Abstract
All models of the RNA world era invoke the presence of ribozymes that can catalyse RNA polymerization. The class I ligase ribozyme selected in vitro 15 years ago from a pool of random RNA sequences catalyses formation of a 3′,5′-phosphodiester linkage analogous to a single step of RNA polymerization. Recently, the three-dimensional structure of the ligase was solved in complex with U1A RNA-binding protein and independently in complex with an antibody fragment. The RNA adopts a tripod arrangement and appears to use a two-metal ion mechanism similar to protein polymerases. Here, we discuss structural implications for engineering a true polymerase ribozyme and describe the use of the antibody framework both as a portable chaperone for crystallization of other RNAs and as a platform for exploring steps in evolution from the RNA world to the RNA–protein world.
Keywords: ligase ribozyme, antibody, RNA world
1. Introduction
Current models of organismal evolution posit the existence of an ancient era when life forms lacked DNA and encoded proteins and relied solely on RNA for information storage and catalysis [1]. Known roles of RNA in catalysis, metabolite sensing and as components of enzymatic cofactors lend indirect support for this hypothesis and further suggest that organisms from this world had made significant evolutionary advances beyond the first protocells. To reproduce, these ‘riboorganisms’ would have required RNA molecules to catalyse RNA polymerization. Generating ribozymes that replicate RNA therefore represents an important milestone in understanding the evolution of life on the Earth. To date, the most promising efforts have commenced with the class I ligase ribozyme [2–4]. The original version of this ribozyme was isolated using in vitro selection methods from a large pool of random RNA sequences and was subsequently improved through mutation and selection [5,6]. The ribozyme catalyses the formation of a 3′,5′-phosphodiester bond whereby the 3′-OH group at the terminus of an oligonucleotide primer attacks the 5′-triphosphate present at the ribozyme's terminus, analogous to a single step in RNA polymerization catalysed by protein polymerases (figure 1).
Figure 1.
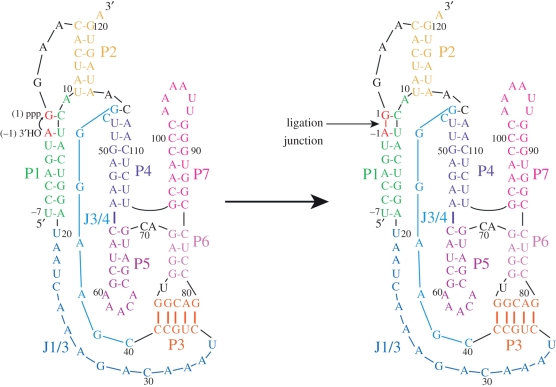
The class I ligase ribozyme catalyses the same reaction chemistry as protein polymerases. The 3′-hydroxyl group of the oligonucleotide primer attacks the alpha phosphorous atom of the triphosphate at the 5′ end of the ribozyme, displacing pyrophosphate and forming a new phosphodiester linkage between the oligonucleotide primer and the ribozyme. Shown is a primary and classical secondary structure representation with helical and secondary structure elements coloured. Adapted from Koldobskaya et al. [7].
The class I ligase has served as a model system for studying ribozyme evolution in vitro [6,8,9]. Additionally, significant effort has been aimed at converting the ligase into a bona fide RNA polymerase that uses nucleotide triphosphates and an external template to synthesize RNA [2–4,6]. Knowledge of the structure of the ligase would not only help in understanding how an RNA active site can catalyse a reaction thought so vital for the evolution of life, but also might aid in attempts to engineer the ribozyme towards greater processivity and ultimately self-replication. This article describes our efforts to develop antibody chaperones for use in RNA crystallization, and its application to the class I ligase. The work started as a collaboration with David Shechner and David Bartel, who first selected the ribozyme as a post-doctoral researcher in Jack Szostak's laboratory [5].
2. The problem of RNA crystallization
Presently, there are roughly 58 000 structures in the Protein Data Bank, but less than 5 per cent of these are experimentally solved RNA structures. One reason for this is that protein crystallography had a head start, and perhaps fewer laboratories work on RNA crystallography than protein crystallography. However, the dearth of RNA structures also reflects the significant challenges associated with RNA crystallization [10,11]. Some of the challenges are essentially the same as those associated with RNA folding: compared with proteins, there are significantly fewer functional groups on RNA to mediate tertiary contacts for folding and lattice contacts for crystal formation, and the repulsive forces from the negative charges on the phosphodiester groups would render such contacts less favourable [10]. Of course, Nature has solved the problem of RNA folding through evolutionary selection, but Nature does not select RNAs for their ability to crystallize. In addition to these problems, RNA molecules are prone to misfolding and adopting alternative conformations [11–15]. Moreover, once crystals are obtained, phasing for RNA crystals remains time-consuming and challenging compared with selenium-assisted phasing of protein crystals [11,16,17].
These problems have led RNA researchers to develop creative approaches to circumvent or ameliorate some of the problems. These include removing peripheral and unstructured domains that are not critical for function, engineering the RNA target for greater stability, including more stable secondary structure and folding at lower divalent metal ion concentrations, and incorporating potential lattice contacts such as tetraloop–tetraloop receptor interactions [4,11,14,18,19]. Another approach involves engineering protein-binding sites into the RNA to enable the formation of a protein–RNA complex. The protein could then help to facilitate crystallization by sequestering counterproductive surface area and by forming lattice contacts. Compared with RNA, proteins have a much greater variety of functional groups for facilitating specific macromolecular contacts [10]. To date, the U1A-binding protein, which binds to a short 10-nucleotide stem-loop structure, has been the approach most frequently used for RNA crystallography [20,21]. All of these approaches to RNA crystallization have met with some success; as yet, there exists no panacea.
(a). Antibody fragments as crystallization chaperones
Several years ago, we embarked on an alternative approach to the problem—the use of antibody fragments as chaperones for RNA crystallization. This strategy appeared attractive for a number of reasons: (i) antibody fragments were already proven as chaperones for the crystallization of recalcitrant membrane proteins [22–26]; (ii) antibodies could bind to an RNA tertiary structure and could potentially eliminate conformational heterogeneity associated with the RNA target (and possibly provide structural information about multiple conformations of the target RNA from structures of the target RNA bound to different antibodies); and (iii) the large size and well-defined architecture of antibodies could help solve the phasing of the crystals using molecular replacement (the U1A protein is three to four times smaller than an antigen-binding fragment (Fab) and possesses less phasing power than a Fab) [22,26].
