Abstract
Aminoacylation of tRNA is an essential event in the translation system. Although in the modern system protein enzymes play the sole role in tRNA aminoacylation, in the primitive translation system RNA molecules could have catalysed aminoacylation onto tRNA or tRNA-like molecules. Even though such RNA enzymes so far are not identified from known organisms, in vitro selection has generated such RNA catalysts from a pool of random RNA sequences. Among them, a set of RNA sequences, referred to as flexizymes (Fxs), discovered in our laboratory are able to charge amino acids onto tRNAs. Significantly, Fxs allow us to charge a wide variety of amino acids, including those that are non-proteinogenic, onto tRNAs bearing any desired anticodons, and thus enable us to reprogramme the genetic code at our will. This article summarizes the evolutionary history of Fxs and also the most recent advances in manipulating a translation system by integration with Fxs.
Keywords: RNA world, ribozyme, flexizyme, translation, genetic code reprogramming
1. Introduction
It has been established indisputably by the X-ray structure of ribosome that its peptidyl transfer centre consists of only RNA [1]. This has given strong support to the hypothesis that primitive translation events could have been catalysed by RNA alone in the RNA world. However, tRNA aminoacylation, another essential reaction of translation, is solely catalysed by protein enzymes, aminoacyl-tRNA synthetases (ARSs), in the modern system. In the RNA world hypothesis, this discrepancy could be explained by the existence of ARS-like ribozymes [2–6]. Although such a ribozyme working in cells has never been discovered so far, over the past decade we and others have isolated such ribozymes by means of in vitro selection [2,6–9]. We herein summarize the evolutionary history of a class of such ribozymes isolated from our laboratory. This class of ribozymes, referred to as flexizymes (Fxs), is capable of charging amino acids onto the 3′-hydroxyl group of tRNA end-like protein ARSs [10]. Moreover, using the unique property of the versatility of Fxs as tRNA acylation catalysts, we and others have developed strategies of manipulating the genetic code, so-called ‘genetic code reprogramming’ [11–13], with the integration of a custom-made reconstituted cell-free translation system [11,14,15]. We also briefly introduce the latest research outcomes for the synthesis of non-standard peptides and perspectives in the selection of novel peptides against protein targets and engineering of the translation machinery.
2. Evolution of aminoacyl-tRNA synthetase-like ribozymes (flexizymes)
Ligation of one of the hydroxyl groups at the tRNA 3′-end with the carboxyl group of amino acid involves two chemistries; (i) activation of the carboxyl group by adenosine triphosphate to yield aminoacyl-adenosine monophosphate (aminoacyl-AMP), and (ii) condensation of aminoacyl-AMP and tRNA to yield aminoacyl-tRNA. In the modern translation system, single ARS species catalyse both chemical reactions. Because of the intrinsic high energy of the aminoacyl-AMP intermediate, step (i) is a highly up-hill reaction, and thus it requires reaction with the tRNA hydroxyl group without long exposure to bulk water. Although certain RNA molecules may be able to catalyse the reaction of step (i) [16], it is however difficult to achieve both steps simultaneously. Alternatively, step (i) may rely on the formation of appropriate esters with prebiotically compatible activating groups, such as cyanomethyl ester [8,17] (the alcohol moiety can be produced by the condensation of cyanide and formaldehyde) or thioester. Thus, primitive ARS-like ribozymes might have catalysed step (ii) in a selective manner where certain amino acids are charged onto the 3′-terminal hydroxyl group of tRNA-like RNA.
