Abstract
Genome-wide association studies (GWAS) have identified several genetic variants associated with coronary heart disease (CHD), and variations in plasma lipoproteins and blood pressure (BP). Loci corresponding to CDKN2A/CDKN2B/ANRIL, MTHFD1L, CELSR2, PSRC1 and SORT1 genes have been associated with CHD, and TMEM57, DOCK7, CELSR2, APOB, ABCG5, HMGCR, TRIB1, FADS2/S3, LDLR, NCAN and TOMM40-APOE with total cholesterol. Similarly, CELSR2-PSRC1-SORT1, PCSK9, APOB, HMGCR, NCAN-CILP2-PBX4, LDLR, TOMM40-APOE, and APOC1-APOE are associated with variations in low-density lipoprotein cholesterol levels. Altogether, forty, forty three and twenty loci have been associated with high-density lipoprotein cholesterol, triglycerides and BP phenotypes, respectively. Some of these identified loci are common for all the traits, some do not map to functional genes, and some are located in genes that encode for proteins not previously known to be involved in the biological pathway of the trait. GWAS have been successful at identifying new and unexpected genetic loci common to diseases and traits, thus rapidly providing key novel insights into disease biology. Since genotype information is fixed, with minimum biological variability, it is useful in early life risk prediction. However, these variants explain only a small proportion of the observed variance of these traits. Therefore, the utility of genetic determinants in assessing risk at later stages of life has limited immediate clinical impact. The future application of genetic screening will be in identifying risk groups early in life to direct targeted preventive measures.
Keywords: Genome-wide association studies, Cardiovascular disease, Lipids, Blood pressure
INTRODUCTION
Cardiovascular disease (CVD) is the leading cause of morbidity and mortality globally[1,2]. There is a concerted effort to reduce this disease burden, particularly that of coronary heart disease (CHD) and cerebrovascular disease in developed countries[3-5]. These range from primary preventive strategies targeted at risk factors through acute management and secondary prevention strategies[6-8]. Kahn et al[9] estimated that aggressive application of nationally recommended prevention activities for CVD would potentially add approximately 224 million quality adjusted life-years to the US adult population over the next 30 years and improve the average lifespan by at least 1.3 years.
CHD is the result of a combination of genetic and environmental factors. More than 200 risk factors have been associated with CHD and, among these low-density lipoprotein cholesterol (LDL-c) and blood pressure (BP) have been shown through randomized controlled trials to be causally related to CHD. A key factor in reducing the global burden of CVD is early prediction of disease to target preventive interventions. More personalised approaches to CVD prevention are attracting increasing interest. Whilst biomarkers and quantitative traits have been extremely useful in targeting primary prevention, the recent advances in genomics offer a smart option for predicting future risk of disease very early in life using the invariant nature of a genotype throughout an individual’s life-span. For example, Cohen et al[10] demonstrated that a genetic variant resulting in a modest 28% reduction in LDL-c from birth results in an 88% reduction in the risk of CHD. Over the last 5 years, genome-wide association studies (GWAS) have revolutionised the discovery of common genetic variants associated with a range of diseases and traits.
There are three key characteristics of a genetic variant that determine its impact on the phenotype studied - (1) the frequency of the variant; (2) the effect size of the variant on the phenotype; and (3) the number of genetic variants acting on the phenotype. The “common disease common variant” hypothesis (CD:CV) is the model invoked to explain how genes influence common traits such as lipids, coronary artery disease (CAD) and BP[11]. This model proposes, using an evolutionary paradigm, that common disease is due to allelic variants with a frequency greater than 5% in the general population and small individual effect size[12]. The CD:CV framework requires population-wide genotyping of very large numbers of common genetic variants (Single Nucleotide Polymorphisms/SNPs) to determine which variants show significant association with the phenotype studied. Technological advances now allow reliable and high-throughput genotyping of hundreds of thousands of SNPs on a genome-wide scale[13]. Such studies employ large scale association mapping using SNPs, making no assumptions about the genomic location or function of the causal variant, and test the hypothesis that allele frequency differs between individuals with differences in phenotype. In most GWAS, emphasis is given to the “P value” for the association of genotype with disease risk, to reduce the potential for false positive association that arises when the association of hundreds of thousands to millions of markers are tested across the whole genome. The current popular method for multiple-test correction is the frequentist approach of adjusting for a number of independent tests - based on this, a significance level of 5 × 10-8 is commonly used, in populations of European ancestry for an overall genome-wide significance threshold of 0.05, adjusted for an estimated 1 million independent SNPs in the genome by the Bonferroni method[14]. It should be noted that the Bonferroni method is a fairly conservative correction method that may increase false negative rate. Other corrections like the False Discovery Rate or permutation testing can be used to set a different threshold. In this context, it is pertinent to recognise that the P-value is an index of a true positive signal and does not in any way reflect the predictive potential of the associated variant. The current gold standard of validity is multiple replication in independent samples. We review the implications of positive GWAS findings in current cardiovascular practice.
GWAS AND CHD
We summarise the GWAS results of CHD from nine case-control studies and three cohort studies[15-26] (Figure 1 and Table 1). The effect sizes (OR) of susceptibility alleles were modest and ranged from 1.05-2.0. Common variants in chromosome 9p21 were implicated in nine independent case-control studies[16-23,25] and in two cohort studies[15,25]. The most replicated SNPs at chromosome 9p21 were rs0757278 and rs13333049. The loci corresponding to MTHFD1L, initially identified in the Wellcome Trust Case Control Consortium (WTCCC) study[17], were later replicated in the German Family MI study[18] with genome-wide statistical significance. However, it did not reach genome-wide statistical significance in the combined analysis of ten different data sets in the study by Kathiresan et al[21]. Genetic loci corresponding to CELSR2, PSRC1 and SORT1 on chromosome 1p13.3 are identified in three independent studies[18,20,21].
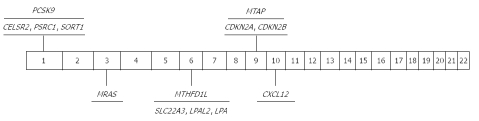
Figure 1.
Significant genome-wide association study findings in coronary heart disease. CELSR2: Cadherin EGF LAG seven-pass G-type receptor 2; PSRC1: Proline/serine-rich coiled-coil 1; SORT1: Sortilin 1; PCSK9: Proprotein convertase subtilisin/kexin type 9; MRAS: Ras-related protein M-Ras; MTHFD1L: Methylenetetrahydrofolate dehydrogenase (NADP+ dependent) 1-like; SLC22A3: Solute carrier family 22 (extraneuronal monoamine transporter), member 3; LPAL2: Lipoprotein, Lp(a)-like 2 pseudogene; LPA: Lipoprotein Lp(a); CDKN2A: Cyclin-dependent kinase inhibitor 2A; CDKN2B: Cyclin-dependent kinase inhibitor 2B; MTAP: Methylthioadenosine phosphorylase; CXCL12: Chemokine (C-X-C motif) ligand 12.
Table 1.
Single Nucleotide Polymorphisms associated with coronary heart disease in genome-wide association studies
| Chromosome | SNP | Position | Sample size | MAF (%) | OR (95% CI) | Pvalue | Proximal gene | Ref. |
| 1 | rs646776 | 109 620 053 | 9746/9746 | 81.0 | 1.17 (1.11-1.24) | CELSR2 | [18,20,21] | |
| rs599839 | 109 623 689 | 2801/4582 | - | 1.29 (1.18-1.40) | 4.05 × 10-9 | PSRC1 | ||
| rs599839 | 109 623 689 | 1926/2938 | 80.8 | 1.20 (1.10-1.31) | 1.30 × 10-5 | SORT1 | ||
| 1 | rs11206510 | 55 268 627 | 25 5381 | 81.0 | 1.15 (1.10-1.21) | PCSK9 | [21] | |
| 1 | rs17465637 | 220 890 152 | 9746/9746 | 72.0 | 1.13 (1.08-1.18) | MIA3 | [18,21] | |
| 2801/4582 | - | 1.20 (1.12-1.30) | 1.27 × 10-6 | |||||
| 2 | rs6725887 | 203 454 130 | 9746/9746 | 14.0 | 1.17 (1.11-1.23) | WDR12 | [21] | |
| 2 | rs2943634 | 226 776 324 | 2801/4582 | 37/32 | 1.21 (1.03-1.30) | 1.60 × 10-7 | Intergenic | [18] |
| 3 | rs9818870 | 139 604 812 | 19 407/21 366 | 17.3/15.4 | 1.15 (1.11-1.19) | 7.44 × 10-13 | MRAS | [23] |
| 6 | rs12526453 | 13 035 530 | 25 5381 | 65.0 | 1.12 (1.08-1.17) | PHACTR1 | [21] | |
| 6 | rs6922269 | 151 294 678 | 2801/4582 | 30.0/26.0 | 1.23 (1.15-1.33) | 2.90 × 10-8 | MTHFD1L | [18] |
| rs6922269 | 151 294 678 | 1926/2938 | 29.4/25.3 | 1.17 (1.04-1.32) | 1.50 × 10-5 | |||
| 62 | rs2048327 | 160 783 522 | 4976/4383 | 4.1/2.1 | 1.82 (1.57-2.12) | 4.20 × 10-15 | SLC22A3 | [22] |
| rs3127599 | 160 827 124 | LPAL2 | ||||||
| rs7767084 | 160 882 493 | LPA | ||||||
| rs10755578 | 160 889 728 | |||||||
| 9 | rs10757278 | 22 114 477 | 1607/6728 | 51.7/45.3 | 1.28 (1.22-1.35) | 3.60 × 10-14 | CDKN2A | [15,16,18,19,21,22,23,25] |
| rs10757274 | 22 086 055 | - | 25.3/20.4 | 1.33 (1.23-1.47) | CDKN2B | |||
| rs1333049 | 22 115 503 | 875/1644 | 54.0/48.0 | 1.33 (1.18-1.51) | 3.40 × 10-6 | |||
| rs1333049 | 22 115 503 | 1926/2938 | 55.4/47.4 | 1.47 (1.27-1.70) | 1.16 × 10-13 | MTAP | ||
| rs1333049 | 22 115 503 | 12 004/28 949 | - | 1.24 (1.20-1.28) | ||||
| - | 9746/9746 | 56.0 | 1.28 (1.23-1.33) | |||||
| rs4977574 | 22 088 574 | - | - | - | ||||
| - | 19 407/21 366 | - | - | |||||
| - | 33 282 | - | 1.20 (1.08-1.34)3 | |||||
| rs1333049 | 22 115 503 | |||||||
| 10 | rs1746048 | 44 095 830 | 9746/9746 | 84.0 | 1.14 (1.08-1.21) | CXCL12 | [18,21] | |
| rs501120 | 44 073 873 | 2801/4582 | -/- | 1.33 (1.20-1.48) | 9.46 × 10-8 | |||
| 12 | rs2259816 | 119 919 970 | 19 407/21 366 | 37.4/35.8 | 1.08 (1.05-1.11) | HNF1A-C12 or f43 | [23] | |
| 16 | rs4329913 | 55 462 933 | 18 245 | - | 1.29 (1.02-1.63)3 | CETP | [26] | |
| rs7202364 | 55 342 891 | 0.76 (0.59-0.99)3 | ||||||
| 19 | rs1122608 | 11 024 601 | 25 5381 | 75.0 | 1.15 (1.10-1.20) | LDLR | [20,21] | |
| rs6511720 | 11 063 306 | 1926/2938 | 90.2 | 1.29 (1.10-1.52) | 6.70 × 10-4 | |||
| 19 | rs4420638 | 50 114 786 | 1926/2938 | 20.9 | 1.17 (1.08-1.28) | 1.00 × 10-4 | APOE/C1/C4 | [20,21] |
| rs4420638 | 50 114 786 | 14 365/30 576 | ||||||
| 21 | rs9982601 | 34 520 998 | 25 5381 | 13.0 | 1.28 (1.23-1.33) | MRPS6 | [21] | |
| SLC5A3 | ||||||||
| KCNE2 |
1WTCC and GerMIFS I and GerMIFS II were added to the total sample; 2Haplotype CCTC; 3Hazard ratio per allele after adjustment for age and multiple risk factors. CELSR2: Cadherin EGF LAG seven-pass G-type receptor 2; PSRC1: Proline/serine-rich coiled-coil 1; SORT1: Sortilin 1; PCSK9: Proprotein convertase subtilisin/kexin type 9; MIA3: Melanoma inhibitory activity family member 3; WDR12: WD repeat protein 12; MRAS: Ras-related protein M-Ras; PHACTR1: Phosphatase and actin regulator 1; MTHFD1L: Methylenetetrahydrofolate dehydrogenase (NADP+ dependent) 1-like; SLC22A3: Solute carrier family 22 (extraneuronal monoamine transporter), member 3; LPAL2: Lipoprotein; Lp(a)-like 2 pseudogene; LPA: Lipoprotein Lp(a); CDKN2A: Cyclin-dependent kinase inhibitor 2A; CDKN2B: Cyclin-dependent kinase inhibitor 2B; MTAP: Methylthioadenosine phosphorylase; CXCL12: Chemokine (C-X-C motif) ligand 12; HNF1A-C12: Hepatocyte nuclear factor-1 homeobox A; CETP: Cholesteryl ester transfer protein plasma; LDLR: Low density lipoprotein receptor; APOE/C1/C4: Apolipoprotein; MRPS6: Mitochondrial ribosomal protein S6; SLC5A3: Solute carrier family 5 (sodium/myo-inositol cotransporter) member 3; KCNE2: Potassium voltage-gated channel subfamily E member 2; SNP: Single nucleotide polymorphisms; MAF: Minor allele frequency; OR: Odds ratio.
GWAS AND LIPIDS
Aulchenko et al[27] studied total cholesterol (TC)-associated genetic markers and identified 11 loci significantly associated with the trait (Figure 2 and Table 2): these corresponded to TMEM57, DOCK7, CELSR2, APOB, ABCG5, HMGCR, TRIB1, FADS2/S3, LDLR, NCAN and TOMM40-APOE. Many of these genes are also implicated in other lipid traits. After screening the genome for common variants associated with plasma lipids in > 100 000 individuals of European ancestry, Teslovich et al[28] identified 39 novel loci associated with TC and replicated several other loci found to be associated with lipid traits in the previous GWAS.
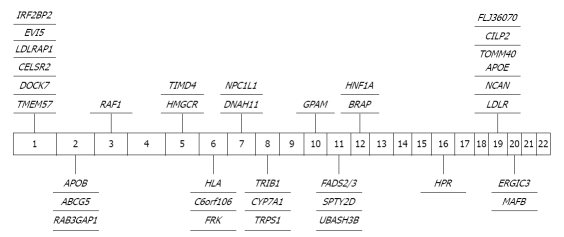
Figure 2.