Beyond the perceived potential applications to RNA structural biology, we envisioned other important applications for antibodies that recognize RNA antigens. The biotechnological and biomedical research communities have built a tremendous research infrastructure around antibodies [27,28]. There are nearly two dozen antibodies currently on the market as drugs, with several hundred more in various stages of clinical trials. Additionally, antibodies provide important diagnostic agents for disease antigens. Finally, as reagents for cell biology, antibodies have been invaluable, serving as research tools to define the components of macromolecular complexes, to investigate macromolecular function, and to establish the cellular locations of proteins. These applications involve predominantly antibodies that bind to protein antigens.
In contrast, we know very little about antibodies that bind to RNA, other than the few isolated from the sera of autoimmune patients [29–32]. For example, antibodies that bind to the U1 small nuclear RNA (and many mammalian small RNA–protein complexes) have been isolated from patients with autoimmune diseases [33]. The reason for the dearth of information is that RNA, when injected into animals, does not trigger the production of antibodies. As a consequence, the traditional hybridoma approaches used for antibody production are unlikely to yield anti-RNA antibodies upon injection of the target antigen into a host animal. This limitation has precluded RNA researchers from exploiting the full potential of immunomethods to the same degree that protein researchers have.
(b). Recombinant antibodies from phage display libraries
It was our vision of tapping into this potential for bringing direct immunomethods to the study of RNA that led us to join forces with our colleagues Shohei Koide and Tony Kossiakoff at the University of Chicago. Kossiakoff brought from Genentech the ability to obtain recombinant antibodies (Fabs) using phage display. Kossiakoff and Koide were heavily engaged in phage display technology to produce antibody and fibronectin domains against protein targets. Jingdong Ye, a post doctoral researcher in my laboratory, spent considerable time in their laboratories getting trained in the ‘art’ of phage display. Although mature and well-established, the phage display technique requires significant expertise to execute. Antibody libraries displayed on filamentous phage yield upto 10 billion different variants. Moreover, because phage display is an in vitro technique, we circumvent the need to inject the RNA target into an animal, thereby minimizing exposure of the target RNA to nucleases.
(i). Platform for phage display
Our display platform uses filamentous phage M13, with the Fab fragment (constant heavy and light chains (CH and CL respectively), and variable heavy and light chains (VH and VL respectively)) fused to pIII, a coat protein present at the pointed end of the phage particle [34,35]. The display fusion point links the C-terminus of the CH domain to the N-terminus of pIII. For RNA targets, we usually engineer a short stretch of 20 or so nucleotides at one of the RNA termini to allow hybridization of a complementary DNA oligonucleotide fused to a biotin tag, allowing immobilization of the RNA target on streptavidin beads [36]. In a typical round of selection, we incubate the phage variants with the RNA target, immobilize the bound phage using streptavidin, wash away unbound phage and elute RNA-bound phage specifically using RNase A [36]. Because we are interested in Fabs that bind with specificity to the RNA structure, subsequent rounds of selection usually include tRNA or another nucleic acid as a competitor to counter the selection of phage carrying Arg- or Lys-enriched Fabs that bind to the RNA by virtue of favourable electrostatic interactions. Following elution with RNase A, individual clones are screened in a competitive phage enzyme-linked immunosorbent assay (ELISA) to identify potential hits. Hits are sequenced and then expressed as soluble proteins from an Escherichia coli periplasmic expression system. We then conduct hydroxyl radical protection assays in the presence and absence of Fab to identify the epitope and determine whether the Fab alters the global fold of the RNA. In purifying the Fab, we take great care to remove any contaminating RNase, a step that can occasionally prove very challenging.
(ii). Reduced codon libraries
Variation of conformation and amino acid sequence in the complementarity determining regions (CDRs) of the VH and VL domains endows the antibody framework with great functional versatility. However, the theoretical number of CDR variants greatly exceeds the 1010 diversity provided in a typical phage display selection. In recent years, efforts have been made to build libraries consisting entirely of human-made diversity, whereby the gap between the theoretical and the practical diversity is reduced [37]. In this view, there is an underlying assumption that the vast majority of potential sequences will not be functional for binding target, and that to enable efficient and robust generation of Fabs against specific targets, the libraries must be biased in some way. Typical variables to be considered in library design include the following: (i) choice of CDR positions to introduce diversity, (ii) inclusion of length variability in specific CDRs, and (iii) choice of amino acids to include in the library. Sachdev Sidhu and his co-workers [34,37–39] at Genentech have constructed synthetic libraries that included diversity in select CDRs and only at solvent accessible positions. To restrict their diversity design to a small subset of amino acids, they turned to the analysis of the Kabat database of antibody–antigen interactions showing amino acid bias in the total CDR composition and in the CDR positions that contact antigen [40]. This analysis showed significant bias towards serine and tyrosine in total CDR composition, and tyrosine and tryptophan in the contact positions. This led Sidhu and co-workers to construct libraries with as little diversity as tyrosine (Y) and serine (S) [38]. Strikingly, using these binary YS libraries they could obtain Fabs that bind to target antigens with high affinity and specificity. Insights from structural analysis of these YS Fab–antigen complexes have shown that tyrosine mediates many of the Fab–antigen interactions, with serine serving as a small, flexible hydrophilic residue that enables the CDRs to arrange the tyrosines in a conformation suitable for binding antigen [41]. These and other studies with these so-called minimalist synthetic-binding proteins have underscored the importance of tyrosine for molecular recognition of protein antigens.
