Abstract
Members of the DAZ (Deleted in AZoospermia) gene family are important players in the process of gametogenesis and their dysregulation accounts for 10% of human male infertility. Boule, the ancestor of the family, is mainly involved in male meiosis in most organisms. With the exception of Drosophila and C. elegans, nothing is known on the function of boule in non-vertebrate animals. In the present study, we report on three boule orthologues in the flatworm Macrostomum lignano. We demonstrate that macbol1 and macbol2 are expressed in testes whilst macbol3 is expressed in ovaries and developing eggs. Macbol1 RNAi blocked spermatocyte differentiation whereas macbol2 showed no effect upon RNAi treatment. Macbol3 RNAi resulted in aberrant egg maturation and led to female sterility. We further demonstrated the evolutionary functional conservation of macbol1 by introducing this gene into Drosophila bol1 mutants. Macbol1 was able to rescue the progression of fly meiotic divisions. In summary, our findings provide evidence for an involvement of boule genes in male and female gamete development in one organism. Furthermore, boule gene function is shown here for the first time in a lophotrochozoan. Our results point to a more diverse functional assignment of boule genes. Therefore, a better understanding of boule function in flatworms can help to elucidate the molecular mechanisms of and concomitant infertility in higher organisms including humans.
Keywords: Boule, Planaria, Flatworms, Platyhelminthes, Germline, Hermaphrodite
Highlights
► The function of boule genes is shown for the first time in a lophotrochozoan. ► There are three distinct macbol orthologues in the flatworm Macrostomum lignano. ► Macbol1 and macbol2 are testis-specific, macbol3 is essential for oogenesis. ► RNAi arrests male meiosis (macbol1) and impedes egg maturation (macbol3). ► Macbol1 is able to rescue meiotic divisions in boule-mutant Drosophila melanogaster.
Introduction
The production of gametes that carry all the genetic information of their genitors by means of meiosis is a crucial step to ensure the generation of fertile offspring. Meiosis is a complex process of which the genetic regulation is not entirely understood (Handel and Schimenti, 2010; Keeney, 2009a, b; Maines and Wasserman, 1998; Orr-Weaver, 1995), despite the remarkable similarity in the generation of male gametes between e.g. Drosophila and mammals (Fuller, 1998).
The Deleted in AZoospermia (DAZ) gene family is known to be crucial for the development of the germline and the generation of male and female gametes (Brook et al., 2009; Kee et al., 2009; Kerr and Cheng, 2010; Takeda et al., 2009; VanGompel and Xu, 2010; Xu et al., 2001). Interested in the background of male infertility, Tiepolo and Zuffardi (1976) proposed the existence of an AZoospermia Factor (AZF) or a spermatogenesis gene that, when not working properly, could lead to a breakdown of sperm production. The first member of the DAZ family was isolated from deleted Y-chromosomal regions in human and was believed to be a strong candidate for AZF (Ma et al., 1993; Reijo et al., 1995). Later on, the three genes belonging to the DAZ family, DAZ, DAZL (DAZ-like) and Boule, proved to be indispensable for gametogenesis (Carani et al., 1997; Cooke et al., 1996; Eberhart et al., 1996; Houston et al., 1998; Houston and King, 2000; Johnson et al., 2001; Karashima et al., 2000; Liu et al., 2009; Maegawa et al., 1999; Ruggiu et al., 1997; Xu et al., 2003; Zhang et al., 2009). The name-giving family member DAZ (Reijo et al., 1995) emerged as a Y-linked duplicate of the autosomal DAZL about 30 million years ago in catarrhine primates (old world monkeys) (Xu et al., 2001). The second family member, DAZL, arose by duplication from boule in an ancestor of vertebrates more than 450 million years ago (Xu et al., 2009). Boule was first identified in Drosophila melanogaster (Castrillon et al., 1993; Eberhart et al., 1996), and is considered the ancestor of the DAZ gene family (Haag, 2001; Xu et al., 2001).
The high evolutionary conservation of orthologues was demonstrated by rescue experiments: Spermatogenesis of sterile fruit flies carrying a bol1-mutation was partially restored by introducing human BOULE (Xu et al., 2003). Interestingly, male sterility in a hybrid bovine species could be caused by reduced boule expression although no direct evidence has been provided (Zhang et al., 2009). Boule was also shown to have an additional function in the nervous system of D. melanogaster. Joiner and Wu (2004) reported a defect in the fly eye when Boule protein was overexpressed. Hoopfer et al. (2008) showed that the protein was a negative regulator of axon pruning. All family members are characterized by a RRM (RNA Recognition Motif) containing RNP-1 and RNP-2 sequences (RiboNucleoProtein binding sites 1 and 2) and a DAZ-motif consisting of one (boule and DAZL) or more (DAZ) DAZ tandem-repeats (Reijo et al., 1995).
Phylogenetic analyses support the existence of two subfamilies, one primarily responsible for early germ cell development (containing DAZ and DAZL), and the other (Boule) with a focus on directing meiosis (Xu et al., 2001). Whereas DAZ can only be found in the male function (Eberhart et al., 1996; Menke et al., 1997; Reijo et al., 1995, 2000), DAZL is required in male and female gonads (Cooke et al., 1996; Saxena et al., 1996). Boule was initially thought to be restricted to spermatogenesis throughout the animal kingdom, until its orthologue in C. elegans, DAZ-1, was shown to be functionally refined to the female part of hermaphroditic worms although it was weakly expressed in the male gonad too (Karashima et al., 2000). This circumstance was interpreted as a switch from an original male to a female function and thought to be causally associated with the invention of amoeboid sperm in the nematode (Haag, 2001). Further, DAZ-1 is pivotal to execute the sperm/oocyte switch (Otori et al., 2006). Recently, the boule-orthologues of medaka and of rainbow trout were shown to be present in both sexes in the same species (Li et al., 2011; Xu et al., 2009).
All studies so far have been performed on vertebrates or in D. melanogaster and C. elegans. In humans, it has been shown that DAZ gene family members are involved in the transition from embryonic stem cells to primordial germ cells (Cauffman et al., 2005; Kee et al., 2009). However, nothing is known on the expression and function of boule-like genes in more basal organisms.
Sexually reproducing flatworms are especially suitable to study germ cell differentiation. The exceptional regenerative capacity of planaria and other flatworms allows them to reconstitute germ cells from somatic stem cells during regeneration (De Mulder et al., 2009, 2010; Egger et al., 2006; Handberg-Thorsager and Salo, 2007; Newmark et al., 2008; Pfister et al., 2008; Sato et al., 2006; Wang et al., 2007; Zayas et al., 2005).
The flatworm M. lignano is an emerging model system in developmental and evolutionary studies (De Mulder et al., 2009, 2010; Egger et al., 2009a, b; Janicke and Schärer, 2009a; Ladurner et al., 2000, 2005a, 2008; Mouton et al., 2009; Pfister et al., 2008; Sekii et al., 2009; Vizoso et al., 2010; Vizoso and Schärer, 2007). As a member of the Platyhelminthes, M. lignano possesses an extraordinary stem cell system that is also responsible for its high power of regeneration (Egger et al., 2006). M. lignano is small in size (up to 1.5 mm), it is highly transparent, can easily be cultured under laboratory conditions, and exhibits comparatively simple organisation of tissues and organs. It is an obligatory cross-fertilising hermaphrodite that produces eggs throughout the whole year in laboratory cultures. Several cell biological tools such as BrdU labelling (Ladurner et al., 2000), monoclonal antibodies (Ladurner et al., 2005a) or molecular methodologies including in situ hybridization and RNA interference are established (Pfister et al., 2007, 2008). More than 20,000 ESTs are available online (Morris et al., 2006) (http://flatworm.uibk.ac.at/macest), and there is an ongoing transcriptome and genome project (http://www.macgenome.org).
Here we report on the isolation and identification of three boule orthologues in the flatworm Macrostomum lignano. We investigated their expression and function and demonstrate that macbol1 RNAi specifically blocked spermatogenesis, whilstmacbol3 RNAi affected oogenesis and led to female sterile animals. Macbol2, however, did not show an RNAi phenotype. In addition, we have performed macbol1 rescue experiments in bol1-mutant flies to demonstrate functional conservation of boule regulation during evolution. In summary, our results provide evidence that boule genes can exhibit a function in male and female gametogenesis. Our findings demonstrate that flatworms are suitable models to study boule function and that these findings are relevant for higher organisms including humans.
Materials and methods
Animal culture
M. lignano was kept in petri dishes with artificial seawater inf/2medium and the diatom Nitzschia curvilineata as food source (see Ladurner et al., 2005b). Climate chambers were set at 60% humidity, 20 °C and a 14:10 day:night cycle (Rieger et al., 1988). D. melanogaster strains were cultured and crossed in 50 ml Eppendorf tubes at RT on standard fly medium (Lewis, 1960).
Identification and cloning of macbol1, macbol2, and macbol3
Macbol1 was identified by screening the M. lignano EST database (ANGU4194, http://flatworm.uibk.ac.at/macest) and confirmed in the M. lignano genome and transcriptome (scaffold scf2520075739037, http://www.macgenome.org), from which also macbol2 (scf2520075752006, RNA328_22493, RNA328_5088) and macbol3 (scf2520075751644, RNA918_15921) sequences were withdrawn. Sequence analyses were done by searching the GenBank database (http://blast.ncbi.nlm.nih.gov/Blast.cgi) with the blastx-tool. Alignments were made with multiple sequence alignment tool implemented in clustalw2 (http://www.ebi.ac.uk/Tools/clustalw2/), employing default parameters. Obvious alignment errors were hand corrected using GeneDoc software. Macbol1 was cloned full length. Its entire open reading frame was amplified with primers containing selected restriction sites: 5′-GACTACTAGTGCGAAATCATGACTGCTGATCTG-3′ and 5′-GACTACTAGTTTCAGTTGCCGTGCGAAGTATGG-3′ (restriction sites underlined). The resulting PCR-product had a size of 1277 bp. Gene-specific primer parts contained a start- or a stop-codon (superscripted letters), and corresponded to aa sequences of EIMTADL and HTSHGN-Stop. The resulting fragment was subcloned into pGEM-T vector (Promega) and sequenced. The gene was excised with enzymes targeting attached restriction sites and cloned into p-element based β2-tubulin testis expression vector (Hoyle and Raff, 1990) in preparation for fly injection. Accession numbers are HM222645 (macbol1), JF911416 (macbol2), and JF911417 (macbol3).
Phylogenetic analyses
For phylogenetic analyses, we downloaded fifteen additional Boule and DAZL sequences from GenBank. Like Xu et al. (2001), we have chosen HRP 1 from Saccharomyces cerevisiae as outgroup because it belongs to the same RNA binding protein family as Boule and DAZL (Shah et al., 2010) (see Fig. 1 for GenBank accession numbers). Sequences were aligned with Mafft version 6 (Katoh and Toh 2008), using the JTT PAM100 matrix. For Bayesian inference of phylogeny, we used MrBayes 3.1 (Ronquist and Huelsenbeck 2003). Four metropolis-coupled Monte Carlo Markov Chains were run in parallel for 2,000,000 generations (burnin = 0.1). Also, a maximum likelihood analysis was performed in Paup* (Swofford 2003), applying the WAG + Γ model; the confidence of the phylogenetic hypotheses was assessed with 100 bootstrap replicates.
Fig. 1.
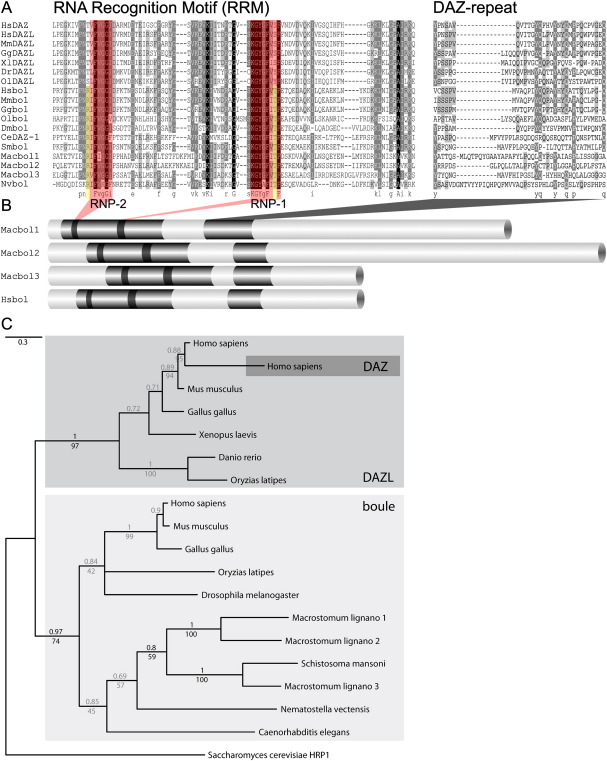
Alignment of DAZ family RRMs and DAZ-repeats of different species, comparative protein domain organisation, and phylogenetic analyses of RRM sequences. (A) Alignment of DAZ family RRM sequences and of DAZ-repeats; the highly conserved motifs RNP-1 and RNP-2 are shaded in pink. The two amino acids specific for all boule orthologues are highlighted in yellow. Species abbreviations: Hs, Homo sapiens (DAZ: AAB02393; DAZL: NP_001342; bol: AAK58689); Mm, Mus musculus (DAZL: AAH99940; bol: AAK69026); Gg, Gallus gallus (DAZL: NP_989549; bol: XP_421917); Xl, Xenopus laevis (AAH97658); Dr, Danio rerio (CAM56544); Ol, Oryzias latipes (DAZL: NP_001098269; bol: ACU31026); Dm, Drosophila melanogaster (Q24207); Ce, Caenorhabditis elegans (AAK68389); Sm, S. mansoni (XM_002575473); Mac, Macrostomum lignano (bol1: HM222645; bol2: JF911416; bol3: JF911417); Nv, N. vectensis (Stellabase ID: SB_53534); accession numbers are IDs for NCBI database unless indicated otherwise. (B) position of conserved motifs within M. lignano Macbol1, Macbol2 and Macbol3 and human Hsbol. (C) Phylogeny of DAZ family members using conserved RRM sequences as depicted in (A); the tree is based on a Bayesian inference analysis with Mr. Bayes. Numbers above the branches indicate Bayesian posterior probabilities, and numbers below the branches are maximum likelihood bootstrap values (100 replicates; WAG + Γ model); note that all three Macbol genes are located within the boule-clade, clustered together with the parasitic flatworm S. mansoni but are split up according to their role in male or female gametogenesis.