Significant genome-wide association study findings in total cholesterol. TMEM57: Transmembrane protein 57; DOCK7: Dedicator of cytokinesis 7; CELSR2: Cadherin, EGF LAG seven-pass G-type receptor 2; LDLRAP1: Low-density lipoprotein receptor adaptor protein 1; EVI5: Ecotropic viral integration site 5; IRF2BP2: Interferon regulatory factor 2 binding protein 2; APOB: Apolipoprotein B; ABCG5: ATP-binding cassette sub-family G member 5; RAB3GAP1: RAB3 GTPase activating protein subunit 1 (catalytic); RAF1: V-raf-1 murine leukemia viral oncogene homolog 1; HMGCR: 3-hydroxy-3-methylglutaryl-CoA reductase; TIMD4: T-cell immunoglobulin and mucin domain containing 4; HLA: Human leukocyte antigen (HLA) complex; C6orf106: Chromosome 6 open reading frame 106; FRK: Fyn-related kinase; DNAH11: Dynein, axonemal, heavy chain 11; NPC1L1: NPC1 (Niemann-Pick disease; type C1, gene)-like 1; TRIB1: Tribbles Homolog-1 (Trib1); CYP7A1: Cytochrome P450, family 7, subfamily A, polypeptide 1; TRPS1: Trichorhinophalangeal syndrome 1; GPAM: Glycerol-3-phosphate acyltransferase, mitochondrial; FADS: Fatty acid desaturase; SPTY2D1: Suppressor of Ty, domain containing 1 (S. cerevisiae); UBASH3B: Ubiquitin associated and SH3 domain containing B; BRAP: BRCA1 associated protein; HNF1A: Hepatocyte nuclear factor-1 α; HPR: Haptoglobin-related protein; LDLR: Low-density lipoprotein receptor; NCAN: Neurocan; TOMM40: Translocase of outer mitochondrial membrane 40 homolog; CILP2: Cartilage intermediate layer protein 2; ERGIC3: Endoplasmic reticulum-Golgi intermediate compartment protein 3; MAFB: V-maf musculoaponeurotic fibrosarcoma oncogene homolog B.
Table 2.
Single nucleotide polymorphisms associated with total cholesterol identified through genome-wide association studies
| Chromosome | Strongest SNP | Chromosome position | Sample size | MAF (average) | β | Pvalue | Proximal gene | Ref. |
| 1 | rs10903129 | 25 641 524 | 22 550 | 54 | 0.061 | 5.4 × 10-10 | TMEM57 | [27] |
| 1 | rs1167998 | 62 704 220 | 17 346 | 32 | -0.073 | 6.4 × 10-10 | DOCK7 | [27] |
| rs108889353 | 19 099 | 32 | -0.079 | 3.7 × 10-12 | [27] | |||
| 1 | rs646776 | 109 620 053 | 17 441 | 22 | -0.128 | 8.5 × 10-22 | CELSR2 | [27] |
| 1 | rs12027135 | 25 648 320 | > 100 000 | 45 | -1.22 | 4.0 × 10-11 | LDLRAP1 | [28] |
| 1 | rs7515577 | 92 782 026 | > 100 000 | 21 | -1.18 | 3.0 × 10-8 | EVI5 | [28] |
| 1 | rs2642442 | 219 040 186 | > 100 000 | 48 | -1.36 | 5.0 × 10-14 | IRF2BP2 | [28] |
| 2 | rs693 | 21 085 700 | 22 500 | 52 | -0.096 | 8.7 × 10-23 | APOB | [27] |
| 2 | rs6756629 | 43 918 594 | 17 472 | 92 | 0.145 | 1.5 × 10-11 | ABCG5 | [27] |
| 2 | rs7570971 | 135 554 376 | > 100 000 | 34 | 1.25 | 2.0 × 10-8 | RAB3GAP1 | [28] |
| 3 | rs2290159 | 12 603 920 | > 100 000 | 22 | -1.42 | 4.0 × 10-9 | RAF1 | [28] |
| 5 | rs384662 | 35 421 429 | 20 873 | 44 | 0.092 | 2.5 × 10-19 | HMGCR | [27] |
| rs12916 | 74 692 295 | > 100 000 | 39 | 2.84 | 9.0 × 10-47 | [28] | ||
| 5 | rs6882076 | 156 322 875 | > 100 000 | 35 | -1.98 | 7.0 × 10-28 | TIMD4 | [28] |
| 6 | rs3177928 | 32 520 413 | > 100 000 | 16 | 2.31 | 4.0 × 10-19 | HLA | [28] |
| 6 | rs2814982 | 34 654 538 | > 100 000 | 11 | -1.86 | 5.0 × 10-11 | C6orf106 | [28] |
| 6 | rs9488822 | 116 419 586 | > 100 000 | 35 | -1.18 | 2.0 × 10-10 | FRK | [28] |
| 7 | rs12670798 | 21 573 877 | > 100 000 | 23 | 1.43 | 9.0 × 10-10 | DNAH11 | [28] |
| 7 | rs2072183 | 44 545 705 | > 100 000 | 25 | 2.01 | 3.0 × 10-11 | NPC1L1 | [28] |
| 8 | rs6987702 | 126 573 908 | 17 413 | 29 | 0.073 | 3.3 × 10-9 | TRIB1 | [27] |
| 8 | rs2081687 | 59 551 119 | > 100 000 | 35 | 1.23 | 2.0 × 10-12 | CYP7A1 | [28] |
| 8 | rs2737229 | 116 717 740 | > 100 000 | 30 | -1.11 | 2.0 × 10-8 | TRPS1 | [28] |
| 10 | rs2255141 | 113 923 876 | > 100 000 | 30 | 1.14 | 2.0 × 10-10 | GPAM | [28] |
| 11 | rs174570 | 61 353 788 | 20 916 | 83 | 0.088 | 1.5 × 10-10 | FADS2/3 | [27] |
| 11 | rs10128711 | 18 589 560 | > 100 000 | 28 | -1.04 | 3.0 × 10-8 | SPTY2D1 | [28] |
| 11 | rs7941030 | 122 027 585 | > 100 000 | 38 | 0.97 | 2.0 × 10-10 | UBASH3B | [28] |
| 12 | rs11065987 | 110 556 807 | > 100 000 | 42 | -0.96 | 7.0 × 10-12 | BRAP | [28] |
| 12 | rs1169288 | 119 901 033 | > 100 000 | 33 | 1.42 | 1.0 × 10-14 | HNF1A | [28] |
| 16 | rs2000999 | 70 665 594 | > 100 000 | 20 | 2.34 | 3.0 × 10-24 | HPR | [28] |
| 19 | rs2228671 | 11 071 912 | 20 910 | 88 | 0.158 | 9.3 × 10-24 | LDLR | [27] |
| 19 | rs2304130 | 19 650 528 | 20 914 | 7 | -0.153 | 2.0 × 10-15 | NCAN | [27] |
| 19 | rs2075650 | 50 087 459 | 17 463 | 15 | 0.138 | 2.9 × 10-19 | TOMM40-APOE | [27] |
| rs157580 | 50 087 106 | 20 903 | 33 | -0.09 | 5.1 × 10-17 | [27] | ||
| 19 | rs10401969 | 19 268 718 | > 100 000 | 7 | -4.74 | 3.0 × 10-38 | CILP2 | [28] |
| 19 | rs492602 | 53 898 229 | > 100 000 | 49 | 1.27 | 2.0 × 10-10 | FLJ36070 | [28] |
| 20 | rs2277862 | > 100 000 | 15 | -1.19 | 4.0 × 10-10 | ERGIC3 | [28] | |
| 20 | rs2902940 | 38 524 901 | > 100 000 | 29 | -1.38 | 6.0 × 10-11 | MAFB | [28] |
TMEM57: Transmembrane protein 57; DOCK7: Dedicator of cytokinesis 7; CELSR2: Cadherin, EGF LAG seven-pass G-type receptor 2; LDLRAP1: Low density lipoprotein receptor adaptor protein 1; EVI5: Ecotropic viral integration site 5; IRF2BP2: Interferon regulatory factor 2 binding protein 2; APOB: Apolipoprotein B; ABCG5: ATP-binding cassette sub-family G member 5; RAB3GAP1: RAB3 GTPase activating protein subunit 1 (catalytic); RAF1: v-raf-1 murine leukemia viral oncogene homolog 1; HMGCR: 3-hydroxy-3-methylglutaryl-CoA reductase; TIMD4: T-cell immunoglobulin and mucin domain containing 4; HLA: Human leukocyte antigen (HLA) complex; C6orf106: Chromosome 6 open reading frame 106; FRK: Fyn-related kinase; DNAH11: Dynein, axonemal, heavy chain 11; NPC1L1: NPC1 (Niemann-Pick disease, type C1, gene)-like 1; TRIB1: Tribbles Homolog-1 (Trib1); CYP7A1: Cytochrome P450, family 7, subfamily A, polypeptide 1; TRPS1: Trichorhinophalangeal syndrome 1; GPAM: Glycerol-3-phosphate acyltransferase; mitochondrial; FADS: Fatty acid desaturase; SPTY2D1: Suppressor of Ty, domain containing 1 (S. cerevisiae); UBASH3B: Ubiquitin associated and SH3 domain containing B; BRAP: BRCA1 associated protein; HNF1A: Hepatocyte nuclear factor-1 α; HPR: Haptoglobin-related protein; LDLR: Low density lipoprotein receptor; NCAN: Neurocan; TOMM40: Translocase of outer mitochondrial membrane 40 homolog; CILP2: Cartilage intermediate layer protein 2; ERGIC3: Endoplasmic reticulum-Golgi intermediate compartment protein 3; MAFB: V-maf musculoaponeurotic fibrosarcoma oncogene homolog B; SNP: Single nucleotide polymorphisms; MAF: Minor allele frequency.
Prior to the publication of the meta-analysis of blood lipids conducted by Teslovich et al[28], 29 loci had been found to be associated with variation in high-density lipoprotein cholesterol (HDL-c) levels[20,27-39]. Teslovich et al[28] identified 31 novel loci associated with HDL-c with genome-wide significance. The most commonly-replicated loci are LPL, LIPC, CETP, ABCA1, LIPG, APOA1/C3/A4/A5 and GALNT2 (Figure 3 and Table 3). The LIPC locus has a set of common variants nearly 50 kb upstream of the gene, strongly associated with HDL-c and appearing to be independent of previously described variants that overlap the transcribed sequence of the gene. SNPs close to the mevalonate kinase-methylmalonic aciduria cblB type (MMAB) locus were found to be associated with HDL-c initially by Willer et al[20] and later confirmed by Kathiresan et al[29].
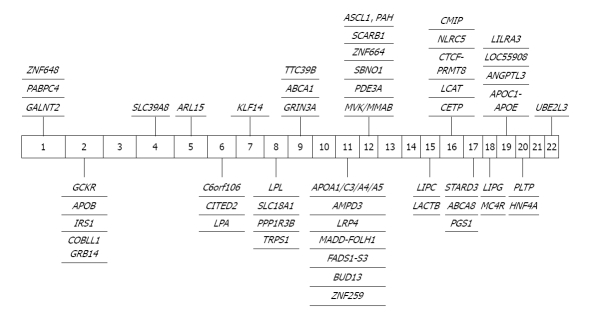
Figure 3.
Significant genome-wide association study findings in high-density lipoprotein cholesterol. GALNT2: N-acetylgalactosaminyltransferase 2; PABPC4: Poly(A) binding protein; cytoplasmic 4 (inducible form); ZNF648: Zinc finger protein 648; GCKR: Glucokinase (hexokinase 4) regulator; APOB: Apolipoprotein B; IRS1: Insulin receptor substrate 1; COBLL1: COBL-like 1; GRB14: Growth factor receptor-bound protein 14; SLC39A8: Solute carrier family 39 (zinc transporter) member 8; ARL15: ADP-ribosylation factor-like 15; C6orf106: Chromosome 6 open reading frame 106; CITED2: Cbp/p300-interacting transactivator, with Glu/Asp-rich carboxy-terminal domain 2; LPA: Lipoprotein, Lp(a); KLF14: Kruppel-like factor 14; LPL: Lipoprotein lipase; SLC18A1: Solute carrier family 18 (vesicular monoamine) member 1; PPP1R3B: Protein phosphatase 1, regulatory (inhibitor) subunit 3B; TRPS1: Trichorhinophalangeal syndrome 1; GRIN3A: Glutamate receptor, ionotropic, N-methyl-D-aspartate 3A; ABCA1: ATP-binding cassette; sub-family A (ABC1) member 1; TTC39B: Tetratricopeptide repeat domain 39B; ABCA1: ATP-binding cassette, sub-family A (ABC1) member 1; APOA1: Apolipoprotein A-I; AMPD3: Adenosine monophosphate deaminase 3; LRP4: Low-density lipoprotein receptor-related protein 4; MADD-FOLH1: MAP-kinase activating death domain- folate hydrolase (prostate-specific membrane antigen) 1; FADS1-S3: Fatty acid desaturase 1; BUD13: BUD13 homolog; ZNF259: Zinc finger protein 259; MVK: Mevalonate kinase; MMAB: Methylmalonic aciduria (cobalamin deficiency) cblB type; PDE3A: Phosphodiesterase 3A; SBNO1: Strawberry notch homolog 1; ZNF664: Zinc finger protein 664; SCARB1: Scavenger receptor class B member 1; ASCL1: Achaete-scute complex homolog 1; PAH: Phenylalanine hydroxylase; LIPC: Hepatic lipase; LACTB: Lactamase β; CETP: Cholesteryl ester transfer protein plasma; LCAT: Lecithin-cholesterol acyltransferase; CTCF: CCCTC-binding factor (zinc finger protein); PRMT8: Protein arginine methyltransferase 8; NLRC5: NLR family CARD domain containing 5; STARD3: StAR-related lipid transfer (START) domain containing 3; ABCA8: ATP-binding cassette; sub-family A (ABC1) member 8; PGS1: Phosphatidylglycerophosphate synthase 1; LIPG: Lipase endothelial; MC4R: Melanocortin 4 receptor; APOC1: Apolipoprotein C-I; APOE: Apolipoprotien E; ANGPTL3: Angiopoietin-like 3; LILRA3: Leukocyte immunoglobulin-like receptor, subfamily A (without TM domain) member 3; PLTP: Phospholipid transfer protein; HNF4A: Hepatocyte nuclear factor 4 α; PLTP: Phospholipid transfer protein; UBE2L3: Ubiquitin-conjugating enzyme E2L 3.
Table 3.