These and other studies in molecular recognition have led to the development of powerful design principles for obtaining antibodies and other synthetic-binding proteins against protein antigens. However, owing to the extremely limited information about RNA interactions with antibodies, the design principles for constructing anti-RNA Fab libraries are completely unknown. A simple guess might be that libraries biased in positively charged amino acids (arginine and lysine) would lead to effective Fab binders against negatively charged RNA; an alternative strategy would be to design the library on the basis of amino acid content found within protein–RNA interfaces. RNA-binding proteins generally are topologically distinct from the antibody framework, so it remains unclear how successful this strategy would be in developing a robust platform for obtaining antibodies. For the two RNA antibody structures we have obtained, analysis of the interfaces shows that the amino acid composition falls within range of that seen in RNA-binding proteins [7,36]. Optimizing an antibody library has a significant iterative component, whereby the sequences from a given library that yield functional binding clones are used to determine sequence composition of the next-generation library. With this in mind, we simply began our RNA selections using the first library we had access to, with little regard for whether the design would be considered suitable for RNA. In our first selections, we used the so-called YSG library (G represents glycine) from Genentech, which was known to be effective against protein antigens.
The YSG library uses a binary code (50% Y and 50% S) in CDR L3, H1 and H2. CDR H3, often the most critical for antigen recognition, contained greater diversity but was still biased towards Y, S and G (20, 15 and 15%, respectively) with 3 per cent of the remaining amino acids except cysteine [34,35]. CDRs L3 and H3 also contained length variability. We chose the independently folding P4–P6 RNA domain as our initial RNA target. This approximately 160 nucleotide RNA is a sequence element found within the Tetrahymena group I intron. This domain maintains its tertiary architecture even when isolated from the remainder of the intron [42–44]. We chose this RNA because it is very well characterized structurally and biochemically. Briefly, we picked seven clones after three rounds of selection. These clones showed enrichment for positively charged amino acids in CDR H3. Some of these clones bound the RNA with affinity in the 30–50 nM range and exhibited excellent specificity for the RNA tertiary structure [36]. We set up crystallization trials and obtained the structure of the Fab–RNA complex at 1.95 Å resolution using the molecular replacement to search for both molecules. However, using only the Fab coordinates, we could begin to build the RNA model into the remaining electron density, illustrating the phasing power of the Fab. Importantly, the Fab had minimal effect on the overall RNA tertiary structure [36].
With this success, we set out to obtain antibodies to other RNA targets using the YSG library. To our surprise and dismay, future selections using the YSG library yielded no positive clones against any of the targets we chose, including the group II intron, domains from RNase P RNA and several riboswitches. Having observed that the positive clones from the P4–P6 selections showed enrichment of positively charged amino acids in CDR H3, we deployed a different Genentech library, termed the YSGR ‘superlibrary’ [7]. This library contained a mixture of four sub-libraries constructed using the same design principles as the YSG library described above but differing in degree of arginine (R) bias in CDR H3. We used this library in subsequent selections against the class I ligase.
3. Antibodies that bind the class I ligase
We targeted our selections against the product of the ligase reaction [7]. After three rounds of selection, we obtained an enrichment ratio of 160, which usually indicates that the selected phage mix contained binders to the RNA target. Following competitive phage ELISA, we sequenced 23 positive clones and obtained three unique sequences. We expressed these as soluble, RNase-free proteins, referred to here as Fab BL1, BL2 and BL3, and used nitrocellose filter binding to determine the affinity of each Fab for the ligase product. In contrast to the 30–50 nM range Kd values that we observed for Fabs targeted against P4–P6, the BL Fabs bound with significantly weaker affinity (BL1, Kd ∼ 500 nM; BL2, Kd ∼ 1000 nM; BL3, Kd ∼ 350 nM). We also tested whether Fab binding affected ligase activity. Fabs BL2 and BL3 had no effect on ligase activity at saturating concentrations, but Fab BL1 inhibited ligase activity in a concentration-dependent manner [7]. The Fab concentration dependence of the inhibition matched the Kd observed by filter binding.
Generally, Fabs that have facilitated structure determination of proteins have bound their targets with Kd values less than 200 nM. Therefore, we attempted to improve affinity before moving on to crystallization trials [7]. We generated another library from the BL3-6 sequence using error-prone PCR to introduce variation throughout the light and heavy chain variable regions, including scaffold regions between the CDRs. For the first round of affinity maturation, we used 500 nM ligase product; in subsequent rounds, we carried out selections in parallel with RNA target concentrations varying from 0.025 to 2.5 nM. If we observed an enrichment ratio greater than 10-fold, we amplified the phage for the next round of selection. This procedure returned clones with no mutations in the heavy chain CDRs, but did yield clones with mutations in CDR L3 and non-CDR (scaffold) regions. These new clones, all derived from Fab BL3, bound with affinities ranging from 35 to 270 nM. The CDR L3 mutation of serine 95 to phenylalanine enhanced the binding most significantly (10-fold) [7]. We viewed the affinity of this Fab (designated hereafter as BL3-6) as sufficient to proceed with crystallization trials.
To determine the location of Fab BL3-6 binding, we used hydroxyl radical footprinting of the RNA product [7]. In the presence of saturating concentrations of Fab, the RNA retained the same protections observed in the absence of Fab, suggesting that the global RNA structure remains unaffected in the presence of Fab. We also observed new areas of protection, one mapping to the P5 loop (5′-AAACA-3′) and the other mapping to the P7 loop (5′-AAAAUU-3′). Regarding these protections, we were struck by the comparison to the footprint for the P4–P6-binding Fabs. P4–P6-binding Fabs protected multiple regions in P4–P6 that mapped to duplex regions that come together in the tertiary structure, and the Fab–P4–P6 structure revealed that the protections reflect Fab interactions in the minor groove of helices P5a and P5c. In contrast, Fab BL3-6 protected the ligase predominantly in single-stranded loop regions, L5 and L7. This difference led us to the idea that perhaps the Fab could bind to one or both of the isolated P5 or P7 hairpins.
We constructed 25-nucleotide hairpins that mimicked the P5 and P7 hairpins and tested them for binding to the suite of BL3-derived Fabs obtained from the affinity maturation protocol [7]. These Fabs had no detectable affinity for the P7 hairpin but bound to P5 with essentially the same affinity as observed for the full-length ligase. This observation allowed us to use the simpler P5 hairpin to address what features of the RNA contributed to the observed binding affinity. Binding assays using Fab BL3-6 revealed that the 5′-AAACA-3′ loop was critical for binding, particularly, the cytidine residue, and with the exception of the closing G-C pair, the identity of the base pairs in the stem were not critical. For the closing base pair, Fab-BL3-6 showed preference for G-C > A-U > C-G [7].
