Abstract
Speed and accuracy of protein synthesis are fundamental parameters for the fitness of living cells, the quality control of translation, and the evolution of ribosomes. The ribosome developed complex mechanisms that allow for a uniform recognition and selection of any cognate aminoacyl-tRNA (aa-tRNA) and discrimination against any near-cognate aa-tRNA, regardless of the nature or position of the mismatch. This review describes the principles of the selection—kinetic partitioning and induced fit—and discusses the relationship between speed and accuracy of decoding, with a focus on bacterial translation. The translational machinery apparently has evolved towards high speed of translation at the cost of fidelity.
Keywords: ribosome, protein synthesis, translation fidelity, tRNA, error frequency, mRNA decoding
1. Speed and accuracy of translation
Protein synthesis on the ribosome is a fundamentally important process that consumes a large part of the energy resources of the cell. Ribosomes are universal macromolecular machines built of two subunits, the small subunit (30S subunit in bacteria), where mRNA decoding takes place, and the large subunit (50S subunit in bacteria), which harbours the catalytic site for peptide bond formation. The decoding and peptidyl transferase centres consist of RNA, suggesting that the ribosome originates from the RNA world. Intuitively, one would expect that the ribosome has evolved to produce proteins with maximum speed and accuracy at minimum metabolic cost. The aim of this review is to discuss the mechanisms by which this is achieved and potential limits to the optimization of the ribosome performance.
Protein synthesis entails four major phases: initiation, elongation, termination and recycling. During initiation, the ribosome selects an mRNA and, assisted by initiation factors, places the initiator tRNA on the appropriate start codon in the P site. In the subsequent elongation phase, amino acids are added to the growing peptide in a cyclic process. Aminoacyl-tRNAs (aa-tRNAs) enter the ribosome in a tight complex with elongation factor Tu (EF-Tu) and guanosine-5′-triphosphate (GTP). Following the recognition of the codon by the anticodon of aa-tRNA and GTP hydrolysis by EF-Tu, aa-tRNA is accommodated in the A site of the 50S subunit and takes part in peptide bond formation. The rate of protein elongation in bacteria is between 4 and 22 amino acids per second at 37°C [1–5]; thus, a protein of an average length of 330 amino acids [6] is completed in about 10–80 s. The times required for initiation, termination and ribosome recycling (around 1 s each [3]) are short enough to make elongation rate-limiting for protein synthesis [7]. Translation of a particular codon depends on both the nature and abundance of the respective tRNAs, particularly on the non-random use of synonymous codons and the availability of the respective isoacceptor tRNAs [8]. The overall rate of translation is limited by the codon-specific rates of cognate ternary complex delivery to the A site and is further attenuated by other factors, such as collisions between individual ribosomes in polysomes [3], controlled ribosome stalling (for a recent review, see [9]), or the cooperation between translating ribosomes and the RNA polymerase machinery [4,10].
Estimations of error frequencies of translation range between 10−5 and 10−3, depending on the type of measurement, concentrations and nature of tRNAs that perform misreading, and the mRNA context [11–13]. Different approaches were taken to measure these values. For instance, error frequencies of amino acid incorporation at a particular position of the protein were estimated based on different physico-chemical properties of native and altered proteins or their fragments (reviewed in [13]). Alternatively, error frequencies were obtained using reporter constructs expressing proteins that gain enzymatic activity upon misincorporation [12,14]. Unfortunately, the number of proteins and types of replacements studied so far are very limited, precluding a comprehensive analysis of error frequencies on particular codons or for different parts of proteins.
The overall measured error rate of protein synthesis reflects the accumulated mistakes from all steps involved in translation, of which tRNA aminoacylation and decoding arguably are the most error-prone. Misincorporation of amino acids into proteins may affect the fitness of an organism by reducing the amount of active proteins, producing proteins that are toxic for the cell, or increasing misfolding. Owing to the limited information available, the consequences of errors on the cellular and organismic levels and the long-term evolutionary responses to errors are not known [11]. Some missense errors may be more readily tolerated than others. Indeed, the common experience in expressing mutant proteins suggests that mutations at many positions in a protein, except within the catalytic site, are often well tolerated, although mild effects on fitness cannot be excluded. Recent estimations of fitness effects of mutations in two ribosomal proteins suggested that most mutations were only weakly detrimental and some were potentially neutral [15]. Furthermore, it was demonstrated that cells tolerated an astonishingly high degree of mistranslation in a case where up to 10 per cent of an enzyme carried a missense error [16]. Under some conditions, increased levels of mistranslation can be advantageous, e.g. upon adaptation of bacterial populations to sub-lethal antibiotic concentrations and the emergence of antibiotic resistance [17] or at conditions of amino acid deprivation (reviewed in [18]).
2. Kinetic discrimination of incorrect tRNAs
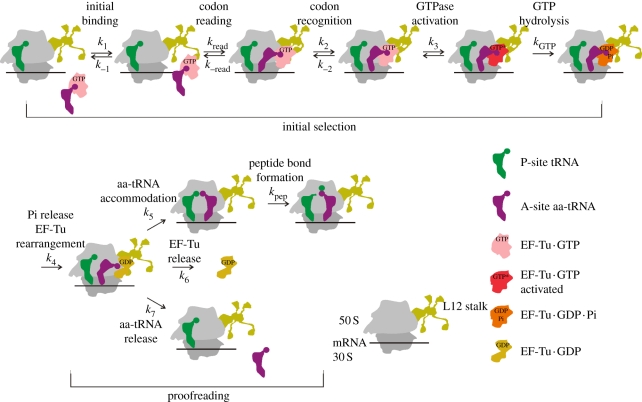
To prevent the incorporation of incorrect amino acids into proteins, aa-tRNA synthetases and ribosomes rigorously control the aminoacylation and the decoding reactions, respectively, employing numerous quality control mechanisms [18]. On the ribosome, the fidelity is controlled at three basic selection stages (figure 1): (i) preferential rejection of incorrect ternary complexes prior to GTP hydrolysis in the initial selection stage, (ii) preferential rejection of incorrect (mostly near-cognate) aa-tRNAs in the proofreading stage after GTP hydrolysis [21–23], and (iii) preferential hydrolysis of erroneous peptidyl-tRNAs by termination factors [24]. The overall missense error frequency of translation depends on the combined efficacy of these selections and on the abundance of the aa-tRNA cognate to a given codon relative to the near-cognate competitors [12].
