Abstract
Ribozymes are RNA molecules that act as chemical catalysts. In contemporary cells, most known ribozymes carry out phosphoryl transfer reactions. The nucleolytic ribozymes comprise a class of five structurally-distinct species that bring about site-specific cleavage by nucleophilic attack of the 2′-O on the adjacent 3′-P to form a cyclic 2′,3′-phosphate. In general, they will also catalyse the reverse reaction. As a class, all these ribozymes appear to use general acid–base catalysis to accelerate these reactions by about a million-fold. In the Varkud satellite ribozyme, we have shown that the cleavage reaction is catalysed by guanine and adenine nucleobases acting as general base and acid, respectively. The hairpin ribozyme most probably uses a closely similar mechanism. Guanine nucleobases appear to be a common choice of general base, but the general acid is more variable. By contrast, the larger ribozymes such as the self-splicing introns and RNase P act as metalloenzymes.
Keywords: ribozymes, catalytic mechanism, general acid–base catalysis, metal ion catalysis
1. RNA catalysis and contemporary ribozymes
Ribozymes are RNA molecules that act as chemical catalysts [1]. In the modern biosphere, the majority of known ribozymes carry out a rather limited range of reactions, mostly involving phosphoryl transfer, notably transesterification or hydrolysis reactions. The discovery that peptidyl transferase is catalysed by the rRNA component of the large ribosomal subunit [2,3] significantly extends the range of chemistry to include the condensation of an amine with an sp2-hybridized carbonyl centre. A greater range of chemical reactions may be catalysed by RNA species selected in the laboratory [4–14], so that ribozymes catalysing a broader set of reactions may conceivably have existed in the past.
Contemporary ribozymes are used for a variety of biological purposes. The nucleolytic ribozymes bring about the site-specific cleavage of RNA by attack of a 2′-hydroxyl group on the adjacent 3′-phosphorus (or by the 5′-hydroxyl group in the reverse reaction). This is used in the processing of replication intermediates, and in the control of gene expression. Ribonuclease P carries out the processing of tRNA in all domains of life, using a hydrolytic reaction [15,16]. A number of introns are spliced out autocatalytically by ribozyme action, initiated either by the attack of a 2′-hydroxyl group located remotely within the intron (group II introns [17]), or by an exogenous guanosine molecule (group I introns [18]). The similarity of the chemistry of mRNA splicing in the spliceosome to that of the group II introns makes it very likely that this too is at least partially RNA catalysed, where snU4/U6 RNA is the ribozyme. Finally, the peptidyl transferase activity of the ribosome catalyses the condensation of amino acids into polypeptides, which is one of the most important reactions of the cell.
In modern biology, most reactions are accelerated by protein enzymes, and it is not hard to see why. The chemical variety of amino acid side chains leads to a wide range of possible catalytic chemistries. Protein enzymes can achieve some extraordinary catalytic rate enhancements. Values of approximately 1018-fold are known. RNA catalysts tend to produce more modest rate enhancements. For example, the nucleolytic ribozymes typically accelerate their transesterification reactions by around a million-fold relative to the uncatalysed reaction in a dinucleotide, with resulting rates of around 1 min−1. For those ribozymes, this may be as fast as it needs to be, so that there is little evolutionary pressure to make them faster. Redesign of some ribozymes has resulted in quite respectable rates of greater than or equal to 10 s−1 [19,20].
Protein structure is based on an electrically neutral backbone, with side chains that introduce a broad range of chemistries, including carboxylic acids, amines, hydroxyl and thiol groups as well as hydrophobic side chains that may be either aliphatic or aromatic. In contrast, RNA comprises four nucleotide bases of similar chemical nature, connected by an electrically-charged ribose-phosphate backbone. So what resources can RNA bring to catalytic chemistry?
First, RNA might exploit its structure. Substrate binding can result in acceleration of reaction rate owing to proximity and orientation, together with structural stabilization of the transition state.
Second, as a polyelectrolyte, RNA can create specific metal ion-binding sites, or pockets that have a high occupancy of atmospherically bound ions. Metal ions can activate nucleophiles, or provide electrostatic stabilization of negative charge such as a phosphorane transition state. Water molecules contained within the inner sphere of coordination can, in principle, participate in general acid–base catalysis.
Third, the nucleobases could participate directly in the catalytic chemistry in a variety of ways. They have hydrogen bond donors and acceptors that can be used to bind the substrate, and potentially to stabilize a transition state. They could also act as general acids and bases, although at first sight their pKa values are not ideally suited to this role at neutral pH values. Adenine N1 and cytosine N3 have low pKa values, whereas those of guanine N1 and uracil N3 are relatively high. To take one example, cytosine (pKa approx. 4) is a relatively strong acid, but only one molecule in 1000 is protonated at neutral pH. Thus, most ribozymes will be in the wrong form to carry out a protonation (having no proton to donate). The great majority of molecules are in the deprotonated form and able to act as a general base, but the conjugate base of a strong acid will be rather unreactive. However, the situation can be improved because nearby anionic phosphate groups may raise the pKa significantly, and values of 5.5–6.5 are quite possible [21,22] making the nucleobase more available as an acid. Similarly, the pKa of guanine might be reduced if it is located close to a bound metal ion, thereby making it basic at a lower pH.
2. How to catalyse phosphoryl transfer reactions—some indications from protein enzymes
Do contemporary protein-based enzymes provide some indications as to how phosphoryl transfer reactions might be catalysed by RNA molecules? Such an analysis suggests two alternative ways in which these reactions can be accelerated.
One solution to the problem is provided by pancreatic ribonuclease A, which brings about cleavage of the RNA backbone via the formation of a cyclic 2′,3′-phosphate equivalent to the nucleolytic ribozymes [23–25]. It uses the imidazole side chains of two histidine residues in general acid–base catalysis, and a lysine side chain to stabilize the phosphorane transition state. Imidazole is well suited to its role, as it has a pKa close to neutrality. The problem for RNA is that there is no equivalent to histidine (although this has been engineered as an experimental tool [26–28]). The nucleobases are aromatic heterocyclic bases, but as discussed above, their pKa values are normally at least two to three units removed from neutrality.
