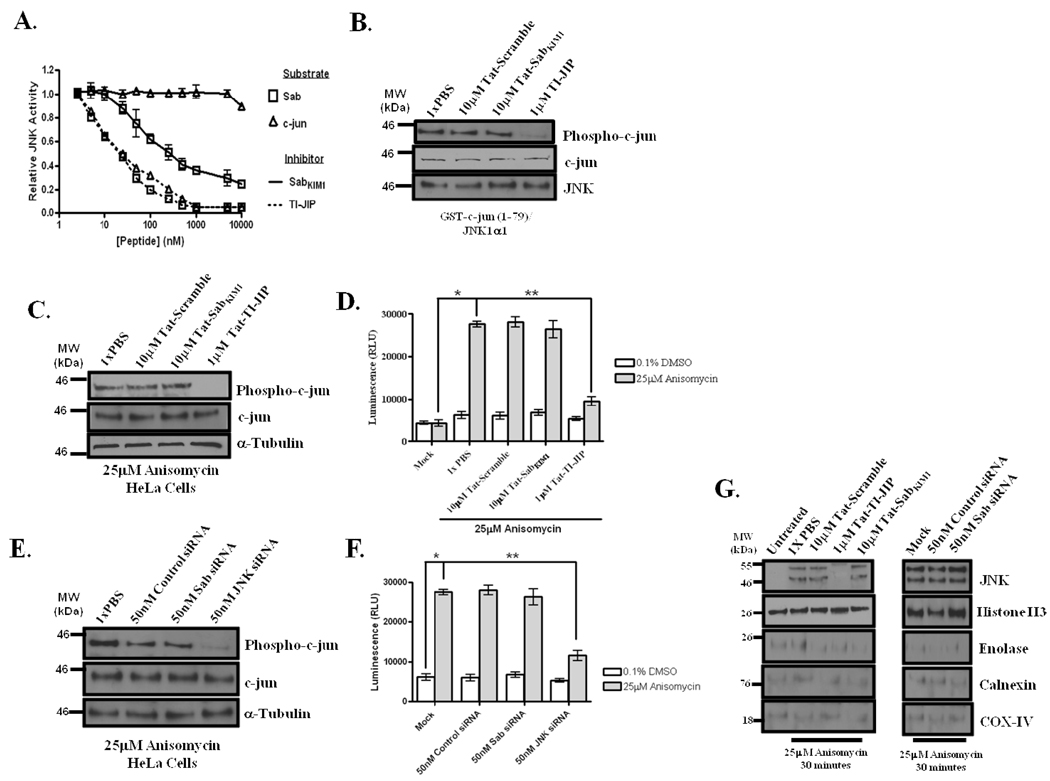
Targeting the JNK/Sab interaction did not perturb JNK-mediated c-Jun phosphorylation or AP-1 transcription
TI-JIP has been shown to be a potent inhibitor of JNK catalytic activity with respect to substrate binding(19–21); however, the Sab KIM1 motif was shown to have little, if any impact on JNK-mediated phosphorylation of transcription factors(15). Based on these data, we examined the impact of Tat-SabKIM1 on c-jun phosphorylation and AP-1 mediated transcription. Using a Kinase-Glo-based activity assay for JNK, we compared Tat-SabKIM1 IC50s for JNK1α1 with either c-jun (1–79) as the substrate or recombinant Sab as the substrate. JNK1α1 was selected over JNK3α1, since the JNK3 isoform is not expressed in HeLa cells(22). Figure 4A, presents data for the inhibition of Sab and c-jun phosphorylation by Tat-SabKIM1. An IC50 of 270±85nM for JNK1α1 phosphorylation of Sab by Tat-SabKIM1 was determined; however, Tat-SabKIM1 only inhibited JNK1α1-mediated c-jun phosphorylation by ~10% at the highest concentration examined (Figure 4A). Similarly Tat-SabKIM1 demonstrated no inhibition with respect to ATF2 (Supplemental Figure S5). The TI-JIP peptide was also used to inhibit JNK1α1. With respect to Sab phosphorylation, TI-JIP had an IC50 = 22±10nM (Figure 4A); TI-JIP also demonstrated inhibition of c-jun phosphorylation by JNK1α1 with an IC50 of 34±8nM (Figure 4A). Unlike the Tat-SabKIM1 peptide, TI-JIP inhibited JNK1α1 phosphorylation of ATF2 with an IC50 of 43±14nM (Supplemental Figure S5). The inhibitory data of each peptide is summarized in Supplemental Table S1.
Figure 4. Interfering with the JNK/Sab interaction did not impact JNK-mediated nuclear events or JNK translocation to the nucleus.