Figure 1.
Schematic of EF-Tu-dependent aa-tRNA binding to the A site. Kinetically resolved steps are indicated by the rate constants k1–k7 (forward reactions) and k−1 and k−2 (backward reactions). The rate of codon reading (presumably a readily reversible step [19]) could not be determined by rapid kinetics; the values available from single-molecule fluorescence resonance energy transfer (FRET) experiments [19,20] are not comparable with the values obtained in bulk experiments owing to differences in experimental conditions. Rate constants of the two chemical steps that are rate-limited by the respective preceding step are designated kGTP and kpep.
The initial interaction of the ternary complex with the ribosome is codon-independent and mainly mediated by contacts between EF-Tu and ribosomal protein L7/12 [25,26]. During subsequent codon reading, the tRNA in the complex undergoes spontaneous structural fluctuations (distortions) that allow it to scan the codon in the decoding site of the 30S subunit, while remaining bound to EF-Tu which is anchored to the 50S subunit [19]. Formation of the correct, fully complementary (cognate) codon–anticodon complex locks the tRNA in the A/T state [27] and induces structural rearrangements of A1492, A1493 and G530 (Escherichia coli numbering is used throughout) in the decoding centre of the 30S subunit [28], establishing a complex network of interactions. At the first and second codon positions, the ribosome recognizes the base pairs according to their universal Watson–Crick geometry, while at the third position, a number of other base pairs are allowed. The rearrangement at the decoding centre is followed by domain closure of the 30S subunit [29], GTPase activation of EF-Tu and GTP hydrolysis [27,30–32]. The release of the reaction product, Pi, leads to an extensive structural change of EF-Tu, which switches from the GTP- to the guanosine-5′-diphosphate GDP-bound form [33,34]. Aa-tRNA has a very low affinity to EF-Tu · GDP and is released to be accommodated in the peptidyl transferase centre, where aa-tRNA takes part in peptide bond formation. Alternatively, the tRNA can be rejected (proofreading). EF-Tu · GDP dissociates from the ribosome and is reactivated by elongation factor Ts (EF-Ts), a guanine nucleotide exchange factor that accelerates the exchange of GDP for GTP [35]. Because of the high cellular concentrations of EF-Tu, practically all aa-tRNA is bound in the ternary complex and immediately available for entering the ribosome.
Kinetic studies have identified four elemental reactions that have different rates for cognate and near-cognate aa-tRNAs, whereas others are very similar (table 1) [27,30,31]. The rates of the ternary complex recruitment to the ribosome and of codon recognition are codon-independent. The codon independence of the latter step at first glance is counterintuitive. We note, however, that the codon-recognition step is likely to entail a large number of substeps comprising a number of conformational rearrangements in the complex, some of which are better resolved by single-molecule techniques [19,20] than by bulk kinetic measurements. The overall rate constant of the codon recognition step is a composite of the elemental rate constants of all these small substeps; it is currently unknown which substep is rate-limiting. The overall k2 value is reduced by antibiotics interfering with the conformational dynamics of the decoding region [38,39]. It is likely, therefore, that the observed k2 value is determined by conformational fluctuations at the decoding site concomitant with codon reading, which precede the formation, and are therefore independent of the geometry, of the codon–anticodon complex.
Table 1.
Rate constants of elemental steps of decoding the cognate UUU and near-cognate CUC codons by Phe-tRNAPhe (HiFi buffer, 20°C) [30,31]. The k6 value (figure 1) was not measured at HiFi conditions (3.5 mM Mg2+, polyamines); at 10 mM Mg2+, k6 = 3 s−1.
| rate constant | UUU | CUC |
|---|---|---|
| k1 (µM−1s−1) | 140 | 140 |
| k−1 (s−1) | 85 | 85 |
| k2 (s−1) | 190 | 190 |
| k−2 (s−1) | 0.2 | 80 |
| k3 (s−1) | 260 | 0.4 |
| k4 (s−1) | ≥10a | n.d. |
| k5 (s−1) | 20 | 0.26b |
| k7 (s−1) | <0.3 | 7b |
aThe value was measured at 1 µM substrate concentration [36].
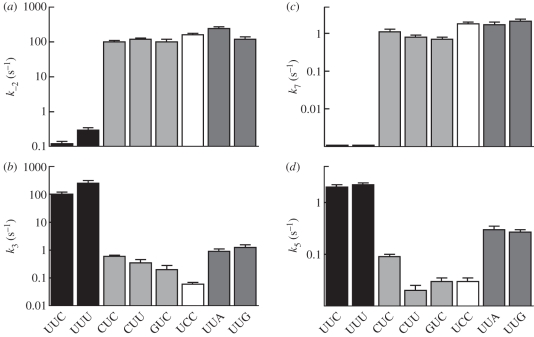
GTPase activation and aa-tRNA accommodation are the two rearrangements that limit the rates of the following chemistry steps, GTP hydrolysis and peptide bond formation, respectively. The rates of these reactions are strongly influenced by codon–anticodon interaction [27,30,38,40,41]. On the other hand, the rates of GTP hydrolysis are remarkably similar (within a factor of 3) for different aa-tRNAs on their respective cognate codons; the same is true for the rates of peptide bond formation [40–44]. This uniformity is achieved by tuning of the tRNA sequence and structure according to the nature of the amino acid [45]. Single mismatches at any position of the codon–anticodon complex result in slower forward reactions and a uniformly 1000-fold faster dissociation of ternary complexes from the ribosome (figure 2). This suggests that the network of interactions in the decoding centre [28] has evolved to yield a similar accuracy threshold, independent of the position of mismatches in the codon–anticodon duplex. Limited data obtained with Phe-tRNAPhe, Trp-tRNATrp and Ala-tRNAAla suggest that error frequencies are also similar for the near-cognate codons of other aa-tRNAs [30,40,44,45]. Uniform rates of decoding cognate and near-cognate codons presumably reflect the selective pressure that maintains similar translational accuracy for all codons.