A second way in which enzymes catalyse phosphoryl transfer reactions is to use bound metal ions. Many polymerases, nucleases, transposases and phospholipases employ variations of a two metal ion mechanism [29], in which one ion activates the nucleophile, while the other acts as Lewis acid to labilize the oxyanion leaving group. Steitz & Steitz [30] made an early prediction that the self-splicing introns of group I and II would operate via two metal ion mechanisms. Clearly, the polyelectrolyte character of RNA makes the coordination of metal ions at specific sites quite feasible.
General acid–base catalysis could operate in parallel with a role for metal ions, for which there are precedents among the protein enzymes [31]. This is quite probable for the nucleolytic ribozymes.
3. General acid–base catalysis in the nucleolytic ribozymes
The nucleolytic ribozymes [1] are a class of five known species that bring about site-specific cleavage of RNA by attack of the 2′-O on its adjacent 3′-P to generate a cyclic 2′,3′-phosphate and a 5′-O (figure 1a). In general, they will also catalyse the reverse (ligation) reaction. The reactions follow an SN2 mechanism, with inversion of configuration at the phosphate, and are accelerated by at least 105-fold when catalysed by the ribozymes. They are the smallest of the ribozymes, of around 70–150 nt in size.
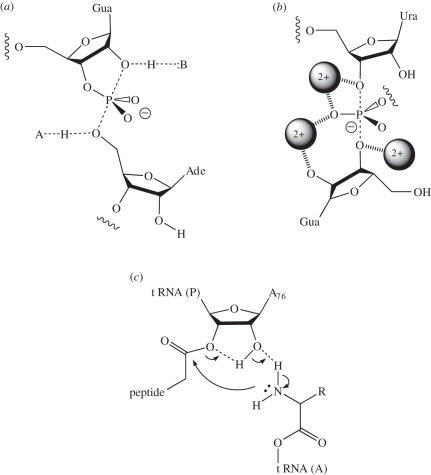
Figure 1.
Three proposed mechanisms for natural ribozymes. (a) General acid–base catalysis in the nucleolytic ribozymes. In the cleavage reaction, B acts as a general base to remove the proton from the 2′-O nucleophile, whereas A donates a proton to the 5′-oxyanion leaving group. In the ligation reaction, the roles are reversed. In the VS (shown) and hairpin ribozymes, B and A are guanine and adenine nucleobases, respectively. (b) RNA metalloenzymes. The self-splicing introns and RNase P act as metalloenzymes. Atomic substitution experiments indicate that three metal ions are bound to the transition state of the group I intron as shown. Two of the metal ion binding sites have been confirmed by crystallography, but that shown bound to the 3′-O of the exogenous guanosine has not been observed in the Azoarcus ribozyme crystal structure. (c) A possible mechanism of peptidyl transferase in the ribosome. The proposed proton shuttle between the nucleophile and the carbonyl target involving the 2′-O of the terminal A76 of the P-site tRNA is shown.
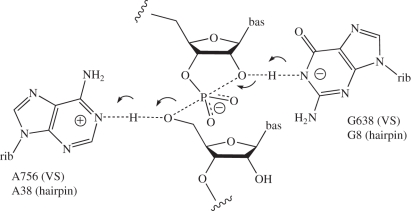
The Varkud satellite (VS) ribozyme [32] is the largest of the nucleolytic ribozymes, and the only one for which there is no crystal structure at present. However, we have used data from small-angle X-ray scattering (SAXS) in solution to propose a model for the complete ribozyme [33] that is broadly in agreement with the earlier studies [34,35]. A trans-acting core of the ribozyme can be released, formed by five helices connected by two three-way junctions. Cleavage and ligation reactions occur within the internal loop—a stem-loop structure that docks into the trans-acting ribozyme. Two key candidate nucleotides important in catalysis have been identified. These are A756 in the body of the ribozyme [26,36–39], and G638 within the internal loop of the substrate stem-loop [40]. Substitution of either nucleotide leads to an impairment of activity by 103- to 104-fold. The combined action of these two nucleotides was demonstrated by complementation experiments in mixtures of cis-acting VS ribozymes [41].
Kinetic isotope effects in the fast, cis-acting form show that proton transfer occurs in the transition state of the cleavage reaction [42]. Present evidence points strongly towards general acid–base catalysis by the nucleobases of A756 and G638. In particular, the pH dependence of the cleavage reaction rate is bell-shaped, corresponding to apparent pKa values of 5.2 and 8.4 [40], and a closely similar bell-shaped pH dependence with pKa values of 5.8 and 8.3 was also observed using the fast-cleaving cis-acting form of the ribozyme [42]. Nucleotide substitution experiments have consistently shown a correlation between the pKa of the nucleobase at position 638 and the observed pKa of the cleavage reaction. 5′-Phosphorothiolate substitution (5′-PS) experiments indicate that A756 is the general acid in the cleavage reaction. While the cleavage activity of VS A756G was impaired 1000-fold on the oxy (5′-PO) substrate, activity was completely restored for the 5′-PS-containing substrate [43]. Thus, cleavage of the 5′-PS substrate is insensitive to substitution at position 756, and therefore A756 is likely to be the general acid for the cleavage reaction. The pH dependence of cleavage reactions on 5′-PS substrates was completely consistent with a sole requirement for the nucleobase at position 638, acting as general base in the cleavage reaction. Thus, all the available data are consistent with a catalytic mechanism for the cleavage reaction in which G638 acts as general base to deprotonate the 2′-O nucleophile, and A756 is the general acid protonating the 5′-oxyanion leaving group. Of course, by the principle of microscopic reversibility, in the ligation reaction, protonated G638 should act as the general acid protonating the 2′-oxyanion leaving group, and unprotonated A756 as the general base deprotonating the 5′-O nucleophile that attacks the cyclic phosphate.