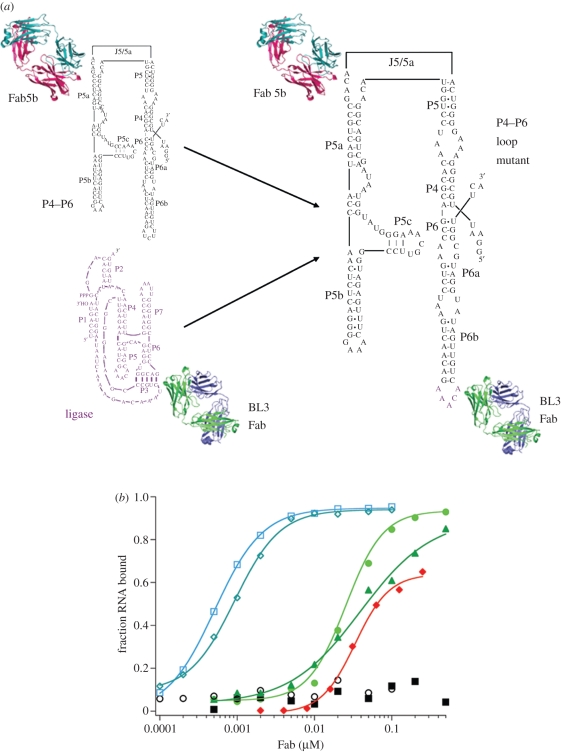
To determine whether the GAAACAC sequence (pentaloop plus closing base pair) could retain binding to Fab BL3-6 in the context of other structured RNA molecules, we replaced stem-loop regions in P4–P6 and the Varkud satellite (VS) ribozyme with the epitope [7]. For P4–P6, we replaced the L6 loop (5′-AUCUU-3′) with 5′-AAACA-3′ (the P6 closing base pair is already G-C) figure 2. For the VS ribozyme, we replaced loop IV and loop VI and closing base pairs with GAAACA. In all three cases, BL3-6 bound with Kd values below 200 nM. These findings raised the possibility of a new portable chaperone system for RNA crystallization that could be used in parallel with the U1A system mentioned above [7]. This has the potential for general use of the Fab chaperone by the broader RNA community as it circumvents the need for direct selection against every RNA target. Of course, because this approach relies on Fab binding to a targeted epitope, the potential advantage of using the Fab to reduce conformational heterogeneity in the RNA target is probably lost. Nevertheless, the approach offers similar advantages as the U1A system, with potential added advantages of the larger size and phasing power provided by the Fab. Our hope is that the Fab BL3-6 system will complement the U1A system. It would be ideal to have a Fab that binds the U1A RNA sequence as such a Fab would eliminate the need to make separate RNA constructs for parallel crystallization trials. As yet, we do not have such a Fab.
Figure 2.
A portable motif for high affinity Fab binding. (a) A variant P4–P6 RNA was constructed in which the antigenic sequence GAAACAAC replaced the L6 loop region (GAUCUUC). (b) This chimeric RNA retained affinity for the P4–P6 binding Fab (Fab 5b; blue curves), showing that the newly constructed RNA retained the P4–P6 global tertiary architecture. Additionally, the newly engineered loop conferred binding of the RNA to BL3 Fab family members with essentially the same affinity as for the class I ligase (red and green curves).
4. Crystal structure of the fab–ligase product complex at 3.1 å resolution
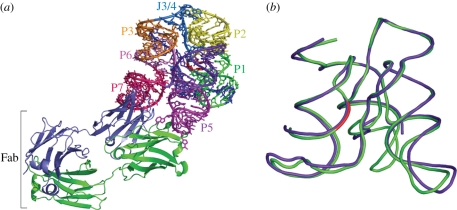
Crystals of the complex were grown using the hanging drop vapour diffusion method. Molecular replacement using Fab coordinates provided sufficient phasing power to resolve the RNA backbone. The asymmetric unit contained two complete Fab–ligase complexes (figure 3a), with the two RNA molecules having an r.m.s.d. of 0.026 Å [7].
Figure 3.
(a) Structure of the class I ligase–BL3-6 Fab complex. (a) Fab binds to RNA in the P5 and P7 regions of the ligase. (b) Overlay of our class I ligase structure with the ligase structure solved by Shechner et al. Purple, Shechner structure; green, our structure, with the ligation junction shown in red. All-atom r.m.s.d. = 1.44 Å. Colour coding in (a) matches that in figure 1. Adapted from Koldobskaya et al. [7].
(a). Global architecture of the class I ligase
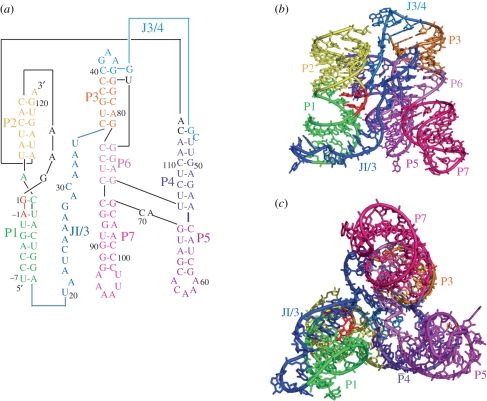
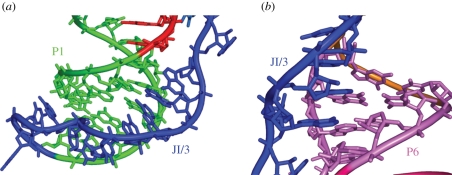
Independently, Shechner et al. [45] engineered the U1A RNA sequence into the P5 region of the ligase and solved the crystal structure of the ligase in complex with the U1A RNA-binding protein. The two structures show excellent overall agreement with an all-atom r.m.s.d. of 1.44 Å [7,45] (figure 3b). The overall ligase structures resemble a tripod, with three sets of coaxially stacked helices, P1/P2, P6/P7 and P4/P5, forming roughly equal legs (figure 4). At the base of the tripod, loops cap the P5 and P7 helices. At the head of the tripod, the three coaxial leg domains are joined by the following elements: the P3 pseudoknot helix, the P4/P5/P6/P7 four helix junction, and two single-stranded regions—J3/4, which connects the P3 pseudoknot to P4, and J1/3, which traverses the entire length of the ribozyme and docks into the minor grooves of P1 and P5 (figure 5) [7,45].