(A) The Tat-SabKIM1 peptide does not interfere with JNK-mediated c-jun phosphorylation in vitro. To determine the IC50s of TI-JIP and SabKIM1 with respect to Sab (open squares) and c-jun (open triangles) substrates, JNK activity was monitored by a Kinase-Glo Assay in the presence of increasing amounts of TI-JIP (dashed line) or Tat-SabKIM1(solid line)peptide. 0.5nM JNK and 1µM substrate were used. JNK activity was determined after 60 minutes of enzyme-substrate interaction. (B) Tat-SabKIM1 peptide does not impact JNK-mediated c-jun phosphorylation. Recombinant JNK1α1 (0.5nM) was incubated with 500nM GST-c-jun (1–79) for 60 minutes at 30°C in a kinase activity assay. Prior to the addition of GST-c-jun (1–79), JNK1α1 was incubated with 10µM Tat-Scramble, 10µM Tat-SabKIM1 or 1µM TI-JIP for 15 minutes. Reactions were then monitored for phospho-c-jun, c-jun, and JNK (loading control) by Western blot. (C) Silencing Sab did not impact JNK-mediated c-jun phosphorylation in anisomycin-treated HeLa cells. Cells were incubated with 10µM Tat-Scramble, 10µM Tat-SabKIM1 or 1µM TI-JIP for 30 minutes prior to the addition of 25µM anisomycin for 45 minutes. Cells were lysed and analyzed by Western blot for the relative abundance of phospho-c-jun, c-jun, and α-tubulin (loading control). (D) Use of the Tat-SabKIM1 peptide did not impact AP-1 transcription in anisomycin-treated HeLa cells. Cells were transfected with empty, or pAP-1:LUC vector for 71 hours prior to anisomycin addition. Cells were incubated with 10µM Tat-Scramble, 10µM Tat-SabKIM1 or 1µM TI-JIP for 30 minutes prior to the addition of 25µM anisomycin for 60 minutes. Cells were then lysed and monitored for luciferase activity. The presence of *’s indicate that the conditions connected by lines are statistically significant with a p-value of less than 0.05. (E) Gene silencing by siRNAs was employed to knockdown Sab and JNK, respectively. 50nM of siRNAs were transfected into cells as described in the methods prior to the addition of 25µM anisomycin. Cells were lysed and monitored for Phospho-c-jun(Ser73), total c-jun, and tubulin by western blotting.(F) Sab knockdown did not impact AP-1 transcription during anisomycin stress in HeLa cells. Cells were transfected with 50nM siRNAs or empty/pAP-1:LUC vector for 71 hours prior to anisomycin addition. Cells were stressed with 25µM anisomycin for 60 minutes. Cells were then lysed and monitored for luciferase activity. The presence of *’s indicate that the conditions connected by lines are statistically significant with a p-value of less than 0.05. (G) HeLa cells were either pretreated with 10µM Tat-Scramble, 1µM TI-JIP, or 10µM Tat-SabKIM1 for 30 minutes prior to incubation with 25µM anisomycin for 30 minutes. Nuclear fractions were prepared as indicated in the experimental procedures. The nuclear fractions were then monitored for the presence of JNK by Western blot analysis. Histone H3 served as the nuclear marker, while calnexin, enolase, and COX-IV were used to determine the relative amount of ER, cytosolic, and mitochondrial contamination, respectively, in the nuclear fractions. Additionally, JNK and Sab were silenced using siRNAs specific for the respective transcripts, JNK and Sab were silenced using 50 nM of their respective siRNAs for 71 hours prior to 30 minute incubation with 25µM anisomycin. Nuclear fractions were prepared as indicated above, and the presence of nuclear JNK was determined by Western blot.
To confirm that the Sab peptide was not able to inhibit JNK phosphorylation of c-jun, we incubated 50ng of active JNK1α1 with 10µM Tat-SabKIM1, 10µM Tat-Scramble, or 1µM Tat-TI-JIP for 15 minutes prior to the addition of GST-c-jun (1–79). Following 60 minutes at 30°C, the samples were examined for c-jun phosphorylation by Western blot analysis. As demonstrated in the IC50 calculation, Tat-SabKIM1 had no impact on JNK-mediated c-jun phosphorylation when compared to PBS-treated or Tat-Scramble treated JNK1α1 (Figure 4B). Moreover, treatment Tat-TI-JIP inhibited nearly all of the JNK-mediated c-jun phosphorylation (Figure 4B). We next evaluated the impact of Tat-SabKIM1 on c-jun phosphorylation in HeLa cells following 45 minutes of anisomycin stress. In cells treated with PBS or 10µM Tat-Scramble prior to anisomycin, JNK phosphorylation of c-jun was not inhibited (Figure 4C). Pre-incubation with 10µM Tat-SabKIM1 also did not prevent JNK-mediated c-jun phosphorylation during anisomycin-induced stress (Figure 4C). In contrast, 1µM Tat-TI-JIP inhibited c-jun phosphorylation completely (Figure 4C). None of the treatments altered total c-jun (Figure 4C). Tubulin was used as a loading control (Figure 4C).
To further confirm Tat-SabKIM1 does not impact JNKs nuclear functions, we monitored JNK-mediated AP-1 transcription during stress using an AP-1 reporter assay. Compared to mock-transfected cells and unstressed cells transfected with pAP1-LUC reporter vector, anisomycin increased AP-1 driven transcription as detected by luminescence (Figure 4D). Treatment with PBS or 10µM Tat-Scramble prior to anisomycin addition did not impact AP-1 transcription (Figure 4D). Conversely, 1 µM Tat-TI-JIP nearly completely inhibited AP-1 mediated transcription during anisomycin stress (Figure 4D); however, 10 µM Tat-SabKIM1 did not inhibit AP-1 driven production of luciferase (Figure 4D). To assure that interfering with the JNK/Sab interaction did not impact JNK-mediated nuclear events, we examined c-jun phosphorylation and AP-1 mediated transcription in cells that had reduced levels of Sab and JNK. Silencing Sab expression did not result in any change in anisomycin-induced c-jun phosphorylation (Figure 4E) or AP-1 transcription (Figure 4F) when compared to mock or control siRNA transfected cells following 45 minutes of stress (Figures 4E and 4F). As expected, reducing JNK expression was sufficient to decrease c-jun phosphorylation (Figure 4E) and AP-1 mediated transcription (Figure 4F) during anisomycin stress.