Whole-mount in situ hybridisation (ISH)
In situ riboprobes were generated using the DIG RNA labelling KIT SP6/T7 (Roche). The PCR templates used for probe synthesis were amplified with the primers 5′-CCTCTGCGACTGAGACAGTAA-3′ and 5′-GCCGGAGCGTAGTAAAGTATG-3′ (macbol1), 5′-TGAGAATCGCATTTTTGTGG-3′ and 5′-TCGAAGTTGACGTGGAAGC-3′ (macbol2) and 5′-AGTCGCCAGCTGTCTCACC-3′ and 5′-CCGCCTAGATCGGAATACC-3′ (macbol3). PCR conditions were initially 94 °C for 3 min, 35 cycles (94 °C: 30 s, Tm: 30 s and 72 °C: 1 min), and 72 °C for 5 min with Tm = 58 °C (macbol1) or Tm = 55 °C (macbol2 and macbol3). The resulting probes had a length of 885 bp, 566 bp and 371 bp, respectively, and were applied in a final concentration of 0.025 ng/μl according to the M. lignano ISH protocol (Pfister et al., 2007). Colour development was done in the dark for 45 min at 37 °C using an NBT/BCIP system (Roche). Specimens were analysed and photographed using DIC optics on a Leica DM5000 light microscope equipped with Pixera Penguin 600CL and Leica DFC 490 digital cameras. Figures were assembled and edited with Photoshop 7.0 software.
Preparation of in situ hybridisation sections
Macbol2 and macbol3 RNAi experiments and consecutive whole mount in situ hybridisations on treated and control animals were performed as described. After visualisation with NBT/BCIP, specimens were washed in PBS and fixed in BOUIN's fluid or 2.5% glutardialdehyde in PBS, dehydrated in standard ethanol series and embedded in Spurr's low viscosity resin (Spurr, 1969). Serial sections were assembled using a Diamond Histo Butler knife (Diatome, Switzerland), mounted, and examined with a Leica DM5000B microscope.
Immunocytochemistry
Macbol1 antibody was raised in rabbit against the peptide CGRGDGRPGLKYMDH (aa 398–412) by GenScript (New Jersey, USA). MSp-1 monoclonal antibody was used to stain spermatids (Ladurner et al., 2005a). Specimens were incubated with both antibodies in single and double staining at 4 °C overnight in diluted 1:1800 (Macbol1) and 1:200 (MSp-1) in BSA-T (PBS with 1% albumin fraction V (Roth) and 0.1% Triton X-100). Secondary antibodies, swine anti-rabbit FITC-labelled (DAKO) and goat anti-mouse Rhodamine-labelled (Rockland), diluted 1:200 and 1:500 in BSA-T, respectively, were applied for 1 h at RT. Photos were taken using either a Leica DM5000 light microscope or a Zeiss LSM 510.
Immunohistochemistry
EnVision + System (Dako) colour development was applied in preparation for semithin sectioning. M. lignano were relaxed in 7.14% MgCl2 for 15 min and fixed in PFA-sucrose (4% paraformaldehyde + 9% sucrose) for 30 min. Specimens were washed 3 × 5 min with PBS-T (PBS with 0.1% Triton X-100) and incubated in peroxidase-blocking reagent (Dako) for 10 min. Blocking of non-specific binding sites was done with BSA-T for 30 min, and primary antibody (Macbol1) was applied and diluted 1:1800 overnight at 4 °C. Visualisation was performed using DAB+ as chromogen (Dako), according to the manufacturer's protocol. Dehydration was done with standard ethanol series. Specimens were then embedded in Spurr's low viscosity resin (Spurr, 1969). Blocks were trimmed and sectioned serially on a Reichert Autocut 2040 microtome with a Diamond Histo Butler knife (Diatome, Switzerland). Sections were counterstained with Richardsons' dye (Richardson et al., 1960), mounted, and examined with a Leica DM5000B light microscope.
Electron microscopy
After three weeks of RNAi, two adult specimens of each treatment group (macbol1, macbol2, and macbol3) and two control worms of the adult group were relaxed in 7.14% MgCl2 and fixed in glutaraldehyde in 0.1 M cacodylate buffer (pH 7.4) for 1 h at 4 °C. Postfixation was performed for 1 h at RT in 1% osmium tetroxide in 0.05 M cacodylate buffer after several washing steps with buffer. Additional washing steps with buffer were done prior to dehydration in ascending ethanol series and embedding in Spurr's low viscosity resin (Spurr, 1969).
Specimens were cut with the prototype of a Butler diamond knife (Diatome), yielding complete series of 1.0 μm sections that were mounted on glass slides. After drying for 2 h at RT, one section series of each experimental group was stained with Richardsons' methylene blue Azur II resin (Richardson et al., 1960), mounted with cedar wood oil and examined for control with a Leica 5000B light microscope. One wild type animal and two animals of each RNAi group were cut with a diamond knife on an Ultracut S ultramicrotome (Leica), yielding complete ultrathin section series. Subsequent double staining with uranyl acetate and lead citrate was performed. Specimens were examined on a ZEISS Libra 120 energy filter electron microscope. Photo-documentation and analysis were done using a 2 k Vario Speed SSCCD camera (Droendle) and the iTEM software (TEM imaging platform, Olympus).
Fly injection, crossing and testis analysis
Injection of wild type Drosophila embryos w1118 with the macbol1 construct was performed by BestGene (USA). Backcrossing into the bol1-mutant background (P{ry[+ t7.2] = PZ}bol[1] ry[506]/TM3, ry[RK] Sb[1] Ser[1]}) and analyses of transformants were performed at the Department of Molecular, Cell and Developmental Biology, UCLA, USA, and at the Department of Ultrastructural Research and Evolutionary Biology at the University of Innsbruck, Austria. For analyses, fly testes were dissected and Macbol1 antibody staining was performed as previously described (Ladurner et al., 2005a). For double staining, DAPI was applied 1:1000 along with the secondary antibody. gDNA was purified with DNeasy® Tissue Kit (QIAGEN) and fragments amplified with the PCR primers 5′-AGCAGATGCGAACAAAAC-3′ and 5′-CCGAAAATAAGCGGAGAGC-3′ for wild type fly boule, 5′-CGCAGTCGTCGTCTCTTTC-3′ and 5′-GCCCAGAAAAGTCAGCAAG-3′ for bol1, 5′-CCTCTGCGACTGAGACAGTAA-3′ and 5′-GCCGGAGCGTAGTAAAGTATG-3′ for macbol1, and 5′-GACCTACGCAGAAAGCTAGCG-3′ and 5′-AAGCGGACGGTAAGATCCAC-3′ for GAPDH.
Postembryonic development and regeneration
To obtain exactly staged worms for analysis of postembryonic development eggs from adult M. lignano were collected and surveyed until the embryos hatched. Hatchlings were kept under regular culture conditions until they reached the desired time points. For regeneration experiments, adult M. lignano were cut between pharynx and testes to remove the gonads completely. Cut specimens were allowed to regenerate missing parts until the defined time points. Specimens were relaxed in 7.14% MgCl2, fixed and frozen in methanol at − 20 °C and then processed for in situ hybridisation and Macbol1 antibody staining. During Macbol1 antibody staining, we analysed 34, 37, 39, 21, 22, 21, 29, 43, and 54 animals at days 1, 2, 3, 4, 5, 6, 7, 10 and 16, respectively, of postembryonic development. At the same time, points 24, 23, 20, 46, 45, 45, 40, 15, and 17 regenerates, respectively, were checked.
RNA interference (RNAi)
RNAi was performed during postembryonic development and regeneration for macbol1 and during homeostasis of M. lignano for macbol1, macbol2, and macbol3 by soaking the animals in a solution of dsRNA (3.0 ng/μl) in f/2 culture medium. dsRNA was generated with an in vitro transcription system (RiboMax™ Large Scale RNA Production System T7, Promega), using the following primer sets: ′5-GGATCCTAATACGACTCACTATAGGCCTCTGCGACTGAGACAGTAA-3′ + ′5-GCCGGAGCGTAGTAAAGTATG-3′ and 5′-CCTCTGCGACTGAGACAGTAA-3′ + 5′-GGATCCTAATACGACTCACTATAGGGCCGGAGCGTAGTAAAGTATG-3′ (macbol1), ′5-GGATCCTAATACGACTCACTATAGGTGAGAATCGCATTTTTGTGG-3′ + ′5-TCGAAGTTGACGTGGAAGC-3′ and 5′-TGAGAATCGCATTTTTGTGG-3′ + 5′-GGATCCTAATACGACTCACTATAGGTCGAAGTTGACGTGGAAGC-3′ (macbol2), and ′5-GGATCCTAATACGACTCACTATAGGAGTCGCCAGCTGTCTCACC-3′ + ′5-CCGCCTAGATCGGAATACC-3′ and 5′-AGTCGCCAGCTGTCTCACC-3′ + 5′-GGATCCTAATACGACTCACTATAGGCCGCCTAGATCGGAATACC-3′ (macbol3). During RNAi, animals were incubated in 250 μl dsRNA in groups of 25, and supplied with food in 24-well plates. RNAi solution was changed once a day. In previous studies, it was shown that luciferase RNAi treated control animals did not show any mock effect (De Mulder et al., 2009; Pfister et al., 2008; Sekii et al., 2009). Therefore, in the present study, control animals were only kept with 250 μl f/2 culture medium. All solutions were treated with ampicillin (1:1000) and kanamycin (1:1000) prior to application. During analyses of macbol1 RNAi, each batch of 25 worms/well was split into 10, 10 and 5 specimen for in situ hybridisation (ISH), antibody/spermatid staining and interference contrast (IC) microscopy, respectively. After three weeks two adult worms each, treated and control, were prepared for transmission electron microscopy and only three analysed by interference contrast imaging. Macbol2- and macbol3-treated batches were only split in 20, 3, and 2 specimen for ISH, IC, and TEM, respectively.
Results
Identification and characterization of macbol genes
Full-length M. lignano boule paralogues macbol1, macbol2 and macbol3 were obtained from the M. lignano EST, genome and transcriptome databases. Macbol1, Macbol2 and Macbol3 have an ORF of 417, 503, and 282 amino acids, respectively, and comprise the two characteristic domains of DAZ family members, an RNA recognition motif (RRM) and the DAZ-repeat (Fig. 1A). The three boule genes show high sequence similarity within the conserved RNP-domains. In our alignment, two amino acids appeared characteristic for all boule proteins (Fig. 1A). In addition, we have performed phylogenetic analyses (Fig. 1C) that revealed two well-supported clades, a DAZL and a boule clade. The three boule sequences retrieved from M. lignano clearly clustered within the boule clade in all analyses (Fig. 1C). Further, there is only one DAZ-repeat present in Boule- and DAZL orthologues but not in DAZ proteins, which contain seven repeats in humans (Reijo et al., 1995). Accordingly, Macbol1, Macbol2, and Macbol3 possess one DAZ-repeat only, notably with only few conserved amino acids (Fig. 1A). However, a modest homology was also observed for the DAZ-repeat when human family members were compared with the fly homologue (Xu et al., 2001). When the cnidarian Nematostella vectensis, the parasitic flatworm Schistosoma mansoni, or the nematode C. elegans were included into the alignment, the DAZ-repeat showed only weak sequence homology (Fig. 1A). However, the overall protein domain organisation of Boule-like proteins is conserved and is consistent with the M. lignano boule domain structures (Fig. 1B). In summary, the conserved amino acids within the RRM, the structural assembly of its conserved domains, and the overall high amino acid identity of the RRM to other Boule proteins, specify M. lignano boule-like genes as true bouleorthologues.