Single nucleotide polymorphisms associated with high-density lipoprotein cholesterol identified through genome-wide association studie
| Chromosome | Strongest SNP | Chromosome position | Sample size | MAF (average) | Change in HDLc/β | Pvalue | Proximal gene | Ref. |
| 1 | rs2144300 | 228 361 539 | 8656 | 40 | - | 6.6 × 10-7 | GALNT2 | [20] |
| rs4846914 | 228 362 314 | 19 794 | 40 | -0.05 SD | 4.0 × 10-8 | [30] | ||
| 1 | rs4660293 | 39 800 767 | > 100 000 | 23 | -0.48 | 4.0 × 10-10 | PABPC4 | [28] |
| 1 | rs1689800 | 180 435 508 | > 100 000 | 35 | -0.47 | 3.0 × 10-10 | ZNF648 | [28] |
| 2 | rs1260326 | 27 584 444 | 16 682 | 41 | 0.93% | < 5 × 10-8 | GCKR | [31] |
| 2 | rs6754295 | 21 059 688 | 17 915 | 25 | 2.63 (z-sc)1 | 4.4 × 10-8 | APOB | [27] |
| 2 | rs2972146 | 226 808 942 | > 100 000 | 37 | 0.46 | 3.0 × 10-9 | IRS1 | [28] |
| 2 | rs10490964 | 51 926 908 | 18 245 | 12 | 1.35 mg/dL | 3.9 × 10-9 | COBLL1, GRB14 | [26] |
| rs12328675 | 165 249 046 | > 100 000 | 13 | 0.68 | 3.0 × 10-10 | COBLL1 | [28] | |
| 4 | rs13107325 | 103 407 732 | > 100 000 | 7 | -0.84 | 7.0 × 10-11 | SLC39A8 | [28] |
| 5 | rs6450176 | 53 333 782 | > 100 000 | 26 | -0.49 | 5.0 × 10-8 | ARL15 | [28] |
| 6 | rs2814944 | 34 660 775 | > 100 000 | 16 | -0.49 | 4.0 × 10-9 | C6orf106 | [28] |
| 6 | rs605066 | 139 871 359 | > 100 000 | 42 | -0.39 | 3.0 × 10-8 | CITED2 | [28] |
| 6 | rs1084651 | 161 009 807 | > 100 000 | 16 | 1.95 | 3.0 × 10-8 | LPA | [28] |
| 7 | rs4731702 | 130 083 924 | > 100 000 | 48 | 0.59 | 1.0 × 10-15 | KLF14 | [28] |
| 8 | rs2083637 | 19 909 455 | 17 922 | 26 | 4.14 (z-sc)1 | 5.5 × 10-18 | LPL | [27] |
| rs10503669 | 19 891 970 | 8656 | 10 | 3.2 × 10-10 | [20] | |||
| rs331 | 19 864 685 | 6382 | 28 | 1.5 mg/dL | 9.1 × 10-7 | [32] | ||
| rs17482753 | 19 876 926 | 8180 | - | 2.8 × 10-11 | [33] | |||
| rs326 | 19 863 719 | 10 536 | 22-30 | 1.8 × 10-8 | [35] | |||
| rs331 | 19 864 685 | 6382 | 28 | 1.5 mg/dL | 9.1 × 10-7 | [32] | ||
| rs12678919 | 19 888 502 | 19 794 | 10 | 0.23 SD | 2.0 × 10-34 | [30] | ||
| rs301 | 19 861 214 | 5592 | 25 | 0.04 | 9.3 × 10-11 | [36] | ||
| 8 | rs3916027 | 19 869 148 | 5592 | 27 | 0.04 | 5.4 × 10-10 | SLC18A1 | [36] |
| 8 | rs331 | 19 864 685 | 16 809 | 27 | 0.43% | < 5 × 10-8 | Intergenic, PPP1R3B, LPL | [31] |
| rs9987289 | 9 220 768 | > 100 000 | 9 | -1.21 | 6.0 × 10-25 | PPP1R3B | [25] | |
| 8 | rs2293889 | 116 668 374 | > 100 000 | 0.41 | -0.44 | 6.0 × 10-11 | TRPS1 | [28] |
| 9 | r1323432 | 103 402 758 | 8656 | 12 | 1.93 mg/dL | 2.5 × 10-8 | GRIN3A | [20] |
| 9 | rs3905000 | 106 696 891 | 17 913 | 14 | -4.37 (z-sc)1 | 8.6 × 10-13 | ABCA1 | [27] |
| rs4149268 | 106 687 041 | 8656 | 36 | 3.3 × 10-7 | [20] | |||
| rs3890182 | 106 687 476 | 21 312 | - | 3.0 × 10-10 | [29] | |||
| rs9282541 | 106 660 656 | 10 536 | 0-9 | 4.8 × 10-8 | [35] | |||
| rs2515614 | 106 724 139 | 16 798 | 34 | 0.20% | < 5 × 10-8 | [31] | ||
| rs1883025 | 106 704 122 | 19 371 | 26 | -0.08 SD | 1.0 × 10-9 | [30] | ||
| 9 | rs471364 | 15 279 578 | 40 414 | 12 | -0.08 SD | 3.0 × 10-10 | TTC39B | [29] |
| rs581080 | 15 295 378 | > 100 000 | 18 | -0.65 | 3.0 × 10-12 | [28] | ||
| 9 | rs1883025 | 106 704 122 | > 100 000 | 25 | -0.94 | 2.0 × 10-33 | ABCA1 | [28] |
| 11 | rs12225230 | 116 233 840 | 6382 | 18 | 1.5 mg/dL | 5.3 × 10-5 | APOA1/C3/A4/A5 | [32] |
| rs618923 | 116 159 369 | 12 111 | 25 | 0.30% | < 5 × 10-8 | [31] | ||
| rs964184 | 116 154 127 | 19 794 | 14 | -0.17 SD | 1.0 × 10-12 | [30 | ||
| rs7350481 | 116 091 493 | 8993 | 28 | 0.62% | 8.8 × 10-10 | [36] | ||
| rs7350481 | 116 091 493 | 18 245 | 2.8 × 10-12 | [26] | ||||
| 11 | rs2923084 | 10 345 358 | > 100 000 | 17 | -0.41 | 5.0 × 10-8 | AMPD3 | [28] |
| 11 | rs3136441 | 46 699 823 | > 100 000 | 15 | 0.78 | 3.0 × 10-18 | LRP4 | [28] |
| 11 | rs7395662 | 17 917 | 39 | 2.82 (z-sc)1 | 6.0 × 10-11 | MADD-FOLH1 | [27] | |
| 11 | rs174547 | 61 327 359 | 40 330 | 33 | -0.09 SD | 2.0 × 10-12 | FADS1-S3 | [30] |
| 11 | rs6589565 | 116 145 447 | 5592 | -0.05 | 4.4 × 10-7 | BUD13 | [36] | |
| 11 | rs2075290 | 116 158 506 | 5592 | 7 | -0.05 | 4.2 × 10-7 | ZNF259 | [36] |
| 12 | rs2338104 | 108 379 551 | 8656 | 45 | 1.9 × 10-6 | MVK/MMAB | [20] | |
| rs2338104 | 108 379 551 | 19 793 | 45 | -0.07 SD | 1.0 × 10-10 | [30] | ||
| rs7134594 | 108 484 574 | > 100 000 | 47 | -0.44 | 7.0 × 10-15 | [28] | ||
| 12 | rs7134375 | 20 365 025 | > 100 000 | 42 | 0.40 | 4.0 × 10-8 | PDE3A | [28] |
| 12 | rs4759375 | 122 362 191 | > 100 000 | 6 | 0.86 | 7.0 × 10-9 | SBNO1 | [28] |
| 12 | rs4765127 | 123 026 120 | > 100 000 | 34 | 0.44 | 3.0 × 10-10 | ZNF664 | [28] |
| 12 | rs838880 | 123 827 546 | > 100 000 | 31 | 0.61 | 3.0 × 10-14 | SCARB1 | [28] |
| 12 | rs1818702 | 102 047 685 | 16 844 | 29 | 0.22% | < 5 × 10-8 | Intergenic, ASCL1, PAH | [31] |
| 15 | rs1532085 | 56 470 658 | 19 736 | 41 | 5.03 (z-sc)1 | 9.7 × 10-36 | LIPC | [27] |
| rs4115041 | 121 186 681 | 8656 | 33 | 2.8 × 10-9 | [20] | |||
| rs1532085 | 56 470 658 | 6382 | 37 | 1.8 mg/dL | 1.3 × 10-10 | [32] | ||
| rs1800588 | 56 510 967 | 21 312 | - | 2.0 × 10-32 | [29] | |||
| rs11858164 | 56 530 023 | 10 536 | 27-55 | 7.0 × 10-8 | [35] | |||
| rs1532085 | 56 470 658 | 6382 | 37 | 1.8 mg/dL | 1.3 × 10-10 | [32] | ||
| rs1800588 | 56 510 967 | 16 811 | 22 | 0.60% | < 5 × 10-8 | [31] | ||
| rs10468017 | 56 465 804 | 19 794 | 30 | 0.10 SD | 8.0 × 10-23 | [30] | ||
| rs1077834 | 56 510 771 | 5987 | 49 | 1.00% | 1.3 × 10-14 | [36] | ||
| rs1077834 | 56 510 771 | 18 245 | 1.4 × 10-23 | [26] | ||||
| rs261342 | 56 518 445 | 5592 | 22 | 0.03 | 6.3 × 10-8 | [36] | ||
| rs1532085 | 56 470 658 | > 100 000 | 39 | 1.45 | 3.0 × 10-96 | [28] | ||
| 15 | rs2652834 | 61 183 920 | > 100 000 | 20 | -0.39 | 9.0 × 10-9 | LACTB | [28] |
| 16 | rs1800775 | 55 552 737 | 2623 | 47 | 2.5 × 10-13 | CETP | [28] | |
| rs1532624 | 55 562 980 | 19 674 | 43 | 8.24 (z-sc)1 | 9.4 × 10-94 | [27] | ||
| rs3764261 | 55 550 825 | 8656 | 31 | 2.42 mg/dL | 2.8 × 10-19 | [20] | ||
| rs3764261 | 55 550 825 | 6382 | 31 | 4.0 mg/dL | 1.0 × 10-41 | [32] | ||
| rs1800775 | 55 552 737 | 2758 | 49 | 2.6 mg/dL | 3.0 × 10-13 | [29] | ||
| rs1800775 | 55 552 737 | 1643 | 47 | 3.99 mg/dL | 6.1 × 10-15 | [33] | ||
| rs9989419 | 55 542 640 | 8216 | - | 8.5 × 10-27 | [33] | |||
| rs7205804 | 55 562 390 | 10 536 | 37-50 | 4.7 × 10-47 | [36] | |||
| rs3764261 | 55 550 825 | 6382 | 31 | 4.0 mg/dL | 1.0 × 10-41 | [32] | ||
| rs1800775 | 55 552 737 | 16 779 | 49 | 2.50% | < 5 × 10-8 | [31] | ||
| rs3764261 | 55 550 825 | 322 | - | 6.2 mg/dL | 3.4 × 10-12 | [39] | ||
| rs3764261 | 55 550 825 | 18 245 | 3.7 × 10-93 | [26] | ||||
| rs173539 | 55 545 545 | 19 794 | 32 | 0.25 SD | 4.0 × 10-75 | [30] | ||
| rs3764261 | 55 550 825 | 5987 | 21 | 2.11% | 4.8 × 10-29 | [37] | ||
| rs3764261 | 55 550 825 | 18 245 | 30-48 | 3.7 × 10-93 | [27] | |||
| rs17231506 | 55 552 029 | 5592 | 32 | 0.07 | 2.3 × 10-36 | [36] | ||
| rs3764261 | 55 550 825 | > 100 000 | 32 | 3.39 | 7.0 × 10-380 | [28] | ||
| 16 | rs255052 | 66 582 496 | 8656 | 17 | 1.5 × 10-6 | LCAT | [20] | |
| rs255052 | 66 582 496 | 8656 + 4534 | - | 1.2 × 10-7 | [20] | |||
| rs2271293 | 66 459 571 | 31 946 | 11 | 0.07 SD | 9.0 × 10-13 | [30] | ||
| rs16942887 | 66 485 543 | > 100 000 | 12 | 1.27 | 8.0 × 10-33 | [28] | ||
| 16 | rs2271293 | 66 459 571 | 17 910 | 13 | 4.99 (z-sc)1 | 8.3 × 10-16 | CTCF-PRMT8 | [27] |
| 16 | rs289743 | 55 575 29 | 5592 | 31 | 0.03 | 8.6 × 10-9 | NLRC5 | [36] |
| 16 | rs2925979 | 80 092 291 | > 100 000 | 30 | -0.45 | 2.0 × 10-11 | CMIP | [28] |
| 17 | rs11869286 | 35 067 382 | > 100 000 | 34 | -0.48 | 1.0 × 10-13 | STARD3 | [28] |
| 17 | rs4148008 | 64 386 889 | > 100 000 | 32 | -0.42 | 2.0 × 10-10 | ABCA8 | [28] |
| 17 | rs4129767 | 73 915 579 | > 100 000 | 49 | -0.39 | 8.0 × 10-9 | PGS1 | [28] |
| 18 | rs4939883 | 45 421 212 | 16 258 | 17 | -3.98 (z-sc)1 | 1.6 × 10-11 | LIPG | [27] |
| rs2156552 | 45 435 666 | 8656 | 16 | 8.4 × 10-7 | [20] | |||
| rs2156552 | 45 435 666 | 21 312 | - | 2.0 × 10-7 | [29] | |||
| rs4939883 | 45 421 212 | 16 648 | 16 | 0.22% | < 5 × 10-8 | [31] | ||
| rs4939883 | 45 421 212 | 19 785 | 17 | -0.14 SD | 7.0 × 10-15 | [30] | ||
| rs4939883 | 45 421 212 | 18 245 | 1.4 × 10-9 | [26] | ||||
| rs7241918 | 45 414 951 | > 100 000 | 17 | -1.31 | 3.0 × 10-49 | [28] | ||
| 18 | rs12967135 | 56 000 003 | > 100 000 | 23 | -0.42 | 7.0 × 10-9 | MC4R | [28] |
| 19 | rs769449 | 50 101 842 | 16 728 | 12 | 0.30% | < 5 × 10-8 | APOC1-APOE | [31] |
| 18 245 | 2.6 × 10-11 | [26] | ||||||
| 19 | rs2967605 | 8 375 738 | 35 151 | 16 | -0.12 SD | 1.0 × 10-8 | ANGPTL3 | [30] |
| rs7255436 | 8 339 196 | > 100 000 | 47 | -0.45 | 3.0 × 10-8 | [28] | ||
| 19 | rs737337 | 11 208 493 | > 100 000 | 8 | -0.64 | 3.0 × 10-9 | LOC55908 | [28] |
| 19 | rs386000 | 59 484 573 | > 100 000 | 20 | 0.83 | 4.0 × 10-16 | LILRA3 | [28] |
| 20 | rs6065906 | 43 987 422 | 16 810 | 48 | 0.40% | < 5 × 10-8 | PLTP | [31] |
| 18 245 | 1.9 × 10-14 | [26] | ||||||
| 20 | rs1800961 | 42 475 778 | 30 714 | 3 | -0.19 SD | 8.0 × 10-10 | HNF4A | [30] |
| rs1800961 | 42 475 778 | > 100 000 | 3 | -1.88 | 1.0 × 10-15 | [28] | ||
| 20 | rs7679 | 44 009 909 | 40 248 | 19 | -0.07 SD | 4.0 × 10-9 | PLTP | [30] |
| rs6065906 | 43 987 422 | > 100 000 | 18 | -0.93 | 2.0 × 10-22 | [28] | ||
| 22 | rs181362 | 20 262 068 | > 100 000 | 20 | -0.46 | 1.0 × 10-8 | UBE2L3 | [28] |
1z-sc: the ENGAGE consortium provided the effect size on the z-scale. GALNT2: N-acetylgalactosaminyltransferase 2; PABPC4: Poly(A) binding protein, cytoplasmic 4 (inducible form); ZNF648: Zinc finger protein 648; GCKR: Glucokinase (hexokinase 4) regulator; APOB: Apolipoprotein B; IRS1: Insulin receptor substrate 1; COBLL1: COBL-like 1; GRB14: Growth factor receptor-bound protein 14; SLC39A8: Solute carrier family 39 (zinc transporter) member 8; ARL15: ADP-ribosylation factor-like 15; C6orf106: Chromosome 6 open reading frame 106; CITED2: Cbp/p300-interacting transactivator, with Glu/Asp-rich carboxy-terminal domain 2; LPA: Lipoprotein, Lp(a); KLF14: Kruppel-like factor 14; LPL: Lipoprotein lipase; SLC18A1: Solute carrier family 18 (vesicular monoamine) member 1; PPP1R3B: Protein phosphatase 1, regulatory (inhibitor) subunit 3B; TRPS1: Trichorhinophalangeal syndrome 1; GRIN3A: Glutamate receptor, ionotropic, N-methyl-D-aspartate 3A; ABCA1: ATP-binding cassette, sub-family A (ABC1) member 1; TTC39B: Tetratricopeptide repeat domain 39B; ABCA1: ATP-binding cassette, sub-family A (ABC1) member 1; APOA1: Apolipoprotein A-I; AMPD3: Adenosine monophosphate deaminase 3; LRP4: Low density lipoprotein receptor-related protein 4; MADD-FOLH1: MAP-kinase activating death domain- folate hydrolase (prostate-specific membrane antigen) 1; FADS1-S3: Fatty acid desaturase 1; BUD13: BUD13 homolog; ZNF259: Zinc finger protein 259; MVK: Mevalonate kinase; MMAB: Methylmalonic aciduria (cobalamin deficiency) cblB type; PDE3A: Phosphodiesterase 3A; SBNO1: Strawberry notch homolog 1; ZNF664: Zinc finger protein 664; SCARB1: Scavenger receptor class B member 1; ASCL1: Achaete-scute complex homolog 1; PAH: Phenylalanine hydroxylase; LIPC: Hepatic lipase; LACTB: Lactamase β; CETP: Cholesteryl ester transfer protein plasma; LCAT: Lecithin-cholesterol acyltransferase; CTCF: CCCTC-binding factor (zinc finger protein); PRMT8: Protein arginine methyltransferase 8; NLRC5: NLR family CARD domain containing 5; STARD3: StAR-related lipid transfer (START) domain containing 3; ABCA8: ATP-binding cassette; sub-family A (ABC1) member 8; PGS1: Phosphatidylglycerophosphate synthase 1; LIPG: Lipase endothelial; MC4R: Melanocortin 4 receptor; APOC1: Apolipoprotein C-I; APOE: Apolipoprotien E; ANGPTL3: Angiopoietin-like 3; LILRA3: Leukocyte immunoglobulin-like receptor, subfamily A (without TM domain) member 3; PLTP: Phospholipid transfer protein; HNF4A: Hepatocyte nuclear factor 4 α; PLTP: Phospholipid transfer protein; UBE2L3: Ubiquitin-conjugating enzyme E2L 3; SNP: Single nucleotide polymorphisms; MAF: Minor allele frequency. HDLc: high-density lipoprotein cholesterol.