Finally, to elucidate if the inability of Sab to alter JNKs nuclear functions was due to failure to inhibit JNK translocation to the nucleus, we examined JNK translocation into the nucleus in the presence and absence of Sab. First, we evaluated JNK nuclear translocation using peptide-mediated interference. Following 30 minutes of anisomycin stress, JNK was found in the nucleus as indicated by co-fractionation with nuclear resident histone H3 (Figure 4G); as described in a previous report(21) and demonstrated in Figure 4G, 1µM Tat-TI-JIP inhibited JNK translocation to the nucleus; whereas 10µM Tat-Scramble peptide did not impact JNK nuclear translocation (Figure 4G). Moreover, treatment with 10µM Tat-SabKIM1 peptide did not prevent JNK migration into the nucleus (Figure 4G). To further demonstrate that interfering with the JNK/Sab interaction did not impact nuclear translocation of JNK, we silenced Sab with siRNAs. In Figure 4G, silencing Sab did not prevent JNK translocation into the nucleus as mock transfected cells, cells transfected with control siRNAs, and cells transfected with Sab-specific siRNAs had the same relative abundance of nuclear JNK. Again, Histone H3 was used as a nuclear loading control (Figure 4G). Nuclear contamination by ER, cytosol, and mitochondria was minimal as demonstrated by Western blot analysis for calnexin, enolase, and COX-IV, respectively (Figure 4G).
Inhibition of JNK or MitoJNK Signaling prevented anisomycin-stress induced phenotypes in HeLa cells
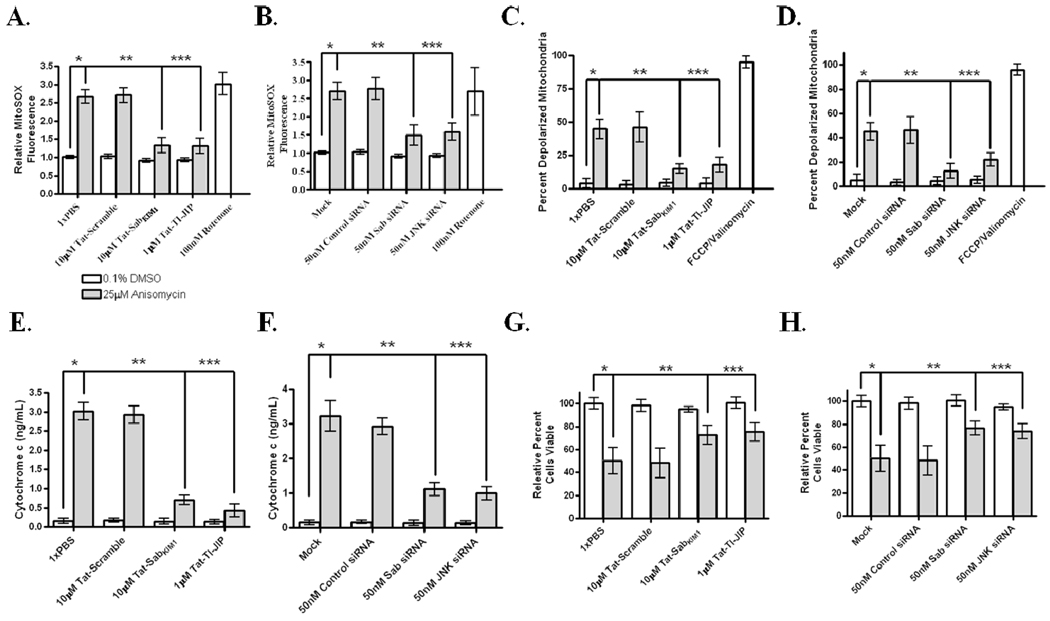
Given that disrupting the JNK/Sab interaction did not disturb nuclear events, we examined the impact of disrupting the JNK mitochondrial localization on stress-related mitochondrial phenotypes. In anisomycin-stressed HeLa cells, 10µM Tat-SabKIM1 prevented JNK induced mitochondrial superoxide production compared to PBS or 10 µM Tat-Scramble treated cells (Figure 5A); similarly, treatment with 1µM Tat-TI-JIP prevented JNK-mediated superoxide generation to the same levels as 10µM Tat-SabKIM1 (Figure 5A). The use of siRNAs was employed to confirm the peptide-based observation. Again, silencing JNK expression statistically significantly decreased mitochondrial superoxide generation compared to mock and control siRNA transfected cells (Figure 5B), and Sab knockdown also prevented JNK-mediated mitochondrial superoxide production (Figure 5B). Rotenone treatment was used as a positive control for mitochondrial superoxide generation (Figure 5A and 5B).
Figure 5. Blocking MitoJNK signaling prevented stress-related mitochondrial dysfunction and cell death.