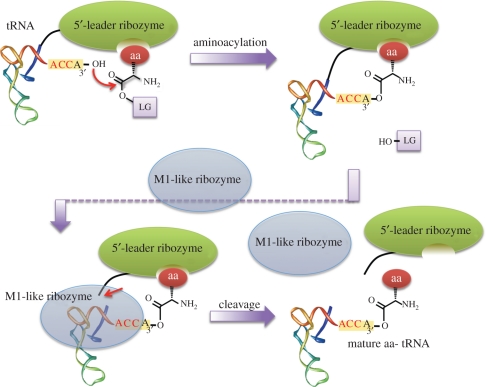
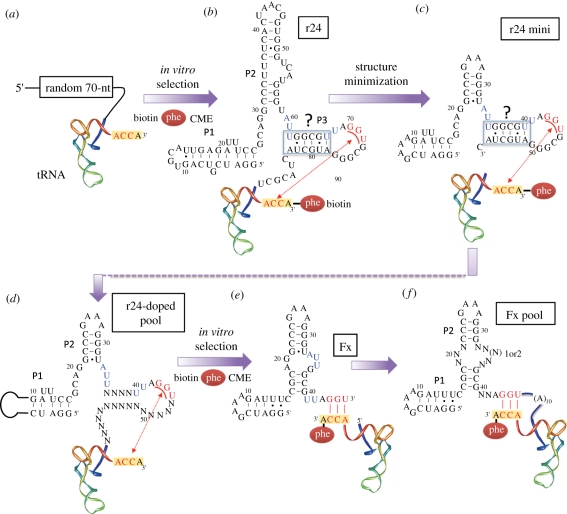
We have hypothesized that primitive ARS-like ribozymes could have evolved as a part of a 5′-leader sequence. This hypothesis seems reasonable in the RNA world hypothesis since M1 RNA of ribonuclease (RNase) P, a known naturally occurring ribozyme [18,19], selectively cleaves the 5′-leader sequence from tRNA independently from tRNA species. Thus, a 5′-leader sequence could have acted as a specific self-aminoacylating catalyst to a cognate tRNA via the covalent linkage, and M1 RNA-like ribozyme would have removed it to yield the mature aminoacyl-tRNA (figure 1). On the basis of this hypothesis, we constructed a RNA library with 70-nucleotide (nt) random sequences attached to the 5′-end of a tRNAGln (figure 2a). In vitro selection was performed using a phenylalanine (Phe) derivative of which the amino group was modified with biotin and the carbonyl group was activated with cyanomethyl ester, and then active species were isolated based on the ability to self-aminoacylate any site of available hydroxyl groups [9]. This selection attempt fortunately yielded a single kind of sequence, referred to as r24, capable of aminoacylating the 3′-terminal hydroxyl group (figure 2b). Importantly, r24 could be removed by the treatment of M1 RNA of RNase P, and thus it fitted well with the hypothetical view of evolution of self-aminoacylating 5′-leader ribozyme (figure 1).
Figure 1.
Hypothetical coevolution of M1-like ribozyme and 5′-leader ribozyme. The 5′-leader ribozyme recognizes a specific amino acid and self-charges to the 3′-end of RNA. In this way, specificity to the tRNA is secured by the direct covalent linkage. M1-like ribozyme is co-evolved with the 5′-leader ribozyme, so that the ribozyme domain is cleaved from the tRNA to release the mature aminoacyl-tRNA. LG, leaving group; aa, aminoacyl.
Figure 2.
The in vitro selection and engineering of flexizymes. (a) RNA pool for in vitro selection of catalytic precursor tRNAs. (b) The secondary structure of r24. Thick lines and dots indicate predicted Watson–Crick and G–U wobble base pairing, respectively. P3 shown in box with question mark is a putative pair region. Proposed bases that form Watson–Crick pairs in r24min (G43–U45) and tRNAs (A73–C75) are underlined and the red-dashed arrow indicates their interactions. Putative bases involving the constitution of the Phe-binding site are highlighted in blue. CME denotes cyanomethyl ester leaving group. (c) The first generation of prototype flexizyme, r24mini. (d) Doped RNA pool for in vitro selection of the second generation of prototype flexizyme. (e) The second generation of prototype flexizyme. Watson–Crick base pairs between the prototype flexizyme and tRNA are in red. (f) Doped RNA pool for in vitro selection of dFx, eFx and aFx that recognize aromatic leaving groups.