Figure 2.
Effect of codon–anticodon mismatches on the crucial steps of (a,b) initial selection (rate constants of dissociation k−2, GTPase activation k3) and (c,d) proofreading (dissociation k7, accommodation k5). Cognate codons are UUC and UUU (black bars). Near-cognate codons contain a single mismatch in the first (CUC, CUU, GUC, light grey), second (UCC, white) or third (UUA, UUG, dark grey) codon position [30].
Although aa-tRNAs have very similar intrinsic decoding properties, not all codons are translated at the same rate. In fact, the rate of GTP hydrolysis by EF-Tu is 2.5 times higher on a cognate codon with a Watson–Crick base pair in the third position compared with a wobble pair [30,46]. Likewise, the rates of GTP hydrolysis on near-cognate codons differ by as much as 20-fold; the same is true for peptide bond formation (figure 2) [30]. Also tRNA modifications play an important role by expanding the repertoire of unconventional base pairs at the third codon position beyond the wobble rules; such codon–anticodon complexes can be treated as ‘almost-cognate’ and are much less disfavoured than the near-cognate ones [44,47]. These variations in the rates of decoding, together with the differences in the concentrations of individual cognate and near-cognate tRNAs, result in different translation rates—and possibly misreading frequencies—on individual codons.
3. The crucial forward steps of decoding
The ribosome controls the differences in the stabilities of the codon–anticodon complexes (k−2, k7) and specifically increases the rate constants of GTPase activation (k3) and accommodation (k5) for correct substrates, implicating both an increased stability of tRNA binding and induced fit as sources of selectivity. Kinetic partitioning between GTPase activation and ternary complex dissociation strongly favours the acceptance of cognate and the rejection of near-cognate ternary complexes. Likewise, cognate aa-tRNA is preferentially accommodated during proofreading, while near-cognate tRNA is largely rejected.
The decoding centre of the ribosome on the 30S subunit is almost 80 Å away from the GTP binding pocket of EF-Tu bound to the 50S subunit, where GTP hydrolysis takes place. A most important unresolved question in decoding is how the correct codon–anticodon interaction results in the acceleration of the forward reactions of GTP hydrolysis (rate-limited by GTPase activation) and peptide bond formation (rate-limited by accommodation). GTP hydrolysis proceeds through the attack of a water molecule on the γ-phosphate of GTP in EF-Tu. His84 in E. coli EF-Tu is the active-site residue that stabilizes the GTPase transition state, but in free EF-Tu is oriented away from the hydrolytic water [34,43,48]. Upon GTPase activation, His84 has to move towards the γ-phosphate, and this movement should be induced only when a correct codon–anticodon complex is formed. A distorted conformation of the tRNA, which is stabilized upon interaction of the cognate codon–anticodon complex with the ribosome, seems to play an important role, because aa-tRNA has to be intact for GTPase activation [49] and because mutations that alter the structure of the tRNA at the site of distortion affect GTP hydrolysis [40,50]. Structures suggest that the tRNA distortion affects the relative orientation of tRNA and EF-Tu [48,51–53]. This leads to subtle rearrangements in EF-Tu and the stabilization of the catalytically active orientation of His84 by an interaction with A2662 of the sarcin–ricin loop of 23S rRNA [48], which ultimately results in GTPase activation. The mechanism of activation must be precisely tuned for each cognate aa-tRNA, as all cognate ternary complexes exhibit similar rates of GTP hydrolysis, despite the structural variations of the tRNAs owing to differences in sequence and post-transcriptional modifications [41]. However, aa-tRNAs are about equally distorted on cognate and near-cognate codons, suggesting that, although the distortion of the tRNA structure is necessary for GTPase activation, it is as such not sufficient to account for the discrimination between cognate and near-cognate ternary complexes [36].
The uniformity of mismatch recognition [30] suggests a global response mechanism, which would be consistent with the idea that all conformational changes that occur upon cognate codon recognition, including domain closure of the 30S subunit, distortions of the tRNA and rearrangements in EF-Tu, are essential for the precise positioning of the GTPase centre of EF-Tu at the sarcin–ricin loop, which is required for GTPase activation [48]. Although the tRNA is distorted also in the near-cognate A/T state [36], subtle changes in the orientation of tRNA and EF-Tu could interfere with GTPase activation by preventing A2662 from properly placing His84 into the active site [48]. In this framework, tRNA mutants that strongly activate GTP hydrolysis on a near-cognate codon [40,45] appear to have found their own unique conformational solution to dock EF-Tu on the sarcin–ricin loop. Nevertheless, other contacts in the codon recognition complex may specifically affect the stringency of decoding, e.g. helix 14 and helix 8 of 16S rRNA, which negatively regulate GTP hydrolysis [54], or the interactions between helix 5 and domain 2 of EF-Tu [55]. Finally, the ribosome may play an active role in monitoring the correct codon–anticodon interaction using a network formed by helices 18 and 44 of 16S rRNA, helices 38, 69 and 89 of 23S rRNA, and proteins S13, S19, L16, L25, L27 and L31 from both ribosomal subunits, as suggested by the crystal structure of a ribosome–tRNA complex in the proofreading stage [56].