Figure 4.
Global architecture of the class I ligase. (a) Secondary structure of the crystallization construct, revised to reflect coaxial stacking and relative orientation of domains. (b) View facing J1/3 single-stranded region. (c) Bottom-up view showing tripod-like architecture of the ribozyme. Adapted from Koldobskaya et al. [7].
Figure 5.
Details of tertiary interactions mediated by J1/3. (a) The 5′ end of J1/3 forms base triples with helix P1; (b) the 3′ end of J1/3 forms base triples with helix P6.
(b). The Fab–RNA interface
The structural features of the Fab–RNA interface show excellent agreement with the biochemical analysis described above [7]. The Fab makes close contact to the P5 and P7 loop regions of the ligase, consistent with the protections observed in the hydroxyl radical footprinting experiments. CDRs L3, H1, H2 and H3 form a deep pocket that mediates recognition of the P5 RNA loop. In contrast, the P7 interaction, mediated by CDR L2 and scaffold residues, appears topologically less rugged. Consistent with our observation that P5 interactions drive binding, the P5 interaction buries significantly more surface area than does the P7 interaction. Our observations from the RNA mutational analysis also conform to structural features. Within CDR H3, three arginine side chains make close contact to the P5 loop. Arg 106 resides within hydrogen-bonding distance of the N7 atom of G59 in the closing base pair, possibly accounting, at least in part, for the Fab's preference for a G-C versus C-G closing base pair. Additionally, G59 stacks onto P5 loop residue A61, which stacks onto a tyrosine residue from CDR-H1. The slightly weaker binding observed for the A-U closing pair, which can still form an Arg–N7 interaction, may reflect increased fraying at the stem-loop junction. The importance of the C-residue in the loop likely comes from interactions with two serine residues in CDR-H2 whose hydroxyl groups are poised to make hydrogen bonds to the Watson–Crick face of the nucleobase. Finally, the Phe95 residue that emerged through affinity maturation to replace Ser95 stacks in-between Tyr62 from CDR-H2 and A62 from the P5 loop, consistent with the ability of the Ser95Phe mutation to enhance binding.
(c). Structural features of the class I ligase active site and implicated mechanism of catalysis
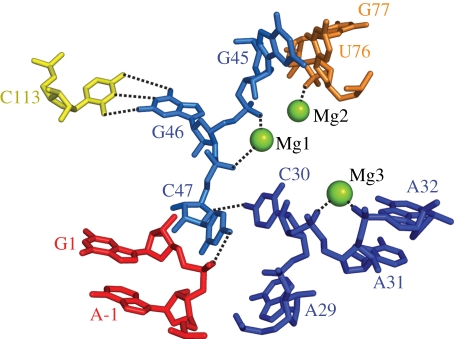
In the ligase product, the phosphate at the ligation junction, located between A−1 and G1, points inward towards the active site (figure 6, ligation junction shown in red). In the immediate vicinity of the ligation junction resides the J3/4 nucleobase, C47, with its exocyclic amine poised to make a hydrogen-bonding interaction to phosphodiester ligation junction. A series of Mg2+-supported backbone turns and base stacking interactions appear to shape the conformation of J3/4 to position C47. The backbone turn between U76 and G77 enables U76 to form a base-pairing interaction with G45. Further downstream, a Watson–Crick base pair in P4 between C113 and G46, together with a backbone turn between G46 and C47, probably contributes to C47 positioning. Further supporting the orientation of C47, the third backbone turn located between C30 and A31 allows C30 to stack with C47.
Figure 6.
Mg2+ supported turns organize tertiary interactions around the ligation junction.
Mutation of C47 decreases ligase activity by a factor of 10−4, strongly suggesting that it plays an important role in catalysis [45,46]. However, further analysis will be required to elucidate the mechanistic role of C47 in catalysis. When considered in the context of how protein enzymes catalyse RNA polymerization, the ligase structure has several interesting implications, as outlined by Shechner et al. [45]. The protein polymerases employ a general acid and two metal ions for catalysis [47,48]. The first metal ion in the protein enzymes interacts with two conserved aspartates and is proposed to activate the nucleophilic 3′-hydroxyl group; the second metal ion interacts with one of the aspartates and forms a chelate interaction with the beta and gamma phosphates of the incoming triphosphate. This chelation putatively serves to stabilize the developing negative charge as does protonation of the oxygen atom that bridges the alpha and beta phosphorous atoms. With respect to the ligase catalytic core, the active site constraints position the A29 and C30 phosphates near one another as well as near the ligation junction [45]. Possibly a metal ion binds in this location to activate the nucleophilic hydroxyl group. Consistent with this possibility, nucleotide analogue interference mapping experiments show a strong phosphorthioate effect for the C30 phosphate [45]. A second metal ion could be brought to the active site via chelation to the triphosphate moiety of the 5′-GTP in the precursor. C47 could supplant the role of the general acid found in protein polymerases, either forming a hydrogen bond with the leaving group or facilitating proton transfer via localized water molecules (figure 7).
Figure 7.
Proposed two-metal ion reaction mechanisms for (a) active site of RNA polymerase, (b) class I ligase active site. Adapted from Shechner et al. [45].
5. Conclusions
(a). A portable chaperone system for RNA crystallization
We have demonstrated the capacity of the Fab scaffold to generate proteins that bind RNA with high affinity and specificity. Reagents derived from this methodology can be used for chaperone assisted crystallography, synthetic biology whereby by the new RNA-binding protein may be used as a regulatory domain, and the application of immunomethods directly to RNA, including, for example, immunoprecipitation, immunohistochemistry, Western blotting and cellular localization studies. Additionally, our discovery of a Fab that binds a seven nucleotide sequence located within a hairpin loop engenders a portable RNA recognition element that can be transplanted into other RNAs to exploit antibody recognition with a wide range of RNAs. Additionally, with respect to crystallography applications, the large size and phasing power of the Fab may impart advantages over the commonly used U1A system.