We performed metal-dependent kinetics and probing, chemical probing, and nucleotide analogue interference mapping of r24, to reveal the sites necessary for Mg2+, tRNA and Phe binding [20,21]. Complementary bases to the tRNA 3′-end region, A73–C75, were found in the 3′-terminal region of r24, and their Watson–Crick base-pair interactions were confirmed by their compensatory mutations albeit tolerance of wobble G73–U* or U73–U* pair (U* denotes the r24 base at position 72). Based on the above knowledge, a shorter version of r24, called r24mini, being 57-nt long, was constructed to confirm the wild-type activity [20] (figure 2c). Accordingly, it was demonstrated that r24mini itself could charge Phe onto tRNA as a trans-acting enzyme like protein enzymes.
To further minimize the structure of r24mini, we maintained the essential bases in the amino acid and tRNA-binding regions but the rest was re-randomized for the second activity selection (figure 2d) [10]. Sequence alignment of the active species revealed that the 3′-terminal region after the tRNA-binding site has no conservation of sequence, suggesting that this region is unimportant for activity. In fact, deletion of this region, giving a new construct only 45-nt long, did not affect the intrinsic catalytic activity of r24mini, but rather increased the fraction of correctly folded active species of the trans-acting ribozyme, i.e. increased the final yield of aminoacyl-tRNA product. Moreover, this short ribozyme was able to charge a variety of aromatic amino acids, such as tyrosine, tryptophan and Phe analogues, onto tRNA as far as they are activated by cyanomethyl ester [22]. Because of the simplicity of the base-pair interactions between this ribozyme and tRNA, it allowed us to prepare a variety of mischarged aminoacyl-tRNAs independent of its body or anticodon sequences. Owing to the flexible utility of this ribozyme, we named it Fx (figure 2e).
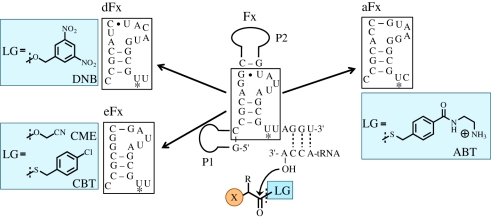
Although the prototype Fx was a useful tool for the preparation of mischarged tRNAs with Phe or Phe analogues, it would become a more versatile and convenient tool if aminoacyl-tRNAs with more variety of kinds of amino acids could be prepared at the user's will. Therefore, we turned our efforts to develop a more versatile and effective Fx tool [11]. A new Fx library was constructed, where the structural scaffold, P1 and P2, was kept the same but bases in putative small loops and junctions were re-randomized along with insertion of a few bases (figure 2f). A drastic change was made in the substrate design. Because during the course of our studies, we learned that the methylene-aromatic ring of the Phe side chain was the major recognition element, we designed such an aromatic structure in the leaving group of the ester. In this way, the substrate recognition of Fx variants would be directed to the common leaving group of the amino acid donor substrate, and therefore any side chain of amino acids would become acceptable. In fact, selection of the above Fx library against lysine with 3,5-dinitrobenzyl ester leaving group successfully yielded a new variant, referred to as dFx (d stands for dinitrobenzyl ester), capable of effectively charging non-aromatic amino acid on tRNAs (figure 3) [11]. Meanwhile, we also repeated selection against Phe-cyanomethyl ester to re-optimize the sequence of the prototype Fx, generating a new variant, referred to as eFx (e stands for enhanced; figure 3) [11]. This eFx turns out to be a good catalyst for not only aromatic amino acids but also non-aromatic amino acids activated with 4-chlorobenzyl thioester. We have also developed another variant aFx that reacts with amino acids activated with an amino-derivatized benzyl thioester group, which is a useful catalyst when the substrate has poor water-solubility owing to a high hydrophobicity of its side chain or certain modifications of amino acids (figure 3) [23].
Figure 3.