(A, B) Interfering with JNK/Sab interaction prevented mitochondrial superoxide generation in anisomycin-stressed HeLa cells. Cells were stressed for 30 minutes with 25µM anisomycin, and then stained with mitochondrial superoxide sensitive dye, MitoSOX Red. Following 10 minutes of staining/stress, the cells were washed, placed in visualizing solution, inspected by microscopy for staining, and then MitoSOX fluorescence was quantitated using a 96-well plated reader. Thirty minutes prior to anisomycin addition cells were either pretreated with 10µM Tat-Scramble, 1µM TI-JIP, or 10µM Tat-SabKIM1, A, or cells were transfected with 50nM siRNAs selective for JNK or Sab for 71 hours prior to anisomycin stress, B. (C, D) Preventing JNK mitochondrial translocation decreased dissipation of mitochondrial membrane potential. Cells were stressed with 25µM anisomycin for 45 minutes prior to staining with the mitochondria membrane sensitive dye, JC-1. Cells were stained as indicated with JC-1, and JC-1 staining was confirmed by microscopy prior to quantitation of JC-1 monomer fluorescence by a 96-well plate reader. Cells were either preincubated with 10µM Tat-Scramble, 1µM TI-JIP, or 10µM Tat-SabKIM1, C, or exposed to 50nM siRNAs selective for JNK or Sab for 71 hours prior to anisomycin stress, D. (E, F) Inhibition of MitoJNK signaling prevented release of cytochrome c from the mitochondria of anisomycin-treated HeLa cells. Thirty minutes prior to anisomycin addition cells were either pretreated with 10µM Tat-Scramble, 1µM TI-JIP, or 10µM Tat-SabKIM1, E, or cells were transfected with 50nM siRNAs selective for JNK or Sab for 71 hours prior to anisomycin stress, F. Cytochrome c levels in the cytosol were quantitated using a cytochrome c ELISA following 4 hours of stress with 25µM anisomycin. (G, H) Disrupting MitoJNK signaling prevented anisomycin-induced cell death in HeLa cells. Cells were either pretreated with 10µM Tat-Scramble, 1µM TI-JIP, or 10µM Tat-SabKIM1 for 30 minutes prior to 25µM anisomycin exposure, G, or cells were transfected with 50nM siRNAs selective for JNK or Sab for 71 hours prior to anisomycin stress, H. Cell viability was measured following 4 hours of anisomycin-induced stress or exposure to DMSO. The presence of *s indicate that the conditions connected by lines are statistically significant with a p-value of less than 0.05.
An early event in cell death responses is loss of mitochondrial membrane potential (MMP)(19). We measured relative cellular MMP dissipation using MMP-sensitive dye JC-1. To demonstrate this dye detected changes in MMP, cells were treated with mitochondrial uncoupler, carbonylcyanide p-trifluoromethoxy-phenylhydrazone (FCCP), and ionophore, valinomycin, a combination which has been shown to induce a near complete loss MMP(19). As seen in Figures 5C and 5D, treatment with FCCP/valinomycin increased the percentage of depolarized mitochondria within HeLa cells. Treatment with 25µM anisomycin also increased the percent depolarized mitochondria compared to DMSO-treated cells showing a 40–50% increase (Figure 5C and 5D). Treatment with 10µM Tat-SabKIM1 or Sab siRNAs decreased the percentage of MMP depolarization when compared to 10µM Tat-scramble and control siRNA transfected cells, respectively (Figures 5C and 5D). Cell pre-treatment with PBS or mock transfected cells had no impact on anisomycin-induced MMP dissipation, while the use of 1 µM Tat-TI-JIP or JNK siRNAs decreased the amount of mitochondria with dissipating MMP (Figures 5C and 5D).
We also monitored the impact of mitochondrial JNK signaling on cytochrome c release from the mitochondria. We found that treatment with 10 µM Tat-SabKIM1 or silencing Sab prevented release of cytochrome c from the mitochondria, as compared to cells treated with 10 µM Tat-Scramble and control siRNAs (Figures 5E and 5F). Additionally, JNK inhibition by1 µM Tat-TI-JIP or JNK knock-down was also capable of reducing cytochrome c release during anisomycin stress (Figure 5E and 5F). Each of these treatments decreased cytochrome c release by 3–5-fold. PBS and mock transfection had no impact on cytochrome c release in response to anisomycin. Finally, we examined if inhibition of mitochondrial JNK signaling by interfering with the JNK/Sab interaction was sufficient to prevent cell death in anisomycin-treated HeLa cells. As stated earlier, treatment with 25µM anisomycin resulted in 50% cell death after 4 hours of stress. The addition of PBS and 10µM Tat-Scramble had no impact on anisomycin-induced cell death (Figure 5G); however, treatment with 10 µM Tat-SabKIM1 peptide rescued cells from anisomycin-induced cell death (Figure 5G). Additionally, silencing Sab also rescued anisomycin-induced cell death (Figure 5H) compared to mock transfection or cells transfected with control siRNAs (Figure 5H). Inhibition of JNK by 1µM Tat-TI-JIP rescued the viability (Figures 1A and 5G); similarly, silencing JNK expression also rescued cells from anisomycin-induced cell death (Figures 1A and 5H). Moreover, siRNA-mediated knockdown of c-jun (Supplemental Figure S6A) did not impact mitochondrial superoxide generation (Supplemental Figures S6B). Silencing c-jun decreased MMP dissipation during anisomycin stress (Supplemental Figure S6C); similarly, silencing c-jun impacted cell viability in response to anisomycin albeit a marginal, but significant increase (Supplemental Figure S6D). However, both the decrease in MMP dissipation and cell death are much less than those changes in the presence of Tat-SabKIM1 peptide.
Discussion
The recent discovery of mitochondrial JNK signaling pathways has revealed that the mechanism of JNK-induced apoptosis may be more dynamic than the mere induction of AP-1-mediated transcription and the modification of pro-apoptotic proteins. Mitochondrial JNK signaling has profound impact on mitochondrial physiology and bioenergetics, and JNK mitochondrial signaling may have a more profound effect than nuclear JNK signaling with regards to the aforementioned JNK-mediated cellular events. Given this concern, we have developed a biochemical probe to selectively evaluate MitoJNK signaling by disrupting the JNK/Sab interaction at the outer mitochondrial membrane.