It is very likely that the VS and hairpin ribozymes are mechanistically closely similar. Although the global architecture of the two ribozymes is quite different, both their active sites appear to be generated by loop–loop interaction, and in both cases an active guanine lies on the opposing strand of the internal loop harbouring the scissile phosphate, whereas an active adenine is provided by the second loop [43,44]. Indeed, if we draw the secondary structures of the two ribozymes to include the loop–loop interactions, then it becomes clear that the polarity of the strands and the relative positioning of the adenine, guanine and sissile phosphate are the same in both cases. In the crystal structure of the hairpin ribozyme [45], the positions of G8 and A38 are consistent with roles of base and acid, respectively, in the cleavage reaction, corresponding to the proposed functions of G638 and A756 in the VS ribozyme. This suggests a fundamental similarity between the active sites of the two ribozymes, perhaps reflecting a deeper similarity in catalytic mechanism [46]. Although there is a degree of confusion in the literature concerning the mechanism of the hairpin ribozyme, all the available experimental data are consistent with the general acid–base catalysis by G8 and A38, as we have critically assessed recently [46]. Furthermore, we are not aware of any data that are inconsistent with this mechanism for either ribozyme, summarized in figure 2.
Figure 2.
The proposed mechanism for the VS and hairpin ribozymes based on general acid–base catalysis by a guanine and an adenine nucleobase.
It seems at this time that all the nucleolytic ribozymes employ general acid–base catalysis to derive their main rate enhancement. Guanine nucleobases appear to be candidates for the role of general base in cleavage reactions catalysed by the hammerhead [47,48] and GlmS [49–51] ribozymes. On the basis of the most recent crystal structure, the hammerhead ribozyme has been proposed to use a 2′-hydroxyl group as the general acid in cleavage reaction [52], but there are no chemical data to support this suggestion at present. The GlmS ribozyme is also a riboswitch, operating with exogenous glucosamine-6-phosphate as its small-molecule effector. Most interestingly, it turns out that the glucosamine-6-phosphate is actually a co-enzyme, its amine acting as the general acid in cleavage reaction [51,53]. This sets an intriguing precedent for the extension of catalytic activity by ribozymes. As noted above, the range of functional groups available to RNA is severely limited, especially when compared with proteins. However, if that can be augmented by the recruitment of other small molecules that are selectively bound to RNA, then this could greatly widen the range of chemistry available to RNA. The riboswitches [54] and laboratory-selected aptamers [55–57] show how selectively and tightly small molecules can bind to RNA, and one can readily speculate on how amino acids and peptides bound to RNA might have been an important step towards the protein-based metabolism of current biology.
The hepatitis delta virus ribozyme is something of an outlier. It is a eukaryotic species, originally viral and now detected in the human genome [58]. Unlike the other nucleolytic ribozymes, it uses a Mg2+ ion to activate the nucleophile, either as general base or Lewis acid [59,60], and a cytosine nucleobase as general acid [59,61–63]. This is the only nucleolytic ribozyme that employs a site-bound metal ion. The other members of this class are active in high concentrations of monovalent metal ions [64], so that the direct participation of a site-bound metal ion as a Lewis acid, or in general acid–base catalysis, is not probable. An electrostatic role of non-site-bound metal ions, however, remains possible in the stabilization of the active site.
We may ask how efficient is nucleobase-mediated general acid–base catalysis in the ribozymes? The fastest observed rate of cleavage for the VS ribozyme acting in trans is approximately 12 min−1, but the intrinsic rate (kcat) is much higher than that because the observed rate (kobs) will be limited by the small fraction of ribozyme molecules that have a protonated A756 (i.e. fA) and a deprotonated G638 (fB), i.e.
We can calculate fAfB from the measured pKa values for A756 and G638 [40] to be 3.8 × 10−4, and hence kcat is approximately 520 s−1. Collins and co-workers have engineered even faster forms of the cis ribozyme [20], so the value we calculate for kcat is likely to be a lower limit. Using two histidine side chains as general acid and base, pancreatic ribonuclease carries out the same cleavage of a phosphodiester linkage in RNA with an observed rate of ≤ 1400 s−1 [65]. Given the pKa for acid and base of 6.2 and 5.8, respectively [66], the maximum kcat is approximately 3700 s−1. Thus, the rate of cleavage by VS ribozyme molecules that are in the correct ionization state is comparable with that of RNase A. The principle limitation on the rates of the RNA-catalysed reaction is the unfavourable pKa values of the nucleobases. Ultimately, this would have given polypeptide-based enzymes a powerful evolutionary advantage.
4. Metal ion catalysis in the larger ribozymes
In contrast to the nucleolytic ribozymes, most of the larger ribozymes such as RNase P and the self-splicing introns appear to act as metalloenzymes (figure 1b). Two metal ions have been found at the active site of the Azoarcus group I intron ribozyme [67], and in the putative active site of a group II intron ribozyme [68]. In these structures, metal ions are coordinated by multiple oxygen atoms of the RNA forming inner sphere complexes. Atomic mutagenesis methods in which the activity of specific sulphur or nitrogen substitutions is restored by soft metal ions [69] have rigorously defined the roles of the ions and their ligands [70] in the transition state of the reaction. Analysis of the first step of a group I intron ribozyme has shown the importance of three ions, bound to the 3′O and pro-S O of the target phosphate (potentially stabilizing developing negative charge on the 3′O leaving group in the transition state), the pro-S O and the 2′O of the guanosine (thereby bridging the two reactants), and the 3′O of the guanosine (probably activating the nucleophile). No ion was observed bound at the latter site in the Azoarcus ribozyme crystal structure [71]. A similar mechanism could occur in the group II intron ribozymes, and there is chemical evidence that this ribozyme acts as a metalloenzyme [72,73]. A closely similar chemistry is performed by the spliceosome in eukaryotic cells. Some domains of spliceosomal RNA show sequence similarity with the catalytic domains of the intron, and there is once again chemical evidence for their participation in catalysis [74]. However, recent structural data offer the suggestion that the key protein Prp8 may assist in the splicing reactions. X-ray crystal structures of a domain of Prp8 reveal a structure that is related to RNase H [75,76], and a plausible model places the splice site adjacent to the vestigial RNase H active site. However, this possible active site lacks two of the four key cation-binding residues observed in RNase H [77], and no metal ion binding has been detected [75]; so it seems unlikely that Prp8 carries out the chemistry unaided. Thus, the spliceosome may be a kind of hybrid RNA–protein enzyme, and perhaps a model of how evolution may have made the transition from the RNA world to the modern protein world.