Macbol1 is located in primary spermatocytes
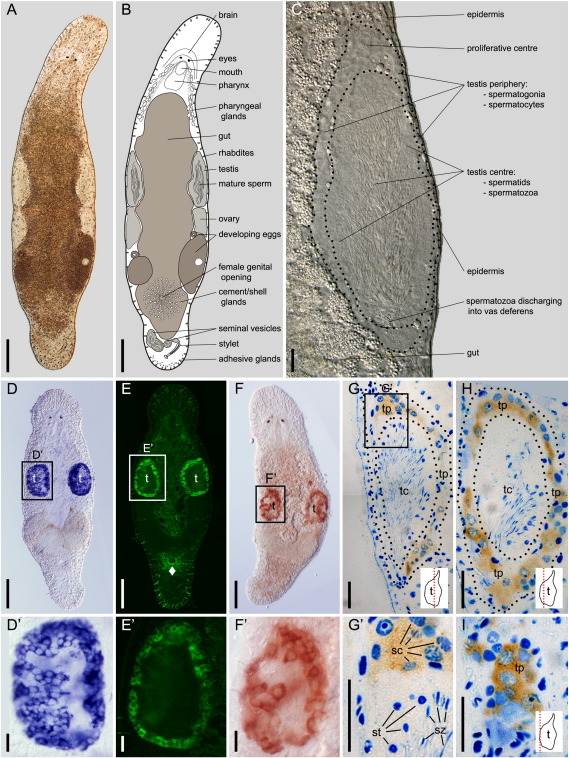
M. lignano (Figs. 2A, B) possesses paired testes and ovaries at the lateral mid-body region. The testes are elongated oval structures (Fig. 2C), surrounded by thin somatic tunica cells and extra cellular matrix. The testis periphery consists of a small number of spermatogonia and numerous spermatocytes I + II (Fig. 2C). This peripheral region is most prominent at the anterior tip (the proliferative centre) that ends in a pointed anterior extension.
Fig. 2.
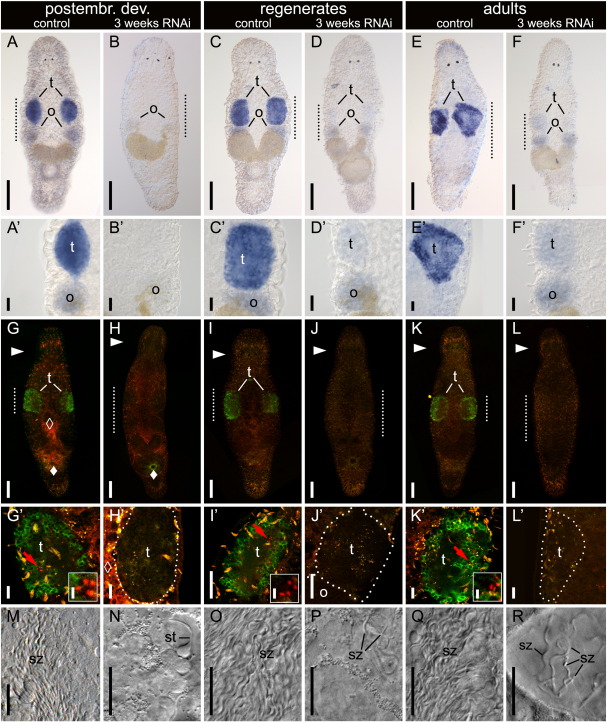
M. lignano morphology and macbol1-expression. (A) interference contrast (IC) image of an adult worm with corresponding scheme in (B); (C) IC image of adult testis with dotted lines bordering the different zones of spermatogenesis; (D, D′) macbol1 in situ hybridization; (E, E′) Macbol1 protein localisation using FITC-labelling; (F, F′) horseradish peroxidase staining; (G–I) histological semi-thin sections of Macbol1 antibody stained testis visualised using horseradish peroxidase (brownish); cell nuclei are counterstained with Richardsons' dye (blue); dotted red lines in the insets indicate cutting planes within the testis; solid diamonds (♦) indicate background in the glands surrounding the female opening; sc, spermatocytes; st, spermatids; sz, spermatozoa; t, testes; tc, testis centre; tp, testis periphery; scale bars 100 μm in A, B, D, E, F; and 20 μm in C, D′, E′, F′–I.
The outer region of the testis centre where maturating spermatids are located (Fig. 2C) and the testis periphery appear rather compact, although towards the lumen vivid mature spermatozoa are present. They discharge into the vas deferens at the posterior end of the testes. Sperm is stored in the seminal vesicles within the tail plate.
To study macbol1 expression during homeostasis, we applied whole-mount in situ hybridisation (ISH) (Figs. 2D, D′) and antibody staining (Figs. 2E–I) in adult animals. Macbol1 ISH staining revealed a strong labelling within distinct cells at the testis periphery (Fig. 2D, D′). Specificity of ISH staining was confirmed by using macbol1 sense probe, which showed no signal.
Macbol1 protein was located in the cytoplasm of cells restricted to the periphery of the testes, as revealed by fluorescence labelling (Figs. 2E, E′) or horseradish peroxidase staining (Figs. 2F, F′). Histological semi-thin sections of the latter showed the presence of Macbol1 in spermatocytes I in the testis periphery (Figs. 2G–I). Spermatocytes I can be recognised by their nuclear morphology showing typical meiotic chromatin organisation representing leptotene or pachytene stages (Fig. 2G′). No signal was detected in spermatogonia or in later stages towards the testis centre.
Macbol1 expression during postembryonic development and regeneration
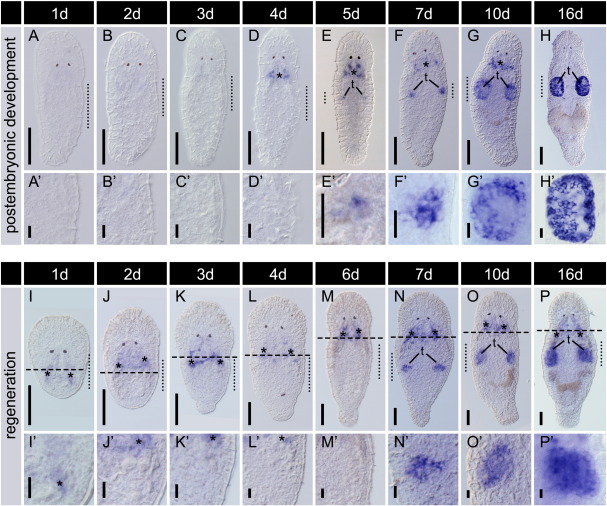
Groups of worms were examined from one to 16 days post hatching (dph). First ISH signals appeared in five days old animals in a group of testis cells (Figs. S1E, E′). The staining intensified after seven dph (Fig. S1F, F′) and positive cells were localised to the testis periphery at 10 days (Figs. S1G, G′). The overall morphology of adult testes was accomplished at day 16 (Figs. S1H, H′).
Fig. S1.
macbol1 mRNA expression during postembryonic development and regeneration. Days after hatching or amputation are indicated above experimental groups; (A–H′) postembryonic development; macbol1 mRNA was first detectable in five day old hatchlings in a cluster of cells (E, E′); (I–P′) regeneration; adult animals were cut behind the pharynx to completely remove the gonads; dashed lines indicate cutting levels (I–P); resulting head fragments were left for regeneration up to 16 days; first macbol1 signals were detected seven days after amputation (N, N′); dotted lines span the lateral region magnified in (A′–P′); unspecific background in pharyngeal glands is marked by asterisks (*); t, testes; scale bars 100 μm (A–P) and 20 μm (A′–P′).
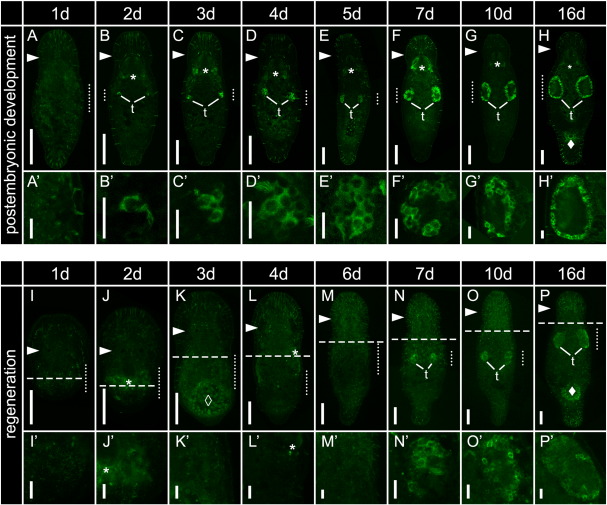
Macbol1 antibody staining revealed that no protein can be found in freshly hatched worms (n = 34) at one dph (Figs. S2A, A′). Macbol1 positive cells could be identified in four of 37 specimens at two dph (Figs. S2B, B′). We assume that the increased sensitivity of immunocytochemistry compared to ISH allowed the identification of the few macbol1 positive cells in these early stages. This observation is in accordance with histological serial sections of two days old hatchlings that exhibit primary spermatocytes in the testis anlage (Salvenmoser, unpubl.). A cluster of two to four Macbol1 positive cells was present in all (39) three days old hatchlings (Figs. S2C, C′). The size of the cluster increased with the progression of development and remained a compact mass of cells until day five (Figs. S2C–E′). From days seven to 10 a central cavity became apparent (Figs. S2F–G′).
Fig. S2.
Macbol1 protein localisation during postembryonic development and regeneration. Days after hatching or amputation are indicated above experimental groups; (A–H′) postembryonic development; Macbol1 labelled cells within testes (t) appeared first at day two post hatching (B, B′); (I–P′) regeneration; adult animals were cut behind the pharynx to completely remove the gonads; dashed lines indicate cutting levels (I–P); resulting head fragments were left for regeneration up to 16 days; note that Macbol1 positive cells within the testes were regenerated seven days after amputation (N, N′); dotted lines span the lateral region magnified in (A′–P′); arrowheads (►) indicate level of the eyes; unspecific background is marked by asterisks (*) in pharyngeal glands, by solid diamonds (♦) in the glands surrounding the female opening and by open diamonds (◊) in the gut; scale bars 100 μm (A–P) and 20 μm (A′–P′).
We next addressed the time course of the reappearance of macbol1 positive cells during regeneration. M. lignano is capable to fully rebuild testes and ovaries from the remaining pool of stem cells after gonads were completely removed by amputation (De Mulder et al., 2009; Egger et al., 2006; Pfister et al., 2008). We analysed macbol1 mRNA expression (Fig. S1I–P′) and Macbol1 protein localization (Figs. S2I–P′) from one to 16 days post amputation (dpa). Macbol1 protein was not detected until day six after amputation. Depending on the level of amputation, the time until complete regeneration varies (Egger et al., 2006). In the present experiment Macbol1 protein and macbol1 mRNA were localised in the testes from day seven on (panels N–P′ in Figs. S1 and S2).
Macbol1 RNAi obstructs meiotic progression
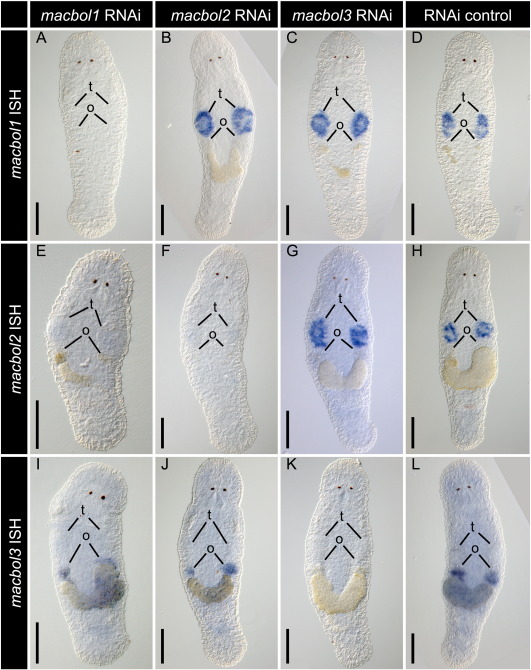
We have performed macbol1 RNA interference knock-down during postembryonic development, regeneration, and in adult animals. We analysed macbol1 RNAi effects using macbol1 ISH (panels A–F′ in Figs. 3, S3, S4), Macbol1 antibody staining (panels G–L in Figs. 3, S3, S4), spermatid labelling (panels G′–L′ in Figs. Figs. 3, S3, S4), and interference contrast microscopy (panels M–R in Figs. 3, S3, S4), after one week (Suppl. Fig. S3), two weeks (Suppl. Fig. S4), and three weeks (Fig. 4) of macbol1 RNAi treatment. In summary, our results demonstrate that macbol1 RNAi in every individual animal of all treatment groups effectively reduced macbol1 mRNA and led to a drastic deficiency in Macbol1 protein and a severe reduction in sperm production (for details see supplemental information). Macbol1 RNAi had no effect on the level of macbol3 expression (Suppl. 7I). Spermatocytes I, however, were arrested in response to macbol1 RNAi prior to the onset of macbol2 expression. Consequently, macbol2 ISH signals were not detected (Suppl. Fig. S7E).