In HeLa cells, anisomycin stress induced cell death in a JNK dependent, mitochondrially localized manner. Here JNK may come into contact with previously identified putative substrates, namely PDH(10, 11) and Bcl-2(4, 16, 17). Inhibition of PDH activity and limitation of pyruvate flux into the mitochondria(10) could explain the decrease in mitochondrial bioenergetics observed in other studies(8–11). While direct phosphorylation of Bcl-2 could initiate signaling resulting in apoptosis by inhibiting Bcl-2 anti-apoptotic functions (4), it could also be responsible for the loss of MMP observed in this study and other work (16, 17). Given that neither JNK nor Sab possess motifs essential for mitochondrial import, one can postulate that JNK mitochondrial signaling begins on the outer membrane (8), and additional downstream signaling events promote the physiological changes that induce cell death. This “outside-in” view of JNK mitochondrial signaling could explain how JNK signaling at the mitochondria could impact the apoptotic and bioenergetic machinery. JNK has the capacity to use mitochondrial localized proteins directly as substrates(4); however, a majority of mitochondrial enzyme activity is regulated by tyrosine phosphorylation(23). One might propose that JNK signaling may activate a protein tyrosine kinase that modulates mitochondrial bioenergetics in conjunction with the serine/threonine kinase activity of JNK.
The observation that catalytically active JNK bound to the mitochondria may suggest that JNK-mediated phosphorylation of Sab was required for mitochondrial docking. Moreover, it implies that there may exist a unique structural conformation in the activated-form of JNK that does not exist in the inactive form, otherwise, JNK might interact with Sab in the absence of stimuli and partly localize to the mitochondria. Additionally there may be a unique conformation of Sab that only binds the active form of JNK. These interpretations of course have many caveats, including the affinity of each of these binding proteins to JNK, as well as the local concentration of each scaffold protein or substrate. Finally, we acknowledge that the presence of the JNK-interacting protein-1 (JIP-1) in the cytosol may also restrict the interactions between JNK and Sab in the absence of stress.
By exploiting the JNK/Sab interaction, we have demonstrated that JNK migration to the mitochondria can be inhibited without impacting nuclear events in JNK signaling, namely c-jun phosphorylation, AP-1-mediated transcription, and JNK nuclear translocation. The inability of the Tat-SabKIM1 peptide to interfere in the nuclear events may be due to the relatively low affinity of Sab for JNK compared to other substrates such as c-jun or ATF-2. For example, TI-JIP can inhibit JNK activity versus ATF-2 at low nanomolar concentrations (Ki= 25nM), or even c-jun (IC50= 38nM) (19, 20), while in our experiments, Tat-SabKIM1 demonstrated essentially no inhibition of c-jun phosphorylation at 10µM. The distinct affinities of JNK for JIP and Sab binding motifs with respect to other substrates, such as ATF-2 and c-Jun, may account for the difference in the mode of action for these two peptides. This is an advantageous characteristic, since our goal was to distinctively target the JNK/Sab interaction.
The observation that silencing Sab or blocking the JNK/Sab interaction prevented cell death and other mitochondrial cell death-associated phenotypes indicated that MitoJNK signaling may have a more pronounced impact on cell death induction than AP-1 mediated transcription. It is interesting to speculate that MitoJNK signaling may be critical to mitochondrial-related cell death. The changes induced by MitoJNK activity could produce a set of changes, both in mitochondrial physiology and signaling, that propagates cell death signaling. It has been suggested that JNK signaling can alter mitochondria in such a manner(4, 5, 8–10, 17, 24–26). In HL-60 cells treated with docetaxel, JNK signaling, induced by early ROS generation and caspase activity, resulted in increased phosphorylation of Bcl-2 and increased ROS production creating a means for cell death through the amplification of mitochondrial dysfunction(24). Our own studies have indicated that mitochondrial JNK is involved in an increase ROS production(27). Thus, the selective inhibition of MitoJNK may provide a selective means to assess JNK-mediated events on the mitochondria contributing to cell death responses.
In this work, we have demonstrated that selectively disrupting the JNK/Sab interaction can be used to inhibit JNK mitochondrial signaling without impacting nuclear events. These tools can now be used to examine the mechanism of JNK-mediated cell death at the mitochondria. Using these techniques we will be able to identify novel JNK substrates on the mitochondria and elucidate new JNK-mediated processes contributing to cell death. The evaluation of this arm of JNK signaling will provide useful information into the necessary mitochondrial perturbations that are required for JNK-induced cell death.
Methods
Materials
Tat-Scramble (LPSVFGDVGAPSRLPEVSLSPPRRRQRRKKRG-NH2) and Tat-SabKIM1 (GFESLSVPSPLDLSGPRVVAPPRRRQRRKKRG-NH2) peptides were purchased from Neo Peptide. Tat-TI-JIP (NH2-YGRKKRRQRRRRPKRPTTLNLF-NH2) and c-jun (1–79) peptide were purchased from Calbiochem. The pAP1-LUC reporter vector was purchased from Clonetech Laboratories, Incorporated. Inactive and active JNK1α1 were purchased from Millipore. Sab siRNAs were purchased from Novus Biologicals, while JNK siRNAs were purchased from Cell Signaling Technologies.
Cell Culture, Transfections, and Gene Silencing
HeLa cells (ATCC) and JNK-null murine embryonic fibroblasts (JNK1−/−, JNK2−/− MEFs; a kind gift from Dr. Roger Davis HHMI, University of Massachusetts) were grown at normal cell culture conditions (37°C and 5% CO2) in DMEM (Invitrogen) supplemented with 10% fetal bovine serum and penicillin/streptomycin. To assure that the cells were actively growing, only cells at ~80% confluency and between passages five and fifteen were used in our experiments. Silencing JNK and Sab expression was achieved by small-interfering RNA (siRNA)-mediated gene silencing. Specific siRNAs for JNK, Sab, or control (nonsense) siRNAs were introduced into HeLa cells using the Qiagen HiPerfect transfection reagent. Briefly, cells were grown to ~50% confluency, and transfected with 50nM of siRNA and 12µL HiPerfect reagent in 100µL medium. The mixture incubated at room temperature for 10 minutes to permit transfection complex formation, and then the complexes were added to cells. After 72 hours post-transfection, knock-down was monitored by western blot analysis.
