Abstract
It is critical to incorporate the process of population dynamics into community genetics studies to identify the mechanisms of the linkage between host plant genetics and associated communities. We studied the effects of plant genotypic diversity of tall goldenrod Solidago altissima on the population dynamics of the aphid Uroleucon nigrotuberculatum. We found genotypic variation in plant resistance to the aphid in our experiments. To determine the impact of plant genotypic diversity on aphid population dynamics, we compared aphid densities under conditions of three treatments: single-genotype plots, mixed-genotype plots and mixed-genotype-with-cages plots. In the latter treatment plants were individually caged to prevent natural enemy attack and aphid movement among plants. The synergistic effects of genotypes on population size were demonstrated by the greater aphid population size in the mixed-genotype treatment than expected from additive effects alone. Two non-exclusive hypotheses are proposed to explain this pattern. First, there is a source–sink relationship among plant genotypes: aphids move from plant genotypes where their reproduction is high to genotypes where their reproduction is low. Second, natural enemy mortality is reduced in mixed plots in a matrix of diverse plant genotypes.
Keywords: community genetics, non-additive effect, plant resistance, source–sink dynamics, Solidago altissima, Uroleucon nigrotuberculatum
1. Introduction
Research on community genetics has demonstrated that intraspecific genetic variation among plants can have important impacts on trophic interactions and the community structure of organisms that interact with the plant [1,2]. Several recent studies have documented that plant genotypic diversity is a key factor shaping the community structure of higher trophic levels [3–6], but the effects of genotypic diversity on the population dynamics of associated species are poorly understood (but see [7]). In order to identify the mechanisms responsible for linking host plant genetics and the community structure of higher trophic levels, it is critical to understand how intraspecific genetic variation among host plants influences the population dynamics of herbivores.
Plant genotypic diversity can affect both the species richness and abundance of herbivore and/or predator communities [3–5,8–10] owing to additive (i.e. a sum of average effects of genotypes) and non-additive (i.e. a synergistic or antagonistic effect of multiple genotypes) effects of plant genotypes [4,5]. Additive effects of plant genotypes on insects would result from the independent influences of plant genotypes on the arthropod community. Diverse patches can have greater abundance and/or richness of arthropods because of the increased probability of including multiple genotypes with distinct communities [11,12]. Alternatively, the effects of genotypes can be non-additive as the result of interactions among plant genotypes, producing higher or lower abundance or diversities of insects than would be predicted by simply adding the effects of individual plant genotypes [4,5,9,10]. Several mechanisms have been proposed by which plant genotypes may interact to produce non-additive effects on herbivore populations, but few studies have tested the existence of these mechanisms [13]. Such studies are needed in order to be able to predict the effects of plant genotype diversity in structuring insect communities [13].
Movement of herbivores among plants influenced by plant genotypic variation may produce non-additive effects on herbivore population size. Root [14] predicted that the abundance of specialist herbivores would be lower in higher diversity plant communities than in lower diversity communities. Diverse plant communities produced a diversity of visual or olfactory cues and this complexity increases the difficulty that specialist herbivores have in locating and moving among host plants [15,16]. Many early studies, which have been motivated by the work of Root [14], have focused on the effects of plant species diversity on herbivore diversity or abundance (e.g. [17]), often without distinguishing non-additive mechanisms, including herbivore movement. On the other hand, theoretical studies predict that local diversity of patch quality can allow for source–sink dynamics of organisms that can increase or decrease population size relative to expectations based on patches without diversity [18–20]. Thus, we predict that non-additive effects of diverse plant genotypes on higher trophic levels will arise from source–sink dynamics of insect populations among plants of different genotypes.
In this study, we examined the effects of genotypic diversity in tall goldenrod, Solidago altissima L., on the population size of the aphid Uroleucon nigrotuberculatum Olive (electronic supplementary material, figure S1). Local populations of Solidago in North America have clones that exhibit considerable trait variation, particularly in resistance to herbivorous insects [21]. Maddox & Root [21] reported that resistance of S. altissima to U. nigrotuberculatum was highly heritable and varied among naturally established genotypes. On more resistant genotypes, both the population growth rate and the density of aphids are lower than on less resistant genotypes. Thus, we hypothesized that intraspecific genotypic variation in plant resistance can influence aphid population size. To test this hypothesis, we addressed the following questions: (i) is there genotypic variation in resistance of S. altissima to the aphid U. nigrotuberculatum? (ii) How does the mixture of genotypes influence the population size of the aphid? (iii) Does plant genotypic diversity effect aphid movement influencing the distribution of aphid densities among plants?