Fig. 3.
Effect of three week macbol1 RNAi treatment on macbol1 expression, Macbol1 protein localisation, spermatid staining, and spermatozoa content in testes during postembryonic development, regeneration and in adult M. lignano. macbol1 expression (A–F′) is present in controls but lacking in animals treated for three weeks with macbol1 RNAi during postembryonic development (A–B′) and regeneration (C–D′), and was greatly reduced in adults (E–F′). Likewise, Macbol1 and spermatid specific MSp-1 antibody doublestaining (G–L′) is present in controls but lacking in macbol1 RNAi treated animals (compare green Macbol1 staining and red MSp-1 labelling in controls (G, G′, I, I′, K, K′) to RNAi affected testes (H, H′, J, J′, L, L′)); arrowheads (►) indicate level of the eyes; dotted lines mark the testes selected for magnification; unspecific background is indicated by solid diamonds (♦) in the gland cells surrounding the female opening and open diamonds (◊) mark autofluorescence of diatoms in the gut; red arrows point to the spermatid clusters enlarged in the respective insets; although the number of spermatozoa (M–R) was large in the testes of control animals (M, O, Q), only single spermatids (N) or spermatozoa (P, R) were observed in testes of macbol1 RNAi treated animals; o, ovaries; st, spermatids; sz, spermatozoa; t, testes; scale bars 100 μm (A–F, G–L), 20 μm (A′–F′, G′–R) and 5 μm (insets).
Fig. S3.
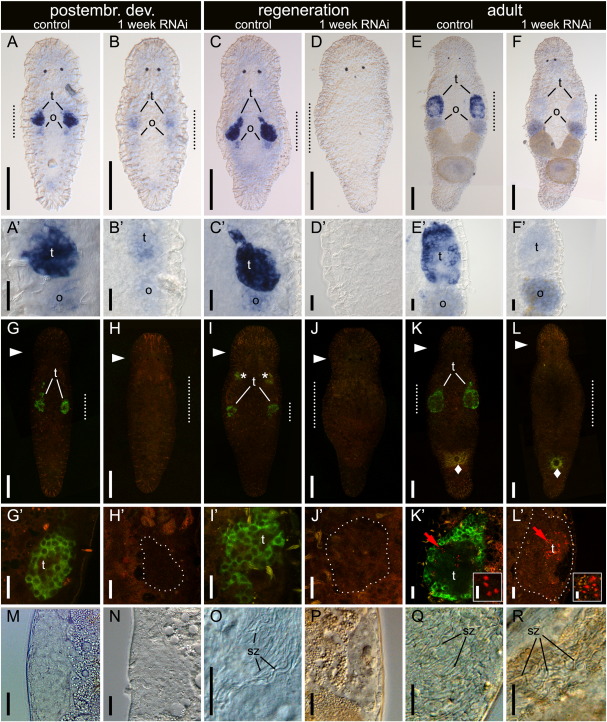
Effect of one week macbol1 RNAi treatment on macbol1 expression, Macbol1 protein level, spermatid staining, and spermatozoa content in testes during postembryonic development, regeneration and in adult M. lignano. macbol1 mRNA (A–F′) was present in controls but greatly reduced in macbol1 RNAi treated animals during postembryonic development (A–B′), almost absent during regeneration (C–D′), and again heavily reduced in adults (E–F′); likewise, Macbol1 antibody staining (G–L′) was present in controls (green staining in G, G′, I, I′, K, K′) but lacking in macbol1 RNAi treated animals (H, H′, J, J′, L, L′); spermatids (red labelling with M. lignano spermatid specific monoclonal antibody MSp-1) could be detected only in adults (K′, L′); arrowheads (►) indicate level of the eyes; unspecific background is indicated by solid diamonds (♦) in the gland cells surrounding the female opening; dotted lines mark the testes selected for magnification; red arrows point to the spermatid clusters enlarged in the respective insets; no spermatozoa were observed in testes of the “postembryonic development” group (M, N) and the macbol1 RNAi treated “regeneration” group (P) but were present in the regeneration control group (O) and in adults (Q, R); o, ovaries; sz, spermatozoa; t, testes; scale bars 100 μm (A–F, G–L), 20 μm (A′–F′, G′–M) and 5 μm (insets).
Fig. S4.
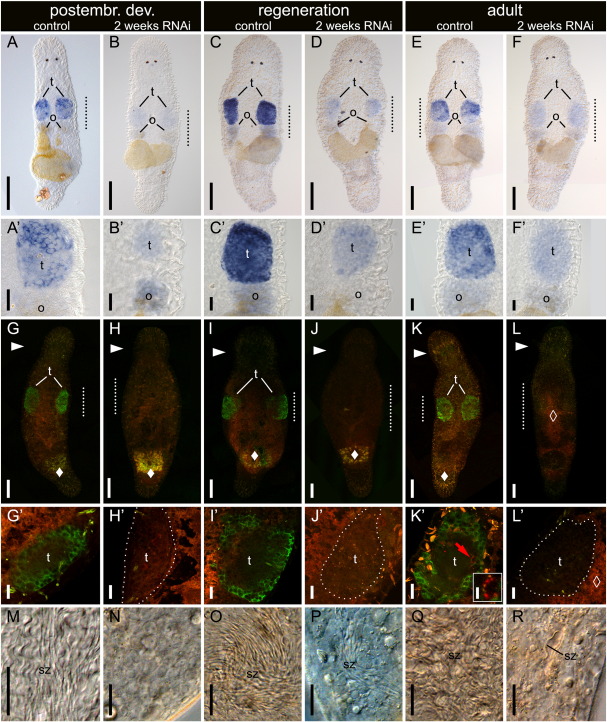
Effect of two week macbol1 RNAi treatment on macbol1 expression, Macbol1 protein level, spermatid staining, and spermatozoa content in testes during postembryonic development, regeneration and in adult M. lignano. macbol1 mRNA (A–F′) was present in controls but greatly reduced in macbol1 RNAi treated animals during postembryonic development (A–B′), during regeneration (C–D′), and in adults (E–F′); likewise, Macbol1 antibody staining (G–L′) was present in controls (green staining in G, G′, I, I′, K, K′) but lacking in macbol1 RNAi treated animals (H, H′, J, J′, L, L′); spermatids (red labelling with M. lignano spermatid specific monoclonal antibody MSp-1) could be detected only in adult controls (K′); arrowheads (►) indicate level of the eyes; unspecific background is indicated by solid diamonds (♦) in the gland cells surrounding the female opening and open diamonds (◊) mark autofluorescence of diatoms in the gut; dotted lines mark the testes selected for magnification; red arrow points to the spermatid cluster enlarged in the inset; although the number of spermatozoa (M–R) was large in the testes of control animals (M, O, Q), only few-to-none (N, P, R) were observed in the testes of macbol1 RNAi treated animals; o, ovaries; sz, spermatozoa; t, testes; scale bars 100 μm (A–F, G–L), 20 μm (A′–F′, G′–M) and 5 μm (insets).
Fig. 4.
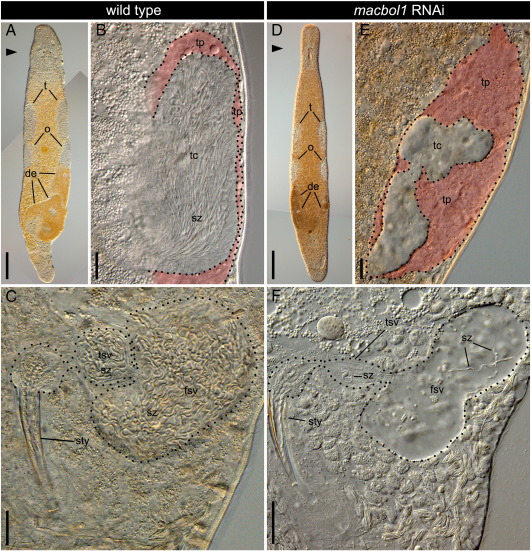
Testes and seminal vesicle comparison of wild type and macbol1 RNAi treated adult M. lignano by interference contrast microscopy. Testes (t) are present in both control (A) and macbol1 RNAi treated (D) animals; o, ovaries; de, developing eggs. (B) detail of wild type testis with a narrow testis periphery (tp) and the testis centre (tc) filled with spermatozoa (sz); (C) detail of seminal vesicles of a control animal; note that the true (tsv) and the false (fsv) seminal vesicle are full of spermatozoa; (E) detail of testis of a macbol1 RNAi treated animal; note the enlarged testis periphery and the empty testis centre; (F) detail of seminal vesicles of a macbol1 RNAi treated animal; note that the true (tsv) and the false (fsv) seminal vesicles are almost empty and contain only very few spermatozoa; scale bars 100 μm in A, D, and 20 μm in B, C, E, F.
Fig. S7.
Cross-effect of three weeks macbol1, macbol2 and macbol3 RNAi on macbol1, macbol2 and macbol3 mRNA expression. RNAi knock-down experiments were carried out in parallel for macbol1 (A, E, I), macbol2 (B, F, J), and macbol3 (C, G, K). Each RNAi-subset as well as an RNAi control group (D, H, L) was analysed by ISH staining for macbol1 (A–D), macbol2 (E–H) and macbol3 (I–L). Consistently, mRNA from the specific gene that was targeted by RNAi could never be visualised (A, F, K), whilst the remaining two riboprobes were able to detect the other genes; note that macbol2 ISH staining is negative in macbol1 RNAi treated specimens because spermatogenesis is blocked before macbol2 expression starts. Controls display the unaffected expression pattern of a gene; scale bars 100 μm.
We next analysed the effect of macbol1 RNAi on testis morphology and seminal vesicle content of adult animals. After three weeks of treatment, the overall size of the testes had not changed dramatically compared to control animals (Figs. 4A, D). However, squeezing preparations (n = 3) using interference contrast (IC) microscopy revealed that the testis periphery of control animals was normally sized, although it was greatly extended in RNAi animals. Furthermore, the testis centre in control animals was large, but it was small in size and devoid of mature spermatozoa after macbol1 RNAi (Figs. 4B, E). In control animals, the false seminal vesicle (basically a widening of the vas deferens) and the true seminal vesicle (which is surrounded by muscles to inject sperm into the stylet during copulation) contained a large number of spermatozoa (Fig. 4C). These structures almost completely lacked sperm in RNAi treated animals (Fig. 4F).
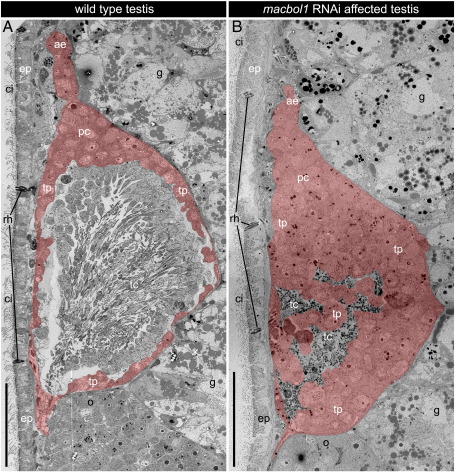
Two adult animals were processed for transmission electron microscopy to further address the question of which cell types were present in the enlarged testis periphery in macbol1 RNAi treated animals. The testis periphery of wild type animals comprised one to two cell layers consisting of spermatogonia and spermatocytes I + II. Only the proliferative centre was expanded and led into the narrow anterior extension (Fig. 5A) containing mostly spermatogonia and few early spermatocytes I. The testis centre comprised spermatids on the outer margin and gradually maturating spermatids further inwards. Mature spermatozoa were in the testis centre (Fig. 5A). In contrast, macbol1 RNAi led to an accumulation of spermatocytes I and a drastically enlarged testis periphery (Fig. 5B). The proliferative centre appeared unaffected. The small-sized testis centre did not contain any differentiated stages of spermatogenesis but debris of disintegrated cells (Fig. 5B and see below). We therefore conclude that macbol1 RNAi interferes with the progression of spermatogenesis.
Fig. 5.
Comparison of testis morphology of wild type and macbol1 RNAi treated adult animals by transmission electron microscopy. (A) The wild type testis centre is filled with late stages of spermatogenesis; The testis periphery is a slim layer containing spermatogonia and spermatocytes I, and only the anterior proliferating centre is expanded; (B) macbol1 RNAi leads to a drastic expansion of the testis periphery; the testis centre contains only degrading cells; ae, anterior extension; ci, cilia; ep, epidermis; g, gut; o, ovary; pc, proliferative centre; rh, rhabdites; tc, testis centre; tp, testis periphery; scale bars 50 μm.