Mitochondria Isolation
Mitochondria were isolated similarly to the method described by Palloti and Lenaz (28). The protocol is included in the Supplemental Methods.
Proteinase K Treatment of Mitochondria
Mitochondria (100µg) isolated as described above were diluted to 2mg/mL in Clark electrode buffer (80mM KCl, 50mM MOPS, 1mM EGTA, 5mM KH2PO4, and 1mg/mL BSA, adjusted to pH 7.4). For recombinant protein studies, JNK1α1 (250ng) was incubated with mitochondria in the presence of 200µM ATP, 2.5mM MgCl2, and 8mM succinate for 40 minutes at 37°C, and then mitochondria were re-obtained by centrifugation at 6000×g for 5 minutes at 4°C. For HeLa cell-based studies, mitochondria were simply diluted in Clark electrode buffer. Next, mitochondria were treated with 50mg/mL Proteinase K for 30 minutes at 4°C. The enzyme reaction was stopped by the addition of 1mM PMSF and Protease Inhibitor Cocktail Set III (Calbiochem). Mitochondria were isolated by centrifugation (6000×g for 5 minutes at 4°C). The supernatant contained proteins cleaved from the outer mitochondrial membrane. The mitochondrial pellet was lysed in RIPA buffer with phosphatase and protease inhibitors. Protein concentration was determined by BCA assay. Samples were resolved by SDS-PAGE, and Western blots were performed to identify proteins found in each mitochondrial subfraction.
Outer Mitochondrial Membrane and Mitoplast Preparation
The outer mitochondrial membrane preparation was obtained by methods described in Schnaitman et al. (29). A detailed description of the protocol can be found in the Supplemental Methods.
Cloning, Expression, and Purification of Recombinant Sab and ATF2
These protocols are described in detail in the Supplemental Methods.
Fluorescence Polarization Assay for Peptide Binding
The binding of JNK3 α1 and JIP, Sab, and Scramble peptides was determined similar to (20). Briefly, binding of the TAMRA-JIP 11-mer peptide (TAMRA-RPKRPTTLNLF) with JNK3α1 was measured in a fluorescence polarization (FP) assay. Under standard assay conditions, different concentrations of unlabeled TI-JIP, TAT-Sab, or Tat-Scramble peptide in assay buffer (10mM HEPES/KOH (pH7.4), 150mM NaCl, 10mM MgCl2, 0.005% Brij-35, 0.1% 2-mercaptoethanol, and 0.05% BSA) were dispensed into a 384-well microtiter plate. Then, JNK3 α1 and TAMRA-JIP peptide were added to the microtiter wells to give a final JNK concentration of 0.8µM and TAMRA-JIP concentration of 5nM. Plates were read on a Perkin Elmer Envision 2104 multilabel plate reader. The Kd for each peptide was calculated as described in the Supplemental Methods.
Kinase-Glo Phosphorylation Assay
Recombinant substrates, Sab and c-jun (1–79), were diluted to 1µM in JNK activity buffer (25mM HEPES, pH 7.4, 10mM MgCl2, 2mM dithiothreotol (DTT), 1mg/mL BSA, and 1µM ATP). The reaction was initiated with the addition of 0.5nM active JNK1α1. The reaction was incubated at 30°C for 60 minutes. The reaction was stopped by the addition of 50mM EDTA. The reaction was combined with the Kinase-Glo reagent at a 1:1 ratio, and then incubated at room temperature for 10 minutes. Luminescence was monitored on a Spectromax M5e plate reader (Molecular Devices) with an integration of 500ms. ATP quantitation was determined based on values interpolated onto an ATP standard curve. Data are reported as percent JNK activity based on uninhibited, active JNK1α1/substrate phosphorylation.
AP-1 Luciferase-based Transcription Assay
The pAP-LUC plasmid or pLUC empty plasmid was transfected into HeLa cells at a 3:1 ratio of plasmid to Fugene6 transfection reagent (Roche) with cells at 60% confluency. Cells were grown for 24 hours, and the media was changed two hours prior to anisomycin stress. The cells were then stressed with 25µM anisomycin for 60 minutes. The luciferase assay was performed with slight modifications (described in Supplemental Methods) from the protocol described by Brasier and Fortin(30).
Nuclear Subcellular Fractionation
The protocol is described in the Supplemental Methods.
Mitochondrial Superoxide Generation
Mitochondrial superoxide production was monitored by utilizing the mitochondrial sensitive dye MitoSOX-Red (Invitrogen)(31). The Supplemental Methods contain a detailed explanation of the procedure.
Mitochondrial Membrane Potential
Mitochondrial membrane potential was determine using the JC-1 stain by methods described in Ryan et al(32). The protocol can be found in the Supplemental Methods.
Cytochrome c Release
Non-mitochondrial cytochrome c levels were monitored by ELISA using Invitrogen’s Cytochrome c Immunoassay Kit protocol. On average, 2.5×107 cells were used per assay. Data are represented as ng/mL cytochrome c based on a cytochrome c standard curve.
Cell Viability
Cell viability was monitored using trypan blue exclusion using the BioRad TC10 Automated Cell Counter. A more detailed protocol is described in the Supplemental Methods.