Compared with GTPase activation and GTP hydrolysis, much less is known about the codon-specific acceleration of peptide bond formation. Based on the available data, the reaction rate is limited by the accommodation of the aa-tRNA in the peptidyl transferase centre on the 50S subunit [37,57]. Computer simulations of accommodation suggested a stepwise movement of the tRNA through a corridor of conserved rRNA bases, which engage in various interactions with the tRNA during this movement [58]. The final accommodation of aa-tRNA in the A site of the peptidyl transferase centre appears to be determined by the interaction between the 3′-CCA end of aa-tRNA and the A loop (helix 92) of 23S rRNA, in particular the universally conserved residues U2492, C2556 and C2573 [59]. The latter residues were proposed to act as a gate that causes the acceptor stem to pause before entrance into the peptidyl transferase centre is allowed [58]. The rigidity and spring-like mechanical properties of the tRNA were suggested to play an important role in the transmission of the decoding signal, enabling the tRNA to precisely position the acceptor arm in the A/T state, the accommodation corridor or the A/A state [58,60]. Correct codon–anticodon alignment results in high rates of accommodation. By contrast, incorrect tRNAs, whose anticodons are misaligned in the decoding site, will also have their acceptor arms misaligned and will therefore be impaired in the movement through the accommodation corridor. Indeed, mutagenesis studies [61] indicated that substitutions of residues U2492 and U2555 at the accommodation gate decreased the fidelity of translation, supporting the view that the accommodation gate may attenuate aa-tRNA binding. However, mutations of C2573 and the neighbouring A2572 did not affect aa-tRNA accommodation, peptide bond formation or the fidelity of aa-tRNA selection, suggesting that the ribosome may allow rapid aa-tRNA accommodation in spite of defects at the accommodation gate, perhaps by using a different pathway [62]. Alternatively, misalignment of the near-cognate tRNA may disturb the network of interactions between tRNA and ribosome [56], thereby disfavouring accommodation and favouring rejection.
4. Trade-off between speed and accuracy
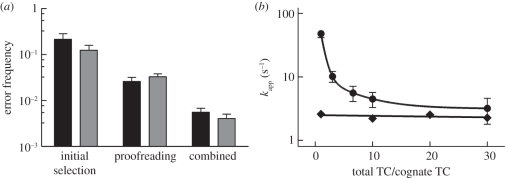
One major question concerns the maximum capacity of the ribosome for tRNA discrimination. Measurements in vitro using reconstituted translation systems working at conditions optimized for speed and fidelity account for an error frequency of about 10−4–10−3 (figure 3a) [24,30,31,37]. The much lower in vitro error frequency of 10−7 reported recently [63] can now be attributed to the use of extremely low concentrations of free Mg2+ that destabilized near-cognate codon recognition complexes much more than cognate ones, leading to an artificially high incorporation of cognate relative to near-cognate amino acids [37]. The about 1000-fold difference between the k−2 values imply a large difference in the thermodynamic stability, ΔΔG°, between cognate and near-cognate codon-recognition complexes [29,31,43,64]. However, this large inherent ΔΔG° of binding does not enter selection as a simple one-step association–dissociation equilibrium. This is because with cognate complexes the following steps of GTPase activation and GTP hydrolysis are rapid, precluding the equilibration of the binding step and leading to high KM values for cognate ternary complexes [31]. In such a case, kinetic partitioning between forward and backward reactions becomes particularly important, because it determines the fate of aa-tRNA on the ribosome. Given the values k−2 = 0.2 s−1 and k3 = 260 s−1 for decoding of the cognate UUU codon (table 1), the cognate ternary complex will preferentially undergo GTP hydrolysis rather than dissociate, whereas on the near-cognate codon (k−2 = 80 s−1, k3 = 0.4 s−1), the ternary complex will be preferentially rejected. Taking into account also the preceding steps of decoding, this results in an error frequency of about 1 : 60 at the initial selection step [31]. Notably, variations in k3 values for near-cognate codons have large effects on the efficiency of tRNA selection; e.g. a 10-fold reduction of the near-cognate k3 reduces the error frequency 10-fold. In contrast, even if the ΔΔG° value between the cognate and near-cognate binding were much less than determined experimentally, this would have little effect on the resulting error frequency; e.g. if the cognate k−2 were 100 times higher than the measured 0.2 s−1, this would reduce the ΔΔG° value dramatically, but would have almost no effect on the error frequency as long as the k−2 value is lower than k3 for the cognate codon (see [27,31] for the respective equations). The same principles of kinetic partitioning apply for proofreading. Such a selection mechanism allows for rapid translation; however, the maximum of intrinsically possible discrimination is not achieved, a phenomenon known as trade-off between speed and accuracy [64,65].
Figure 3.
Accuracy and speed of decoding in vitro. (a) Contributions of initial selection and proofreading to combined error frequency at optimized conditions in polymix (black bars) and HiFi (grey bars) buffers (adapted from Wohlgemuth et al. [37]). (b) Rates (kapp) of GTP hydrolysis (circles) and dipeptide formation (diamonds) were measured upon addition of increasing concentrations of competing near- and non-cognate ternary complexes (TC).
As a consequence of high rates of GTP hydrolysis and lack of equilibration, the apparent affinity of the ternary complex in the cognate codon recognition complex, expressed as KM, is much less than its true thermodynamic affinity represented by Kd. The consequence of the high cognate GTPase rate (which at first glance feels counterintuitive) becomes apparent when the KM value for the cognate complex is compared with those expected for the near- or non-cognate complexes: the KM values for the cognate and non-cognate ternary complexes are quite similar [21], suggesting that ternary complexes are selected solely on the basis of differences in kcat. On the other hand, it has been argued that similar KM values for the cognate and non-cognate complexes would lead to slow cognate amino acid incorporation owing to competition by the large excess of incorrect ternary complexes over correct ones [65,66]. However, competition experiments showed that, although the presence of excess bulk ternary complexes reduced the rate of GTP hydrolysis for the cognate ternary complex about 10-fold, this is not reflected in a decrease of the rate of cognate peptide bond formation, because the rate of GTP hydrolysis even at high concentrations of inhibitory ternary complexes is higher than the rate of peptide bond formation, buffering the inhibition by competition (figure 3b) [37]. Thus, the high speed of GTP hydrolysis on cognate codons in fact causes a loss of intrinsically available selectivity, but at the same time precludes that the rate of amino acid incorporation is decreased by competition with bulk ternary complexes. This suggests that it is the rate of GTP hydrolysis, and not that of peptide bond formation, which governs the evolution and the optimization of both speed and accuracy of translation, and explains why the maximum accuracy intrinsic in the system is not achieved.