GWAS have identified several genetic loci associated with LDL-c (Figure 4 and Table 4)[20,27-32,34,36,40], such as the study by Teslovich et al[28] which identified 22 novel and 25 previously implicated loci. CELSR2-PSRC1-SORT1 and PCSK9 loci on chromosome 1, APOB, HMGCR, NCAN-CILP2-PBX4, LDLR, TOMM40-APOE, and APOC1-APOE were the most commonly-replicated loci in LDL-c. Several of these loci were also associated with CHD in the WTCCC study[17].
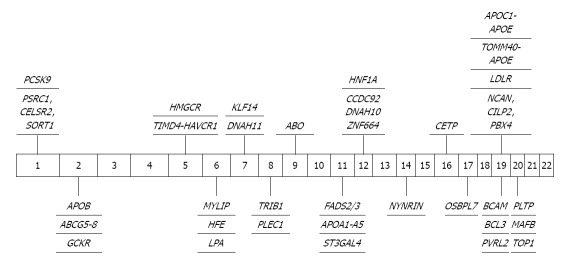
Figure 4.
Significant genome-wide association study findings in low-density lipoprotein cholesterol. CELSR2: Cadherin EGF LAG seven-pass G-type receptor 2; PSRC1: Proline/serine-rich coiled-coil 1; SORT1: Sortilin 1; PCSK9: Proprotein convertase subtilisin/kexin type 9; APOB: Apolipoprotein B; ABCG: ATP-binding cassette sub-family G; GCKR: Glucokinase (hexokinase 4) regulator; TIMD4: T-cell immunoglobulin and mucin domain containing 4; HAVCR1: Hepatitis A virus cellular receptor 1; HMGCR: 3-hydroxy-3-methylglutaryl-CoA reductase; MYLIP: Myosin regulatory light chain interacting protein; HFE: Human hemochromatosis; LPA: Lipoprotein, Lp(a); DNAH11: Dynein axonemal heavy chain 11; KLF14: Kruppel-like factor 14; TRIB1: Tribbles homolog 1; PLEC1: Plectin; ABO: ABO blood group (transferase A, α 1-3-N-acetylgalactosaminyltransferase, transferase B, α 1-3-galactosyltransferase); FADS: Fatty acid desaturase; APOA1: Apolipoprotien A1; ST3GAL4: ST3 β-galactoside α-2;3-sialyltransferase 4; CCDC92: Coiled-coil domain containing 92; DNAH10: Dynein axonemal heavy chain 10; ZNF664: Zinc finger protein 664; HNF1A: HNF1 homeobox A; NYNRIN: NYN domain and retroviral integrase containing; CETP: Cholesteryl ester transfer protein plasma; OSBPL7: Oxysterol binding protein-like 7; NCAN: Nucleoporin 214kDa; CILP2: Cartilage intermediate layer protein 2; PBX4: Pre-B-cell leukemia homeobox 4; LDLR: Low-density lipoprotein receptor; TOMM40: Translocase of outer mitochondrial membrane 40 homolog; APOE: Apolipoprotien E; APOC2: Apolipoprotein C-II; APOE: Apolipoprotien E; BCAM: Basal cell adhesion molecule; BCL3: B-cell CLL/lymphoma 3; PVRL2: Poliovirus receptor-related 2 (herpesvirus entry mediator B); PLPT: Proteolipid protein; MAFB: V-maf musculoaponeurotic fibrosarcoma oncogene homolog B; TOP1: Topoisomerase (DNA) 1.
Table 4.
Single nucleotide polymorphisms associated with low-density lipoprotein cholesterol identified through genome-wide association studies
| Chromosome | Strongest SNP | Chromosome position | Sample size | MAF (average) | β | Pvalue | Proximal gene | Ref. |
| 1 | rs1167998 | 62 704 220 | 12 685 | 22 | -0.155 | 7.8 × 10-23 | PSRC1, CELSR2, SORT1 | [27] |
| rs646776 | 109 620 053 | 6382 | 22 | - | 4.9 × 10-19 | [32] | ||
| rs599839 | 109 623 689 | 1636 | 24 | - | 1.1 × 10-7 | [34] | ||
| rs602633 | 109 623 034 | 8589 | 20 | - | 4.8 × 10-14 | [20] | ||
| rs646776 | 109 620 053 | 16 791 | 22 | -0.04 | < 5 × 10-8 | CELSR2 | [31] | |
| rs646776 | 109 620 053 | 21 312 | 24 | -0.16 | 5 × 10-42 | PSRC1 | [29] | |
| rs12740374 | 109 619 113 | 19 648 | 21 | -0.23 | 2.0 × 10-42 | [30] | ||
| rs599839 | 109 623 689 | 11 685 | 21 | -0.05 | 1.7 × 10-15 | [40] | ||
| rs646776 | 109 620 053 | 4337 | 21 | -0.16 | 4.3 × 10-9 | [40] | ||
| rs12740374 | 109 619 113 | 5592 | 21 | -0.15 | 1.8 × 10-9 | SORT1 | [36] | |
| rs646776 | 109 620 053 | 5592 | 22 | -0.14 | 3.8 × 10-8 | [36] | ||
| rs629301 | 109 619 829 | > 100 000 | 22 | -5.65 | 1.0 × 10-170 | [28] | ||
| 1 | rs11591147 | 55 278 235 | 16 826 | 2 | -0.12 | < 5 × 10-8 | PCSK9 | [31] |
| rs11591147 | 55 278 235 | 12 167 | 2 | -0.13 | < 5 × 10-8 | [31] | ||
| rs11591147 | 55 278 235 | 21 312 | 1 | -0.26 | 2.0 × 10-24 | [30] | ||
| rs11206510 | 55 268 627 | 19394 | 3.5 × 10-11 | [20] | ||||
| rs11206510 | 55 268 627 | 19 629 | 19 | -0.09 | 4.0 × 10-8 | [30] | ||
| rs11591147 | 55 278 235 | 5592 | -0.55 | 9.3 × 10-12 | [36] | |||
| rs2479409 | 55 277 238 | > 100 000 | 30 | 2.01 | 2.0 × 10-28 | [28] | ||
| 2 | rs693 | 21 085 700 | 2601 | 22 | 7.1 × 10-7 | APOB | [38] | |
| rs562338 | 21 141 826 | 1636 + 2631 | 17 | 8.6 × 10-13 | [34] | |||
| rs515135 | 21 139 562 | 8589 | 83 | 3.1 × 10-14 | [20] | |||
| rs562338 | 21 141 826 | 8589 + 10 849 | 5.6 × 10-22 | [20] | ||||
| rs693 | 21 085 700 | 16 112 | 52 | -0.098 | 3.6 × 10-17 | [28] | ||
| rs506585 | 21 250 687 | 6382 | 20 | -4.6 | 9.3 × 10-9 | [32] | ||
| rs506585 | 21 250 687 | 16 842 | 20 | -0.04 | < 5 × 10-8 | [31] | ||
| rs693 | 21 085 700 | 21 312 | 48 | 0.12 | 1.0 × 10-21 | [29] | ||
| rs515135 | 21 139 562 | 19 648 | 20 | -0.16 | 5.0 × 10-29 | [30] | ||
| rs562338 | 21 141 826 | 11 685 | 20 | -0.04 | 1.4 × 10-9 | [40] | ||
| rs1713222 | 21 124 828 | 4337 | 16 | -0.17 | 1.0 × 10-8 | [40] | ||
| rs562338 | 21 141 826 | 5592 | 18 | -0.18 | 1.2 × 10-11 | [36] | ||
| rs1367117 | 21 117 405 | > 100 000 | 30 | 4.05 | 4.0 × 10-114 | [28] | ||
| 2 | rs6756629 | 43 918 594 | 12 706 | 92 | 0.157 | 2.6 × 10-10 | ABCG5 | [27] |
| 2 | rs6544713 | 43 927 385 | 23 456 | 32 | 0.15 | 2 × 10-20 | ABCG8 | [30] |
| rs4299376 | 43 926 080 | > 100 000 | 30 | 2.75 | 2.0 × 10-47 | ABCG5/8 | [28] | |
| 2 | rs780094 | 27 594 741 | 16 841 | 40 | 0.03 | < 5 × 10-8 | GCKR | [31] |
| 5 | rs1501908 | 156 330 747 | 27 280 | 37 | -0.07 | 1 × 10-11 | TIMD4-HAVCR1 | [30] |
| 5 | rs3846662 | 74 686 840 | 16 135 | 44 | 0.079 | 1.5 × 10-11 | HMGCR | [27] |
| rs12654264 | 74 684 359 | 2758 + 18 554 | 39 | 0.1 | 1.0 × 10-20 | [29] | ||
| rs3846663 | 74 691 482 | 19 648 | 38 | 0.07 | 8.0 × 10-12 | [30] | ||
| rs12654264 | 74 684 359 | 5592 | 38 | 0.11 | 5.8 × 10-8 | [36] | ||
| 6 | rs3757354 | 88 570 980 | > 100 000 | 22 | -1.43 | 1.0 × 10-11 | MYLIP | [28] |
| 6 | rs1800562 | 26 201 120 | > 100 000 | 6 | -2.22 | 6.0 × 10-10 | HFE | [28] |
| 6 | rs1564348 | 160 498 850 | > 100 000 | 17 | -0.56 | 2.0 × 10-17 | LPA | [28] |
| 7 | rs12670798 | 21 573 877 | 12 695 | 24 | 0.089 | 6.1 × 10-9 | DNAH11 | [27] |
| 7 | rs4731702 | 130 083 924 | 16 747 | 49 | -0.02 | < 5 × 10-8 | KLF14 | [31] |
| 8 | rs6982636 | 126 548 497 | 16 798 | 47 | -0.02 | < 5 × 10-8 | TRIB1 | [31] |
| 8 | rs11136341 | 145 115 531 | > 100 000 | 40 | 1.4 | 4.0 × 10-13 | PLEC1 | [28] |
| 9 | rs9411489 | 135 144 821 | > 100 000 | 20 | 2.24 | 6.0 × 10-13 | ABO | [28] |
| 11 | rs174570 | 61 353 788 | 16 153 | 83 | 0.11 | 4.4 × 10-13 | FADS2/3 | [31] |
| 11 | rs3135506 | 116 167 617 | 16 837 | 6 | -0.13 | < 5 × 10-8 | APOA1-A5 | [31] |
| rs2072560 | 116 167 036 | 5592 | 6 | 0.22 | 2.4 × 10-7 | [36] | ||
| 11 | rs11220462 | 125 749 162 | > 100 000 | 14 | 1.95 | 1.0 × 10-15 | ST3GAL4 | [28] |
| 12 | rs7307277 | 123 041 109 | 16 804 | 34 | -0.02 | < 5 × 10-8 | CCDC92/DNAH10/ZNF664 | [31] |
| 12 | rs2650000 | 119 873 345 | 39 340 | 36 | 0.07 | 2.0 × 10-8 | HNF1A | [30] |
| 14 | rs8017377 | 51 667 587 | > 100 000 | 47 | 1.14 | 5.0 × 10-1 | NYNRIN | [28] |
| 16 | rs708272 | 55 553 789 | 16 843 | 43 | -0.04 | < 5 × 10-8 | CETP | [31] |
| rs17231506 | 55 552 029 | 5592 | 32 | -0.11 | 5.0 × 10-7 | [36] | ||
| 17 | rs7206971 | 42 780 114 | > 100 000 | 49 | 0.78 | 2.0 × 10-8 | OSBPL7 | [28] |
| 19 | rs1699614 | 19 519 472 | 21 312 | 10 | -0.1 | 3 × 10-8 | NCAN, CILP2, PBX4 | [29] |
| rs2228603 | 19 190 924 | 8589 | 7 | 1.8 × 10-7 | [20] | |||
| rs16996148 | 19 519 472 | 19 394 | 2.7 × 10-9 | [20] | ||||
| rs10401969 | 19 268 718 | 19 648 | 6 | -0.05 | 2.0 × 10-8 | [30] | ||
| 19 | rs688 | 11 088 602 | 4267 | 45 | 7.3 × 10-7 | LDLR | [34] | |
| rs6511720 | 11 063 306 | 8589 | 9 | 6.8 × 10-10 | [20] | |||
| rs6511720 | 11 063 306 | 19 394 | 4.2 × 10-23 | [20] | ||||
| rs2228671 | 11 071 912 | 16 148 | 82 | 0.136 | 4.2 × 10-14 | [27] | ||
| rs6511720 | 11 063 306 | 6382 | 12 | -7.7 | 5.2 × 10-15 | [32] | ||
| rs6511720 | 11 063 306 | 16 843 | 12 | -0.04 | < 5 × 10-8 | [31] | ||
| rs6511720 | 11 063 306 | 21 312 | 10 | -0.26 | 2 × 10-51 | [29] | ||
| rs6511720 | 11 063 306 | 19 648 | 10 | -0.26 | 2.0 × 10-26 | [30] | ||
| rs2228671 | 11 071 912 | 4337 | 12 | -0.18 | 1.1 × 10-8 | [40] | ||
| rs17248720 | 11 059 187 | 5592 | 13 | -0.31 | 7.8 × 10-25 | [36] | ||
| rs6511720 | 11 063 306 | > 100 000 | 11 | 6.99 | 4.0 × 10-117 | [28] | ||
| 19 | rs2075650 | 50 087 459 | 12 697 | 15 | 0.16 | 9.3 × 10-19 | TOMM40-APOE | [27] |
| rs157580 | 50 087 106 | 16 160 | 68 | -0.111 | 2.1 × 10-19 | [27] | ||
| rs2075650 | 50 087 459 | 4337 | 13 | 0.23 | 7.1 × 10-14 | [40] | ||
| rs2075650 | 50 087 459 | 5592 | 14 | 0.23 | 1.1 × 10-14 | [36] | ||
| 19 | rs4420638 | 50 114 786 | 2601 | 22 | 3.4 × 10-13 | APOC1-APOE | [38] | |
| rs4420638 | 50 114 786 | 4267 | 19 | 8.3 × 10-14 | [34] | |||
| rs4420638 | 50 114 786 | 8589 | 12 | 1.5 × 10-21 | [20] | |||
| rs4420638 | 50 114 786 | 19 394 | 3.0 × 10-43 | [20] | ||||
| rs4803750 | 49 939 467 | 6382 | 7 | -9.6 | 3.6 × 10-14 | [32] | ||
| rs4803750 | 49 939 467 | 16 616 | 7 | -9.3 | < 5 × 10-8 | [32] | ||
| rs4420638 | 50 114 786 | 21 312 | 20 | 0.19 | 1.0 × 10-60 | [29] | ||
| rs4420638 | 50 114 786 | 11 881 | 16 | 0.29 | 4.0 × 10-27 | [30] | ||
| rs4420638 | 50 114 786 | 11 685 | 18 | 0.06 | 1.2 × 10-20 | APOC2 | [40] | |
| rs12721046 | 50 113 094 | 5592 | 15 | 0.21 | 7.6 × 10-14 | [36] | ||
| rs12721109 | 50 139 061 | 5592 | 2 | -0.54 | 5.1 × 10-14 | [36] | ||
| rs4420638 | 50 114 786 | > 100 000 | 17 | 7.14 | 9.0 × 10-147 | APOE | [28] | |
| 19 | rs10402271 | 50 021 054 | 11 685 | 33 | 0.03 | 4.1 × 10-8 | BCAM | [40] |
| rs4605275 | 50 030 333 | 4337 | 31 | -0.13 | 4.7 × 10-8 | [40] | ||
| 19 | rs4803750 | 49 939 467 | 4337 | 7 | -0.28 | 2.4 × 10-11 | BCL3 | [40] |
| rs1531517 | 49 934 013 | 5592 | 7 | -0.22 | 5.3 × 10-8 | [36] | ||
| 19 | rs10402271 | 50 021 054 | 5592 | 33 | 0.15 | 2.1 × 10-12 | PVRL2 | [36] |
| 20 | rs6065906 | 43 987 422 | 16 843 | 48 | 0.02 | < 5 × 10-8 | PLPT | [31] |
| 20 | rs6102059 | 38 662 198 | 28 895 | 32 | -0.06 | 4.0 × 10-9 | MAFB | [30] |
| 20 | rs6029526 | 39 106 032 | > 100 000 | 47 | 1.39 | 4.0 × 10-19 | TOP1 | [28] |