Flexizymes and their cognate leaving groups in substrates. Chemical structures of a benzyl ester and thioester leaving group. R represents amino acid side chains including non-proteinogenic ones. Each flexizyme recognizes the specific leaving group and charges the acyl group onto the 3′-hydroxyl group at the tRNA 3′-end. LG, leaving group; DNB, dinitrobenzyl ester; CME, cyanomethyl ester; CBT, p-chlorobenzyl thioester; ABT, amino-derivatized benzyl thioester.
To this end, we have devised a versatile ‘Fx’ system that enables us to perform aminoacylation of any tRNAs with a wide array of proteinogenic amino acids and non-proteinogenic ones, such as d-amino acids [24], β-amino acids [11], N-methyl-amino acids [25] and N-acyl-amino acids [26]. Having this system, we initiated a new project to manipulate the genetic code, so-called genetic code reprogramming, leading us to express non-standard peptides containing multiple non-proteinogenic amino acids.
3. Genetic code reprogramming facilitated by flexible in vitro translation system
Genetic code reprogramming [11–13] is a concept in which some codons are reassigned from proteinogenic amino acids to non-proteinogenic ones, allowing expression of non-standard polypeptides by using an in vitro translation apparatus. Despite the simplicity of the concept, the codon reassignments require two major technical challenges; (i) preparation of aminoacyl-tRNAs with non-proteinogenic amino acids, and (ii) making vacant codons to which the above amino acids are reassigned. Although there are several different approaches available for (i), such as semi-chemical synthetic method [12,27–32] or ARS-mischarging method [13,33–37], Fxs are far superior to others in terms of flexibility of the choices of non-proteinogenic amino acids and tRNAs as well as facility and reliability [11]. In the ordinary reconstituted cell-free translation, 30 pre-purified translation factors are essential for normal protein synthesis. Removing specific amino acids and/or their cognate ARSs from the translation system pauses the expression or induces peptidyl-tRNA drop-off at the vacant codons in (ii). When the Fx system is integrated with the custom-made reconstituted translation apparatus, non-proteinogenic amino acids charged on specific tRNAs reoccupy the vacant codons. We referred to this Fx–custom-made translation integrated system as the flexible in vitro translation (FIT) system [14].
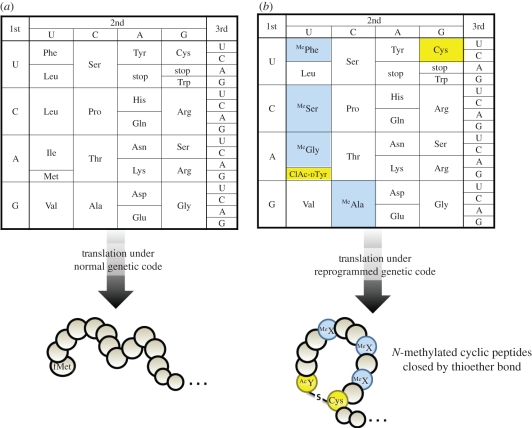
In the last few years, we have made major efforts to express a wide variety of non-standard polypeptides containing multiple non-proteinogenic amino acids in FIT systems. Non-proteinogenic amino acids (resulting peptides are in parentheses) used in these studies (table 1) are (i) N-methyl-amino acids in elongation (N-methyl-peptides) [25,38], (ii) N-alkyl glycine in elongation (peptoid–peptide hybrid) [39], (iii) N-acyl-amino acids and exotic peptides in initiation (N-terminal modified peptides) [26], (iv) α-hydroxy acids in elongation (polyesters or backbone cyclic peptides) [38,40], and (v) those with unique side chains in initiation and elongation, which can react with specific partner amino acids (e.g. cyclic peptides closed by non-reducible covalent bond) [26]. Combination of any of these methods yields structurally diverse non-standard peptides such as natural product-like peptides and artificial cyclic peptides, allowing us to express unique non-standard peptide libraries from mRNA templates under reprogrammed genetic codes (figure 4).