The ribosome is an ancient RNA catalyst which in many respects differs from the specialized enzymes consisting of protein. It was argued that owing to the conditions at the onset of life, the evolution of the protein enzymes was driven by optimization of the activation enthalpy, ΔH≠, in such a way that for most modern enzymes it is around 12 kcal mol−1 [67]. This value is significantly lower than the corresponding values for the ribosome-catalysed reactions. Although the mechanisms of GTP hydrolysis and peptide bond formation on the ribosome are quite different from one another, the activation enthalpy for both GTP hydrolysis and peptide bond formation is 17–20 kcal mol−1 [63,68]. For the GTPase reaction on the ribosome, one possible reason for this difference to the enzyme-catalysed reactions is the necessity to optimize speed and fidelity at the same time; the optimization of subsequent peptide bond formation beyond the efficiency of the GTPase would be unnecessary. In fact, the ribosome accelerates the peptidyl transfer reaction by a factor of about 107–108 [37,42,68], i.e. it is much less efficient than many protein enzymes that accelerate reactions by up to 1023-fold [69].
Another peculiar feature of the ribosome is the necessity to deal with different peptidyl-tRNAs and aa-tRNAs with each codon combination in P and A sites. Because the chemical properties of these substrates can be different, the precise adjustment of the catalytic groups that could potentially contribute to the enthalpic effect in the peptidyl transferase centre may not be feasible and thus catalysis is predominantly entropic [68]. This may also explain why the ribosome did not evolve to contain proteins, with their large repertoire of chemical groups suitable for efficient catalysis, at its catalytic sites. Apparently, the evolutionary pressure favoured the optimization of speed and accuracy of the rate-limiting steps of protein synthesis, while at the same time retaining the ability to accept a large number of substrates with potentially different properties. This might have allowed the ribosome to retain its RNA-based catalytic strategy during the evolution from a prebiotic translational ribozyme into a modern ribosome, which thus appears to be a living fossil of a primitive catalyst of the RNA world.
Acknowledgements
We thank Wolfgang Wintermeyer for critical reading of the manuscript. The work was supported by the Deutsche Forschungsgemeinschaft.
References
- 1.Bremer H., Dennis P. P. 1987. Modulation of chemical composition and other parameters of the cell by growth rate. In Escherichia coli and Salmonella typhimurium: cellular and molecular biology (ed. Neidhardt F. C.), pp. 1553–1569 Washington, DC: American Society for Microbiology [Google Scholar]
- 2.Liang S. T., Xu Y. C., Dennis P., Bremer H. 2000. mRNA composition and control of bacterial gene expression. J. Bacteriol. 182, 3037–3044 10.1128/JB.182.11.3037-3044.2000 (doi:10.1128/JB.182.11.3037-3044.2000) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Mitarai N., Sneppen K., Pedersen S. 2008. Ribosome collisions and translation efficiency: optimization by codon usage and mRNA destabilization. J. Mol. Biol. 382, 236–245 10.1016/j.jmb.2008.06.068 (doi:10.1016/j.jmb.2008.06.068) [DOI] [PubMed] [Google Scholar]
- 4.Proshkin S., Rahmouni A. R., Mironov A., Nudler E. 2010. Cooperation between translating ribosomes and RNA polymerase in transcription elongation. Science 328, 504–508 10.1126/science.1184939 (doi:10.1126/science.1184939) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Sorensen M. A., Pedersen S. 1991. Absolute in vivo translation rates of individual codons in Escherichia coli. The two glutamic acid codons GAA and GAG are translated with a threefold difference in rate. J. Mol. Biol. 222, 265–280 10.1016/0022-2836(91)90211-N (doi:10.1016/0022-2836(91)90211-N) [DOI] [PubMed] [Google Scholar]
- 6.Netzer W. J., Hartl F. U. 1997. Recombination of protein domains facilitated by co-translational folding in eukaryotes. Nature 388, 343–349 10.1038/41024 (doi:10.1038/41024) [DOI] [PubMed] [Google Scholar]
- 7.Mehra A., Hatzimanikatis V. 2006. An algorithmic framework for genome-wide modeling and analysis of translation networks. Biophys. J. 90, 1136–1146 10.1529/biophysj.105.062521 (doi:10.1529/biophysj.105.062521) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Buchan J. R., Stansfield I. 2007. Halting a cellular production line: responses to ribosomal pausing during translation. Biol. Cell 99, 475–487 10.1042/BC20070037 (doi:10.1042/BC20070037) [DOI] [PubMed] [Google Scholar]
- 9.Ito K., Chiba S., Pogliano K. 2010. Divergent stalling sequences sense and control cellular physiology. Biochem. Biophys. Res. Commun. 393, 1–5 10.1016/j.bbrc.2010.01.073 (doi:10.1016/j.bbrc.2010.01.073) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Burmann B. M., Schweimer K., Luo X., Wahl M. C., Stitt B. L., Gottesman M. E., Rosch P. 2010. A NusE : NusG complex links transcription and translation. Science 328, 501–504 10.1126/science.1184953 (doi:10.1126/science.1184953) [DOI] [PubMed] [Google Scholar]