We next determined which cell types in the testes were specifically affected by macbol1 RNAi treatment. Consequently, we compared the ultrastructure of successive stages of spermatogenesis (Fig. 6). It became apparent that spermatogonia (Figs. 6A, B) and early spermatocytes I (Figs. 6C, D) showed an unaffected morphology in macbol1 RNAi treated animals. Late spermatocytes I, however, exhibited a modified fine structure and showed clear signs of degeneration (Figs. 6E–H). They acquired a change in nuclear morphology with lobes and foldings. Therefore, in ultrastructural sections, cytoplasmic inclusions can be seen within the nucleus (Fig. 6F), marking the onset of morphological degeneration. Cells beyond the spermatocyte I stage appeared disintegrated and carried a highly aberrant cellular content with membrane fragments (Fig. 6H) and a fragmented nucleus.
Fig. 6.
Effect of macbol1 RNAi on successive stages of spermatogenesis examined by transmission electron microscopy. Spermatogonia (A) and early primary spermatocytes (C) appear unaffected in macbol1 RNAi treated animals (B, D). Later stages of spermatogenesis begin to change their nuclear shape (compare E, F) and are finally degraded (compare G, H); ad, advanced degradation; dc, degrading cell; sc I, primary spermatocytes; sc II, secondary spermatocytes; sg, spermatogonia; st, spermatids; arrowheads (►) point at cell membranes (black) or nuclear envelopes (white); scale bars 2 μm.
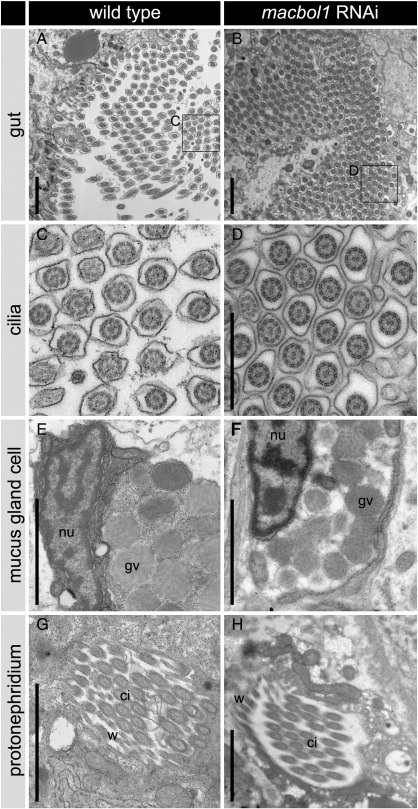
Detailed electron microscopical analyses revealed that macbol1 RNAi had no detectable effect on the morphology of cells and tissues such as nerve cells (Suppl. Figs. S5A–H), oocytes (Suppl. Figs. S5I, J), epidermal cells and muscle cells (Suppl. Figs. S5K, L), rhabdite forming cells (Suppl. Figs. S5M, N), gut cells and their associated cilia (Suppl. Figs. S6A–D), mucus gland cells (Suppl. Figs. S6E, F), or protonephridial cells (Suppl. Figs. S6G, H). Finally, we determined reproductive success of the adult control versus RNAi-treated animals (n = 25/group) by counting freshly hatched worms. After three weeks of macbol1 RNAi, controls had fathered 30 but treated animals only nine offspring.
Fig. S5.
macbol1 RNAi treatment has no effect on the ultrastructural morphology of other different cell types. (A–H) nervous tissue; (I, J) oocyte; (K, L) epidermis with muscle fibres; (M, N), rhabdite forming cell; ci, cilia; cm, circular muscles; dcv, dense core vesicles; ldv, large dense vesicles; lm, longitudinal muscles; lv, lucent vesicles; mrv, membrane retrieval vesicles; nu, nucleus; nuo, nucleolus; rh, rhabdites; urh, ultrarhabdites; scale bars 100 nm (A–H) and 2 μm (I–N).
Fig. S6.
macbol1 RNAi treatment has no effect on the ultrastructural morphology of other different cell types, continued. (A, B) gut; (C, D) cilia; (E, F) mucus gland cell; ci, cilia; gv, gland vesicles; nu, nucleus; scale bars 2 μm (A, B, E, F) and 1 μm (C, D).
Macbol1 shows partial functional equivalency to Drosophila boule
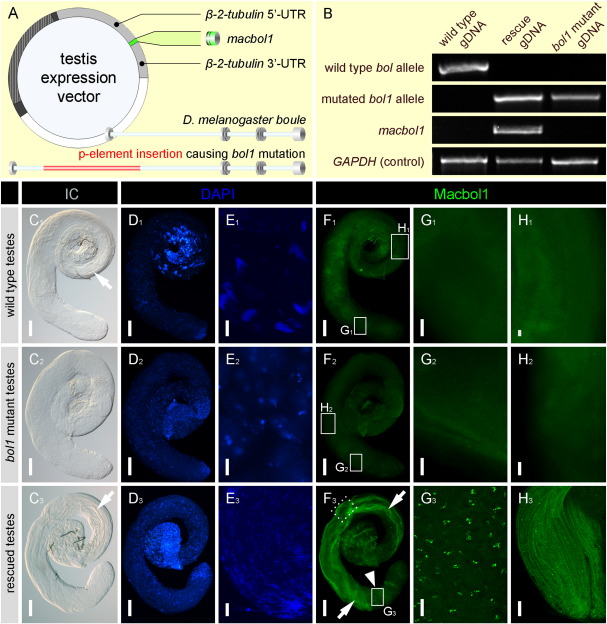
To address the evolutionary conservation of macbol1 function we introduced macbol1 into bol1-mutant Drosophila. We prepared a macbol1 carrying construct (Fig. 7A) for injection and generated a macbol1-transgenic bol1-mutant Drosophila strain. Testes (n = 10 each) from wild type, mutant and rescued flies were dissected and analysed by PCR, DAPI and Macbol1 antibody staining. PCR experiments with primers specifically detecting macbol1, D. melanogaster wild type boule, the bol1 mutation, or GAPDH were performed. These data corroborated that the rescue-phenotype could only result from the introduced flatworm gene (Fig. 7B). In wild type testes, interference contrast microscopy (Fig. 7C1) and DAPI staining (Figs. 7D1, E1) revealed numerous bundles of elongated spermatozoa and single sperm in the seminal vesicle. Macbol1 protein was not detected in wild type testes (Figs. 7F1–H1). In bol1 mutant testes no stages of spermatogenesis were observed (Figs. 7C2–E2) and Macbol1 was absent (Figs. 7F2–H2). In macbol1 rescue flies, bundles of sperm were observed in the testes (Fig. 7C3). However, the number was lower compared to wild type animals. Furthermore, in the proximal region of the testis tubule sperm nuclei were observed (Fig. 7D3). The shape of sperm nuclei was different from the wild type condition in that they were broader and not as sharply pointed (Fig. 7E3). Rescued testes stained with the Macbol1 antibody revealed a signal within the tubule (Fig. 7F3). In a distal testis area, Macbol1 positive cells were identified (Figs. 7F3 arrowhead; G3), probably accounting for spermatocytes. Tails of maturating spermatids and sperm were also labelled (Figs. 7F3 arrows; H3). When macbol1 carrying bol1-males were crossed to a wild type female, however, they were not able to produce offspring (n = 75). Therefore we conclude that macbol1 rescued the meiotic defect and sperm could be produced. Nevertheless, the final steps of spermatogenesis could not be completed and flies were infertile, a situation that was also observed when fly or human boulewas used to rescue bol1 mutant flies (Eberhart et al.; 1996, Xu et al., 2003).
Fig. 7.
Evolutionary conservation of boule function between platyhelminths and insects. macbol1 was introduced into male sterile Drosophila bol1-mutant; (A) scheme of the expression vector carrying macbol1; below: schematic drawing of the genomic organisation of Drosophila boule (exons are indicated as discs); in the bol1-mutant strain wild type fly boule is disrupted by a p-element insertion; (B) PCR from gDNA of wild type, mutant and rescued Drosophila with primers detecting wild type fly boule, bol1, macbol1 or GAPDH (positive control); wild type, rescued and mutant testes were analysed by interference contrast images (C1–C3), DAPI (D1–E3), and Macbol1 antibody staining (F1–H3); details E1–E3 show proximal region of testis tubules; arrowhead points at spermatocytes; arrows highlight elongated spermatids and sperm; solid squares frame regions enlarged from F1–F3; dotted square marks relative position of detail H3 which was taken from a testis other than F3; scale bars 100 μm (C1–D3, F1–F3) and 10 μm (E1–E3, G1–H3).
Macbol2 is expressed in spermatocytes II and spermatids
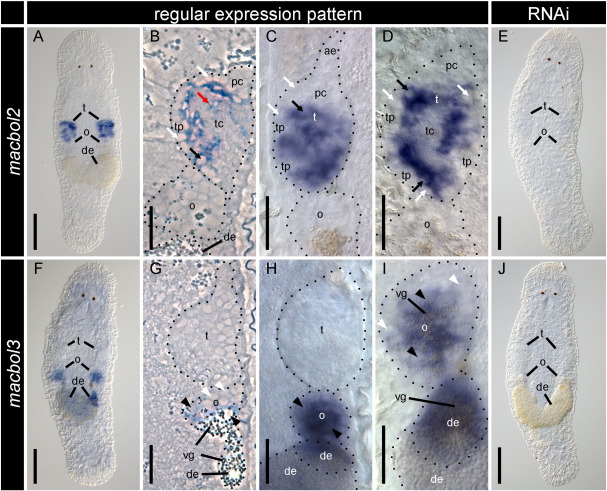
Whole mount ISH and histological sections revealed that the second M. lignano testis-specific gene, macbol2, was localised to spermatocytes II and spermatids (Figs. 8A–D). Spermatocytes II can be recognised by a large nucleus surrounded by a broad rim of cytoplasm (Fig. 8B). Spermatids are located more to the centre of the testis and show nuclear condensation and elongation of the cell. The proliferating centre and the anterior extension – regions that contain spermatogonia and spermatocytes I – lack any ISH signal (Figs. 8B–D). Likewise, the testis periphery and the testis centre did not show any macbol2 staining (Fig. 8D). After three weeks of macbol2 RNAi treatment, no macbol2 expression could be detected (Figs. 8E, S7F); however, no phenotype was observed. Accordingly, after three weeks, 20 offspring were counted for both, adult RNAi-treated and control worms. Macbol2 RNAi had no effect on the expression of macbol1 and macbol3 (Suppl. Figs. S7B, J).
Fig. 8.
Macbol2 and macbol3 expression pattern and RNAi treatment of adult M. lignano. macbol2 ISH was performed on wildtype (A–D) and macbol2 RNAi treated (E) adult specimens; macbol2 mRNA is confined to the testes (t) (A); as shown in semithin sections of ISH stained testes (B) and higher magnifications of the gonads (C, D) macbol2 is expressed in secondary spermatocytes (black arrows) and early spermatids (red arrows) but not in the testis centre (tc) and the testis periphery (tp) with its primary spermatocytes (white arrows), proliferative centre (pc), and anterior extension (ae); after three weeks of macbol2 RNAi mRNA was not detected (E); likewise macbol3 ISH on wildtype individuals as depicted in an overview (F), a semithin section (G) and a higher magnification of the gonads (H, I) revealed respective mRNA to be expressed in vitellogenic oocytes (black arrowheads) and developing eggs (de) but not in previtellogenic oocytes (white arrowheads); the staining fades with progression of egg development; macbol3 RNAi results in undetectable macbol3 ISH staining; o), ovaries; vg) vitelline granules; all images were taken using IC optics except for phase contrast panels B) and G); scale bars 100 μm in overviews, 20 μm in gonad magnifications.
Macbol3 regulates oocyte maturation
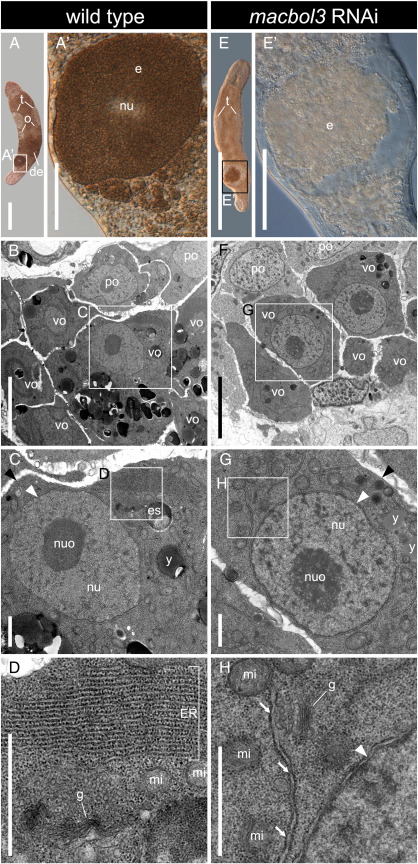
We found the third M. lignano boule ortholog to be expressed in oocytes and developing eggs (Figs. 8F–I). Macbol3 expression was lacking in oogonia and early oocytes, although it was present in later oocytes that commenced vitellogenesis (Figs. 8G–I). Macbol3 RNAi resulted in macbol3 knock-down (Fig. 8J, S7K), but did not show any cross effects on macbol1 and macbol2 (Suppl. Figs. S7C, G). Macbol3 RNAi treatment resulted in aberrant egg maturation and caused female sterility with complete penetrance within the treatment group (Figs. 9A, E). After three weeks of macbol3 RNAi, treated animals had not produced any offspring compared to 27 in the control group. Early stages of oogenesis like oogonia and early oocytes I seemed not to be affected by macbol3 RNAi treatment compared to the wild type control. The oocytes of macbol3 treated animals showed typical meiotic figures including synaptonemal complexes. Later stages such as oocytes I in vitellogenesis, however, showed distinct changes in their ultrastructure. As a striking difference, these oocytes lacked the prominent rER compartments (Figs. 9B–H). In addition, the cells appeared smaller and less developed in their cytoplasmic maturation (such as yolk and egg-shell granule production). Finally, egg maturation could not be completed and eggs exhibited an abnormal phenotype. Animals ceased egg laying and they became female sterile hermaphrodites.