β: Estimated mean; CELSR2: Cadherin EGF LAG seven-pass G-type receptor 2; PSRC1: Proline/serine-rich coiled-coil 1; SORT1: Sortilin 1; PCSK9: Proprotein convertase subtilisin/kexin type 9; APOB: Apolipoprotein B; ABCG: ATP-binding cassette sub-family G; GCKR: Glucokinase (hexokinase 4) regulator; TIMD4: T-cell immunoglobulin and mucin domain containing 4; HAVCR1: Hepatitis A virus cellular receptor 1; HMGCR: 3-hydroxy-3-methylglutaryl-CoA reductase; MYLIP: Myosin regulatory light chain interacting protein; HFE: Human hemochromatosis; LPA: Lipoprotein, Lp(a); DNAH11: Dynein axonemal heavy chain 11; KLF14: Kruppel-like factor 14; TRIB1: Tribbles homolog 1; PLEC1: Plectin; ABO: ABO blood group (transferase A, α 1-3-N-acetylgalactosaminyltransferase, transferase B, α 1-3-galactosyltransferase); FADS: Fatty acid desaturase; APOA1: Apolipoprotien A1; ST3GAL4: ST3 β-galactoside α-2,3-sialyltransferase 4; CCDC92: Coiled-coil domain containing 92; DNAH10: Dynein axonemal heavy chain 10; ZNF664: Zinc finger protein 664; HNF1A: HNF1 homeobox A; NYNRIN: NYN domain and retroviral integrase containing; CETP: Cholesteryl ester transfer protein plasma; OSBPL7: Oxysterol binding protein-like 7; NCAN: Nucleoporin 214 kDa; CILP2: Cartilage intermediate layer protein 2; PBX4: Pre-B-cell leukemia homeobox 4; LDLR: Low density lipoprotein receptor; TOMM40: Translocase of outer mitochondrial membrane 40 homolog; APOE: Apolipoprotien E; APOC2: Apolipoprotein C-II; APOE: Apolipoprotien E; BCAM: Basal cell adhesion molecule; BCL3: B-cell CLL/lymphoma 3; PVRL2: Poliovirus receptor-related 2 (herpesvirus entry mediator B); PLPT: Proteolipid protein; MAFB: V-maf musculoaponeurotic fibrosarcoma oncogene homolog B; TOP1: Topoisomerase (DNA) 1; SNP: Single nucleotide polymorphisms; MAF: Minor allele frequency.
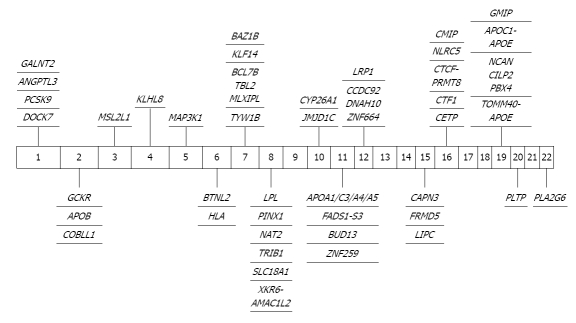
In total, 43 different loci have been found to be associated with triglycerides (TAG) in GWAS (Figure 5 and Table 5). SNPs in proximity to ANGPTL3, APOB, GCKR, MLXIPL, LPL, TRIB1, APOA1/A4/A5/C3, and NCAN-CILP2-PBX4 have been associated with TAG in several GWAS.
Figure 5.
Significant genome-wide association study findings in triglycerides. DOCK7: Dedicator of cytokinesis 7; PCSK9: Proprotein convertase subtilisin/kexin type 9; GALNT2: N-acetylgalactosaminyltransferase 2; ANGPTL3: Angiopoietin-like 3; APOB: Apolipoprotien B; GCKR: Glucokinase (hexokinase 4) regulator; COBLL1: COBL-like 1; MSL2L1: Male-specific lethal 2 homolog; KLHL8: Kelch-like 8; MAP3K1: Mitogen-activated protein kinase kinase kinase 1; BTNL2: Butyrophilin-like 2 (MHC class II associated); HLA: Major histocompatibility complex; TYW1B: tRNA-yW synthesizing protein 1 homolog B; TBL2: Transducin (β)-like 2; BCL7B: B-cell CLL/lymphoma 7B; TBL2: Transducin (β)-like 2; MLXIPL: MLX interacting protein-like; KLF14: Kruppel-like factor 14; BAZ1B: Bromodomain adjacent to zinc finger domain 1B; PINX1: PIN2/TERF1 interacting, telomerase inhibitor 1; NAT2: N-acetyltransferase 2 (arylamine N-acetyltransferase); LPL: Lipoprotein lipase; PP1R3B: Protein phosphatase 1, regulatory (inhibitor) subunit 3B; TRIB1: Tribbles homolog 1; SLC18A1: Solute carrier family 18 (vesicular monoamine) member 1; XKR6: XK Kell blood group complex subunit-related family member 6; AMAC1L2: Acyl-malonyl condensing enzyme 1-like 2; JMJD1C: Jumonji domain containing 1C; CYP26A1: Cytochrome P450 family 26 subfamily A polypeptide 1; APOA1: Apolipoprotein A-I; FADS: Fatty acid desaturase; CCDC92: Coiled-coil domain containing 92; DNAH10: Dynein axonemal heavy chain 10; ZNF664: Zinc finger protein 664; LRP1: Low density lipoprotein receptor-related protein 1; CAPN3: Calpain 3, (p94); FRMD5: FERM domain containing 5; LIPC: Hepatic lipase; CTF1: Cardiotrophin 1; CETP: Cholesteryl ester transfer protein plasma; TOMM40: Translocase of outer mitochondrial membrane 40 homolog; APOE: Apolipoprotien E; NCAN: Nucleoporin 214kDa; CILP2: Cartilage intermediate layer protein 2; PBX4: Pre-B-cell leukemia homeobox 4; GMIP: GEM interacting protein; PLPT: Palmitoyl-protein thioesterase 1; PLA2G6: Phospholipase A2, group VI (cytosolic; calcium-independent).
Table 5.
Single nucleotide polymorphisms associated with triglycerides identified through genome-wide association studies
| Chromosome | Strongest SNP | Studies | Sample size | MAF (average) | β | Pvalue | Proximal gene | Ref. |
| 1 | rs1167998 | 62 704 220 | 14 268 | 32 | -0.091 | 2.0 × 10-12 | DOCK7 | [27] |
| rs10889353 | 62 890 784 | 14 337 | 32 | -0.085 | 8.2 × 10-11 | [27] | ||
| 1 | rs11591147 | 55 278 235 | 16 826 | 2 | -0.09 | < 5 × 10-8 | PCSK9 | [31] |
| 1 | rs12042319 | 62 822 407 | 4267 | 34 | 3.2 × 10-7 | ANGPTL3 | [34] | |
| rs10889353 | 62 890 784 | 16 831 | 33 | -0.03 | < 5 × 10-8 | [31] | ||
| rs10889353 | 62 890 784 | 8993 | 14 | -0.13 | 2.0 × 10-9 | [37] | ||
| rs12130333 | 62 964 365 | 21 312 | 22 | -0.11 | 2.0 × 10-8 | [29] | ||
| rs10889353 | 62 890 784 | 19 834 | 33 | -0.05 | 3.0 × 10-7 | [30] | ||
| rs1748195 | 62 822 181 | 18 243 | 1.7 × 10-10 | [20] | ||||
| rs2131925 | 62 798 530 | > 100 000 | 32 | -4.94 | 9.0 × 10-43 | [28] | ||
| 1 | rs4846914 | 228 362 314 | 21 312 | 40 | 0.08 | 7.0 × 10-15 | GALNT2 | [28] |
| 2 | rs6754295 | 21 059 688 | 14 338 | 25 | -0.077 | 2.5 × 10-8 | APOB | [27] |
| rs673548 | 21 091 049 | 12 694 | 76 | 0.086 | 1.1 × 10-8 | [27] | ||
| rs673548 | 21 091 049 | 16 797 | 21 | -0.04 | < 5 × 10-8 | [31] | ||
| rs693 | 21 085 700 | 21 312 | 48 | 0.12 | 1.0 × 10-21 | [29] | ||
| rs7557067 | 21 061 717 | 19 840 | 22 | -0.08 | 9.0 × 10-12 | [30] | ||
| rs1042034 | 21 078 786 | > 100 000 | 22 | -5.99 | 1.0 × 10-45 | [28] | ||
| 2 | rs780094 | 27 594 741 | 2659 | 35 | 3.7 × 10-8 | GCKR | [38] | |
| rs780094 | 27 594 741 | 4267 | 39 | 8.1 × 10-14 | [34] | |||
| rs1260326 | 27 584 444 | 8684 | 40 | 1.5 × 10-15 | [20] | |||
| rs780094 | 27 594 741 | 18 243 | 6.1 × 10-32 | [20] | ||||
| rs780094 | 27 594 741 | 17 790 | 63 | -0.103 | 3.1 × 10-20 | [27] | ||
| rs1260326 | 27 584 444 | 6382 | 41 | 0.07 | 1.3 × 10-16 | [32] | ||
| rs1260326 | 27 584 444 | 16 650 | 41 | 0.07 | < 5 × 10-8 | [31] | ||
| rs1260326 | 27 584 444 | 8993 | 45 | -0.101 | 1.1 × 10-11 | [37] | ||
| rs780094 | 27 594 741 | 21 312 | 34 | 0.13 | 3.0 × 10-14 | [29] | ||
| rs1260326 | 27 584 444 | 19 840 | 45 | 0.12 | 2.0 × 10-31 | [30] | ||
| rs1260326 | 27 584 444 | 5592 | 40 | 0.06 | 1.8 × 10-7 | [36] | ||
| rs1260326 | 27 584 444 | > 100 000 | 41 | 8.76 | 6.0 × 10-133 | [28] | ||
| 2 | rs10195252 | 165 221 337 | > 100 000 | 40 | -2.01 | 2.0 × 10-10 | COBLL1 | [28] |
| 3 | rs645040 | 137 409 312 | > 100 000 | 22 | -2.22 | 3.0 × 10-8 | MSL2L1 | [28] |
| 4 | rs442177 | 88 249 285 | > 100 000 | 41 | -2.25 | 9.0 × 10-12 | KLHL8 | [28] |
| 5 | rs9686661 | 55 897 543 | > 100 000 | 20 | 2.57 | 1.0 × 10-10 | MAP3K1 | [28] |
| 6 | rs2076530 | 32 471 794 | 16 829 | 43 | 0.03 | < 5 × 10-8 | BTNL2 | [31] |
| 6 | rs2247056 | 31 373 469 | > 100 000 | 25 | -2.99 | 2.0 × 10-15 | HLA | [28] |
| 7 | rs13238203 | 71 767 603 | > 100 000 | 4 | -7.91 | 1.0 × 10-9 | TYW1B | [28] |
| 7 | rs17145738 | 72 620 810 | 2758 + 18 554 | 13 | -0.14 | 7.0 × 10-22 | BCL7B, TBL2, MLXIPL | [29] |
| TBL2 | ||||||||
| rs11974409 | 72 627 326 | 5592 | 20 | -0.08 | 5.7 × 10-9 | MLXIPL | [36] | |
| rs10551921 | 107 998 852 | 5592 | 20 | -0.08 | 1.3 × 10-8 | [36] | ||
| 7 | rs2240466 | 72 494 205 | 12 680 | 87 | 0.137 | 1.1 × 10-12 | MLXIPL | [27] |
| rs11974409 | 72 627 326 | 16 839 | 19 | -0.04 | < 5 × 10-8 | [31] | ||
| rs714052 | 72 502 805 | 19 840 | 12 | -0.16 | 3.0 × 10-15 | [30] | ||
| rs17145738 | 72 620 810 | 18 243 | 2.0 × 10-12 | [20] | ||||
| rs17145738 | 72 620 810 | > 100 000 | 12 | -9.32 | 6.0 × 10-58 | [28] | ||
| 7 | rs4731702 | 130 083 924 | 16 714 | 49 | -0.03 | < 5 × 10-8 | KLF14 | [31] |
| 7 | rs17145713 | 72 542 746 | 5592 | 20 | -0.09 | 5.3 × 10-10 | BAZ1B | [36] |
| 8 | rs11776767 | 10 721 339 | > 100 000 | 37 | 2.01 | 1.0 × 10-8 | PINX1 | [28] |
| 8 | rs1495741 | 18 317 161 | > 100 000 | 22 | 2.85 | 5.0 × 10-14 | NAT2 | [28] |
| 8 | rs2083637 | 19 909 455 | 14 344 | 26 | -0.107 | 1.0 × 10-14 | LPL | [27] |
| rs10096633 | 19 875 201 | 12 708 | 88 | 0.174 | 1.9 × 10-18 | [27] | ||
| rs12678919 | 19 888 502 | 19 840 | 10 | -0.25 | 2.0 × 10-41 | [30] | ||
| rs10096633 | 19 875 201 | 8993 | 12 | -0.169 | 9.3 × 10-14 | [37] | ||
| rs331 | 19 864 685 | 5592 | 25 | -0.08 | 1.7 × 10-11 | [36] | ||
| rs12678919 | 19 888 502 | > 100 000 | 12 | -13.64 | 2.0 × 10-115 | [28] | ||
| 8 | rs17482753 | 19 876 926 | 2652 | 11 | 4.9 × 10-7 | LPL | [38] | |
| rs17482753 | 19 876 926 | 1636 | 10 | 1.2 × 10-9 | [34] | |||
| rs17482753 | 19 876 926 | 1636 + 2631 | 5.2 × 10-15 | [34] | ||||
| rs6993414 | 19 947 198 | 8684 | 46 | 1.4 × 10-13 | [20] | |||
| rs10503669 | 19 891 970 | 4267 | 3.9 × 10-22 | [20] | ||||
| rs328 | 19 864 004 | 6382 | 11 | -0.09 | 4.7 × 10-11 | Intergenic, PPP1R3B, LPL | [32] | |
| rs331 | 19 864 685 | 6382 | 28 | -0.06 | 1.7 × 10-9 | [32] | ||
| rs328 | 19 864 685 | 16 812 | 11 | -0.09 | < 5 × 10-8 | LPL | [31] | |
| rs328 | 19 864 004 | 21 242 | 9 | -0.19 | 2.0 × 10-28 | [29] | ||
| 8 | rs6982636 | 126 548 497 | 16 765 | 47 | -0.03 | < 5 × 10-8 | TRIB1 | [31] |
| rs17321515 | 12 655 591 | 21 242 | 49 | -0.08 | 4.0 × 10-17 | [29] | ||
| rs2954029 | 126 560 154 | 8684 | 56 | 2.8 × 10-8 | [20] | |||
| rs17321515 | 12 655 591 | 14 176 | 7.0 × 10-13 | [20] | ||||
| rs2954029 | 126 560 154 | 19 840 | 44 | -0.11 | 3.0 × 10-19 | [30] | ||
| rs2954029 | 126 560 154 | > 100 000 | 47 | -5.64 | 3.0 × 10-55 | [28] | ||
| 8 | rs3916027 | 19 869 148 | 5592 | 27 | -0.08 | 1.0 × 10-10 | SLC18A1 | [36] |
| 8 | rs7819412 | 11 082 571 | 33 336 | 48 | -0.04 | 3.0 × 10-8 | XKR6-AMAC1L2 | [30] |
| 10 | rs10761731 | 64 697 616 | > 100 000 | 43 | -2.38 | 3.0 × 10-12 | JMJD1C | [28] |
| 10 | rs2068888 | 94 829 632 | > 100 000 | 46 | -2.28 | 2.0 × 10-8 | CYP26A1 | [28] |
| 11 | rs12272004 | 116 108 934 | 12 622 | 7 | -0.181 | 5.4 × 10-13 | APO (A1/A4/A5/C3) | [27] |
| rs6589566 | 116 157 633 | 1636 | 6 | 1.5 × 10-11 | [34] | |||