Biological Replicates and Statistics
A minimum of four biological replicates were considered for all cell-based studies. For mitochondrial enrichments, a minimum of three biological replicates were used. Recombinant protein studies were performed in quadruplicate at minimum. Biochemical assays, fluorometric detection of superoxide, and other cellular measures were done with a minimum of five experimental replicates. To determine statistical significance a student’s paired t-test was employed. Statistical significance is indicated by (*) in figures when the p-value is less than 0.05.
Supplementary Material
Acknowledgments
JNK1−/−/JNK2−/− MEFs were a kind gift from Dr. Roger Davis HHMI, University of Massachusetts. This work was supported by NIH grant NS057153 awarded to P.L. and support from the Saul and Theresa Esman Foundation.
Footnotes
Supporting Information: Supporting information associated with this manuscript is available online.
Author Contributions
JWC designed and conducted the experiments with the exception of the determination of Kds for peptide binding, which were performed by LC. JDL and MFL produced recombinant Sab and ATF2, respectively. JWC and PVL wrote the manuscript with methods contributed from LC, JDL, and MFL.
References
- 1.Weston CR, Davis RJ. The JNK signal transduction pathway. Curr Opin Cell Biol. 2007;19:142–149. doi: 10.1016/j.ceb.2007.02.001. [DOI] [PubMed] [Google Scholar]
- 2.Martin JH, Mohit AA, Miller CA. Developmental expression in the mouse nervous system of the p493F12 SAP kinase. Brain Res Mol Brain Res. 1996;35:47–57. doi: 10.1016/0169-328x(95)00181-q. [DOI] [PubMed] [Google Scholar]
- 3.Barr RK, Bogoyevitch MA. The c-Jun N-terminal protein kinase family of mitogen-activated protein kinases (JNK MAPKs) Int J Biochem Cell Biol. 2001;33:1047–1063. doi: 10.1016/s1357-2725(01)00093-0. [DOI] [PubMed] [Google Scholar]
- 4.Dhanasekaran DN, Reddy EP. JNK signaling in apoptosis. Oncogene. 2008;27:6245–6251. doi: 10.1038/onc.2008.301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kim MJ, Lee KH, Lee SJ. Ionizing radiation utilizes c-Jun N-terminal kinase for amplification of mitochondrial apoptotic cell death in human cervical cancer cells. FEBS J. 2008;275:2096–2108. doi: 10.1111/j.1742-4658.2008.06363.x. [DOI] [PubMed] [Google Scholar]
- 6.Kharbanda S, Saxena S, Yoshida K, Pandey P, Kaneki M, Wang Q, Cheng K, Chen YN, Campbell A, Sudha T, Yuan ZM, Narula J, Weichselbaum R, Nalin C, Kufe D. Translocation of SAPK/JNK to mitochondria and interaction with Bcl-x(L) in response to DNA damage. J Biol Chem. 2000;275:322–327. doi: 10.1074/jbc.275.1.322. [DOI] [PubMed] [Google Scholar]
- 7.Ito Y, Mishra NC, Yoshida K, Kharbanda S, Saxena S, Kufe D. Mitochondrial targeting of JNK/SAPK in the phorbol ester response of myeloid leukemia cells. Cell Death Differ. 2001;8:794–800. doi: 10.1038/sj.cdd.4400886. [DOI] [PubMed] [Google Scholar]
- 8.Hanawa N, Shinohara M, Saberi B, Gaarde WA, Han D, Kaplowitz N. Role of JNK translocation to mitochondria leading to inhibition of mitochondria bioenergetics in acetaminophen-induced liver injury. J Biol Chem. 2008;283:13565–13577. doi: 10.1074/jbc.M708916200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Aoki H, Kang PM, Hampe J, Yoshimura K, Noma T, Matsuzaki M, Izumo S. Direct activation of mitochondrial apoptosis machinery by c-Jun N-terminal kinase in adult cardiac myocytes. J Biol Chem. 2002;277:10244–10250. doi: 10.1074/jbc.M112355200. [DOI] [PubMed] [Google Scholar]
- 10.Zhou Q, Lam PY, Han D, Cadenas E. c-Jun N-terminal kinase regulates mitochondrial bioenergetics by modulating pyruvate dehydrogenase activity in primary cortical neurons. J Neurochem. 2008;104:325–335. doi: 10.1111/j.1471-4159.2007.04957.x. [DOI] [PubMed] [Google Scholar]
- 11.Zhou Q, Lam PY, Han D, Cadenas E. Activation of c-Jun-N-terminal kinase and decline of mitochondrial pyruvate dehydrogenase activity during brain aging. FEBS Lett. 2009;583:1132–1140. doi: 10.1016/j.febslet.2009.02.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zhao Y, Herdegen T. Cerebral ischemia provokes a profound exchange of activated JNK isoforms in brain mitochondria. Mol Cell Neurosci. 2009;41:186–195. doi: 10.1016/j.mcn.2009.02.012. [DOI] [PubMed] [Google Scholar]
- 13.Wiltshire C, Gillespie DA, May GH. Sab (SH3BP5), a novel mitochondria-localized JNK-interacting protein. Biochem Soc Trans. 2004;32:1075–1077. doi: 10.1042/BST0321075. [DOI] [PubMed] [Google Scholar]