Fig. 9.
Effect of macbol3 RNAi on egg development. Eggs stuffed with yolk (A, A′) are the result of wild type oogenesis (A–D), which requires granulogenesis in vitellogenic oocytes (B); yolk granules (C) are generated based on massive stacks of endoplasmic reticulum (D); Eggs produced during macbol3 RNAi (E–H) contain less yolk (E, E′); granulogenesis is scaled down (F) whilst cellular morphology is like in the wild type (compare G to C) except for heavily reduced endoplasmic reticulum, which is not present in stacks but in single layers (compare H to D); de, developing egg; e, egg; ER, endoplasmic reticulum; es, egg shell granule; g, golgi; mi, mitochondria; nu, nucleus; nuo, nucleolus; o, ovaries; po, previtellogenic oocytes; t, testes; vo, vitellogenic oocytes; y, yolk granules; black arrowhead, cell membrane; white arrowhead, nuclear envelope; white arrows, ER lamella; all images were taken using transmission electron microscopy except for interference contrast panels A, A′, E and E′; scale bars 500 μm (A, E), 100 μm (A′, E′), 5 μm (B, F), and 1 μm (C, D, G, H).
Discussion
Evolution of DAZ family proteins
Up to now, there is a complete lack of knowledge on the expression and function of boule-like genes in Lophotrochozoa. In M. lignano, we were able to identify three distinct boule paralogues. Previously, two potential boule paralogues have also been reported from the sea anemone N. vectensis. One of them perfectly matched a Boule protein consensus sequence (Shah et al., 2010), and the other showed both similarities and differences in critical positions. Nevertheless, there is the possibility that multiple boule paralogues – just as in our flatworm model – do exist already in Cnidaria. However, there are no indications that multiple boule genes are present in Deuterostomia. In M. lignano, all three reported boule paralogues match the consensus sequence of Shah et al. (2010). Our findings demonstrate that the paralogues corroborate the well-known function in male meiosis but also act during female gametogenesis. Nevertheless, the single and differently employed boule orthologues that we can identify today in various taxa like vertebrates, insects and nematodes do not necessarily represent the same ancestral gene. They could be the result of preservation of particular ancient paralogues and loss of the others. To follow up this hypothesis, more invertebrate species of different phyla, especially deuterostome invertebrates but also other cnidarian and flatworm species, should be investigated and potential boule sequences should be analysed and compared in detail.
Functional conservation of boule-like genes
Human BOULE's importance in female embryos and the notion that it may have a role in primordial germ cell (PGC) migration (Kee et al., 2009), together with expression of fish boule in PGCs (Xu et al., 2009), suggest an early role of boule in female germ cell development of vertebrates. As in D. melanogaster and C. elegans, we could neither detect PGC-specific boule expression for any of the paralogues of M. lignano, nor an additional paralogue in its genome. Taken together, these findings could either hint at a loss of DAZ family mediated regulation of PGCs in Protostomia or its evolution in Deuterostomia. In the latter scenario, however, the question of how an upstream regulatory function has been acquired remains elusive. However, although in fish and mice boule shows a certain degree of functional redundancy with Dazl (Li et al., 2011; VanGompel and Xu, 2010; Xu et al., 2009), in invertebrates, boule genes are indispensable for first meiotic divisions (Eberhart et al., 1996; Karashima et al., 2000). Molecular evolution may be rapid in reproductive genes (Wyckoff et al., 2000), which also seems to be the case in DAZ family members that have undergone two consecutive duplication events (Bielawski and Yang, 2001; Haag, 2001; Tung et al., 2006). The cause may be positive Darwinian selection or a relaxation of functional constraints (Bielawski and Yang, 2001). Overall, DAZ family proteins act during gametogenesis in all organisms studied so far. They promote translation initiation of target mRNAs with only a short poly-A-tail upon binding to their 3′-UTR. It has been shown that twine mRNA, the cdc25 homologue in Drosophila is a potential substrate for boule (Maines and Wasserman, 1999). Twine or stringshows differential expression in male and female gonads of D. melanogaster (Alphey et al., 1992; Courtot et al., 1992; Sigrist et al., 1995). In the M. lignano transcriptome (build 100918) we have identified four twine or string cdc25 phosphatases (RNA328_6352, RNA328_80531, RNA328_24244, RNA328_2512) that could be potential Macbol interaction partners. Furthermore, proteins such as PUM2 and PABP (Poly-Adenylation Binding Protein) in humans have also been shown to interact with Boule (Brook et al., 2009; Collier et al., 2005; Moore et al., 2003; Urano et al., 2005). Brook et al. (2009) gives a complete list of DAZ family protein interaction partners. According to a proposed model, interaction is mediated between the DAZ family protein's RRM and DAZ repeat and the response elements in the 3′-UTR of targeted mRNAs (Collier et al., 2005; Meagawa et al., 2002). Like poly-A tails, DAZ-family proteins are able to recruit PABPs that would not be attracted sufficiently by short poly-A tailed mRNAs alone. As a consequence, the 5′- and the 3′-UTR can be bridged by the numerous proteins of the initiation complex, form a “closed loop”, and can be joined by the ribosomal subunits. In general, the role of boule orthologues, i.e. to participate in the proper propagation of first meiotic divisions, seems to be conserved throughout the animal kingdom (Luetjens et al., 2004). This is underlined by the fact that both human BOULE (Xu et al., 2003) and macbol1 are able to substitute to the same extent for the defect allele in Drosophila. These cross-species rescues could indeed reflect conservation of boule function. Alternatively, biochemical interactions that are not based on evolutionary relation of boule genes could be responsible for the partial rescue. However, to fundamentally understand such rescue experiments, a detailed knowledge on the molecular function of boule proteins in these species would be required. When introduced into the invertebrate background, vertebrate Xdazl can also act during meiosis (Houston et al., 1998). It is thought that meiosis represents an evolutionary bottleneck during male gamete production (Xu et al., 2003), and boule and DAZL seem to be under similar functional constraints (Tung et al., 2006). Despite the number of identified interacting molecules, virtually nothing is known about factors that specifically direct DAZ family mediated stimulation into male and/or female gametogenesis in M. lignano and other organisms.
Comparison of expression and function of macbol genes
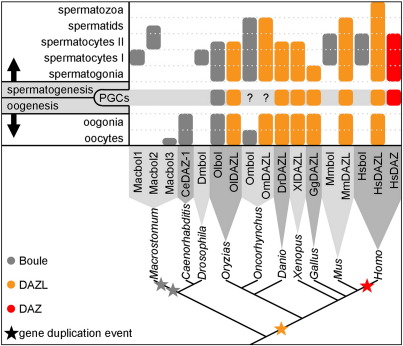
Concerning macbol1 and macbol2 expression, our data line up M. lignano with most other animals investigated so far, since boule genes are thought to be mainly involved in male meiosis (Cheng et al., 1998; Eberhart et al., 1996). Macbol1 mRNA and Macbol1 protein were exclusively found in primary spermatocytes whilstmacbol2 was expressed in secondary spermatocytes and early spermatids (Fig. 10). Likewise, boule-like genes have been shown to be vital players in the process of male gamete production in fly (Eberhart et al., 1996), bovinae (Zhang et al., 2009), mouse (VanGompel and Xu, 2010) and human (Luetjens et al., 2004; Xu et al., 2001). Moreover, BOULE isoforms were detected in the testes of four bat species (Yuan et al., 2009). However, in mice, boule is essential in the wake of meiosis for spermatid differentiation (VanGompel and Xu, 2010).
Fig. 10.
Comparison of DAZ family gene expression patterns during gametogenesis of diverse genera. So far, DAZ family genes were not detected in PGCs of organisms other than vertebrates; only genes with expression patterns confirmed by staining were included in this figure; expression data after Xu et al. (2009) and Brook et al. (2009) were complemented with data from Macrostomum and chicken; note that the depicted phylogenetic tree only displays the relationship between genera but not exact evolutionary distances.
We now found macbol3 to be expressed in the ovaries and developing eggs of M. lignano. From the three boule genes that so far have been identified during oogenesis – the orthologues of medaka, rainbow trout, and C. elegans (Fig. 10) – only the nematode's gene was demonstrated to be functionally refined to egg production. However, there was also faint staining in male gonads (Karashima et al., 2000; Otori et al., 2006). Thus, macbol3 represents the first boule homologue that, in expression and function, is specific to oogenesis only. It is therefore evident that different boule genes regulate male and female meiosis in M. lignano.
Germline specificity of macbol paralogues
The experimental accessibility of germ cell differentiation in flatworms (De Mulder et al., 2009, 2010; Egger et al., 2006; Handberg-Thorsager and Salo, 2007; Pfister et al., 2008; Sato et al., 2006; Sekii et al., 2009; Wang et al., 2007; Zayas et al., 2005) during regeneration and postembryonic development renders these animals as model systems to study the function of DAZ family genes during the stem cell — germ line transition and germ cell differentiation. Due to M. lignano's translucent body, its germline is amenable to experimentation and perfectly visible even in living adult specimen. Hence, the animal is used as a model for sex allocation studies — the differential investment into male and/or female function (Brauer et al., 2007; Janicke and Schärer, 2009b; Schärer, 2009; Vizoso and Schärer, 2007). It has previously been shown in M. lignano that an RNAi knock-down of melav2, a gene involved in spermatid differentiation, can be used to alter gonadal development (Sekii et al., 2009). In contrast, genes such as vasa and piwi, although well characterized in germline development in many organisms, cannot serve as gonad specific markers in flatworms because they have also been shown to be present in somatic stem cells (De Mulder et al., 2009; Palakodeti et al., 2008; Pfister et al., 2007, 2008; Reddien et al., 2005; Rossi et al., 2006; Shibata et al., 1999). In M. lignano, our findings demonstrate the tight interdependency of macbol1 and meiotic divisions during spermatogenesis in adults during postembryonic development and regeneration. Our detailed ultrastructural analyses of germ cells and various other cells and structures did not reveal any role of macbol1 other than male meiotic control. Macbol1 RNAi led to male sterility whilstmacbol3 RNAi resulted in female sterile animals. Therefore, macbol1 and macbol3 would provide versatile tools to artificially alter the amount of energy dedicated to gametogenic cascades and to address reproductive biology in M. lignano. As a corollary, this will also change our understanding of boule's significance for general animal reproduction.
Conclusions
The recent success in sequencing the genomes of diverse invertebrate species will be the next step in answering important evolutionary questions. As we have demonstrated, the DAZ family homologues of M. lignano are true boule genes by primary sequence, domain structure and function. Further, we report for the first time different boule homologues that direct oogenesis and spermatogenesis in the same animal. Because M. lignano is the only invertebrate species where boule genes are essential for both gametogenic pathways, the animal is amenable for genetic manipulation, and there is an ongoing genome and transcriptome project that will provide access to potential Boule interaction partners, and the M. lignano boule paralogues are especially suited to study DAZ gene family evolution and the role of boule during male and female reproduction and (in)fertility.
The following are the supplementary materials related to this article.
Supplementary material.
Acknowledgments
The authors especially want to thank L. Mandal for help with the fly rescue experiment. Further, we want to thank F. Marx for support in gene isolation, A. Lusser for help with fly biology. This work was supported by FWF grant 18099 to P. L. (Austria), an FWO grant to K. D. M. (Belgium), and SPA/02 - 81/Plattwürmer to G.K.
Contributor Information
Georg Kuales, Email: Georg.Kuales@uibk.ac.at.
Katrien De Mulder, Email: katrien.demulder@ugent.be.
Jade Glashauser, Email: Jade.Glashauser@student.uibk.ac.at.
Willi Salvenmoser, Email: willi.salvenmoser@uibk.ac.at.
Shigeo Takashima, Email: stakashima@ucla.edu.
Volker Hartenstein, Email: volkerh@mcdb.ucla.edu.
Walter Salzburger, Email: walter.salzburger@unibas.ch.
Peter Ladurner, Email: Peter.Ladurner@uibk.ac.at.