2. Material and methods
(a). Plant resistance to the aphid
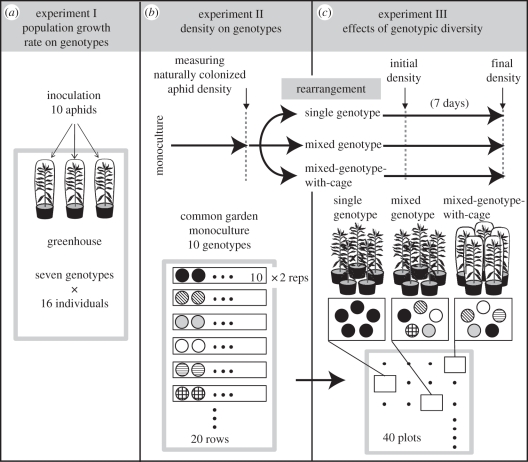
To examine genotypic variation in Solidago resistance to the aphid U. nigrotuberculatum, we conducted two experiments. Plants used in the experiment were collected as rhizomes from six sites in Minnesota, USA, in 2003, and each plant genotype was propagated by repeatedly dividing the rhizomes into new pots at 2 year intervals at the University of Minnesota Duluth Research and Field Studies Center, Duluth, Minnesota, USA (47° N, 92° W; electronic supplementary material, figure S1). Rhizomes collected from a single plant were divided into rhizome segments to obtain genotypic replicates. To examine the difference in population growth rate of aphids among plant genotypes, we conducted an aphid-inoculation experiment in 2009 (figure 1a). Rhizomes from seven genotypes were transported to Center for Ecological Research (CER), Kyoto University, Japan (35° N, 136° E), with the permission of the US and Japanese governments. Rhizome segments were grown in pots (18 cm in diameter) and placed in randomly assigned positions in a greenhouse covered with 1 mm mesh net from early May to late July 2009. Then each plant was covered with a fine non-woven fibre cloth with high light transmissibility that is used to protect vegetable crops from pests (Marsol Co. Ltd, Okayama, Japan). We collected adult aphids from wild populations of U. nigrotuberculatum on S. altissima growing on the grounds of CER. Ten adult aphids were inoculated on each plant on 30 July. On 9 August 2009, we counted the number of aphids to determine aphid population growth rate on each covered plant. We calculated population growth rates (dN/dt) as a function of ln(N1 − N0)/day; N0 is the number of inoculated aphids on each plant and N1 is the final number of aphids. We performed a one-way ANOVA to examine the effects of Solidago genotypes on population growth rate of the aphid. Although the genetic composition of Japanese populations of the aphid might differ from US populations, population growth rates of Japanese aphids on other imported US Solidago genotypes in the CER greenhouse were similar to the pattern in Minnesota (Y. Ando, S. Utsumi & T. Ohgushi 2009, unpublished data).
Figure 1.
A graphical summary of the experimental design.
To examine genotype effects on the number of natural long-term aphids, we conducted a common garden experiment at the University of Minnesota Duluth Research and Field Studies Center (figure 1b). We used 10 plant genotypes, including seven genotypes used for the above experiment in Japan. In April 2008, rhizomes from each genotype were divided into 20 rhizome segments to obtain replicates. All replicates were randomly assigned placement in the 20-row common garden. Ten pots of each genotype were closely arranged in a single row. Two rows were created for each genotype. All rows were randomly assigned in the 20-row common garden and adjacent rows were separated by 90 cm. Aphids were allowed to colonize naturally on the experimental plants. We randomly selected five plants from each row (i.e. 10 replicates for each genotype) and counted the number of the aphids on the plants on 20 August 2008. The data were log(n + 1)-transformed because assumptions of normality and equal variance were not met. To examine the effects of Solidago genotypes on aphid density, we first performed a nested-ANOVA with factors of genotype and row nested within genotype. Because the row effect was not significant (F10,80 = 0.78, p = 0.65), we performed a one-way ANOVA with genotype effect. To examine differences in plant size of Solidago genotypes, we measured the height of the experimental plants on 20 August 2008. The data were analysed using one-way ANOVA. To examine variation in the genotypes' resistance to the aphid while accounting for the differences in plant size, we used residuals from a regression of aphid number on plant height in one-way ANOVA of effects of genotypes on aphid densities. We also compared the results between the two experiments using a correlation analysis for final densities in the first experiment and densities in the second experiment.