| rs6589566 | 116 157 633 | 1636 + 2631 | 3.7 × 10-12 | [34] | ||||
| rs964184 | 116 154 127 | 8684 | 12 | 1.5 × 10-16 | [20] | |||
| rs12286037 | 116 157 417 | 18 422 | 1.0 × 10-26 | [20] | ||||
| rs3135506 | 116 167 617 | 6382 | 6 | 0.13 | 5.5 × 10-12 | [32] | ||
| rs662799 | 116 168 917 | 6382 | 6 | 0.14 | 2.9 × 10-15 | [32] | ||
| rs3135506 | 116 167 617 | 16 804 | 6 | 0.14 | < 5 × 10-8 | [31] | ||
| rs7350481 | 116 091 493 | 8993 | 43 | 0.24 | 1.4 × 10-49 | [37] | ||
| rs28927680 | 116 124 283 | 21 312 | 7 | 0.26 | 2.0 × 10-17 | [29] | ||
| rs964184 | 116 154 127 | 19 840 | 14 | 0.3 | 4.0 × 10-62 | [30] | ||
| rs651821 | 116 167 789 | 5592 | 6 | 0.21 | 8.8 × 10-21 | APOA1 | [36] | |
| rs964184 | 116 154 127 | > 100 000 | 13 | 16.95 | 7.0 × 10-240 | [28] | ||
| 11 | rs174547 | 61 327 359 | 38 846 | 33 | 0.06 | 2.0 × 10-14 | FADS1-S3 | [30] |
| rs174546 | 61 326 406 | > 100 000 | 34 | 3.82 | 5.0 × 10-24 | [28] | ||
| 11 | rs6589565 | 116 145 447 | 5592 | 7 | 0.19 | 4.5 × 10-20 | BUD13 | [36] |
| 11 | rs2075290 | 116 158 506 | 5592 | 7 | 0.19 | 6.6 × 10-20 | ZNF259 | [36] |
| 12 | rs7307277 | 123 041 109 | 16 771 | 34 | -0.04 | < 5 × 10-8 | CCDC92/DNAH10/ | [31] |
| ZNF664 | ||||||||
| 12 | rs11613352 | - | > 100 000 | 23 | -2.7 | 4.0 × 10-10 | LRP1 | [28] |
| 15 | rs2412710 | 40 471 079 | > 100 000 | 2 | 7 | 2.0 × 10-8 | CAPN3 | [28] |
| 15 | rs2929282 | 42 033 223 | > 100 000 | 5 | 5.13 | 2.0 × 10-11 | FRMD5 | [28] |
| 15 | rs4775041 | 56 461 987 | 8684 | 67 | 7.3 × 10-5 | LIPC | [20] | |
| rs4775041 | 56 461 987 | 17 104 | 1.6 × 10-8 | [20] | ||||
| 16 | Rs11649653 | 30 825 988 | > 100 000 | 40 | -2.13 | 3.0 × 10-8 | CTF1 | [28] |
| 16 | rs1800775 | 55 552 737 | 16 779 | 49 | -0.03 | < 5 × 10-8 | CETP | [31] |
| 19 | rs157580 | 50 087 106 | 16 160 | 33 | -0.069 | 1.2 × 10-8 | TOMM40-APOE | [27] |
| rs439401 | 50 106 291 | 11 885 | 68 | 0.086 | 1.8 × 10-9 | [27] | ||
| 19 | rs16996148 | 19 519 472 | 21 312 | 10 | -0.1 | 4.0 × 10-9 | NCAN, CILP2, PBX4 | [29] |
| rs10401969 | 19 268 718 | 8684 | 8 | 2.3 × 10-7 | [20] | |||
| rs16996148 | 19 519 472 | 18 391 | 2.5 × 10-9 | [20] | ||||
| rs17216525 | 46 471 516 | 19 840 | 7 | -0.11 | 4.0 × 10-11 | [30] | ||
| rs12610185 | 19 582 722 | 5592 | 9 | -0.1 | 5.6 × 10-7 | [36] | ||
| 19 | rs439401 | 50 106 291 | 16 638 | 35 | -0.04 | < 5 × 10-8 | APOC1-APOE | [31] |
| rs439401 | 50 106 291 | > 100 000 | 36 | -5.5 | 1.0 × 10-30 | APOE | [28] | |
| 19 | rs2304128 | 19 607 151 | 5592 | 9 | -0.1 | 3.2 × 10-7 | GMI | [36] |
| 20 | rs6065906 | 43 987 422 | 16 810 | 48 | 0.04 | < 5 × 10-8 | PLPT | [31] |
| rs7679 | 44 009 909 | 38 561 | 19 | 0.07 | 7.0 × 10-11 | [30] | ||
| 22 | rs5756931 | 36 875 979 | > 100 000 | 40 | -1.54 | 4.0 × 10-8 | PLA2G6 | [28] |
β: Estimated mean; DOCK7: Dedicator of cytokinesis 7; PCSK9: Proprotein convertase subtilisin/kexin type 9; GALNT2: N-acetylgalactosaminyltransferase 2; ANGPTL3: Angiopoietin-like 3; APOB: Apolipoprotien B; GCKR: Glucokinase (hexokinase 4) regulator; COBLL1: COBL-like 1; MSL2L1: Male-specific lethal 2 homolog; KLHL8: Kelch-like 8; MAP3K1: Mitogen-activated protein kinase kinase kinase 1; BTNL2: Butyrophilin-like 2 (MHC class II associated); HLA: Major histocompatibility complex; TYW1B: tRNA-yW synthesizing protein 1 homolog B; TBL2: Transducin (β)-like 2; BCL7B: B-cell CLL/lymphoma 7B; TBL2: Transducin (β)-like 2; MLXIPL: MLX interacting protein-like; KLF14: Kruppel-like factor 14; BAZ1B: Bromodomain adjacent to zinc finger domain 1B; PINX1: PIN2/TERF1 interacting, telomerase inhibitor 1; NAT2: N-acetyltransferase 2 (arylamine N-acetyltransferase); LPL: Lipoprotein lipase; PP1R3B: Protein phosphatase 1, regulatory (inhibitor) subunit 3B; TRIB1: Tribbles homolog 1; SLC18A1: Solute carrier family 18 (vesicular monoamine) member 1; XKR6: XK Kell blood group complex subunit-related family member 6; AMAC1L2: Acyl-malonyl condensing enzyme 1-like 2; JMJD1C: Jumonji domain containing 1C; CYP26A1: Cytochrome P450 family 26 subfamily A polypeptide 1; APOA1: Apolipoprotein A-I; FADS: Fatty acid desaturase; CCDC92: Coiled-coil domain containing 92; DNAH10: Dynein axonemal heavy chain 10; ZNF664: Zinc finger protein 664; LRP1: Low density lipoprotein receptor-related protein 1; CAPN3: Calpain 3, (p94); FRMD5: FERM domain containing 5; LIPC: Hepatic lipase; CTF1: Cardiotrophin 1; CETP: Cholesteryl ester transfer protein plasma; TOMM40: Translocase of outer mitochondrial membrane 40 homolog; APOE: Apolipoprotien E; NCAN: Nucleoporin 214 kDa; CILP2: Cartilage intermediate layer protein 2; PBX4: Pre-B-cell leukemia homeobox 4; GMIP: GEM interacting protein; PLPT: Palmitoyl-protein thioesterase 1; PLA2G6: Phospholipase A2, group VI (cytosolic, calcium-independent); SNP: Single nucleotide polymorphisms; MAF: Minor allele frequency.
GWAS AND BP
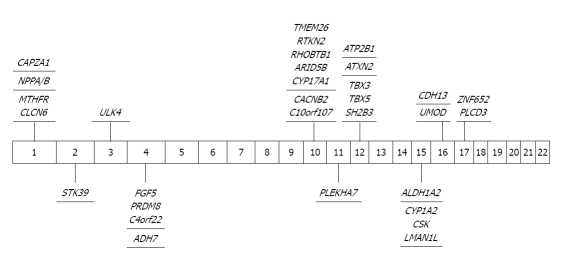
In 2007, the Framingham Heart Study[41] reported on 1327 individuals whose BP had been sampled longitudinally in the Framingham Community project. In the same year, the WTCCC[17] reported results from 2000 Northern European subjects with HTN. Although a few SNPs did reach a statistical significance of P < 10-5, none of them achieved genome-wide significance (P < 5 × 10-8). The most significant GWAS findings in blood pressure are summarized in Table 6 and Figure 6 [42-50].
Table 6.
Single nucleotide polymorphisms associated with hypertension and blood pressure in genome-wide association studies
| Chr | SNP | Position | Ancestry | N (discovery) | Phenotype | Risk allele | Risk allele frequency | OR/β | P | Nearest gene | Ref. |
| 1 | rs17367504 | 11 785 365 | E | 34 433 | SBP | G | 0.14 | -0.85 | 2 × 10-13 | MTHFR, CLCN6, NPPA, NPPB, AGTRAP | [42,43] |
| 2 | rs6749447 | 168 749 632 | E | 542 | SBP | G | 0.28 | 1.90 | 8 × 10-5 | STK39 | [47] |
| 3 | rs9815354 | 41 887 655 | E | 29 136 | DBP | A | 0.17 | 0.49 | 3 × 10-9 | ULK4 | [42,43] |
| 4 | rs16998073 | 81 403 365 | E | 34 433 | DBP | T | 0.21 | 0.50 | 1 × 10-21 | FGF5, PRDM8, C4orf22 | [42,43] |
| 4 | rs991316 | 100 541 468 | AA | 1017 | SBP | T | 0.45 | 1.62 | 5 × 10-6 | ADH7 | [44] |
| 10 | rs11014166 | 18 748 804 | E | 29 136 | DBP | A | 0.66 | 0.37 | 1 × 10-8 | CACNB2 | [42,43] |
| 10 | rs1530440 | 63 194 597 | E | 34 433 | DBP | T | 0.19 | -0.39 | 1 × 10-9 | C10orf107, TMEM26, RTKN2, RHOBTB1, ARID5B, CYP17A1 | [42,43] |
| 10 | rs1004467 | 104 584 497 | E | 29 136 | SBP | A | 0.90 | 1.05 | 1 × 10-10 | TMEM26, RTKN2, RHOBTB1, ARID5B, CYP17A1 | [42,43] |
| 10 | rs11191548 | 104 836 168 | E | 34 433 | SBP | T | 0.91 | 1.16 | 3 × 10-7 | CYP17A1, AS3MT, CNNM2, NT5C2 | [42,43] |
| 11 | rs381815 | 16 858 844 | E | 29 136 | SBP | T | 0.26 | 0.65 | 2 × 10-9 | PLEKHA7 | [42,43] |
| 12 | rs17249754 | 88584 | EA | 8842 | SBP, DBP | A | 0.37 | 1.06 | 9 × 10-7 | ATP2B1 | [49] |
| 12 | rs2681472 | 88 533 090 | E | 29 136 | SBP, DBP, HTN | A | 0.83 | 0.50 | 2 × 10-9 | ATP2B1 | [42,43] |
| 12 | rs2681492, | 88 537 220 | E | 29 136 | SBP, DBP, HTN | T | 0.80 | 0.85 | 4 × 10-11 | ATP2B1 | [42,43] |
| 12 | rs3184504 | 110 368 991 | E | 29 136 | SBP, DBP | T | 0.49 | 0.48 | 3 × 10-14 | ATXN2, SH2B3 | [42,43] |
| 12 | rs653178 | 110 492 139 | E | 34 433 | DBP | T | 0.53 | -0.46 | 3 × 10-18 | ATXN2, SH2B3 | [42,43] |
| 12 | rs2384550 | 113 837 114 | E | 29 136 | DBP | A | 0.35 | 0.43 | 4 × 10-8 | TBX3, TBX5 | [42,43] |
| 15 | rs1550576 | 56 000 706 | AA | 1017 | SBP | C | 0.86 | 1.92 | 3 × 10-6 | ALDH1A2 | [44] |
| 15 | rs1378942 | 72 865 396 | E | 34 433 | DBP | C | 0.36 | 0.43 | 1 × 10-23 | CSK, CYP1A1, CYP1A2, LMAN1L, CPLX3, ARID3B, ULK3 | [42,43] |
| 15 | rs6495122 | 72 912 698 | E | 29 136 | DBP | A | 0.42 | 0.40 | 2 × 10-10 | CSK, CYP1A1, CYP1A2, LMAN1L, CPLX3, ARID3B, ULK3 | [42,43] |
| 16 | rs13333226 | 20 273 155 | E | 3320 | HTN | A | 0.81 | 1.15 | 4 × 10-11 | UMOD | [50] |
| 16 | rs11646213 | 81 200 152 | E | 1977 | HTN | T | 0.60 | 1.28 | 8 × 10-6 | CDH13 | [48] |
| 17 | rs12946454 | 40 563 647 | E | 34 433 | SBP | T | 0.28 | 0.57 | 1 × 10-8 | PLCD3, ACBD4, HEXIM1, HEXIM2 | [42,43] |
| 17 | rs16948048 | 44 795 465 | E | 34 433 | DBP | G | 0.39 | 0.31 | 5 × 10-9 | ZNF652, PHB | [42,43] |
E: European; AA: African American; EA: East Asians; SBP: Systolic blood pressure; DBP: Diastolic blood pressure; HTN: Hypertension; ACBD4: Acyl-CoA binding domain containing 4; ADH7: Alcohol dehydrogenase 7 (class IV), mu or sigma polypeptide; AGTRAP: Angiotensin II receptor-associated protein; ALDH1A2: Aldehyde dehydrogenase 1 family, member A2; ARID5B: AT rich interactive domain 5B (MRF1-like); AS3MT: Arsenic (+3 oxidation state) methyltransferase; ATP2B1: ATPase; Ca++ transporting; plasma membrane 1; ATXN2: Ataxin 2; C10orf107: Chromosome 10 open reading frame 107; C4orf22: Chromosome 4 open reading frame 22; CACNB2: Calcium channel, voltage-dependent, β 2 subunit; CDH13: Cadherin 13, H-cadherin (heart); CLCN6: Chloride channel 6; CNNM2: Cyclin M2; CPLX3: Complexin 3; CSK: C-src tyrosine kinase; CYP17A1: Cytochrome P450, family 17, subfamily A, polypeptide 1; CYP1A1: Cytochrome P450; family 1, subfamily A, polypeptide 1; CYP1A2: Cytochrome P450, family 1, subfamily A, polypeptide 2; FGF5: Fibroblast growth factor 5; HEXIM1: Hexamethylene bis-acetamide inducible 1; HEXIM2: Hexamthylene bis-acetamide inducible 2; LMAN1L: Lectin, mannose-binding, 1 like; MTHFR: Methylenetetrahydrofolate reductase (NAD(P)H); NPPA: Natriuretic peptide A; NPPB: Natriuretic peptide B; NT5C2: 5'-nucleotidase, cytosolic II; PHB: Prohibitin; PLCD3: Phospholipase C, Δ 3; PLEKHA7: Pleckstrin homology domain containing, family A member 7; PRDM8: PR domain containing 8; RHOBTB1: Rho-related BTB domain containing 1; RTKN2: Rhotekin 2; SH2B3: SH2B adaptor protein 3; STK39: Serine threonine kinase 39; TBX3: T-box 3; TBX5: T-box 5; TMEM26: Transmembrane protein 26; ULK3: Unc-51-like kinase 3 (C. elegans); ULK4: Unc-51-like kinase 4 (C. elegans); UMOD: Uromodulin, ZNF652: Zinc finger protein 652; SNP: Single nucleotide polymorphisms; OR: Odds ratio.