- 14.Wiltshire C, Matsushita M, Tsukada S, Gillespie DA, May GH. A new c-Jun N-terminal kinase (JNK)-interacting protein, Sab (SH3BP5), associates with mitochondria. Biochem J. 2002;367:577–585. doi: 10.1042/BJ20020553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Barr RK, Boehm I, Attwood PV, Watt PM, Bogoyevitch MA. The critical features and the mechanism of inhibition of a kinase interaction motif-based peptide inhibitor of JNK. J Biol Chem. 2004;279:36327–36338. doi: 10.1074/jbc.M402181200. [DOI] [PubMed] [Google Scholar]
- 16.Lee JJ, Lee JH, Ko YG, Hong SI, Lee JS. Prevention of premature senescence requires JNK regulation of Bcl-2 and reactive oxygen species. Oncogene. 2010;29:561–575. doi: 10.1038/onc.2009.355. [DOI] [PubMed] [Google Scholar]
- 17.Lei K, Nimnual A, Zong WX, Kennedy NJ, Flavell RA, Thompson CB, Bar-Sagi D, Davis RJ. The Bax subfamily of Bcl2-related proteins is essential for apoptotic signal transduction by c-Jun NH(2)-terminal kinase. Mol Cell Biol. 2002;22:4929–4942. doi: 10.1128/MCB.22.13.4929-4942.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Barr RK, Kendrick TS, Bogoyevitch MA. Identification of the critical features of a small peptide inhibitor of JNK activity. J Biol Chem. 2002;277:10987–10997. doi: 10.1074/jbc.M107565200. [DOI] [PubMed] [Google Scholar]
- 19.Ember B, Kamenecka T, LoGrasso P. Kinetic Mechanism and Inhibitor Characterization for c-jun-N-Terminal Kinase 3α1. Biochemistry. 2008;47:3076–3084. doi: 10.1021/bi701852z. [DOI] [PubMed] [Google Scholar]
- 20.Ember B, LoGrasso P. Mechanistic characterization for c-jun-N-Terminal Kinase 1[alpha]1. Archives of Biochemistry and Biophysics. 2008;477:324–329. doi: 10.1016/j.abb.2008.06.001. [DOI] [PubMed] [Google Scholar]
- 21.Dickens M, Rogers JS, Cavanagh J, Raitano A, Xia Z, Halpern JR, Greenberg ME, Sawyers CL, Davis RJ. A Cytoplasmic Inhibitor of the JNK Signal Transduction Pathway. Science. 1997;277:693–696. doi: 10.1126/science.277.5326.693. [DOI] [PubMed] [Google Scholar]
- 22.Ventura JJ, Hubner A, Zhang C, Flavell RA, Shokat KM, Davis RJ. Chemical genetic analysis of the time course of signal transduction by JNK. Mol Cell. 2006;21:701–710. doi: 10.1016/j.molcel.2006.01.018. [DOI] [PubMed] [Google Scholar]
- 23.Pagliarini DJ, Dixon JE. Mitochondrial modulation: reversible phosphorylation takes center stage? Trends Biochem Sci. 2006;31:26–34. doi: 10.1016/j.tibs.2005.11.005. [DOI] [PubMed] [Google Scholar]
- 24.Cao D, Qiao B, Ge Z, Yuan Y. Amplification loop cascade for increasing caspase activity induced by docetaxel. J Cell Biochem. 2005;96:810–820. doi: 10.1002/jcb.20563. [DOI] [PubMed] [Google Scholar]
- 25.Shen HM, Liu ZG. JNK signaling pathway is a key modulator in cell death mediated by reactive oxygen and nitrogen species. Free Radic Biol Med. 2006;40:928–939. doi: 10.1016/j.freeradbiomed.2005.10.056. [DOI] [PubMed] [Google Scholar]
- 26.Ventura JJ, Cogswell P, Flavell RA, Baldwin AS, Jr, Davis RJ. JNK potentiates TNF-stimulated necrosis by increasing the production of cytotoxic reactive oxygen species. Genes Dev. 2004;18:2905–2915. doi: 10.1101/gad.1223004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kamenecka T, Jiang R, Song X, Duckett D, Chen W, Ling YY, Habel J, Laughlin JD, Chambers J, Figuera-Losada M, Cameron MD, Lin L, Ruiz CH, LoGrasso PV. Synthesis, biological evaluation, X-ray structure, and pharmacokinetics of aminopyrimidine c-jun-N-terminal kinase (JNK) inhibitors. J Med Chem. 2010;53:419–431. doi: 10.1021/jm901351f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Pallotti F, Lenaz G. Isolation and subfractionation of mitochondria from animal cells and tissue culture lines. Methods Cell Biol. 2007;80:3–44. doi: 10.1016/S0091-679X(06)80001-4. [DOI] [PubMed] [Google Scholar]
- 29.Schnaitman C, Erwin VG, Greenawalt JW. The submitochondrial localization of monoamine oxidaseAn enzymatic marker for the outer membrane of rat liver mitochondria. J Cell Biol. 1967;32:719–735. doi: 10.1083/jcb.32.3.719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Brasier AR, Fortin JJ. Nonisotopic Assays for Reporter Gene Activity. John Wiley & Sons, Inc.; 2001. [DOI] [PubMed] [Google Scholar]
- 31.Mukhopadhyay P, Rajesh M, Hasko G, Hawkins BJ, Madesh M, Pacher P. Simultaneous detection of apoptosis and mitochondrial superoxide production in live cells by flow cytometry and confocal microscopy. Nat Protoc. 2007;2:2295–2301. doi: 10.1038/nprot.2007.327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ryan JA, Brunelle JK, Letai A. Heightened mitochondrial priming is the basis for apoptotic hypersensitivity of CD4+ CD8+ thymocytes. Proceedings of the National Academy of Sciences. 2010;107:12895–12900. doi: 10.1073/pnas.0914878107. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.