References
- Alphey L., Jimenez J., White-Cooper H., Dawson I., Nurse P., Glover D.M. twine, a cdc25 homolog that functions in the male and female germline of Drosophila. Cell. 1992;69:977–988. doi: 10.1016/0092-8674(92)90616-k. [DOI] [PubMed] [Google Scholar]
- Bielawski J.P., Yang Z. Positive and negative selection in the DAZ gene family. Mol. Biol. Evol. 2001;18:523–529. doi: 10.1093/oxfordjournals.molbev.a003831. [DOI] [PubMed] [Google Scholar]
- Brauer V.S., Schärer L., Michiels N.K. Phenotypically flexible sex allocation in a simultaneous hermaphrodite. Evolution. 2007;61:216–222. doi: 10.1111/j.1558-5646.2007.00018.x. [DOI] [PubMed] [Google Scholar]
- Brook M., Smith J.W., Gray N.K. The DAZL and PABP families: RNA-binding proteins with interrelated roles in translational control in oocytes. Reproduction. 2009;137:595–617. doi: 10.1530/REP-08-0524. [DOI] [PubMed] [Google Scholar]
- Carani C., Gromoll J., Brinkworth M.H., Simoni M., Weinbauer G.F., Nieschlag E. cynDAZLA: a cynomolgus monkey homologue of the human autosomal DAZ gene. Mol. Hum. Reprod. 1997;3:479–483. doi: 10.1093/molehr/3.6.479. [DOI] [PubMed] [Google Scholar]
- Castrillon D.H., Gonczy P., Alexander S., Rawson R., Eberhart C.G., Viswanathan S., DiNardo S., Wasserman S.A. Toward a molecular genetic analysis of spermatogenesis in Drosophila melanogaster: characterization of male-sterile mutants generated by single P element mutagenesis. Genetics. 1993;135:489–505. doi: 10.1093/genetics/135.2.489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cauffman G., Van de Velde H., Liebaers I., Van Steirteghem A. DAZL expression in human oocytes, preimplantation embryos and embryonic stem cells. Mol. Hum. Reprod. 2005;11:405–411. doi: 10.1093/molehr/gah167. [DOI] [PubMed] [Google Scholar]
- Cheng M.H., Maines J.Z., Wasserman S.A. Biphasic subcellular localization of the DAZL-related protein boule in Drosophila spermatogenesis. Dev. Biol. 1998;204:567–576. doi: 10.1006/dbio.1998.9098. [DOI] [PubMed] [Google Scholar]
- Collier B., Gorgoni B., Loveridge C., Cooke H.J., Gray N.K. The DAZL family proteins are PABP-binding proteins that regulate translation in germ cells. EMBO J. 2005;24:2656–2666. doi: 10.1038/sj.emboj.7600738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cooke H.J., Lee M., Kerr S., Ruggiu M. A murine homologue of the human DAZ gene is autosomal and expressed only in male and female gonads. Hum. Mol. Genet. 1996;5:513–516. doi: 10.1093/hmg/5.4.513. [DOI] [PubMed] [Google Scholar]
- Courtot C., Fankhauser C., Simanis V., Lehner C.F. The Drosophila cdc25 homolog twine is required for meiosis. Development. 1992;116:405–416. doi: 10.1242/dev.116.2.405. [DOI] [PubMed] [Google Scholar]
- De Mulder K., Pfister D., Kuales G., Egger B., Salvenmoser W., Willems M., Steger J., Fauster K., Micura R., Borgonie G., Ladurner P. Stem cells are differentially regulated during development, regeneration and homeostasis in flatworms. Dev. Biol. 2009;334:198–212. doi: 10.1016/j.ydbio.2009.07.019. [DOI] [PubMed] [Google Scholar]
- De Mulder K., Kuales G., Pfister D., Egger B., Seppi T., Eichberger P., Borgonie G., Ladurner P. Potential of Macrostomum lignano to recover from γ-ray irradiation. Cell Tissue Res. 2010;339:527–542. doi: 10.1007/s00441-009-0915-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eberhart C.G., Maines J.Z., Wasserman S.A. Meiotic cell cycle requirement for a fly homologue of human Deleted in Azoospermia. Nature. 1996;381:783–785. doi: 10.1038/381783a0. [DOI] [PubMed] [Google Scholar]
- Egger B., Ladurner P., Nimeth K., Gschwentner R., Rieger R. The regeneration capacity of the flatworm Macrostomum lignano — on repeated regeneration, rejuvenation, and the minimal size needed for regeneration. Dev. Genes Evol. 2006;216:565–577. doi: 10.1007/s00427-006-0069-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Egger B., Gschwentner R., Hess M.W., Nimeth K.T., Adamski Z., Willems M., Rieger R., Salvenmoser W. The caudal regeneration blastema is an accumulation of rapidly proliferating stem cells in the flatworm Macrostomum lignano. BMC Dev. Biol. 2009;9:41. doi: 10.1186/1471-213X-9-41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Egger B., Steinke D., Tarui H., De Mulder K., Arendt D., Borgonie G., Funayama N., Gschwentner R., Hartenstein V., Hobmayer B., Hooge M., Hrouda M., Ishida S., Kobayashi C., Kuales G., Nishimura O., Pfister D., Rieger R., Salvenmoser W., Smith J., Technau U., Tyler S., Agata K., Salzburger W., Ladurner P. To be or not to be a flatworm: the acoel controversy. PLoS One. 2009;4:e5502. doi: 10.1371/journal.pone.0005502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fuller M.T. Genetic control of cell proliferation and differentiation in Drosophila spermatogenesis. Semin. Cell Dev. Biol. 1998;9:433–444. doi: 10.1006/scdb.1998.0227. [DOI] [PubMed] [Google Scholar]
- Haag E.S. Rolling back to BOULE. Proc. Natl. Acad. Sci. USA. 2001;98:6983–6985. doi: 10.1073/pnas.141237898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Handberg-Thorsager M., Salo E. The planarian nanos-like gene Smednos is expressed in germline and eye precursor cells during development and regeneration. Dev. Genes Evol. 2007;217:403–411. doi: 10.1007/s00427-007-0146-3. [DOI] [PubMed] [Google Scholar]
- Handel M.A., Schimenti J.C. Genetics of mammalian meiosis: regulation, dynamics and impact on fertility. Nat. Rev. Genet. 2010;11:124–136. doi: 10.1038/nrg2723. [DOI] [PubMed] [Google Scholar]
- Hoopfer E.D., Penton A., Watts R.J., Luo L. Genomic analysis of Drosophila neuronal remodeling: a role for the RNA-binding protein Boule as a negative regulator of axon pruning. J. Neurosci. 2008;28:6092–6103. doi: 10.1523/JNEUROSCI.0677-08.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Houston D.W., King M.L. A critical role for Xdazl, a germ plasm-localized RNA, in the differentiation of primordial germ cells in Xenopus. Development. 2000;127:447–456. doi: 10.1242/dev.127.3.447. [DOI] [PubMed] [Google Scholar]
- Houston D.W., Zhang J., Maines J.Z., Wasserman S.A., King M.L. A Xenopus DAZ-like gene encodes an RNA component of germ plasm and is a functional homologue of Drosophila boule. Development. 1998;125:171–180. doi: 10.1242/dev.125.2.171. [DOI] [PubMed] [Google Scholar]
- Hoyle H.D., Raff E.C. Two Drosophila beta tubulin isoforms are not functionally equivalent. J. Cell Biol. 1990;111:1009–1026. doi: 10.1083/jcb.111.3.1009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Janicke T., Schärer L. Determinants of mating and sperm-transfer success in a simultaneous hermaphrodite. J. Evol. Biol. 2009;22:405–415. doi: 10.1111/j.1420-9101.2008.01660.x. [DOI] [PubMed] [Google Scholar]
- Janicke T., Schärer L. Sex allocation predicts mating rate in a simultaneous hermaphrodite. Proc. Biol. Sci. 2009;276:4247–4253. doi: 10.1098/rspb.2009.1336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson A.D., Bachvarova R.F., Drum M., Masi T. Expression of axolotl DAZL RNA, a marker of germ plasm: widespread maternal RNA and onset of expression in germ cells approaching the gonad. Dev. Biol. 2001;234:402–415. doi: 10.1006/dbio.2001.0264. [DOI] [PubMed] [Google Scholar]
- Joiner M.L., Wu C.F. Nervous system function for the testis RNA-binding protein BOULE in Drosophila. J. Neurogenet. 2004;18:341–363. doi: 10.1080/01677060490477435. [DOI] [PubMed] [Google Scholar]
- Karashima T., Sugimoto A., Yamamoto M. Caenorhabditis elegans homologue of the human azoospermia factor DAZ is required for oogenesis but not for spermatogenesis. Development. 2000;127:1069–1079. doi: 10.1242/dev.127.5.1069. [DOI] [PubMed] [Google Scholar]
- Katoh K., Toh H. Recent developments in the MAFFT multiple sequence alignment program. Brief. Bioinform. 2008;9:286–298. doi: 10.1093/bib/bbn013. [DOI] [PubMed] [Google Scholar]
- Kee K., Angeles V.T., Flores M., Nguyen H.N., Reijo Pera R.A. Human DAZL, DAZ and BOULE genes modulate primordial germ-cell and haploid gamete formation. Nature. 2009;462:222–225. doi: 10.1038/nature08562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keeney S., editor. Meiosis: Cytological Methods. Humana Press; Berlin: 2009. [Google Scholar]
- Keeney S., editor. Meiosis: Molecular and Genetic Methods. Humana Press; Berlin: 2009. [DOI] [PubMed] [Google Scholar]
- Kerr C.L., Cheng L. The dazzle in germ cell differentiation. J. Mol. Cell Biol. 2010;2:26–29. doi: 10.1093/jmcb/mjp041. [DOI] [PubMed] [Google Scholar]
- Ladurner P., Rieger R., Baguña J. Spatial distribution and differentiation potential of stem cells in hatchlings and adults in the marine platyhelminth Macrostomum sp.: a bromodeoxyuridine analysis. Dev. Biol. 2000;226:231–241. doi: 10.1006/dbio.2000.9867. [DOI] [PubMed] [Google Scholar]
- Ladurner P., Pfister D., Seifarth C., Schärer L., Mahlknecht M., Salvenmoser W., Gerth R., Marx F., Rieger R. Production and characterization of cell- and tissue-specific monoclonal antibodies for the flatworm Macrostomum sp. Histochem. Cell Biol. 2005;123:89–104. doi: 10.1007/s00418-004-0722-9. [DOI] [PubMed] [Google Scholar]
- Ladurner P., Schärer L., Salvenmoser W., Rieger R.M. A new model organism among the lower Bilateria and the use of digital microscopy in taxonomy of meiobenthic Platyhelminthes: Macrostomum lignano, n. sp (Rhabditophora, Macrostomorpha) J. Zool. Syst. Evol. Res. 2005;43:114–126. [Google Scholar]
- Ladurner P., Egger B., De Mulder K., Pfister D., Kuales G., Salvenmoser W., Schärer L. The stem cell system of the basal flatworm Macrostomum lignano. In: Bosch T.C.G., editor. Stem Cells from Hydra to Man, Vol. 1. Springer; Netherlands: 2008. pp. 75–94. [Google Scholar]
- Lewis E.B. A new standard food medium. Drosoph. Inf. Serv. 1960;34:117–118. [Google Scholar]
- Li M., Shen Q., Xu H., Wong F.M., Cui J., Li Z., Hong N., Wang L., Zhao H., Ma B., Hong Y. Differential conservation and divergence of fertility genes boule and dazl in the rainbow trout. PLoS One. 2011;6:e15910. doi: 10.1371/journal.pone.0015910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J., Linher K., Li J. Porcine DAZL messenger RNA: its expression and regulation during oocyte maturation. Mol. Cell. Endocrinol. 2009;311:101–108. doi: 10.1016/j.mce.2009.06.003. [DOI] [PubMed] [Google Scholar]
- Luetjens C.M., Xu E.Y., Reijo Pera R.A., Kamischke A., Nieschlag E., Gromoll J. Association of meiotic arrest with lack of BOULE protein expression in infertile men. J. Clin. Endocrinol. Metab. 2004;89:1926–1933. doi: 10.1210/jc.2003-031178. [DOI] [PubMed] [Google Scholar]
- Ma K., Inglis J.D., Sharkey A., Bickmore W.A., Hill R.E., Prosser E.J., Speed R.M., Thomson E.J., Jobling M., Taylor K. A Y chromosome gene family with RNA-binding protein homology: candidates for the azoospermia factor AZF controlling human spermatogenesis. Cell. 1993;75:1287–1295. doi: 10.1016/0092-8674(93)90616-x. [DOI] [PubMed] [Google Scholar]
- Maegawa S., Yasuda K., Inoue K. Maternal mRNA localization of zebrafish DAZ-like gene. Mech. Dev. 1999;81:223–226. doi: 10.1016/s0925-4773(98)00242-1. [DOI] [PubMed] [Google Scholar]
- Maegawa S., Yamashita M., Yasuda K., Inoue K. Zebrafish DAZ-like protein controls translation via the sequence ‘GUUC’. Genes Cells. 2002;7:971–984. doi: 10.1046/j.1365-2443.2002.00576.x. [DOI] [PubMed] [Google Scholar]