Figure 6.
Significant genome-wide association study findings in blood pressure. ACBD4: Acyl-CoA binding domain containing 4; ADH7: Alcohol dehydrogenase 7 (class IV), mu or sigma polypeptide; AGTRAP: Angiotensin II receptor-associated protein; ALDH1A2: Aldehyde dehydrogenase 1 family, member A2; ARID5B: AT rich interactive domain 5B (MRF1-like); AS3MT: Arsenic (+3 oxidation state) methyltransferase; ATP2B1: ATPase, Ca++ transporting; plasma membrane 1; ATXN2: Ataxin 2; C10orf107: Chromosome 10 open reading frame 107; C4orf22: Chromosome 4 open reading frame 22; CACNB2: Calcium channel, voltage-dependent, β 2 subunit; CDH13: Cadherin 13, H-cadherin (heart); CLCN6: Chloride channel 6; CNNM2: Cyclin M2; CPLX3: Complexin 3; CSK: C-src tyrosine kinase; CYP17A1: Cytochrome P450, family 17, subfamily A; polypeptide 1; CYP1A1: Cytochrome P450, family 1, subfamily A, polypeptide 1; CYP1A2: Cytochrome P450, family 1, subfamily A, polypeptide 2; FGF5: Fibroblast growth factor 5; HEXIM1: Hexamethylene bis-acetamide inducible 1; HEXIM2: Hexamthylene bis-acetamide inducible 2; LMAN1L: Lectin, mannose-binding, 1 like; MTHFR: Methylenetetrahydrofolate reductase (NAD(P)H); NPPA: Natriuretic peptide A; NPPB: Natriuretic peptide B; NT5C2: 5'-nucleotidase, cytosolic II; PHB: Prohibitin; PLCD3: Phospholipase C, Δ 3; PLEKHA7: Pleckstrin homology domain containing, family A member 7; PRDM8: PR domain containing 8; RHOBTB1: Rho-related BTB domain containing 1; RTKN2: Rhotekin 2; SH2B3: SH2B adaptor protein 3; STK39: Serine threonine kinase 39; TBX3: T-box 3; TBX5: T-box 5; TMEM26: Transmembrane protein 26; ULK3: Unc-51-like kinase 3 (C. elegans); ULK4: Unc-51-like kinase 4 (C. elegans); UMOD: Uromodulin; ZNF652: Zinc finger protein 652.
The global BPgen consortium[42] studied 34 433 subjects of European ancestry, subsequently followed up the findings with direct genotyping of 71 225 individuals of European ancestry and 12 889 individuals of Indian Asian ancestry and conducted a joint analysis. They identified an association between systolic or diastolic BP (SBP/DBP) and common variants in eight regions near the CYP17A1 (intergenic CNNM2/NT5C2), CYP1A2 (intron CSK), FGF5, SH2B3 (intron ATXN2), MTHFR, c10orf107, ZNF652 and intron PLCD3. Furthermore, three of these common variants (MTHFR, CYP17A1 and CYP17A2 or CSK) were associated with HTN (P < 5 × 10-8). The CHARGE consortium study (n = 29 136) identified 13, 20 and 10 SNPs for SBP, DBP and HTN respectively[43].
In a joint meta-analysis of CHARGE consortium data with BPgen consortium data (n = 34 433)[43], four CHARGE loci attained genome-wide significance for SBP (ATP2B1, CYP17A1, PLEKHA7, SH2B3), six for DBP (ATP2B1, CACNB2, CSK-ULK3, SH2B3, TBX3-TBX5, ULK4) and one for HTN (ATP2B1). The KORA study by Org et al[48] in a South German Cohort identified a SNP upstream of T-cadherin adhesion molecule (CDH13) gene on chromosome 16 (rs11646213) as significantly associated with HTN at a genome-wide level. Finally, in a population of African origin, Adeyemo et al[44] identified four common variants (MYLIP, chr 6; YWHAZ, chr 8; IPO7, chr 11 and SLC24A4, chr 14) associated with SBP with genome-wide significance.
Wang et al[47] identified STK39, SPAK (STE20/SPS1-related proline and alanine rich kinase; a serine/threonine kinase) with a P value of 1.6 × 10-7 in an Amish cohort. Several other studies also identified potentially important genetic loci associated with BP traits with borderline genome-wide significance. These include ATP2B1[43,51] (ATPase, Ca++ transporting, plasma membrane 1) on chromosome 12, FOXD3[41] (fork head box D3) on chromosome 1, CCNG1 (cyclin G1)[48] on chromosome 5, BCAT1 (branched chain aminotransferase 1, cytosolic)[17] on chromosome 12, ATXN2 (ataxin 2)[42,43] on chromosome 12 and TBX3 (T-box3)[43] on chromosome 12 (Figure 6 and Table 6). However, none of these loci were replicated in other studies. Using an extreme case-control design, Padmanabhan et al[50] identified a novel HTN locus on chromosome 16 in the promoter region of uromodulin (UMOD; rs13333226, combined P value 3.6 × 10-11). The minor G allele of this SNP is associated with a lower risk of HTN [OR (95% CI): 0.87 (0.84-0.91)], reduced urinary UMOD excretion and increased estimated glomerular filtration rate (3.6 mL/min per minor-allele, P = 0.012), and borderline association with renal sodium balance.
CLINICAL IMPLICATIONS
GWAS are a useful tool in the identification of new and unexpected genetic loci of common diseases and traits, thus providing key novel insights into disease biology. But the clinical utility of these discoveries is negligible at this stage. The comparatively small numbers of variants which have been successfully replicated in several independent studies explain only a small proportion of the observed variation of these traits and explain in aggregate less than 20% of disease heritability. For example, the loci underpinning LDL-C levels[28] and BP account for < 20% of the variance of these quantitative traits. The variants associated with CHD increase disease risk by up to 20% per allele[51,52]. Next generation sequencing is now used to study low-frequency and rare variants that may potentially explain some of the missing heritabilities; however it is likely that studies designed to test for gene-environment interactions and gene-gene interactions may hold the answer. There were attempts to develop genetic profiles using the results from GWAS studies, but these have very limited value in personalised risk prediction as the genotype-phenotype effect sizes are very small. In the few studies that have evaluated the ability of a panel of genetic markers to discriminate CHD cases, the area under the receiver operating characteristic curve has been small indicating that conventional risk factors and family history are better at predicting risk and the incremental advantage of adding genetic markers is negligible. A few studies have attempted reclassification based on incorporation of SNPs from GWAS of CAD, lipids, etc.[52-58], and while they showed some improvement in net reclassification, the interpretation of these are still controversial and not translatable into general use[59]. Many companies are providing direct-to-consumer genetic tests that provide a “genetic risk profile” for an individual using risk alleles of small-to-moderate effects despite the clinical utility of genetic screening not being established. None of the major healthcare providers in Europe and USA have adopted these tests for CHD risk prediction, and the FDA has advised that direct-to-consumer genetic tests should be considered to be medical devices requiring FDA approval for commercial use. The future application of genetic screening will be in identifying risk groups early in life to direct targeted preventive measures and potentially pharmacogenetic tests to identify those at higher risk for adverse events. While technology is not a barrier to achieving this, the discovery, evaluation and deployment of these tests will require the same standards as non-genetic tests[60].
Footnotes
Peer reviewer: Boris Z Simkhovich, MD, PhD, The Heart Institute, Good Samaritan Hospital, 1225 Wilshire Boulevard, Los Angeles, CA 90017, United States
Supported by A Wellcome Trust Capacity Strengthening Strategic Award to the Public Health Foundation of India and a consortium of UK universities (to Jeemon P); Research grants from National Heart Lung and Blood Institute, United States of America (HHSN286200900026C) and National Institute of Health, United States of America (1D43HD065249) (to Prabhakaran D)
S- Editor Tian L L- Editor O’Neill M E- Editor Zheng XM
References
- 1.World Health Organization. The World Health Report, 2002: reducing risks, promoting healthy life. Geneva, 2002. doi: 10.1080/1357628031000116808. [DOI] [PubMed] [Google Scholar]
- 2.Ezzati M, Lopez AD, Rodgers A, Vander Hoorn S, Murray CJ. Selected major risk factors and global and regional burden of disease. Lancet. 2002;360:1347–1360. doi: 10.1016/S0140-6736(02)11403-6. [DOI] [PubMed] [Google Scholar]
- 3.Unal B, Critchley JA, Capewell S. Explaining the decline in coronary heart disease mortality in England and Wales between 1981 and 2000. Circulation. 2004;109:1101–1107. doi: 10.1161/01.CIR.0000118498.35499.B2. [DOI] [PubMed] [Google Scholar]
- 4.Ford ES, Capewell S. Coronary heart disease mortality among young adults in the U.S. from 1980 through 2002: concealed leveling of mortality rates. J Am Coll Cardiol. 2007;50:2128–2132. doi: 10.1016/j.jacc.2007.05.056. [DOI] [PubMed] [Google Scholar]
- 5.Peeters A, Nusselder WJ, Stevenson C, Boyko EJ, Moon L, Tonkin A. Age-specific trends in cardiovascular mortality rates in the Netherlands between 1980 and 2009. Eur J Epidemiol. 2011;26:369–373. doi: 10.1007/s10654-011-9546-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ford ES, Capewell S. Proportion of the decline in cardiovascular mortality disease due to prevention versus treatment: public health versus clinical care. Annu Rev Public Health. 2011;32:5–22. doi: 10.1146/annurev-publhealth-031210-101211. [DOI] [PubMed] [Google Scholar]
- 7.Unal B, Critchley JA, Fidan D, Capewell S. Life-years gained from modern cardiological treatments and population risk factor changes in England and Wales, 1981-2000. Am J Public Health. 2005;95:103–108. doi: 10.2105/AJPH.2003.029579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kabir Z, Bennett K, Shelley E, Unal B, Critchley J, Feely J, Capewell S. Life-years-gained from population risk factor changes and modern cardiology treatments in Ireland. Eur J Public Health. 2007;17:193–198. doi: 10.1093/eurpub/ckl090. [DOI] [PubMed] [Google Scholar]
- 9.Kahn R, Robertson RM, Smith R, Eddy D. The impact of prevention on reducing the burden of cardiovascular disease. Circulation. 2008;118:576–585. doi: 10.1161/CIRCULATIONAHA.108.190186. [DOI] [PubMed] [Google Scholar]
- 10.Cohen JC, Boerwinkle E, Mosley TH, Hobbs HH. Sequence variations in PCSK9, low LDL, and protection against coronary heart disease. N Engl J Med. 2006;354:1264–1272. doi: 10.1056/NEJMoa054013. [DOI] [PubMed] [Google Scholar]
- 11.Lander ES. The new genomics: global views of biology. Science. 1996;274:536–539. doi: 10.1126/science.274.5287.536. [DOI] [PubMed] [Google Scholar]
- 12.McCarthy MI, Abecasis GR, Cardon LR, Goldstein DB, Little J, Ioannidis JP, Hirschhorn JN. Genome-wide association studies for complex traits: consensus, uncertainty and challenges. Nat Rev Genet. 2008;9:356–369. doi: 10.1038/nrg2344. [DOI] [PubMed] [Google Scholar]
- 13.Ku CS, Loy EY, Pawitan Y, Chia KS. The pursuit of genome-wide association studies: where are we now? J Hum Genet. 2010;55:195–206. doi: 10.1038/jhg.2010.19. [DOI] [PubMed] [Google Scholar]
- 14.Pe'er I, Yelensky R, Altshuler D, Daly MJ. Estimation of the multiple testing burden for genomewide association studies of nearly all common variants. Genet Epidemiol. 2008;32:381–385. doi: 10.1002/gepi.20303. [DOI] [PubMed] [Google Scholar]
- 15.McPherson R, Pertsemlidis A, Kavaslar N, Stewart A, Roberts R, Cox DR, Hinds DA, Pennacchio LA, Tybjaerg-Hansen A, Folsom AR, et al. A common allele on chromosome 9 associated with coronary heart disease. Science. 2007;316:1488–1491. doi: 10.1126/science.1142447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Helgadottir A, Thorleifsson G, Manolescu A, Gretarsdottir S, Blondal T, Jonasdottir A, Jonasdottir A, Sigurdsson A, Baker A, Palsson A, et al. A common variant on chromosome 9p21 affects the risk of myocardial infarction. Science. 2007;316:1491–1493. doi: 10.1126/science.1142842. [DOI] [PubMed] [Google Scholar]
- 17.Wellcome Trust Case Control Consortium. Genome-wide association study of 14,000 cases of seven common diseases and 3,000 shared controls. Nature. 2007;447:661–678. doi: 10.1038/nature05911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Samani NJ, Erdmann J, Hall AS, Hengstenberg C, Mangino M, Mayer B, Dixon RJ, Meitinger T, Braund P, Wichmann HE, et al. Genomewide association analysis of coronary artery disease. N Engl J Med. 2007;357:443–453. doi: 10.1056/NEJMoa072366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Schunkert H, Götz A, Braund P, McGinnis R, Tregouet DA, Mangino M, Linsel-Nitschke P, Cambien F, Hengstenberg C, Stark K, et al. Repeated replication and a prospective meta-analysis of the association between chromosome 9p21.3 and coronary artery disease. Circulation. 2008;117:1675–1684. doi: 10.1161/CIRCULATIONAHA.107.730614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Willer CJ, Sanna S, Jackson AU, Scuteri A, Bonnycastle LL, Clarke R, Heath SC, Timpson NJ, Najjar SS, Stringham HM, et al. Newly identified loci that influence lipid concentrations and risk of coronary artery disease. Nat Genet. 2008;40:161–169. doi: 10.1038/ng.76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kathiresan S, Voight BF, Purcell S, Musunuru K, Ardissino D, Mannucci PM, Anand S, Engert JC, Samani NJ, Schunkert H, et al. Genome-wide association of early-onset myocardial infarction with single nucleotide polymorphisms and copy number variants. Nat Genet. 2009;41:334–341. doi: 10.1038/ng.327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Trégouët DA, König IR, Erdmann J, Munteanu A, Braund PS, Hall AS, Grosshennig A, Linsel-Nitschke P, Perret C, DeSuremain M, et al. Genome-wide haplotype association study identifies the SLC22A3-LPAL2-LPA gene cluster as a risk locus for coronary artery disease. Nat Genet. 2009;41:283–285. doi: 10.1038/ng.314. [DOI] [PubMed] [Google Scholar]