(b). Exploring the transition from the RNA to the ribonucleoprotein world
We have little insight about the evolutionary steps by which protein enzymes came to supplant the majority of RNA enzymes present in the RNA world. One possibility for this takeover could entail gradual evolution of protein–RNA complexes to ultimately shed their RNA. Evolutionary intermediates in this process might include ribozyme–protein complexes in which the protein engages in substrate binding or provides elements of the ribozyme's catalytic apparatus. For example, both protein and RNA could contribute ligands that coordinate a catalytic metal ion. Our methodology may provide an avenue for developing proof-of-concept systems to explore the evolutionary transition from RNA catalysis to protein catalysis via ribonucleoprotein complexes. For example, the U2 and U6 small nuclear RNAs can catalyse splicing chemistry inefficiently when isolated from the spliceosome [49,50]. Possibly the phage display platform could be used to identify Fabs that enhance RNA activity. Although this approach would certainly not yield protein descendents from spliceosome evolution, it might reveal possible fundamental mechanisms by which proteins could synergize with and ultimately supplant the RNA component.
(c). RNA-catalysed RNA polymerization
The structure of the catalytic core of the class I ligase ribozyme provides a powerful starting point for investigating the mechanism of RNA polymerization catalysed by a ribozyme and the postulated similarity to protein polymerases. We note that the current structure represents only one product of the ligation reaction and lacks the pyrophosphate leaving group. Moreover, the ligation event extends the P1 helix by an additional base pair, which could provide a thermodynamic driving force for formation of a catalytically inactive conformation. Deeper understanding of the mechanism will certainly require structural analysis of the pre-ligation state. In recent years, at least for the small endonucleolytic ribozymes, nucleobase catalysis has become the norm rather than the exception [51,52]. For the sophisticated metabolism postulated to have existed in the RNA world, ribozymes would probably have had to rely on nucleobases for catalysis of reactions other than endonucleolytic cleavage. The ligase structure suggests that such catalysis is possible. Finally, obtaining polymerase ribozymes able to replicate RNA remains an important goal for origin of life research. The structural data revealing interactions with the template–primer duplex may guide future efforts towards engineering the class I ligase into self-replicating polymerase ribozyme.
Acknowledgements
The work described herein involved collaboration among multiple laboratories. David Shechner and David Bartel supplied the ligase ribozyme, provided insight into the structure determination, and shared coordinates of their U1A bound ligase. We thank Shohei Koide, Tony Kossiakoff and the members of their laboratories for assistance with all aspects of the projects. We thank Erica Duguid for invaluable assistance in solving the structure of the Fab–ligase complex and refining the model. The VS ribozyme experiments were carried out by Nikolai Suslov in collaboration with David Lilley and Tim Wilson. We are grateful to Jon Sutherland and David Lilley for their work in organizing the Royal Society Discussion Meeting on the Chemical Origins of Life and Its Early Evolution.
References
- 1.Joyce G. F., Orgel L. E. () 1999. The RNA world. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press [Google Scholar]
- 2.Johnston W. K., Unrau P. J., Lawrence M. S., Glasner M. E., Bartel D. P. 2001. RNA-catalyzed RNA polymerization: accurate and general RNA-templated primer extension. Science 292, 1319–1325 10.1126/science.1060786 (doi:10.1126/science.1060786) [DOI] [PubMed] [Google Scholar]
- 3.Lawrence M. S., Bartel D. P. 2005. New ligase-derived RNA polymerase ribozymes. RNA 11, 1173–1180 10.1261/rna.2110905 (doi:10.1261/rna.2110905) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Zaher H. S., Unrau P. J. 2007. Selection of an improved RNA polymerase ribozyme with superior extension and fidelity. RNA 13, 1017–1026 10.1261/rna.548807 (doi:10.1261/rna.548807) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bartel D. P., Szostak J. W. 1993. Isolation of new ribozymes from a large pool of random sequences (see comment). Science 261, 1411–1418 10.1126/science.7690155 (doi:10.1126/science.7690155) [DOI] [PubMed] [Google Scholar]
- 6.Ekland E. H., Szostak J. W., Bartel D. P. 1995. Structurally complex and highly active RNA ligases derived from random RNA sequences. Science 269, 364–370 10.1126/science.7618102 (doi:10.1126/science.7618102) [DOI] [PubMed] [Google Scholar]
- 7.Koldobskaya Y., et al. 2011. A portable RNA sequence whose recognition by a synthetic antibody facilitates structural determination. Nat. Struct. Mol. Biol. 18, 100–106 10.1038/nsmb.1945 (doi:10.1038/nsmb.1945) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wright M. C., Joyce G. F. 1997. Continuous in vitro evolution of catalytic function. Science 276, 614–617 10.1126/science.276.5312.614 (doi:10.1126/science.276.5312.614) [DOI] [PubMed] [Google Scholar]
- 9.Joyce G. F. 2004. Directed evolution of nucleic acid enzymes. Annu. Rev. Biochem. 73, 791–836 10.1146/annurev.biochem.73.011303.073717 (doi:10.1146/annurev.biochem.73.011303.073717) [DOI] [PubMed] [Google Scholar]
- 10.Doudna J. A., Cate J. H. 1997. RNA structure: crystal clear? Curr. Opin. Struct. Biol. 7, 310–316 10.1016/S0959-440X(97)80045-0 (doi:10.1016/S0959-440X(97)80045-0) [DOI] [PubMed] [Google Scholar]