5. The mechanism of peptidyl transferase on the ribosome
The peptidyltransferase centre of the ribosome accelerates amino acid condensation by 107 fold [78]. The reaction contrasts with those of the preceding ribozymes in that the nucleophile is an amine and the target an sp2-hybridized carbonyl centre, forming and resolving a tetrahedral intermediate. There are no data supporting metal ion participation, and none has been observed in the active centre by crystallography, so at present it seems that the ribosome is not a metalloenzyme. The reaction is pH-insensitive and neither nucleobases nor metal ions have been shown to be critical for catalysis. However, the key role of the A76 2′-hydroxyl of the P site tRNA has been established [79], leading to a proposed mechanism wherein a proton is shuttled from the nucleophile via the 2′O to the 3′O leaving group [80] (figure 1c). The Brønsted coefficient of the nucleophile is close to zero [81], consistent with a fully concerted proton shuttle mechanism in which there is simultaneous making and breaking of all bonds, obviating a shift in the pKa of the 2′ hydroxyl. The ribosome achieves the significant acceleration over the uncatalysed reaction through a favourable change in entropy [78]. While no ribosomal groups directly participate in catalysis, an extensive network of hydrogen bonds serves to position and align the substrates. Furthermore, the ribosome provides a desolvated environment in which the reaction can proceed, shielded from the unfavourable entropic effects of solvent reorganization by the large structure of the ribosome.
6. Conclusion
Most contemporary ribozymes accelerate phosphoryl transfer reactions of various kinds—transesterification, nucleotidyl transfer or hydrolysis reactions. Moreover, in echoes of the protein enzymes, the majority operate either by general acid–base catalysis or as metalloenzymes. All the available evidence points towards the (generally smaller) nucleolytic ribozymes using general acid–base catalysis, whereas the (mostly larger) self-splicing introns and RNase P employ specifically bound metal ions.
The nucleolytic ribozymes often but not exclusively exploit their nucleobases in general acid–base catalysis, and guanine nucleobases are used as the general base in cleavage by four of the five known ribozymes. The acid used is more variable, including adenine (in two cases), cytosine and glucosamine-6-phosphate. General acid–base catalysis is potentially efficient, but severely limited by the pKa values of the nucleobases. There may be rather few solutions to the problem of how to design such an activity in RNA alone. The fact that only five distinct kinds of nucleolytic ribozyme have been discovered (the last of which, GlmS, is already 6 years old [82]) suggests that we might have discovered all that there are. Indeed, although there are a number of examples of hairpin and hammerhead ribozymes, there has been only a single isolate of the VS ribozyme indicating that it is a very rare species.
While most cellular reactions are catalysed by proteins, just a few ribozymes are present in the modern world. Most of these do not require to turn over, which is perhaps why these roles have not been usurped by protein enzymes. Thus perhaps only the least demanding reactions remain from an RNA world. Might a greater range of RNA-based chemistry have been available in early evolution of life on the planet? In vitro selection of RNA species capable of catalysing reactions such as Michael addition [10] and Diels–Alder cycloaddition [7] suggest this should be the case. However, no trace of these remains in current metabolism. Moreover, structures of in vitro-selected ribozymes where known suggest that they operate in a relatively crude way. The Diels–Alder ribozyme essentially puts the two reactants together to promote reaction [83], which is probably all that is required for this electrocyclic reaction. The structure of a selected RNA ligase reveals no indication of nucleobase involvement in the chemistry [84], but rather may function as a metalloenzyme. Of course, another limitation of RNA catalysts is the very limited range of functional groups that they can bring to bear on the chemistry. However, the GlmS ribozyme and the group I intron ribozyme show that small molecules (glucosamine-6-phosphate and guanosine, respectively) can be bound to participate in the chemistry. Recruitment of other small molecules could easily be envisaged, thus rapidly widening the available chemistries, and might have led to a protein enzyme-based metabolism relatively swiftly.