Table 1.
Representative examples for the incorporation of non-proteinogenic amino acids into a peptide chain using the FIT system.
| non-proteinogenic amino acids | amino acid position | peptides | references |
|---|---|---|---|
| N-methyl amino acids | elongation | N-methyl peptides | [25,38] |
| N-alkyl glycine | elongation | peptoid–peptide hybrid | [39] |
| N-acyl amino acids | initiation | N-terminal modified peptides | [26] |
| α-hydroxy acids | elongation | polyesters or backbone cyclic peptides | [38,40] |
| chloro-acetyl amino acid | initiation | thioether-based cyclic peptides | [26] |
Figure 4.
Reprogrammed genetic code using the FIT system. (a) Normal codon table and (b) reprogrammed codon table. In the reprogrammed genetic code, codons of methionine (Met), phenylalanine (Phe), leucine (Leu), isoleucine (Ile) and alanine (Ala) are reassigned to N-(2-chloroacetyl)-d-tyrosine (ClAc-dTyr), N-methyl-phenylalanine (MePhe), N-methyl-serine (MeSer), N-methyl-glycine (MeGly) and N-methyl-alanine (MeAla), respectively. Expression of a certain mRNA under the reprogrammed genetic code yields a specific sequence of peptides containing the above non-proteinogenic amino acids, where the N-terminal ClAc group on d-Tyr reacts with the sulfhydryl group of the side chain of cysteine (Cys) at the C-terminus, giving a cyclic peptide closed by thioether bond.
4. Perspectives
Having the methodology that enables us to readily construct non-standard peptide libraries, it is a logical next step to integrate it with an appropriate in vitro display technology such as mRNA display. Such an approach will allow us to select non-standard peptide aptamers against therapeutic targets from a highly diverse library with the complexity of over 1012. In fact, we have recently built cyclic non-standard peptide libraries and selected desired molecules with a dozen distinct targets with dissociation constants of low nanomolar to sub nanomolar. Moreover, some of these molecules exhibited inhibitory activities of enzymes as well as protein–protein interactions. Such results will be reported in the near future.
Another direction that we have been taking with Fx technology in our laboratory is to investigate the ribosomal translation machinery by using aminoacyl-tRNAs with a non-CCA-3′-end. We recently found that Fxs are able to catalyse aminoacylation on non-CCA-3′ tRNAs efficiently when the Fx's tRNA-binding bases were mutated to make base pairs with the tRNA 3′-end (G. Hayashi and H. Suga 2011, unpublished data). Furthermore, in a combination of engineered ribosomes having compensatory mutations at rRNA against non-CCA-3′-end tRNAs, we have developed an artificial ribosome–tRNAs pair that acts orthogonal to the naturally occurring ribosome–tRNAs pair, i.e. we have created an ‘orthogonal genetic code’.
In conclusion, Fxs are a unique tool to open many new doors for not only basic science but also applications involving the translation apparatus. More new ideas should follow in the future.
Acknowledgements
This work was supported by grants from the Japan Society for the Promotion of Science Specially Promoted Research (21000005), research and development projects of the Industrial Science and Technology Program in the New Energy and Industrial Technology Development Organization (NEDO), and the World Class University project of the MEST and the NRF (R31-2008-000-10103-0). G.H. was supported by a Japan Society for the Promotion of Science Research Fellowship for Young Scientists.