- 11.Golden B. L., Kundrot C. E. 2003. RNA crystallization. J. Struct. Biol. 142, 98. 10.1016/S1047-8477(03)00042-X (doi:10.1016/S1047-8477(03)00042-X) [DOI] [PubMed] [Google Scholar]
- 12.Edwards A. L., Garst A. D., Batey R. T. 2009. Determining structures of RNA aptamers and riboswitches by X-ray crystallography. Nucleic Acid Pept. Aptamers 535, 1–29 10.1007/978-1-59745-557-2_9 (doi:10.1007/978-1-59745-557-2_9) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ke A., Doudna J. A. 2004. Crystallization of RNA and RNA-protein complexes. Methods 34, 408–414 10.1016/j.ymeth.2004.03.027 (doi:10.1016/j.ymeth.2004.03.027) [DOI] [PubMed] [Google Scholar]
- 14.Ferre-D'Amare A. R., Zhou K., Doudna J. A. 1998. A general module for RNA crystallization. J. Mol. Biol. 279, 621–631 10.1006/jmbi.1998.1789 (doi:10.1006/jmbi.1998.1789) [DOI] [PubMed] [Google Scholar]
- 15.Edwards A. L., Garst A. D., Batey R. T. 2009. Determining structures of RNA aptamers and riboswitches by X-ray crystallography. Methods Mol. Biol. 535, 135–163 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Carrasco N., Buzin Y., Tyson E., Halpert E., Huang Z. 2004. Selenium derivatization and crystallization of DNA and RNA oligonucleotides for X-ray crystallography using multiple anomalous dispersion. Nucl. Acids Res. 32, 1638–1646 10.1093/nar/gkh325 (doi:10.1093/nar/gkh325) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hobartner C., Rieder R., Kreutz C., Puffer B., Lang K., Polonskaia A., Serganov A., Micura R. 2005. Syntheses of RNAs with up to 100 nucleotides containing site-specific 2′-methylseleno labels for use in X-ray crystallography. J. Am. Chem. Soc. 127, 12 035–12 045 10.1021/ja051694k (doi:10.1021/ja051694k) [DOI] [PubMed] [Google Scholar]
- 18.Keel A. Y., Rambo R. P., Batey R. T., Kieft J. S. 2007. A general strategy to solve the phase problem in RNA crystallography. Structure 15, 761–772 10.1016/j.str.2007.06.003 (doi:10.1016/j.str.2007.06.003) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Robertson M. P., Scott W. G. 2008. A general method for phasing novel complex RNA crystal structures without heavy-atom derivatives. Acta Crystallogr. D Biol. Crystallogr. D64, 738–744 10.1107/S0907444908011578 (doi:10.1107/S0907444908011578) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ferre-D'Amare A. R., Doudna J. A. 2000. Crystallization and structure determination of a hepatitis delta virus ribozyme: use of the RNA-binding protein U1A as a crystallization module. J. Mol. Biol. 295, 541–556 10.1006/jmbi.1999.3398 (doi:10.1006/jmbi.1999.3398) [DOI] [PubMed] [Google Scholar]
- 21.Ferré-D'Amaré A. R., Zhou K., Doudna J. A. 1998. Crystal structure of a hepatitis delta virus ribozyme. Nature 395, 567–574 10.1038/26912 (doi:10.1038/26912) [DOI] [PubMed] [Google Scholar]
- 22.Koide S. 2009. Engineering of recombinant crystallization chaperones. Curr. Opin. Struct. Biol. 19, 449–457 10.1016/j.sbi.2009.04.008 (doi:10.1016/j.sbi.2009.04.008) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Dutzler R., Campbell E. B., Cadene M., Chait B. T., MacKinnon R. 2002. X-ray structure of a ClC chloride channel at 3.0 Å reveals the molecular basis of anion selectivity. Nature 415, 287–294 10.1038/415287a (doi:10.1038/415287a) [DOI] [PubMed] [Google Scholar]
- 24.Tereshko V., Uysal S., Koide A., Margalef K., Koide S., Kossiakoff A. A. 2008. Toward chaperone-assisted crystallography: protein engineering enhancement of crystal packing and X-ray phasing capabilities of a camelid single-domain antibody (VHH) scaffold. Protein Sci. 17, 1175–1187 10.1110/ps.034892.108 (doi:10.1110/ps.034892.108) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Iwata S., Ostermeier C., Ludwig B., Michel H. 1995. Structure at 2.8 Å resolution of cytochrome c oxidase from Paracoccus denitrificans. Nature 376, 660–669 10.1038/376660a0 (doi:10.1038/376660a0) [DOI] [PubMed] [Google Scholar]
- 26.Koide S. 2009. Engineering of recombinant crystallization chaperones. Curr. Opin. Struct. Biol. 19, 449–457 10.1016/j.sbi.2009.04.008 (doi:10.1016/j.sbi.2009.04.008) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Carter P. J. 2006. Potent antibody therapeutics by design. Nat. Rev. Immunol. 6, 343–357 10.1038/nri1837 (doi:10.1038/nri1837) [DOI] [PubMed] [Google Scholar]
- 28.Schrama D., Reisfeld R. A., Becker J. C. 2006. Antibody targeted drugs as cancer therapeutics. Nat. Rev. Drug Discov. 5, 147–159 10.1038/nrd1957 (doi:10.1038/nrd1957) [DOI] [PubMed] [Google Scholar]
- 29.van Venrooij W. J., Hoet R., Castrop J., Hageman B., Mattaj I. W., van de Putte L. B. 1990. Anti-(U1) small nuclear RNA antibodies in anti-small nuclear ribonucleoprotein sera from patients with connective tissue diseases. J. Clin. Invest. 86, 2154–2160 10.1172/JCI114954 (doi:10.1172/JCI114954) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Gilliam A. C., Steitz J. A. 1993. Rare scleroderma autoantibodies to the U11 small nuclear ribonucleoprotein and to the trimethylguanosine cap of U small nuclear RNAs. Proc. Natl Acad. Sci. USA 90, 6781–6785 10.1073/pnas.90.14.6781 (doi:10.1073/pnas.90.14.6781) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kalmakoff J., Maskill W. J., Thongkrajai P., Palmer D. G. 1981. Antibodies against double-stranded RNA in patients with rheumatoid arthritis, osteoarthrosis and Paget's disease of bone. Aust. N. Z. J. Med. 11, 173–178 10.1111/j.1445-5994.1981.tb04227.x (doi:10.1111/j.1445-5994.1981.tb04227.x) [DOI] [PubMed] [Google Scholar]