References
- 1.Lilley D. M. J., Eckstein F. (eds) 2008. Ribozymes and RNA catalysis. Cambridge, UK: Royal Society of Chemistry [Google Scholar]
- 2.Noller H. F., Hoffarth V., Zimniak L. 1992. Unusual resistance of peptidyl transferase to protein extraction procedures. Science 256, 1416–1419 10.1126/science.1604315 (doi:10.1126/science.1604315) [DOI] [PubMed] [Google Scholar]
- 3.Nissen P., Hansen J., Ban N., Moore P. B., Steitz T. A. 2000. The structural basis of ribosome activity in peptide bond synthesis. Science 289, 920–930 10.1126/science.289.5481.920 (doi:10.1126/science.289.5481.920) [DOI] [PubMed] [Google Scholar]
- 4.Ekland E. H., Szostak J. W., Bartel D. P. 1995. Structurally complex and highly active RNA ligases derived from random RNA sequences. Science 269, 364–370 10.1126/science.7618102 (doi:10.1126/science.7618102) [DOI] [PubMed] [Google Scholar]
- 5.Pan T. 1997. Novel and variant ribozymes obtained through in vitro selection. Curr. Opin. Chem. Biol. 1, 17–25 10.1016/S1367-5931(97)80104-4 (doi:10.1016/S1367-5931(97)80104-4) [DOI] [PubMed] [Google Scholar]
- 6.Unrau P. J., Bartel D. P. 1998. RNA-catalysed nucleotide synthesis. Nature 395, 260–263 10.1038/26193 (doi:10.1038/26193) [DOI] [PubMed] [Google Scholar]
- 7.Seelig B., Jäschke A. 1999. A small catalytic RNA motif with Diels–Alderase activity. Chem. Biol. 6, 167–176 10.1016/S1074-5521(99)89008-5 (doi:10.1016/S1074-5521(99)89008-5) [DOI] [PubMed] [Google Scholar]
- 8.Johnston W. K., Unrau P. J., Lawrence M. S., Glasner M. E., Bartel D. P. 2001. RNA-catalyzed RNA polymerization: accurate and general RNA-templated primer extension. Science 292, 1319–1325 10.1126/science.1060786 (doi:10.1126/science.1060786) [DOI] [PubMed] [Google Scholar]
- 9.Robertson M. P., Hesselberth J. R., Ellington A. D. 2001. Optimization and optimality of a short ribozyme ligase that joins non-Watson–Crick base pairings. RNA 7, 513–523 10.1017/S1355838201002199 (doi:10.1017/S1355838201002199) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sengle G., Eisenfuh R. A., Arora P. S., Nowick J. S., Famulok M. 2001. Novel RNA catalysts for the Michael reaction. Chem. Biol. 8, 459–473 10.1016/S1074-5521(01)00026-6 (doi:10.1016/S1074-5521(01)00026-6) [DOI] [PubMed] [Google Scholar]
- 11.Paul N., Joyce G. F. 2002. A self-replicating ligase ribozyme. Proc. Natl Acad. Sci. USA 99, 12 733–12 740 10.1073/pnas.202471099 (doi:10.1073/pnas.202471099) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Voytek S. B., Joyce G. F. 2007. Emergence of a fast-reacting ribozyme that is capable of undergoing continuous evolution. Proc. Natl Acad. Sci. USA 104, 15 288–15 293 10.1073/pnas.0707490104 (doi:10.1073/pnas.0707490104) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Xiao H., Murakami H., Suga H., Ferre-D'Amare A. R. 2008. Structural basis of specific tRNA aminoacylation by a small in vitro selected ribozyme. Nature 454, 358–361 10.1038/nature07033 (doi:10.1038/nature07033) [DOI] [PubMed] [Google Scholar]
- 14.Lincoln T. A., Joyce G. F. 2009. Self-sustained replication of an RNA enzyme. Science 323, 1229–1232 10.1126/science.1167856 (doi:10.1126/science.1167856) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Guerrier-Takada C., Gardiner K., Marsh T., Pace N., Altman S. 1983. The RNA moiety of ribonuclease P is the catalytic subunit of the enzyme. Cell 35, 849–857 10.1016/0092-8674(83)90117-4 (doi:10.1016/0092-8674(83)90117-4) [DOI] [PubMed] [Google Scholar]
- 16.Kazantsev A. V., Pace N. R. 2006. Bacterial RNase P: a new view of an ancient enzyme. Nat. Rev. Microbiol. 4, 729–740 10.1038/nrmicro1491 (doi:10.1038/nrmicro1491) [DOI] [PubMed] [Google Scholar]
- 17.Peebles C. L., Perlman P. S., Mecklenburg K. L., Petrillo M. L., Tabor J. H., Jarrell K. A., Cheng H.-L. 1986. A self-splicing RNA excises an intron lariat. Cell 44, 213–223 10.1016/0092-8674(86)90755-5 (doi:10.1016/0092-8674(86)90755-5) [DOI] [PubMed] [Google Scholar]
- 18.Cech T. R., Zaug A. J., Grabowski P. J. 1981. In vitro splicing of the ribosomal RNA precursor of Tetrahymena: involvement of a guanosine nucleotide in the excision of the intervening sequence. Cell 27, 487–496 10.1016/0092-8674(81)90390-1 (doi:10.1016/0092-8674(81)90390-1) [DOI] [PubMed] [Google Scholar]
- 19.Canny M. D., Jucker F. M., Kellogg E., Khvorova A., Jayasena S. D., Pardi A. 2004. Fast cleavage kinetics of a natural hammerhead ribozyme. J. Am. Chem. Soc. 126, 10 848–10 849 10.1021/ja046848v (doi:10.1021/ja046848v) [DOI] [PubMed] [Google Scholar]
- 20.Zamel R., Poon A., Jaikaran D., Andersen A., Olive J., De Abreu D., Collins R. A. 2004. Exceptionally fast self-cleavage by a Neurospora Varkud satellite ribozyme. Proc. Natl Acad. Sci. USA 101, 1467–1472 10.1073/pnas.0305753101 (doi:10.1073/pnas.0305753101) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Legault P., Pardi A. 1997. Unusual dynamics and pKa shift at the active site of a lead-dependent ribozyme. J. Am. Chem. Soc. 119, 6621–6628 10.1021/ja9640051 (doi:10.1021/ja9640051) [DOI] [Google Scholar]
- 22.Ravindranathan S., Butcher S. E., Feigon J. 2000. Adenine protonation in domain B of the hairpin ribozyme. Biochemistry 39, 16 026–16 032 10.1021/bi001976r (doi:10.1021/bi001976r) [DOI] [PubMed] [Google Scholar]