- Maines J., Wasserman S. Regulation and execution of meiosis in Drosophila males. Curr. Top. Dev. Biol. 1998;37:301–332. doi: 10.1016/s0070-2153(08)60178-7. [DOI] [PubMed] [Google Scholar]
- Maines J.Z., Wasserman S.A. Post-transcriptional regulation of the meiotic Cdc25 protein Twine by the Dazl orthologue Boule. Nat. Cell Biol. 1999;1:171–174. doi: 10.1038/11091. [DOI] [PubMed] [Google Scholar]
- Menke D.B., Mutter G.L., Page D.C. Expression of DAZ, an azoospermia factor candidate, in human spermatogonia. Am. J. Hum. Genet. 1997;60:237–241. [PMC free article] [PubMed] [Google Scholar]
- Moore F.L., Jaruzelska J., Fox M.S., Urano J., Firpo M.T., Turek P.J., Dorfman D.M., Pera R.A. Human Pumilio-2 is expressed in embryonic stem cells and germ cells and interacts with DAZ (Deleted in AZoospermia) and DAZ-like proteins. Proc. Natl. Acad. Sci. USA. 2003;100:538–543. doi: 10.1073/pnas.0234478100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morris J., Ladurner P., Rieger R., Pfister D., Del Mar De Miguel-Bonet M., Jacobs D., Hartenstein V. The Macrostomum lignano database as a molecular resource for studying platyhelminth development and phylogeny. Dev. Genes Evol. 2006;216:695–707. doi: 10.1007/s00427-006-0098-z. [DOI] [PubMed] [Google Scholar]
- Mouton S., Willems M., Braeckman B.P., Egger B., Ladurner P., Schärer L., Borgonie G. The free-living flatworm Macrostomum lignano: a new model organism for ageing research. Exp. Gerontol. 2009;44:243–249. doi: 10.1016/j.exger.2008.11.007. [DOI] [PubMed] [Google Scholar]
- Newmark P.A., Wang Y., Chong T. Germ cell specification and regeneration in planarians. Cold Spring Harb. Symp. Quant. Biol. 2008;73:573–581. doi: 10.1101/sqb.2008.73.022. [DOI] [PubMed] [Google Scholar]
- Orr-Weaver T.L. Meiosis in Drosophila: seeing is believing. Proc. Natl. Acad. Sci. USA. 1995;92:10443–10449. doi: 10.1073/pnas.92.23.10443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Otori M., Karashima T., Yamamoto M. The Caenorhabditis elegans homologue of deleted in azoospermia is involved in the sperm/oocyte switch. Mol. Biol. Cell. 2006;17:3147–3155. doi: 10.1091/mbc.E05-11-1067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palakodeti D., Smielewska M., Lu Y.C., Yeo G.W., Graveley B.R. The PIWI proteins SMEDWI-2 and SMEDWI-3 are required for stem cell function and piRNA expression in planarians. RNA. 2008;14:1174–1186. doi: 10.1261/rna.1085008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfister D., De Mulder K., Philipp I., Kuales G., Hrouda M., Eichberger P., Borgonie G., Hartenstein V., Ladurner P. The exceptional stem cell system of Macrostomum lignano: screening for gene expression and studying cell proliferation by hydroxyurea treatment and irradiation. Front. Zool. 2007;4:9. doi: 10.1186/1742-9994-4-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfister D., De Mulder K., Hartenstein V., Kuales G., Borgonie G., Marx F., Morris J., Ladurner P. Flatworm stem cells and the germ line: developmental and evolutionary implications of macvasa expression in Macrostomum lignano. Dev. Biol. 2008;319:146–159. doi: 10.1016/j.ydbio.2008.02.045. [DOI] [PubMed] [Google Scholar]
- Reddien P.W., Oviedo N.J., Jennings J.R., Jenkin J.C., Sanchez Alvarado A. SMEDWI-2 is a PIWI-like protein that regulates planarian stem cells. Science. 2005;310:1327–1330. doi: 10.1126/science.1116110. [DOI] [PubMed] [Google Scholar]
- Reijo R., Lee T.Y., Salo P., Alagappan R., Brown L.G., Rosenberg M., Rozen S., Jaffe T., Straus D., Hovatta O. Diverse spermatogenic defects in humans caused by Y chromosome deletions encompassing a novel RNA-binding protein gene. Nat. Genet. 1995;10:383–393. doi: 10.1038/ng0895-383. [DOI] [PubMed] [Google Scholar]
- Reijo R.A., Dorfman D.M., Slee R., Renshaw A.A., Loughlin K.R., Cooke H., Page D.C. DAZ family proteins exist throughout male germ cell development and transit from nucleus to cytoplasm at meiosis in humans and mice. Biol. Reprod. 2000;63:1490–1496. doi: 10.1095/biolreprod63.5.1490. [DOI] [PubMed] [Google Scholar]
- Richardson K.C., Jarett L., Finke E.H. Embedding in epoxy resins for ultrathin sectioning in electron microscopy. Stain Technol. 1960;35:313–323. doi: 10.3109/10520296009114754. [DOI] [PubMed] [Google Scholar]
- Rieger R.M., Gehlen M., Haszprunar G., Holmlund M., Legniti A., Salvenmoser W., Tyler S. Laboratory cultures of marine Macrostomida (Turbellaria) Forts. Zool. 1988;36:523–525. [Google Scholar]
- Ronquist F., Huelsenbeck J.P. MrBayes 3: Bayesian phylogenetic inference under mixed models. Bioinformatics. 2003;19:1572–1574. doi: 10.1093/bioinformatics/btg180. [DOI] [PubMed] [Google Scholar]
- Rossi L., Salvetti A., Lena A., Batistoni R., Deri P., Pugliesi C., Loreti E., Gremigni V. DjPiwi-1, a member of the PAZ-Piwi gene family, defines a subpopulation of planarian stem cells. Dev. Genes Evol. 2006;216:335–346. doi: 10.1007/s00427-006-0060-0. [DOI] [PubMed] [Google Scholar]
- Ruggiu M., Speed R., Taggart M., McKay S.J., Kilanowski F., Saunders P., Dorin J., Cooke H.J. The mouse Dazla gene encodes a cytoplasmic protein essential for gametogenesis. Nature. 1997;389:73–77. doi: 10.1038/37987. [DOI] [PubMed] [Google Scholar]
- Sato K., Shibata N., Orii H., Amikura R., Sakurai T., Agata K., Kobayashi S., Watanabe K. Identification and origin of the germline stem cells as revealed by the expression of nanos-related gene in planarians. Dev. Growth Differ. 2006;48:615–628. doi: 10.1111/j.1440-169X.2006.00897.x. [DOI] [PubMed] [Google Scholar]
- Saxena R., Brown L.G., Hawkins T., Alagappan R.K., Skaletsky H., Reeve M.P., Reijo R., Rozen S., Dinulos M.B., Disteche C.M., Page D.C. The DAZ gene cluster on the human Y chromosome arose from an autosomal gene that was transposed, repeatedly amplified and pruned. Nat. Genet. 1996;14:292–299. doi: 10.1038/ng1196-292. [DOI] [PubMed] [Google Scholar]
- Schärer L. Tests of sex allocation theory in simultaneously hermaphroditic animals. Evolution. 2009;63:1377–1405. doi: 10.1111/j.1558-5646.2009.00669.x. [DOI] [PubMed] [Google Scholar]
- Sekii K., Salvenmoser W., De Mulder K., Schärer L., Ladurner P. Melav2, an elav-like gene, is essential for spermatid differentiation in the flatworm Macrostomum lignano. BMC Dev. Biol. 2009;9:62. doi: 10.1186/1471-213X-9-62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shah C., Vangompel M.J., Naeem V., Chen Y., Lee T., Angeloni N., Wang Y., Xu E.Y. Widespread presence of human BOULE homologs among animals and conservation of their ancient reproductive function. PLoS Genet. 2010;6:e1001022. doi: 10.1371/journal.pgen.1001022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shibata N., Umesono Y., Orii H., Sakurai T., Watanabe K., Agata K. Expression of vasa(vas)-related genes in germline cells and totipotent somatic stem cells of planarians. Dev. Biol. 1999;206:73–87. doi: 10.1006/dbio.1998.9130. [DOI] [PubMed] [Google Scholar]
- Sigrist S., Ried G., Lehner C.F. Dmcdc2 kinase is required for both meiotic divisions during Drosophila spermatogenesis and is activated by the Twine/cdc25 phosphatase. Mech. Dev. 1995;53:247–260. doi: 10.1016/0925-4773(95)00441-3. [DOI] [PubMed] [Google Scholar]
- Spurr A.R. A low-viscosity epoxy resin embedding medium for electron microscopy. J. Ultrastruct. Res. 1969;26:31–43. doi: 10.1016/s0022-5320(69)90033-1. [DOI] [PubMed] [Google Scholar]
- Swofford D.L. 4.0. Sinauer; Sunderland, MA: 2003. PAUP* — Phylogenetic Analyses Using Parsimony and Other Methods, Version 4.0. [Google Scholar]
- Takeda Y., Mishima Y., Fujiwara T., Sakamoto H., Inoue K. DAZL relieves miRNA-mediated repression of germline mRNAs by controlling poly(A) tail length in zebrafish. PLoS One. 2009;4:e7513. doi: 10.1371/journal.pone.0007513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tiepolo L., Zuffardi O. Localization of factors controlling spermatogenesis in the nonfluorescent portion of the human Y chromosome long arm. Hum. Genet. 1976;34:119–124. doi: 10.1007/BF00278879. [DOI] [PubMed] [Google Scholar]
- Tung J.Y., Luetjens C.M., Wistuba J., Xu E.Y., Reijo Pera R.A., Gromoll J. Evolutionary comparison of the reproductive genes, DAZL and BOULE, in primates with and without DAZ. Dev. Genes Evol. 2006;216:158–168. doi: 10.1007/s00427-005-0039-2. [DOI] [PubMed] [Google Scholar]
- Urano J., Fox M.S., Reijo Pera R.A. Interaction of the conserved meiotic regulators, BOULE (BOL) and PUMILIO-2 (PUM2) Mol. Reprod. Dev. 2005;71:290–298. doi: 10.1002/mrd.20270. [DOI] [PubMed] [Google Scholar]
- VanGompel M.J., Xu E.Y. A novel requirement in mammalian spermatid differentiation for the DAZ-family protein Boule. Hum. Mol. Genet. 2010:1–10. doi: 10.1093/hmg/ddq109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vizoso D.B., Schärer L. Resource-dependent sex-allocation in a simultaneous hermaphrodite. J. Evol. Biol. 2007;20:1046–1055. doi: 10.1111/j.1420-9101.2007.01294.x. [DOI] [PubMed] [Google Scholar]
- Vizoso D.B., Rieger G., Schärer L. Goings-on inside a worm: functional hypotheses derived from sexual conflict thinking. Biol. J. Linn. Soc. 2010;99:370–383. [Google Scholar]
- Wang Y., Zayas R.M., Guo T., Newmark P.A. nanos function is essential for development and regeneration of planarian germ cells. Proc. Natl. Acad. Sci. USA. 2007;104:5901–5906. doi: 10.1073/pnas.0609708104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wyckoff G.J., Wang W., Wu C.I. Rapid evolution of male reproductive genes in the descent of man. Nature. 2000;403:304–309. doi: 10.1038/35002070. [DOI] [PubMed] [Google Scholar]
- Xu E.Y., Moore F.L., Pera R.A. A gene family required for human germ cell development evolved from an ancient meiotic gene conserved in metazoans. Proc. Natl. Acad. Sci. USA. 2001;98:7414–7419. doi: 10.1073/pnas.131090498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu E.Y., Lee D.F., Klebes A., Turek P.J., Kornberg T.B., Reijo Pera R.A. Human BOULE gene rescues meiotic defects in infertile flies. Hum. Mol. Genet. 2003;12:169–175. doi: 10.1093/hmg/ddg017. [DOI] [PubMed] [Google Scholar]
- Xu H., Li Z., Li M., Wang L., Hong Y. Boule is present in fish and bisexually expressed in adult and embryonic germ cells of medaka. PLoS One. 2009;4:e6097. doi: 10.1371/journal.pone.0006097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan L., Zuo X., He L., Racey P., Levin E., Zhang S. Cloning and characterization of novel isoforms of the BOULE gene in bats. Biochem. Genet. 2009;48:173–180. doi: 10.1007/s10528-009-9299-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zayas R.M., Hernandez A., Habermann B., Wang Y., Stary J.M., Newmark P.A. The planarian Schmidtea mediterranea as a model for epigenetic germ cell specification: analysis of ESTs from the hermaphroditic strain. Proc. Natl. Acad. Sci. USA. 2005;102:18491–18496. doi: 10.1073/pnas.0509507102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Q., Li J., Li Q., Li X., Liu Z., Song D., Xie Z. Cloning and characterization of the gene encoding the bovine BOULE protein. Mol. Genet. Genomics. 2009;281:67–75. doi: 10.1007/s00438-008-0394-6. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary material.