- 32.Ohosone Y., Matsumura M., Chiba J., Nagaoka S., Matsuoka Y., Irimajiri S., Mimori T. 1998. Anti-transfer RNA antibodies in two patients with pulmonary fibrosis, Raynaud's phenomenon and polyarthritis. Clin. Rheumatol. 17, 144–147 10.1007/BF01452262 (doi:10.1007/BF01452262) [DOI] [PubMed] [Google Scholar]
- 33.Steitz J. A. 1989. [32] Immunoprecipitation of ribonucleoproteins using autoantibodies. Methods Enzymol. 180, 468–481 10.1016/0076-6879(89)80118-1 (doi:10.1016/0076-6879(89)80118-1) [DOI] [PubMed] [Google Scholar]
- 34.Fellouse F. A., et al. 2007. High-throughput generation of synthetic antibodies from highly functional minimalist phage-displayed libraries. J. Mol. Biol. 373, 924–940 10.1016/j.jmb.2007.08.005 (doi:10.1016/j.jmb.2007.08.005) [DOI] [PubMed] [Google Scholar]
- 35.Koide A., Gilbreth R. N., Esaki K., Tereshko V., Koide S. 2007. High-affinity single-domain binding proteins with a binary-code interface. Proc. Natl Acad. Sci. USA 104, 6632–6637 10.1073/pnas.0700149104 (doi:10.1073/pnas.0700149104) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ye J.-D., Tereshko V., Frederiksen J. K., Koide A., Fellouse F. A., Sidhu S. S., Koide S., Kossiakoff A. A., Piccirilli J. E. 2008. Synthetic antibodies for specific recognition and crystallization of structured RNA. Proc. Natl Acad. Sci. USA 105, 82–87 10.1073/pnas.0709082105 (doi:10.1073/pnas.0709082105) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Michnick S. W., Sidhu S. S. 2008. Submitting antibodies to binding arbitration. Nat. Chem. Biol. 4, 326–329 10.1038/nchembio0608-326 (doi:10.1038/nchembio0608-326) [DOI] [PubMed] [Google Scholar]
- 38.Fellouse F. A., Li B., Compaan D. M., Peden A. A., Hymowitz S. G., Sidhu S. S. 2005. Molecular recognition by a binary code. J. Mol. Biol. 348, 1153–1162 10.1016/j.jmb.2005.03.041 (doi:10.1016/j.jmb.2005.03.041) [DOI] [PubMed] [Google Scholar]
- 39.Fellouse F. A., Wiesmann C., Sidhu S. S. 2004. Synthetic antibodies from a four-amino-acid code: a dominant role for tyrosine in antigen recognition. Proc. Natl Acad. Sci. USA 101, 12 467–12 472 10.1073/pnas.0401786101 (doi:10.1073/pnas.0401786101) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kabat E. A., Wu T. T. 1971. Attempts to locate complementarity-determining residues in the variable positions of light and heavy chains. Ann. N. Y. Acad. Sci. 190, 382–393 10.1111/j.1749-6632.1971.tb13550.x (doi:10.1111/j.1749-6632.1971.tb13550.x) [DOI] [PubMed] [Google Scholar]
- 41.Koide S., Sidhu S. S. 2009. The importance of being tyrosine: lessons in molecular recognition from minimalist synthetic binding proteins. ACS Chem. Biol. 4, 325–334 10.1021/cb800314v (doi:10.1021/cb800314v) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Murphy F. L., Cech T. R. 1993. An independently folding domain of RNA tertiary structure within the Tetrahymena ribozyme. Biochemistry 32, 5291–5300 10.1021/bi00071a003 (doi:10.1021/bi00071a003) [DOI] [PubMed] [Google Scholar]
- 43.Cate J. H., Gooding A. R., Podell E., Zhou K., Golden B. L., Kundrot C. E., Cech T. R., Doudna J. A. 1996. Crystal structure of a group I ribozyme domain: principles of RNA packing. Science 273, 1678–1685 10.1126/science.273.5282.1678 (doi:10.1126/science.273.5282.1678) [DOI] [PubMed] [Google Scholar]
- 44.Juneau K., Podell E., Harrington D. J., Cech T. R. 2001. Structural basis of the enhanced stability of a mutant ribozyme domain and a detailed view of RNA–solvent interactions. Structure 9, 221–231 10.1016/S0969-2126(01)00579-2 (doi:10.1016/S0969-2126(01)00579-2) [DOI] [PubMed] [Google Scholar]
- 45.Shechner D. M., Grant R. A., Bagby S. C., Koldobskaya Y., Piccirilli J. A., Bartel D. P. 2009. Crystal structure of the catalytic core of an RNA-polymerase ribozyme. Science 326, 1271–1275 10.1126/science.1174676 (doi:10.1126/science.1174676) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Bagby S. C., Bergman N. H., Shechner D. M., Yen C., Bartel D. P. 2009. A class I ligase ribozyme with reduced Mg2+ dependence: selection, sequence analysis, and identification of functional tertiary interactions. RNA 15, 2129–2146 10.1261/rna.1912509 (doi:10.1261/rna.1912509) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Castro C., Smidansky E. D., Arnold J. J., Maksimchuk K. R., Moustafa I., Uchida A., Götte M., Konigsberg W., Cameron C. E. 2009. Nucleic acid polymerases use a general acid for nucleotidyl transfer. Nat. Struct. Mol. Biol. 16, 212–218 10.1038/nsmb.1540 (doi:10.1038/nsmb.1540) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Sträter N., Lipscomb W. N., Klabunde T., Krebs B. 1996. Two-metal ion catalysis in enzymatic acyl- and phosphoryl-transfer reactions. Angew. Chem. Int. Ed. Engl. 35, 2024–2055 10.1002/anie.199620241 (doi:10.1002/anie.199620241) [DOI] [Google Scholar]
- 49.Valadkhan S., Mohammadi A., Jaladat Y., Geisler S. 2009. Protein-free small nuclear RNAs catalyze a two-step splicing reaction. Proc. Natl Acad. Sci. USA 106, 11 901–11 906 10.1073/pnas.0902020106 (doi:10.1073/pnas.0902020106) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Valadkhan S., Manley J. L. 2001. Splicing-related catalysis by protein-free snRNAs. Nature 413, 701–707 10.1038/35099500 (doi:10.1038/35099500) [DOI] [PubMed] [Google Scholar]
- 51.Wilson T. J., McLeod A. C., Lilley D. M. 2007. A guanine nucleobase important for catalysis by the VS ribozyme. EMBO J. 26, 2489–2500 10.1038/sj.emboj.7601698 (doi:10.1038/sj.emboj.7601698) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Wilson T. J., Lilley D. M. 2011. Do the hairpin and VS ribozymes share a common catalytic mechanism based on general acid–base catalysis? A critical assessment of available experimental data. RNA 17, 213–221 10.1261/rna.2473711 (doi:10.1261/rna.2473711) [DOI] [PMC free article] [PubMed] [Google Scholar]