- 23.Wyckoff H. W., Tsernoglou D., Hanson A. W., Knox J. R., Lee B., Richards F. M. 1970. The three-dimensional structure of ribonuclease-S. Interpretation of an electron density map at a nominal resolution of 2 Å. J. Biol. Chem. 245, 305–328 [PubMed] [Google Scholar]
- 24.Richards F. M., Wyckoff H. W. 1971. Bovine pancreatic ribonuclease. In The enzymes (ed. Boyer P. D.), pp. 647–806 New York, NY: Academic Press [Google Scholar]
- 25.Wodak S. Y., Liu M. Y., Wyckoff H. W. 1977. The structure of cytidilyl (2′,5′) adenosine when bound to pancreatic ribonuclease S. J. Mol. Biol. 116, 855–875 10.1016/0022-2836(77)90275-3 (doi:10.1016/0022-2836(77)90275-3) [DOI] [PubMed] [Google Scholar]
- 26.Zhao Z., McLeod A., Harusawa S., Araki L., Yamaguchi M., Kurihara T., Lilley D. M. J. 2005. Nucleobase participation in ribozyme catalysis. J. Am. Chem. Soc. 127, 5026–5027 10.1021/ja0502775 (doi:10.1021/ja0502775) [DOI] [PubMed] [Google Scholar]
- 27.Wilson T. J., Ouellet J., Zhao Z. Y., Harusawa S., Araki L., Kurihara T., Lilley D. M. J. 2006. Nucleobase catalysis in the hairpin ribozyme. RNA 12, 980–987 10.1261/rna.11706 (doi:10.1261/rna.11706) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Araki L., Morita K., Yamaguchi M., Zhao Z., Wilson T. J., Lilley D. M. J., Harusawa S. 2009. Synthesis of novel C4-linked C2-imidazole ribonucleoside phosphoramidite and its application to probing the catalytic mechanism of a ribozyme. J. Org. Chem. 74, 2350–2356 10.1021/jo802556s (doi:10.1021/jo802556s) [DOI] [PubMed] [Google Scholar]
- 29.Yang W., Lee J. Y., Nowotny M. 2006. Making and breaking nucleic acids: two-Mg2+-ion catalysis and substrate specificity. Mol. Cell 22, 5–13 10.1016/j.molcel.2006.03.013 (doi:10.1016/j.molcel.2006.03.013) [DOI] [PubMed] [Google Scholar]
- 30.Steitz T. A., Steitz J. A. 1993. A general 2-metal-ion mechanism for catalytic RNA. Proc. Natl Acad. Sci. USA 90, 6498–6502 10.1073/pnas.90.14.6498 (doi:10.1073/pnas.90.14.6498) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Suck D., Lahm A., Oefner C. 1988. Structure refined to 2A of a nicked DNA octanucleotide complex with DNase I. Nature 332, 464–468 10.1038/332464a0 (doi:10.1038/332464a0) [DOI] [PubMed] [Google Scholar]
- 32.Beattie T. L., Olive J. E., Collins R. A. 1995. A secondary-structure model for the self-cleaving region of Neurospora VS RNA. Proc. Natl Acad. Sci. USA 92, 4686–4690 10.1073/pnas.92.10.4686 (doi:10.1073/pnas.92.10.4686) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lipfert J., Ouellet J., Norman D. G., Doniach S., Lilley D. M. J. 2008. The complete VS ribozyme in solution studied by small-angle X-ray scattering. Structure 16, 1357–1367 10.1016/j.str.2008.07.007 (doi:10.1016/j.str.2008.07.007) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lafontaine D. A., Norman D. G., Lilley D. M. J. 2001. Structure, folding and activity of the VS ribozyme: importance of the 2-3-6 helical junction. EMBO J. 20, 1415–1424 10.1093/emboj/20.6.1415 (doi:10.1093/emboj/20.6.1415) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lafontaine D. A., Norman D. G., Lilley D. M. J. 2002. The global structure of the VS ribozyme. EMBO J. 21, 2461–2471 10.1093/emboj/21.10.2461 (doi:10.1093/emboj/21.10.2461) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lafontaine D. A., Wilson T. J., Norman D. G., Lilley D. M. J. 2001. The A730 loop is an important component of the active site of the VS ribozyme. J. Mol. Biol. 312, 663–674 10.1006/jmbi.2001.4996 (doi:10.1006/jmbi.2001.4996) [DOI] [PubMed] [Google Scholar]
- 37.Lafontaine D. A., Wilson T. J., Zhao Z.-Y., Lilley D. M. J. 2002. Functional group requirements in the probable active site of the VS ribozyme. J. Mol. Biol. 323, 23–34 10.1016/S0022-2836(02)00910-5 (doi:10.1016/S0022-2836(02)00910-5) [DOI] [PubMed] [Google Scholar]
- 38.Hiley S. L., Sood V. D., Fan J., Collins R. A. 2002. 4-thio-U cross-linking identifies the active site of the VS ribozyme. EMBO J. 21, 4691–4698 10.1093/emboj/cdf462 (doi:10.1093/emboj/cdf462) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Jones F. D., Strobel S. A. 2003. Ionization of a critical adenosine residue in the Neurospora Varkud Satellite ribozyme active site. Biochemistry 42, 4265–4276 10.1021/bi020707t (doi:10.1021/bi020707t) [DOI] [PubMed] [Google Scholar]
- 40.Wilson T. J., McLeod A. C., Lilley D. M. J. 2007. A guanine nucleobase important for catalysis by the VS ribozyme. EMBO J. 26, 2489–2500 10.1038/sj.emboj.7601698 (doi:10.1038/sj.emboj.7601698) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ouellet J., Byrne M., Lilley D. M. J. 2009. Formation of an active site in trans by interaction of two complete Varkud Satellite ribozymes. RNA 15, 1822–1826 10.1261/rna.1759009 (doi:10.1261/rna.1759009) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Smith M. D., Collins R. A. 2007. Evidence for proton transfer in the rate-limiting step of a fast-cleaving Varkud satellite ribozyme. Proc. Natl Acad. Sci. USA 104, 5818–5823 10.1073/pnas.0608864104 (doi:10.1073/pnas.0608864104) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wilson T. J., Li N.-S., Lu J., Frederiksen J. K., Piccirilli J. A., Lilley D. M. J. 2010. Nucleobase-mediated general acid-base catalysis in the Varkud satellite ribozyme. Proc. Natl Acad. Sci. USA 107, 11 751–11 756 10.1073/pnas.1004255107 (doi:10.1073/pnas.1004255107) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Rupert P. B., Ferré-D'Amaré A. R. 2001. Crystal structure of a hairpin ribozyme–inhibitor complex with implications for catalysis. Nature 410, 780–786 10.1038/35071009 (doi:10.1038/35071009) [DOI] [PubMed] [Google Scholar]