References
- 1.Nissen P., Hansen J., Ban N., Moore P., Steitz T. 2000. The structural basis of ribosome activity in peptide bond synthesis. Science 289, 920–930 10.1126/science.289.5481.920 (doi:10.1126/science.289.5481.920) [DOI] [PubMed] [Google Scholar]
- 2.Illangasekare M., Sanchez G., Nickles T., Yarus M. 1995. Aminoacyl-RNA synthesis catalyzed by an RNA. Science 267, 643–647 10.1126/science.7530860 (doi:10.1126/science.7530860) [DOI] [PubMed] [Google Scholar]
- 3.Schimmel P., Giegé R., Moras D., Yokoyama S. 1993. An operational RNA code for amino acids and possible relationship to genetic code. Proc. Natl Acad. Sci. USA 90, 8763–8768 10.1073/pnas.90.19.8763 (doi:10.1073/pnas.90.19.8763) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Piccirilli J., McConnell T., Zaug A., Noller H., Cech T. 1992. Aminoacyl esterase activity of the Tetrahymena ribozyme. Science 256, 1420–1424 10.1126/science.1604316 (doi:10.1126/science.1604316) [DOI] [PubMed] [Google Scholar]
- 5.De Pouplana L., Turner R., Steer B., Schimmel P. 1998. Genetic code origins: tRNAs older than their synthetases? Proc. Natl Acad. Sci. USA 95, 11 295–11 300 10.1073/pnas.95.19.11295 (doi:10.1073/pnas.95.19.11295) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Illangasekare M., Yarus M. 1999. Specific, rapid synthesis of Phe-RNA by RNA. Proc. Natl Acad. Sci. USA 96, 5470–5475 10.1073/pnas.96.10.5470 (doi:10.1073/pnas.96.10.5470) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Jenne A., Famulok M. 1998. A novel ribozyme with ester transferase activity. Chem. Biol. 5, 23–34 10.1016/S1074-5521(98)90084-9 (doi:10.1016/S1074-5521(98)90084-9) [DOI] [PubMed] [Google Scholar]
- 8.Lee N., Bessho Y., Wei K., Szostak J., Suga H. 2000. Ribozyme-catalyzed tRNA aminoacylation. Nat. Struct. Biol. 7, 28–33 10.1038/71225 (doi:10.1038/71225) [DOI] [PubMed] [Google Scholar]
- 9.Saito H., Kourouklis D., Suga H. 2001. An in vitro evolved precursor tRNA with aminoacylation activity. EMBO J. 20, 1797–1806 10.1093/emboj/20.7.1797 (doi:10.1093/emboj/20.7.1797) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Murakami H., Saito H., Suga H. 2003. A versatile tRNA aminoacylation catalyst based on RNA. Chem. Biol. 10, 655–662 10.1016/S1074-5521(03)00145-5 (doi:10.1016/S1074-5521(03)00145-5) [DOI] [PubMed] [Google Scholar]
- 11.Murakami H., Ohta A., Ashigai H., Suga H. 2006. A highly flexible tRNA acylation method for non-natural polypeptide synthesis. Nat. Methods 3, 357–359 10.1038/nmeth877 (doi:10.1038/nmeth877) [DOI] [PubMed] [Google Scholar]
- 12.Forster A., Tan Z., Nalam M., Lin H., Qu H., Cornish V., Blacklow S. 2003. Programming peptidomimetic syntheses by translating genetic codes designed de novo. Proc. Natl Acad. Sci. USA 100, 6353–6357 10.1073/pnas.1132122100 (doi:10.1073/pnas.1132122100) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Josephson K., Hartman M., Szostak J. 2005. Ribosomal synthesis of unnatural peptides. J. Am. Chem. Soc. 127, 11 727–11 735 10.1021/ja0515809 (doi:10.1021/ja0515809) [DOI] [PubMed] [Google Scholar]
- 14.Goto Y., Katoh T., Suga H. 2011. Flexizymes for genetic code reprogramming. Nat. Protoc. 6, 779–790 10.1038/nprot.2011.331 (doi:10.1038/nprot.2011.331) [DOI] [PubMed] [Google Scholar]