- 23.Erdmann J, Grosshennig A, Braund PS, König IR, Hengstenberg C, Hall AS, Linsel-Nitschke P, Kathiresan S, Wright B, Trégouët DA, et al. New susceptibility locus for coronary artery disease on chromosome 3q22.3. Nat Genet. 2009;41:280–282. doi: 10.1038/ng.307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Elliott P, Chambers JC, Zhang W, Clarke R, Hopewell JC, Peden JF, Erdmann J, Braund P, Engert JC, Bennett D, et al. Genetic Loci associated with C-reactive protein levels and risk of coronary heart disease. JAMA. 2009;302:37–48. doi: 10.1001/jama.2009.954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Karvanen J, Silander K, Kee F, Tiret L, Salomaa V, Kuulasmaa K, Wiklund PG, Virtamo J, Saarela O, Perret C, et al. The impact of newly identified loci on coronary heart disease, stroke and total mortality in the MORGAM prospective cohorts. Genet Epidemiol. 2009;33:237–246. doi: 10.1002/gepi.20374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ridker PM, Paré G, Parker AN, Zee RY, Miletich JP, Chasman DI. Polymorphism in the CETP gene region, HDL cholesterol, and risk of future myocardial infarction: Genomewide analysis among 18 245 initially healthy women from the Women's Genome Health Study. Circ Cardiovasc Genet. 2009;2:26–33. doi: 10.1161/CIRCGENETICS.108.817304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Aulchenko YS, Ripatti S, Lindqvist I, Boomsma D, Heid IM, Pramstaller PP, Penninx BW, Janssens AC, Wilson JF, Spector T, et al. Loci influencing lipid levels and coronary heart disease risk in 16 European population cohorts. Nat Genet. 2009;41:47–55. doi: 10.1038/ng.269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Teslovich TM, Musunuru K, Smith AV, Edmondson AC, Stylianou IM, Koseki M, Pirruccello JP, Ripatti S, Chasman DI, Willer CJ, et al. Biological, clinical and population relevance of 95 loci for blood lipids. Nature. 2010;466:707–713. doi: 10.1038/nature09270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kathiresan S, Melander O, Guiducci C, Surti A, Burtt NP, Rieder MJ, Cooper GM, Roos C, Voight BF, Havulinna AS, et al. Six new loci associated with blood low-density lipoprotein cholesterol, high-density lipoprotein cholesterol or triglycerides in humans. Nat Genet. 2008;40:189–197. doi: 10.1038/ng.75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kathiresan S, Willer CJ, Peloso GM, Demissie S, Musunuru K, Schadt EE, Kaplan L, Bennett D, Li Y, Tanaka T, et al. Common variants at 30 loci contribute to polygenic dyslipidemia. Nat Genet. 2009;41:56–65. doi: 10.1038/ng.291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Chasman DI, Paré G, Mora S, Hopewell JC, Peloso G, Clarke R, Cupples LA, Hamsten A, Kathiresan S, Mälarstig A, et al. Forty-three loci associated with plasma lipoprotein size, concentration, and cholesterol content in genome-wide analysis. PLoS Genet. 2009;5:e1000730. doi: 10.1371/journal.pgen.1000730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Chasman DI, Paré G, Zee RY, Parker AN, Cook NR, Buring JE, Kwiatkowski DJ, Rose LM, Smith JD, Williams PT, et al. Genetic loci associated with plasma concentration of low-density lipoprotein cholesterol, high-density lipoprotein cholesterol, triglycerides, apolipoprotein A1, and Apolipoprotein B among 6382 white women in genome-wide analysis with replication. Circ Cardiovasc Genet. 2008;1:21–30. doi: 10.1161/CIRCGENETICS.108.773168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Heid IM, Boes E, Müller M, Kollerits B, Lamina C, Coassin S, Gieger C, Döring A, Klopp N, Frikke-Schmidt R, et al. Genome-wide association analysis of high-density lipoprotein cholesterol in the population-based KORA study sheds new light on intergenic regions. Circ Cardiovasc Genet. 2008;1:10–20. doi: 10.1161/CIRCGENETICS.108.776708. [DOI] [PubMed] [Google Scholar]
- 34.Wallace C, Newhouse SJ, Braund P, Zhang F, Tobin M, Falchi M, Ahmadi K, Dobson RJ, Marçano AC, Hajat C, et al. Genome-wide association study identifies genes for biomarkers of cardiovascular disease: serum urate and dyslipidemia. Am J Hum Genet. 2008;82:139–149. doi: 10.1016/j.ajhg.2007.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kooner JS, Chambers JC, Aguilar-Salinas CA, Hinds DA, Hyde CL, Warnes GR, Gómez Pérez FJ, Frazer KA, Elliott P, Scott J, et al. Genome-wide scan identifies variation in MLXIPL associated with plasma triglycerides. Nat Genet. 2008;40:149–151. doi: 10.1038/ng.2007.61. [DOI] [PubMed] [Google Scholar]
- 36.Talmud PJ, Drenos F, Shah S, Shah T, Palmen J, Verzilli C, Gaunt TR, Pallas J, Lovering R, Li K, et al. Gene-centric association signals for lipids and apolipoproteins identified via the HumanCVD BeadChip. Am J Hum Genet. 2009;85:628–642. doi: 10.1016/j.ajhg.2009.10.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kamatani Y, Matsuda K, Okada Y, Kubo M, Hosono N, Daigo Y, Nakamura Y, Kamatani N. Genome-wide association study of hematological and biochemical traits in a Japanese population. Nat Genet. 2010;42:210–215. doi: 10.1038/ng.531. [DOI] [PubMed] [Google Scholar]
- 38.Saxena R, Voight BF, Lyssenko V, Burtt NP, de Bakker PI, Chen H, Roix JJ, Kathiresan S, Hirschhorn JN, Daly MJ, et al. Genome-wide association analysis identifies loci for type 2 diabetes and triglyceride levels. Science. 2007;316:1331–1336. doi: 10.1126/science.1142358. [DOI] [PubMed] [Google Scholar]
- 39.Hiura Y, Shen CS, Kokubo Y, Okamura T, Morisaki T, Tomoike H, Yoshida T, Sakamoto H, Goto Y, Nonogi H, et al. Identification of genetic markers associated with high-density lipoprotein-cholesterol by genome-wide screening in a Japanese population: the Suita study. Circ J. 2009;73:1119–1126. doi: 10.1253/circj.cj-08-1101. [DOI] [PubMed] [Google Scholar]
- 40.Sandhu MS, Waterworth DM, Debenham SL, Wheeler E, Papadakis K, Zhao JH, Song K, Yuan X, Johnson T, Ashford S, et al. LDL-cholesterol concentrations: a genome-wide association study. Lancet. 2008;371:483–491. doi: 10.1016/S0140-6736(08)60208-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Levy D, Larson MG, Benjamin EJ, Newton-Cheh C, Wang TJ, Hwang SJ, Vasan RS, Mitchell GF. Framingham Heart Study 100K Project: genome-wide associations for blood pressure and arterial stiffness. BMC Med Genet. 2007;8 Suppl 1:S3. doi: 10.1186/1471-2350-8-S1-S3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Newton-Cheh C, Johnson T, Gateva V, Tobin MD, Bochud M, Coin L, Najjar SS, Zhao JH, Heath SC, Eyheramendy S, et al. Genome-wide association study identifies eight loci associated with blood pressure. Nat Genet. 2009;41:666–676. doi: 10.1038/ng.361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Levy D, Ehret GB, Rice K, Verwoert GC, Launer LJ, Dehghan A, Glazer NL, Morrison AC, Johnson AD, Aspelund T, et al. Genome-wide association study of blood pressure and hypertension. Nat Genet. 2009;41:677–687. doi: 10.1038/ng.384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Adeyemo A, Gerry N, Chen G, Herbert A, Doumatey A, Huang H, Zhou J, Lashley K, Chen Y, Christman M, et al. A genome-wide association study of hypertension and blood pressure in African Americans. PLoS Genet. 2009;5:e1000564. doi: 10.1371/journal.pgen.1000564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kato N, Miyata T, Tabara Y, Katsuya T, Yanai K, Hanada H, Kamide K, Nakura J, Kohara K, Takeuchi F, et al. High-density association study and nomination of susceptibility genes for hypertension in the Japanese National Project. Hum Mol Genet. 2008;17:617–627. doi: 10.1093/hmg/ddm335. [DOI] [PubMed] [Google Scholar]
- 46.Yang HC, Liang YJ, Wu YL, Chung CM, Chiang KM, Ho HY, Ting CT, Lin TH, Sheu SH, Tsai WC, et al. Genome-wide association study of young-onset hypertension in the Han Chinese population of Taiwan. PLoS One. 2009;4:e5459. doi: 10.1371/journal.pone.0005459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Wang Y, O'Connell JR, McArdle PF, Wade JB, Dorff SE, Shah SJ, Shi X, Pan L, Rampersaud E, Shen H, et al. From the Cover: Whole-genome association study identifies STK39 as a hypertension susceptibility gene. Proc Natl Acad Sci U S A. 2009;106:226–231. doi: 10.1073/pnas.0808358106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Org E, Eyheramendy S, Juhanson P, Gieger C, Lichtner P, Klopp N, Veldre G, Döring A, Viigimaa M, Sõber S, et al. Genome-wide scan identifies CDH13 as a novel susceptibility locus contributing to blood pressure determination in two European populations. Hum Mol Genet. 2009;18:2288–2296. doi: 10.1093/hmg/ddp135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Cho YS, Go MJ, Kim YJ, Heo JY, Oh JH, Ban HJ, Yoon D, Lee MH, Kim DJ, Park M, et al. A large-scale genome-wide association study of Asian populations uncovers genetic factors influencing eight quantitative traits. Nat Genet. 2009;41:527–534. doi: 10.1038/ng.357. [DOI] [PubMed] [Google Scholar]
- 50.Padmanabhan S, Melander O, Johnson T, Di Blasio AM, Lee WK, Gentilini D, Hastie CE, Menni C, Monti MC, Delles C, et al. Genome-wide association study of blood pressure extremes identifies variant near UMOD associated with hypertension. PLoS Genet. 2010;6:e1001177. doi: 10.1371/journal.pgen.1001177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Ioannidis JP. Prediction of cardiovascular disease outcomes and established cardiovascular risk factors by genome-wide association markers. Circ Cardiovasc Genet. 2009;2:7–15. doi: 10.1161/CIRCGENETICS.108.833392. [DOI] [PubMed] [Google Scholar]
- 52.Morrison AC, Bare LA, Chambless LE, Ellis SG, Malloy M, Kane JP, Pankow JS, Devlin JJ, Willerson JT, Boerwinkle E. Prediction of coronary heart disease risk using a genetic risk score: the Atherosclerosis Risk in Communities Study. Am J Epidemiol. 2007;166:28–35. doi: 10.1093/aje/kwm060. [DOI] [PubMed] [Google Scholar]
- 53.Paynter NP, Chasman DI, Buring JE, Shiffman D, Cook NR, Ridker PM. Cardiovascular disease risk prediction with and without knowledge of genetic variation at chromosome 9p21.3. Ann Intern Med. 2009;150:65–72. doi: 10.7326/0003-4819-150-2-200901200-00003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Kathiresan S, Melander O, Anevski D, Guiducci C, Burtt NP, Roos C, Hirschhorn JN, Berglund G, Hedblad B, Groop L, et al. Polymorphisms associated with cholesterol and risk of cardiovascular events. N Engl J Med. 2008;358:1240–1249. doi: 10.1056/NEJMoa0706728. [DOI] [PubMed] [Google Scholar]
- 55.Talmud PJ, Cooper JA, Palmen J, Lovering R, Drenos F, Hingorani AD, Humphries SE. Chromosome 9p21.3 coronary heart disease locus genotype and prospective risk of CHD in healthy middle-aged men. Clin Chem. 2008;54:467–474. doi: 10.1373/clinchem.2007.095489. [DOI] [PubMed] [Google Scholar]
- 56.Paynter NP, Chasman DI, Paré G, Buring JE, Cook NR, Miletich JP, Ridker PM. Association between a literature-based genetic risk score and cardiovascular events in women. JAMA. 2010;303:631–637. doi: 10.1001/jama.2010.119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Rosenberg S, Elashoff MR, Beineke P, Daniels SE, Wingrove JA, Tingley WG, Sager PT, Sehnert AJ, Yau M, Kraus WE, et al. Multicenter validation of the diagnostic accuracy of a blood-based gene expression test for assessing obstructive coronary artery disease in nondiabetic patients. Ann Intern Med. 2010;153:425–434. doi: 10.7326/0003-4819-153-7-201010050-00005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Ripatti S, Tikkanen E, Orho-Melander M, Havulinna AS, Silander K, Sharma A, Guiducci C, Perola M, Jula A, Sinisalo J, et al. A multilocus genetic risk score for coronary heart disease: case-control and prospective cohort analyses. Lancet. 2010;376:1393–1400. doi: 10.1016/S0140-6736(10)61267-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Pepe MS, Feng Z, Gu JW. Comments on 'Evaluating the added predictive ability of a new marker: From area under the ROC curve to reclassification and beyond' by M. J. Pencina et al., Statistics in Medicine (DOI: 10.1002/sim.2929) Stat Med. 2008;27:173–181. doi: 10.1002/sim.2991. [DOI] [PubMed] [Google Scholar]
- 60.Hlatky MA, Greenland P, Arnett DK, Ballantyne CM, Criqui MH, Elkind MS, Go AS, Harrell FE, Hong Y, Howard BV, et al. Criteria for evaluation of novel markers of cardiovascular risk: a scientific statement from the American Heart Association. Circulation. 2009;119:2408–2416. doi: 10.1161/CIRCULATIONAHA.109.192278. [DOI] [PMC free article] [PubMed] [Google Scholar]