- 45.Rupert P. B., Massey A. P., Sigurdsson S. T., Ferré-D'Amaré A. R. 2002. Transition state stabilization by a catalytic RNA. Science 298, 1421–1424 10.1126/science.1076093 (doi:10.1126/science.1076093) [DOI] [PubMed] [Google Scholar]
- 46.Wilson T. J., Lilley D. M. J. 2011. Do the hairpin and VS ribozymes share a common catalytic mechanism based on general acid-base catalysis? A critical assessment of available experimental data. RNA 17, 213–221 10.1261/rna.2473711 (doi:10.1261/rna.2473711) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Han J., Burke J. M. 2005. Model for general acid-base catalysis by the hammerhead ribozyme: pH–activity relationships of G8 and G12 variants at the putative active site. Biochemistry 44, 7864–7870 10.1021/bi047941z (doi:10.1021/bi047941z) [DOI] [PubMed] [Google Scholar]
- 48.Martick M., Horan L. H., Noller H. F., Scott W. G. 2008. A discontinuous hammerhead ribozyme embedded in a mammalian messenger RNA. Nature 454, 899–902 10.1038/nature07117 (doi:10.1038/nature07117) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Klein D. J., Been M. D., Ferré-D'Amaré A. R. 2007. Essential role of an active-site guanine in glmS ribozyme catalysis. J. Am. Chem. Soc. 129, 14 858–14 859 10.1021/ja0768441 (doi:10.1021/ja0768441) [DOI] [PubMed] [Google Scholar]
- 50.Cochrane J. C., Lipchock S. V., Strobel S. A. 2007. Structural investigation of the GlmS ribozyme bound to its catalytic cofactor. Chem. Biol. 14, 97–105 10.1016/j.chembiol.2006.12.005 (doi:10.1016/j.chembiol.2006.12.005) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Cochrane J. C., Lipchock S. V., Smith K. D., Strobel S. A. 2009. Structural and chemical basis for glucosamine 6-phosphate binding and activation of the glmS ribozyme. Biochemistry 48, 3239–3246 10.1021/bi802069p (doi:10.1021/bi802069p) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Martick M., Scott W. G. 2006. Tertiary contacts distant from the active site prime a ribozyme for catalysis. Cell 126, 309–320 10.1016/j.cell.2006.06.036 (doi:10.1016/j.cell.2006.06.036) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Klein D. J., Ferré-D'Amaré A. R. 2006. Structural basis of glmS ribozyme activation by glucosamine-6-phosphate. Science 313, 1752–1756 10.1126/science.1129666 (doi:10.1126/science.1129666) [DOI] [PubMed] [Google Scholar]
- 54.Mandal M., Boese B., Barrick J. E., Winkler W. C., Breaker R. R. 2003. Riboswitches control fundamental biochemical pathways in Bacillus subtilis and other bacteria. Cell 113, 577–586 10.1016/S0092-8674(03)00391-X (doi:10.1016/S0092-8674(03)00391-X) [DOI] [PubMed] [Google Scholar]
- 55.Macaya R. F., Schultze P., Smith F. W., Roe J. A., Feigon J. 1993. Thrombin-binding DNA aptamer forms a unimolecular quadruplex structure in solution. Proc. Natl Acad. Sci. USA. 90, 3745–3749 10.1073/pnas.90.8.3745 (doi:10.1073/pnas.90.8.3745) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Huizenga D. E., Szostak J. W. 1995. A DNA aptamer that binds adenosine and ATP. Biochemistry 34, 656–665 10.1021/bi00002a033 (doi:10.1021/bi00002a033) [DOI] [PubMed] [Google Scholar]
- 57.Robertson M. P., Ellington A. D. 2000. Design and optimization of effector-activated ribozyme ligases. Nucleic Acids Res. 28, 1751–1759 10.1093/nar/28.8.1751 (doi:10.1093/nar/28.8.1751) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Salehi-Ashtiani K., Luptak A., Litovchick A., Szostak J. W. 2006. A genomewide search for ribozymes reveals an HDV-like sequence in the human CPEB3 gene. Science 313, 1788–1792 10.1126/science.1129308 (doi:10.1126/science.1129308) [DOI] [PubMed] [Google Scholar]
- 59.Nakano S., Chadalavada D. M., Bevilacqua P. C. 2000. General acid-base catalysis in the mechanism of a hepatitis delta virus ribozyme. Science 287, 1493–1497 10.1126/science.287.5457.1493 (doi:10.1126/science.287.5457.1493) [DOI] [PubMed] [Google Scholar]
- 60.Chen J. H., Yajima R., Chadalavada D. M., Chase E., Bevilacqua P. C., Golden B. L. 2010. A 1.9 A crystal structure of the HDV ribozyme precleavage suggests both Lewis acid and general acid mechanisms contribute to phosphodiester cleavage. Biochemistry 49, 6508–6518 10.1021/bi100670p (doi:10.1021/bi100670p) [DOI] [PubMed] [Google Scholar]
- 61.Ferré-d'Amaré A. R., Zhou K., Doudna J. A. 1998. Crystal structure of a hepatitis delta virus ribozyme. Nature 395, 567–574 10.1038/26912 (doi:10.1038/26912) [DOI] [PubMed] [Google Scholar]
- 62.Ke A., Zhou K., Ding F., Cate J. H., Doudna J. A. 2004. A conformational switch controls hepatitis delta virus ribozyme catalysis. Nature 429, 201–205 10.1038/nature02522 (doi:10.1038/nature02522) [DOI] [PubMed] [Google Scholar]
- 63.Das S. R., Piccirilli J. A. 2005. General acid catalysis by the hepatitis delta virus ribozyme. Nat. Chem. Biol. 1, 45–52 10.1038/nchembio703 (doi:10.1038/nchembio703) [DOI] [PubMed] [Google Scholar]