- 11.Drummond D. A., Wilke C. O. 2009. The evolutionary consequences of erroneous protein synthesis. Nat. Rev. Genet. 10, 715–724 10.1038/nrg2662 (doi:10.1038/nrg2662) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kramer E. B., Farabaugh P. J. 2007. The frequency of translational misreading errors in E. coli is largely determined by tRNA competition. RNA 13, 87–96 10.1261/rna.294907 (doi:10.1261/rna.294907) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Parker J. 1989. Errors and alternatives in reading the universal genetic code. Microbiol. Rev. 53, 273–298 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sergiev P. V., Lesnyak D. V., Kiparisov S. V., Burakovsky D. E., Leonov A. A., Bogdanov A. A., Brimacombe R., Dontsova O. A. 2005. Function of the ribosomal E-site: a mutagenesis study. Nucl. Acids Res. 33, 6048–6056 10.1093/nar/gki910 (doi:10.1093/nar/gki910) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lind P. A., Berg O. G., Andersson D. I. 2010. Mutational robustness of ribosomal protein genes. Science 330, 825–827 10.1126/science.1194617 (doi:10.1126/science.1194617) [DOI] [PubMed] [Google Scholar]
- 16.Ruan B., Palioura S., Sabina J., Marvin-Guy L., Kochhar S., Larossa R. A., Soll D. 2008. Quality control despite mistranslation caused by an ambiguous genetic code. Proc. Natl Acad. Sci. USA 105, 16 502–16 507 10.1073/pnas.0809179105 (doi:10.1073/pnas.0809179105) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lee H. H., Molla M. N., Cantor C. R., Collins J. J. 2010. Bacterial charity work leads to population-wide resistance. Nature 467, 82–85 10.1038/nature09354 (doi:10.1038/nature09354) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Reynolds N. M., Lazazzera B. A., Ibba M. 2010. Cellular mechanisms that control mistranslation. Nat. Rev. Microbiol. 8, 849–856 10.1038/nrmicro2472 (doi:10.1038/nrmicro2472) [DOI] [PubMed] [Google Scholar]
- 19.Geggier P., Dave R., Feldman M. B., Terry D. S., Altman R. B., Munro J. B., Blanchard S. C. 2010. Conformational sampling of aminoacyl-tRNA during selection on the bacterial ribosome. J. Mol. Biol. 399, 576–595 10.1016/j.jmb.2010.04.038 (doi:10.1016/j.jmb.2010.04.038) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Blanchard S. C., Gonzalez R. L., Kim H. D., Chu S., Puglisi J. D. 2004. tRNA selection and kinetic proofreading in translation. Nat. Struct. Mol. Biol. 11, 1008–1014 10.1038/nsmb831 (doi:10.1038/nsmb831) [DOI] [PubMed] [Google Scholar]
- 21.Rodnina M. V., Wintermeyer W. 2001. Fidelity of aminoacyl-tRNA selection on the ribosome: kinetic and structural mechanisms. Annu. Rev. Biochem. 70, 415–435 10.1146/annurev.biochem.70.1.415 (doi:10.1146/annurev.biochem.70.1.415) [DOI] [PubMed] [Google Scholar]
- 22.Ruusala T., Ehrenberg M., Kurland C. G. 1982. Is there proofreading during polypeptide synthesis? EMBO J. 1, 741–745 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Thompson R. C., Stone P. J. 1977. Proofreading of the codon–anticodon interaction on ribosomes. Proc. Natl Acad. Sci. USA 74, 198–202 10.1073/pnas.74.1.198 (doi:10.1073/pnas.74.1.198) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zaher H. S., Green R. 2009. Quality control by the ribosome following peptide bond formation. Nature 457, 161–166 10.1038/nature07582 (doi:10.1038/nature07582) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Diaconu M., Kothe U., Schlunzen F., Fischer N., Harms J. M., Tonevitsky A. G., Stark H., Rodnina M. V., Wahl M. C. 2005. Structural basis for the function of the ribosomal L7/12 stalk in factor binding and GTPase activation. Cell 121, 991–1004 10.1016/j.cell.2005.04.015 (doi:10.1016/j.cell.2005.04.015) [DOI] [PubMed] [Google Scholar]
- 26.Kothe U., Wieden H. J., Mohr D., Rodnina M. V. 2004. Interaction of helix D of elongation factor Tu with helices 4 and 5 of protein L7/12 on the ribosome. J. Mol. Biol. 336, 1011–1021 10.1016/j.jmb.2003.12.080 (doi:10.1016/j.jmb.2003.12.080) [DOI] [PubMed] [Google Scholar]
- 27.Pape T., Wintermeyer W., Rodnina M. 1999. Induced fit in initial selection and proofreading of aminoacyl-tRNA on the ribosome. EMBO J. 18, 3800–3807 10.1093/emboj/18.13.3800 (doi:10.1093/emboj/18.13.3800) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ogle J. M., Brodersen D. E., Clemons W. M., Jr, Tarry M. J., Carter A. P., Ramakrishnan V. 2001. Recognition of cognate transfer RNA by the 30S ribosomal subunit. Science 292, 897–902 10.1126/science.1060612 (doi:10.1126/science.1060612) [DOI] [PubMed] [Google Scholar]
- 29.Ogle J. M., Murphy F. V., Tarry M. J., Ramakrishnan V. 2002. Selection of tRNA by the ribosome requires a transition from an open to a closed form. Cell 111, 721–732 10.1016/S0092-8674(02)01086-3 (doi:10.1016/S0092-8674(02)01086-3) [DOI] [PubMed] [Google Scholar]
- 30.Gromadski K. B., Daviter T., Rodnina M. V. 2006. A uniform response to mismatches in codon–anticodon complexes ensures ribosomal fidelity. Mol. Cell 21, 369–377 10.1016/j.molcel.2005.12.018 (doi:10.1016/j.molcel.2005.12.018) [DOI] [PubMed] [Google Scholar]