- 15.Shimizu Y., Inoue A., Tomari Y., Suzuki T., Yokogawa T., Nishikawa K., Ueda T. 2001. Cell-free translation reconstituted with purified components. Nat. Biotechnol. 19, 751–755 10.1038/90802 (doi:10.1038/90802) [DOI] [PubMed] [Google Scholar]
- 16.Kumar R. K., Yarus M. 2001. RNA-catalyzed amino acid activation. Biochemistry 40, 6998–7004 10.1021/bi010710x (doi:10.1021/bi010710x) [DOI] [PubMed] [Google Scholar]
- 17.Suga H., Lohse P., Szostak J. 1998. Structural and kinetic characterization of an acyl transferase ribozyme. J. Am. Chem. Soc. 120, 1151–1156 10.1021/ja972472s (doi:10.1021/ja972472s) [DOI] [PubMed] [Google Scholar]
- 18.Guerrier-Takada C., Gardiner K., Marsh T., Pace N., Altman S. 1983. The RNA moiety of ribonuclease P is the catalytic subunit of the enzyme. Cell 35, 849–857 10.1016/0092-8674(83)90117-4 (doi:10.1016/0092-8674(83)90117-4) [DOI] [PubMed] [Google Scholar]
- 19.Frank D., Pace N. 1998. Ribonuclease P: unity and diversity in a tRNA processing ribozyme. Annu. Rev. Biochem. 67, 153–180 10.1146/annurev.biochem.67.1.153 (doi:10.1146/annurev.biochem.67.1.153) [DOI] [PubMed] [Google Scholar]
- 20.Saito H., Watanabe K., Suga H. 2001. Concurrent molecular recognition of the amino acid and tRNA by a ribozyme. RNA 7, 1867–1878 10.1017/S1355838201013164 (doi:10.1017/S1355838201013164) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Saito H., Suga H. 2002. Outersphere and innersphere coordinated metal ions in an aminoacyl-tRNA synthetase ribozyme. Nucleic Acids Res. 30, 5151–5159 10.1093/nar/gkf641 (doi:10.1093/nar/gkf641) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Murakami H., Kourouklis D., Suga H. 2003. Using a solid-phase ribozyme aminoacylation system to reprogram the genetic code. Chem. Biol. 10, 1077–1084 10.1016/j.chembiol.2003.10.010 (doi:10.1016/j.chembiol.2003.10.010) [DOI] [PubMed] [Google Scholar]
- 23.Niwa N., Yamagishi Y., Murakami H., Suga H. 2009. A flexizyme that selectively charges amino acids activated by a water-friendly leaving group. Bioorg. Med. Chem. Lett. 19, 3892–3894 10.1016/j.bmcl.2009.03.114 (doi:10.1016/j.bmcl.2009.03.114) [DOI] [PubMed] [Google Scholar]
- 24.Goto Y., Murakami H., Suga H. 2008. Initiating translation with D-amino acids. RNA 14, 1390–1398 10.1261/rna.1020708 (doi:10.1261/rna.1020708) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kawakami T., Murakami H., Suga H. 2008. Messenger RNA-programmed incorporation of multiple N-methyl-amino acids into linear and cyclic peptides. Chem. Biol. 15, 32–42 10.1016/j.chembiol.2007.12.008 (doi:10.1016/j.chembiol.2007.12.008) [DOI] [PubMed] [Google Scholar]
- 26.Goto Y., Ohta A., Sako Y., Yamagishi Y., Murakami H., Suga H. 2008. Reprogramming the translation initiation for the synthesis of physiologically stable cyclic peptides. ACS Chem. Biol. 3, 120–129 10.1021/cb700233t (doi:10.1021/cb700233t) [DOI] [PubMed] [Google Scholar]
- 27.Noren C., Anthony-Cahill S., Griffith M., Schultz P. 1989. A general method for site-specific incorporation of unnatural amino acids into proteins. Science 244, 182–188 10.1126/science.2649980 (doi:10.1126/science.2649980) [DOI] [PubMed] [Google Scholar]