- 64.Murray J. B., Seyhan A. A., Walter N. G., Burke J. M., Scott W. G. 1998. The hammerhead, hairpin and VS ribozymes are catalytically proficient in monovalent cations alone. Chem. Biol. 5, 587–595 10.1016/S1074-5521(98)90116-8 (doi:10.1016/S1074-5521(98)90116-8) [DOI] [PubMed] [Google Scholar]
- 65.delCardayré S. B., Raines R. T. 1994. Structural determinants of enzymatic processivity. Biochemistry 33, 6031–6037 10.1021/bi00186a001 (doi:10.1021/bi00186a001) [DOI] [PubMed] [Google Scholar]
- 66.Raines R. T. 1998. Ribonuclease A. Chem. Rev. 98, 1045–1066 10.1021/cr960427h (doi:10.1021/cr960427h) [DOI] [PubMed] [Google Scholar]
- 67.Adams P. L., Stahley M. R., Wang J., Strobel S. A. 2004. Crystal structure of a self-splicing group I intron with both exons. Nature 430, 45–50 10.1038/nature02642 (doi:10.1038/nature02642) [DOI] [PubMed] [Google Scholar]
- 68.Toor N., Keating K. S., Taylor S. D., Pyle A. M. 2008. Crystal structure of a self-spliced group II intron. Science 320, 77–82 10.1126/science.1153803 (doi:10.1126/science.1153803) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Shan S. O., Yoshida A., Sun S. G., Piccirilli J. A., Herschlag D. 1999. Three metal ions at the active site of the Tetrahymena group I ribozyme. Proc. Natl Acad. Sci. USA 96, 12 299–12 304 10.1073/pnas.96.22.12299 (doi:10.1073/pnas.96.22.12299) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Forconi M., Lee J., Lee J. K., Piccirilli J. A., Herschlag D. 2008. Functional identification of ligands for a catalytic metal ion in group I introns. Biochemistry 47, 6883–6894 10.1021/bi800519a (doi:10.1021/bi800519a) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Stahley M. R., Strobel S. A. 2005. Structural evidence for a two-metal-ion mechanism of group I intron splicing. Science 309, 1587–1590 10.1126/science.1114994 (doi:10.1126/science.1114994) [DOI] [PubMed] [Google Scholar]
- 72.Gordon P. M., Piccirilli J. A. 2001. Metal ion coordination by the AGC triad in domain 5 contributes to group II intron catalysis. Nat. Struct. Biol. 8, 893–898 10.1038/nsb1001-893 (doi:10.1038/nsb1001-893) [DOI] [PubMed] [Google Scholar]
- 73.Gordon P. M., Fong R., Piccirilli J. A. 2007. A second divalent metal ion in the group II intron reaction center. Chem. Biol. 14, 607–612 10.1016/j.chembiol.2007.05.008 (doi:10.1016/j.chembiol.2007.05.008) [DOI] [PubMed] [Google Scholar]
- 74.Gordon P. M., Sontheimer E. J., Piccirilli J. A. 2000. Metal ion catalysis during the exon-ligation step of nuclear pre-mRNA splicing: extending the parallels between the spliceosome and group II introns. RNA 6, 199–205 10.1017/S1355838200992069 (doi:10.1017/S1355838200992069) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Pena V., Rozov A., Fabrizio P., Luhrmann R., Wahl M. C. 2008. Structure and function of an RNase H domain at the heart of the spliceosome. EMBO J. 27, 2929–2940 10.1038/emboj.2008.209 (doi:10.1038/emboj.2008.209) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Ritchie D. B., Schellenberg M. J., Gesner E. M., Raithatha S. A., Stuart D. T., Macmillan A. M. 2008. Structural elucidation of a PRP8 core domain from the heart of the spliceosome. Nat. Struct. Mol. Biol. 15, 1199–1205 10.1038/nsmb.1505 (doi:10.1038/nsmb.1505) [DOI] [PubMed] [Google Scholar]
- 77.Nowotny M., Gaidamakov S. A., Crouch R. J., Yang W. 2005. Crystal structures of RNase H bound to an RNA/DNA hybrid: substrate specificity and metal-dependent catalysis. Cell 121, 1005–1016 10.1016/j.cell.2005.04.024 (doi:10.1016/j.cell.2005.04.024) [DOI] [PubMed] [Google Scholar]
- 78.Sievers A., Beringer M., Rodnina M. V., Wolfenden R. 2004. The ribosome as an entropy trap. Proc. Natl Acad. Sci. USA 101, 7897–7901 10.1073/pnas.0402488101 (doi:10.1073/pnas.0402488101) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Weinger J. S., Parnell K. M., Dorner S., Green R., Strobel S. A. 2004. Substrate-assisted catalysis of peptide bond formation by the ribosome. Nat. Struct. Mol. Biol. 11, 1101–1106 10.1038/nsmb841 (doi:10.1038/nsmb841) [DOI] [PubMed] [Google Scholar]
- 80.Schmeing T. M., Huang K. S., Kitchen D. E., Strobel S. A., Steitz T. A. 2005. Structural insights into the roles of water and the 2′ hydroxyl of the P site tRNA in the peptidyl transferase reaction. Mol. Cell 20, 437–448 10.1016/j.molcel.2005.09.006 (doi:10.1016/j.molcel.2005.09.006) [DOI] [PubMed] [Google Scholar]
- 81.Kingery D. A., Pfund E., Voorhees R. M., Okuda K., Wohlgemuth I., Kitchen D. E., Rodnina M. V., Strobel S. A. 2008. An uncharged amine in the transition state of the ribosomal peptidyl transfer reaction. Chem. Biol. 15, 493–500 10.1016/j.chembiol.2008.04.005 (doi:10.1016/j.chembiol.2008.04.005) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Winkler W. C., Nahvi A., Roth A., Collins J. A., Breaker R. R. 2004. Control of gene expression by a natural metabolite-responsive ribozyme. Nature 428, 281–286 10.1038/nature02362 (doi:10.1038/nature02362) [DOI] [PubMed] [Google Scholar]
- 83.Serganov A., et al. 2005. Structural basis for Diels–Alder ribozyme-catalyzed carbon-carbon bond formation. Nat. Struct. Mol. Biol. 12, 218–224 10.1038/nsmb906 (doi:10.1038/nsmb906) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Robertson M. P., Scott W. G. 2007. The structural basis of ribozyme-catalyzed RNA assembly. Science 315, 1549–1553 10.1126/science.1136231 (doi:10.1126/science.1136231) [DOI] [PubMed] [Google Scholar]