- 31.Gromadski K. B., Rodnina M. V. 2004. Kinetic determinants of high-fidelity tRNA discrimination on the ribosome. Mol. Cell 13, 191–200 10.1016/S1097-2765(04)00005-X (doi:10.1016/S1097-2765(04)00005-X) [DOI] [PubMed] [Google Scholar]
- 32.Rodnina M. V., Fricke R., Kuhn L., Wintermeyer W. 1995. Codon-dependent conformational change of elongation factor Tu preceding GTP hydrolysis on the ribosome. EMBO J. 14, 2613–2619 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kothe U., Rodnina M. V. 2006. Delayed release of inorganic phosphate from elongation factor Tu following GTP hydrolysis on the ribosome. Biochemistry 45, 12 767–12 774 10.1021/bi061192z (doi:10.1021/bi061192z) [DOI] [PubMed] [Google Scholar]
- 34.Berchtold H., Reshetnikova L., Reiser C. O., Schirmer N. K., Sprinzl M., Hilgenfeld R. 1993. Crystal structure of active elongation factor Tu reveals major domain rearrangements. Nature 365, 126–132 10.1038/365126a0 (doi:10.1038/365126a0) [DOI] [PubMed] [Google Scholar]
- 35.Gromadski K. B., Wieden H. J., Rodnina M. V. 2002. Kinetic mechanism of elongation factor Ts-catalyzed nucleotide exchange in elongation factor Tu. Biochemistry 41, 162–169 10.1021/bi015712w (doi:10.1021/bi015712w) [DOI] [PubMed] [Google Scholar]
- 36.Mittelstaet J., Konevega A. L., Rodnina M. V. 2011. Distortion of tRNA upon near-cognate codon recognition on the ribosome. J. Biol. Chem. 286, 8158–8164 10.1074/jbc.M110.210021 (doi:10.1074/jbc.M110.210021) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wohlgemuth I., Pohl C., Rodnina M. V. 2010. Optimization of speed and accuracy of decoding in translation. EMBO J. 29, 3701–3709 10.1038/emboj.2010.229 (doi:10.1038/emboj.2010.229) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Gromadski K. B., Rodnina M. V. 2004. Streptomycin interferes with conformational coupling between codon recognition and GTPase activation on the ribosome. Nat. Struct. Mol. Biol. 11, 316–322 10.1038/nsmb742 (doi:10.1038/nsmb742) [DOI] [PubMed] [Google Scholar]
- 39.Pape T., Wintermeyer W., Rodnina M. V. 2000. Conformational switch in the decoding region of 16S rRNA during aminoacyl-tRNA selection on the ribosome. Nat. Struct. Biol. 7, 104–107 10.1038/72364 (doi:10.1038/72364) [DOI] [PubMed] [Google Scholar]
- 40.Cochella L., Green R. 2005. An active role for tRNA in decoding beyond codon:anticodon pairing. Science 308, 1178–1180 10.1126/science.1111408 (doi:10.1126/science.1111408) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ledoux S., Uhlenbeck O. C. 2008. Different aa-tRNAs are selected uniformly on the ribosome. Mol. Cell 31, 114–123 10.1016/j.molcel.2008.04.026 (doi:10.1016/j.molcel.2008.04.026) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Johansson M., Ieong K. W., Trobro S., Strazewski P., Aqvist J., Pavlov M. Y., Ehrenberg M. 2011. pH-sensitivity of the ribosomal peptidyl transfer reaction dependent on the identity of the A-site aminoacyl-tRNA. Proc. Natl Acad. Sci. USA 108, 79–84 10.1073/pnas.1012612107 (doi:10.1073/pnas.1012612107) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Daviter T., Gromadski K. B., Rodnina M. V. 2006. The ribosome's response to codon–anticodon mismatches. Biochimie 88, 1001–1011 10.1016/j.biochi.2006.04.013 (doi:10.1016/j.biochi.2006.04.013) [DOI] [PubMed] [Google Scholar]
- 44.Kothe U., Rodnina M. V. 2007. Codon reading by tRNAAla with modified uridine in the wobble position. Mol. Cell 25, 167–174 10.1016/j.molcel.2006.11.014 (doi:10.1016/j.molcel.2006.11.014) [DOI] [PubMed] [Google Scholar]
- 45.Ledoux S., Olejniczak M., Uhlenbeck O. C. 2009. A sequence element that tunes Escherichia coli tRNA(Ala)(GGC) to ensure accurate decoding. Nat. Struct. Mol. Biol. 16, 359–634 10.1038/nsmb.1581 (doi:10.1038/nsmb.1581) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Thomas L. K., Dix D. B., Thompson R. C. 1988. Codon choice and gene expression: synonymous codons differ in their ability to direct aminoacylated-transfer RNA binding to ribosomes in vitro. Proc. Natl Acad. Sci. USA 85, 4242–4246 10.1073/pnas.85.12.4242 (doi:10.1073/pnas.85.12.4242) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Murphy IV F. V., Ramakrishnan V. 2004. Structure of a purine–purine wobble base pair in the decoding center of the ribosome. Nat. Struct. Mol. Biol. 11, 1251–1252 10.1038/nsmb866 (doi:10.1038/nsmb866) [DOI] [PubMed] [Google Scholar]
- 48.Voorhees R. M., Schmeing T. M., Kelley A. C., Ramakrishnan V. 2010. The mechanism for activation of GTP hydrolysis on the ribosome. Science 330, 835–838 10.1126/science.1194460 (doi:10.1126/science.1194460) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Piepenburg O., Pape T., Pleiss J. A., Wintermeyer W., Uhlenbeck O. C., Rodnina M. V. 2000. Intact aminoacyl-tRNA is required to trigger GTP hydrolysis by elongation factor Tu on the ribosome. Biochemistry 39, 1734–1738 10.1021/bi992331y (doi:10.1021/bi992331y) [DOI] [PubMed] [Google Scholar]