- 28.Bain J., Diala E., Glabe C., Dix T., Chamberlin A. 1989. Biosynthetic site-specific incorporation of a non-natural amino acid into a polypeptide. J. Am. Chem. Soc. 111, 8013–8014 10.1021/ja00202a052 (doi:10.1021/ja00202a052) [DOI] [Google Scholar]
- 29.Frankel A., Millward S., Roberts R. 2003. Encodamers: unnatural peptide oligomers encoded in RNA. Chem. Biol. 10, 1043–1050 10.1016/j.chembiol.2003.11.004 (doi:10.1016/j.chembiol.2003.11.004) [DOI] [PubMed] [Google Scholar]
- 30.Tan Z., Forster A., Blacklow S., Cornish V. 2004. Amino acid backbone specificity of the Escherichia coli translation machinery. J. Am. Chem. Soc. 126, 12 752–12 753 10.1021/ja0472174 (doi:10.1021/ja0472174) [DOI] [PubMed] [Google Scholar]
- 31.Heckler T., Chang L., Zama Y., Naka T., Chorghade M., Hecht S. 1984. T4 RNA ligase mediated preparation of novel ‘chemically misacylated’ tRNAPheS. Biochemistry 23, 1468–1473 10.1021/bi00302a020 (doi:10.1021/bi00302a020) [DOI] [PubMed] [Google Scholar]
- 32.Robertson S., Ellman J., Schultz P. 1991. A general and efficient route for chemical aminoacylation of transfer RNAs. J. Am. Chem. Soc. 113, 2722–2729 10.1021/ja00007a055 (doi:10.1021/ja00007a055) [DOI] [Google Scholar]
- 33.Hendrickson T., de Crecy-Lagard V., Schimmel P. 2004. Incorporation of nonnatural amino acids into proteins. Annu. Rev. Biochem. 73, 147–176 10.1146/annurev.biochem.73.012803.092429 (doi:10.1146/annurev.biochem.73.012803.092429) [DOI] [PubMed] [Google Scholar]
- 34.Wang L., Schultz P. 2005. Expanding the genetic code. Angew. Chem. Int. Ed. 44, 34–66 10.1002/anie.200460627 (doi:10.1002/anie.200460627) [DOI] [PubMed] [Google Scholar]
- 35.Santoro S., Wang L., Herberich B., King D., Schultz P. 2002. An efficient system for the evolution of aminoacyl-tRNA synthetase specificity. Nat. Biotechnol. 20, 1044–1048 10.1038/nbt742 (doi:10.1038/nbt742) [DOI] [PubMed] [Google Scholar]
- 36.Kiga D., et al. 2002. An engineered Escherichia coli tyrosyl-tRNA synthetase for site-specific incorporation of an unnatural amino acid into proteins in eukaryotic translation and its application in a wheat germ cell-free system. Proc. Natl Acad. Sci. USA 99, 9715–9720 10.1073/pnas.142220099 (doi:10.1073/pnas.142220099) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Link A., Tirrell D. 2005. Reassignment of sense codons in vivo. Methods 36, 291–298 10.1016/j.ymeth.2005.04.005 (doi:10.1016/j.ymeth.2005.04.005) [DOI] [PubMed] [Google Scholar]
- 38.Kawakami T., Ohta A., Ohuchi M., Ashigai H., Murakami H., Suga H. 2009. Diverse backbone-cyclized peptides via codon reprogramming. Nat. Chem. Biol. 5, 888–890 10.1038/nchembio.259 (doi:10.1038/nchembio.259) [DOI] [PubMed] [Google Scholar]
- 39.Kawakami T., Murakami H., Suga H. 2008. Ribosomal synthesis of polypeptoids and peptoid–peptide hybrids. J. Am. Chem. Soc. 130, 16 861–16 863 10.1021/ja806998v (doi:10.1021/ja806998v) [DOI] [PubMed] [Google Scholar]
- 40.Ohta A., Murakami H., Higashimura E., Suga H. 2007. Synthesis of polyester by means of genetic code reprogramming. Chem. Biol. 14, 1315–1322 10.1016/j.chembiol.2007.10.015 (doi:10.1016/j.chembiol.2007.10.015) [DOI] [PubMed] [Google Scholar]