- 50.Pan D., Zhang C. M., Kirillov S., Hou Y. M., Cooperman B. S. 2008. Perturbation of the tRNA tertiary core differentially affects specific steps of the elongation cycle. J. Biol. Chem. 283, 18 431–18 440 10.1074/jbc.M801560200 (doi:10.1074/jbc.M801560200) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Schmeing T. M., Voorhees R. M., Kelley A. C., Gao Y. G., Murphy VI F. V., Weir J. R., Ramakrishnan V. 2009. The crystal structure of the ribosome bound to EF-Tu and aminoacyl-tRNA. Science 326, 688–694 10.1126/science.1179700 (doi:10.1126/science.1179700) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Schuette J. C., et al. 2009. GTPase activation of elongation factor EF-Tu by the ribosome during decoding. EMBO J. 28, 755–765 10.1038/emboj.2009.26 (doi:10.1038/emboj.2009.26) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Villa E., et al. 2009. Ribosome-induced changes in elongation factor Tu conformation control GTP hydrolysis. Proc. Natl Acad. Sci. USA 106, 1063–1068 10.1073/pnas.0811370106 (doi:10.1073/pnas.0811370106) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.McClory S. P., Leisring J. M., Qin D., Fredrick K. 2010. Missense suppressor mutations in 16S rRNA reveal the importance of helices h8 and h14 in aminoacyl-tRNA selection. RNA 16, 1925–1934 10.1261/rna.2228510 (doi:10.1261/rna.2228510) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Vorstenbosch E., Pape T., Rodnina M. V., Kraal B., Wintermeyer W. 1996. The G222D mutation in elongation factor Tu inhibits the codon-induced conformational changes leading to GTPase activation on the ribosome. EMBO J. 15, 6766–6774 [PMC free article] [PubMed] [Google Scholar]
- 56.Jenner L., Demeshkina N., Yusupova G., Yusupov M. 2010. Structural rearrangements of the ribosome at the tRNA proofreading step. Nat. Struct. Mol. Biol. 17, 1072–1078 10.1038/nsmb.1880 (doi:10.1038/nsmb.1880) [DOI] [PubMed] [Google Scholar]
- 57.Pape T., Wintermeyer W., Rodnina M. V. 1998. Complete kinetic mechanism of elongation factor Tu-dependent binding of aminoacyl-tRNA to the A site of the E. coli ribosome. EMBO J. 17, 7490–7497 10.1093/emboj/17.24.7490 (doi:10.1093/emboj/17.24.7490) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Sanbonmatsu K. Y., Joseph S., Tung C. S. 2005. Simulating movement of tRNA into the ribosome during decoding. Proc. Natl Acad. Sci. USA 102, 15 854–15 859 10.1073/pnas.0503456102 (doi:10.1073/pnas.0503456102) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Kim D. F., Green R. 1999. Base-pairing between 23S rRNA and tRNA in the ribosomal A site. Mol. Cell 4, 859–864 10.1016/S1097-2765(00)80395-0 (doi:10.1016/S1097-2765(00)80395-0) [DOI] [PubMed] [Google Scholar]
- 60.Whitford P. C., Geggier P., Altman R. B., Blanchard S. C., Onuchic J. N., Sanbonmatsu K. Y. 2010. Accommodation of aminoacyl-tRNA into the ribosome involves reversible excursions along multiple pathways. RNA 16, 1196–1204 10.1261/rna.2035410 (doi:10.1261/rna.2035410) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.O'Connor M., Dahlberg A. E. 1995. The involvement of two distinct regions of 23 S ribosomal RNA in tRNA selection. J. Mol. Biol. 254, 838–847 10.1006/jmbi.1995.0659 (doi:10.1006/jmbi.1995.0659) [DOI] [PubMed] [Google Scholar]
- 62.Burakovsky D. E., Sergiev P. V., Steblyanko M. A., Kubarenko A. V., Konevega A. L., Bogdanov A. A., Rodnina M. V., Dontsova O. A. 2010. Mutations at the accommodation gate of the ribosome impair RF2-dependent translation termination. RNA 16, 1848–1853 10.1261/rna.2185710 (doi:10.1261/rna.2185710) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Johansson M., Bouakaz E., Lovmar M., Ehrenberg M. 2008. The kinetics of ribosomal peptidyl transfer revisited. Mol. Cell 30, 589–598 10.1016/j.molcel.2008.04.010 (doi:10.1016/j.molcel.2008.04.010) [DOI] [PubMed] [Google Scholar]
- 64.Thompson R. C., Karim A. M. 1982. The accuracy of protein biosynthesis is limited by its speed: high fidelity selection by ribosomes of aminoacyl-tRNA ternary complexes containing GTPγS. Proc. Natl Acad. Sci. USA 79, 4922–4926 10.1073/pnas.79.16.4922 (doi:10.1073/pnas.79.16.4922) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Lovmar M., Ehrenberg M. 2006. Rate, accuracy and cost of ribosomes in bacterial cells. Biochimie 88, 951–961 10.1016/j.biochi.2006.04.019 (doi:10.1016/j.biochi.2006.04.019) [DOI] [PubMed] [Google Scholar]
- 66.Bilgin N., Ehrenberg M., Kurland C. 1988. Is translation inhibited by noncognate ternary complexes? FEBS Lett. 233, 95–99 10.1016/0014-5793(88)81362-0 (doi:10.1016/0014-5793(88)81362-0) [DOI] [PubMed] [Google Scholar]
- 67.Stockbridge R. B., Lewis C. A., Jr, Yuan Y., Wolfenden R. 2010. Impact of temperature on the time required for the establishment of primordial biochemistry, and for the evolution of enzymes. Proc. Natl Acad. Sci. USA 107, 22 102–22 105 10.1073/pnas.1013647107 (doi:10.1073/pnas.1013647107) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Sievers A., Beringer M., Rodnina M. V., Wolfenden R. 2004. The ribosome as an entropy trap. Proc. Natl Acad. Sci. USA 101, 7897–7901 10.1073/pnas.0402488101 (doi:10.1073/pnas.0402488101) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Radzicka A., Wolfenden R. 1995. A proficient enzyme. Science 267, 90–93 10.1126/science.7809611 (doi:10.1126/science.7809611) [DOI] [PubMed] [Google Scholar]