(b). Effect of genotype mixture on aphid population size
To measure how plant genotypic diversity, aphid movement among plants and natural enemy attack affected aphid population size, we conducted a third common garden experiment. We spatially rearranged potted plants from the second experiment into three treatments: single genotype, mixed genotype and mixed-genotype-with-cage where each plant was individually caged (figure 1c). The cage treatment prevented aphids from moving among plants and prevented natural enemies from attacking aphids. On 21 August 2008, we set up the following common garden experiment. We established forty 2 × 1.5 m plots and randomly assigned each to one of the three treatments. We moved the plants to these plots, leaving plants harbouring all of the insects naturally colonized in the second experiment. However, those plants were not heavily colonized by herbivores other than aphids. Each plot contained five individual plants, with a total of 200 plants in the experiment. Twenty plots were assigned to the single-genotype treatment where each plot consisted of five plants of a single genotype. There were two replicates in this treatment for each of the 10 genotypes. In the 10 plots in the mixed-genotype treatment and in the 10 plots in the mixed-genotype-with-cage treatment, each plot consisted of one plant each of five genotypes randomly selected from the 10 genotypes with the constraint that no two plots could have the same composition. In the mixed-genotype-with-cage treatment, we removed all predators on plants and covered each plant completely with Reemay, a fine non-woven fibre cloth. In each plot, we placed the plant individuals pentagonally and randomly with a 30 cm space between adjacent pots. Plots were separated by 1.5 m. We counted the number of aphids on 21 August 2008 to determine the initial densities on all experimental plants.
On 28 August, we counted the number of aphids on the experimental plants to determine the final density and the total number of aphids in each plot. The data were log(n + 1)-transformed because assumptions of normality and equal variance were not met. We performed a repeated-measures ANOVA on aphid population size per plot at the beginning versus population size at the end of the experiment to examine treatment effects. Next, we performed a paired t-test to examine changes in densities of aphids on each plot for each treatment. To identify non-additive effects of the increasing genotypic diversity, we applied the statistical approach described by Johnson et al. [5]. We first calculated the mean final density per plant of the aphid on each genotype in the single-genotype treatment, and then calculated the expected density per plot for the mixed-genotype treatment by summing the appropriate mean values from the single-genotype plots for each genotype in the mixed-genotype plots. No randomization was required for such density data [5]. We compared the observed density for each plot to this expected density in the absence of non-additive effects using a paired t-test.
We calculated population growth rate (dN/dt) on each plant during the third experiment as a function of ln(N1 − N0)/day and performed separate ANCOVA with genotypes as a main factor and initial density as a covariate for each of the data of the three treatment plots to examine the effects of genotypes on population growth rates. Also, a regression analysis was conducted to compare the effects of initial aphid density (N0) on population growth rate in the single genotype, mixed genotype and mixed-genotype-with-cage treatments. Furthermore, in the mixed-genotype plots, to examine the relationship between combinations of genotype resistance and aphid population growth rates, we ranked resistance levels of 10 genotypes from 1 (lowest resistance) to 10 (highest) according to average aphid densities in the second experiments, and then performed ANCOVA with plots as a main factor and ranked resistance level as a covariate.
3. Results
(a). Plant resistance to the aphid
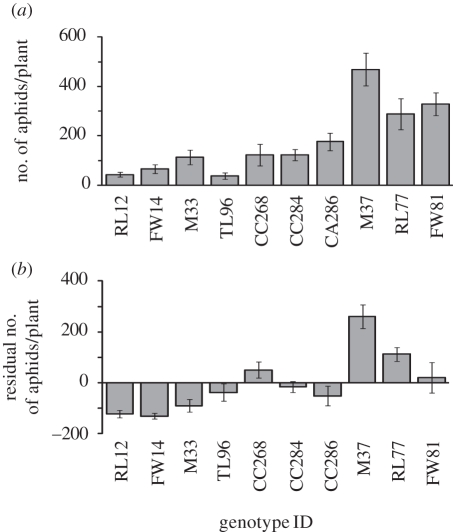
The first experiment in Japan demonstrated that plant genotype significantly influenced the population growth rate of the aphids (F6,103 = 42.89, p < 0.0001; figure 2). We also found significant differences in natural long-term aphid densities among Solidago genotypes in the second experiment in the USA (F9,90 = 8.18, p < 0.0001; figure 3a). There was a twelvefold variation in aphid density among genotypes, ranging from 37.6 to 469.6 individuals per plant. Final aphid densities on plant genotypes in the first experiment were strongly correlated with densities of the aphid on these genotypes in the second experiment (rPearson = 0.81, p = 0.028). These results indicate that genotypic variation in plant resistance to the aphid was consistent in Japan and USA.
Figure 2.
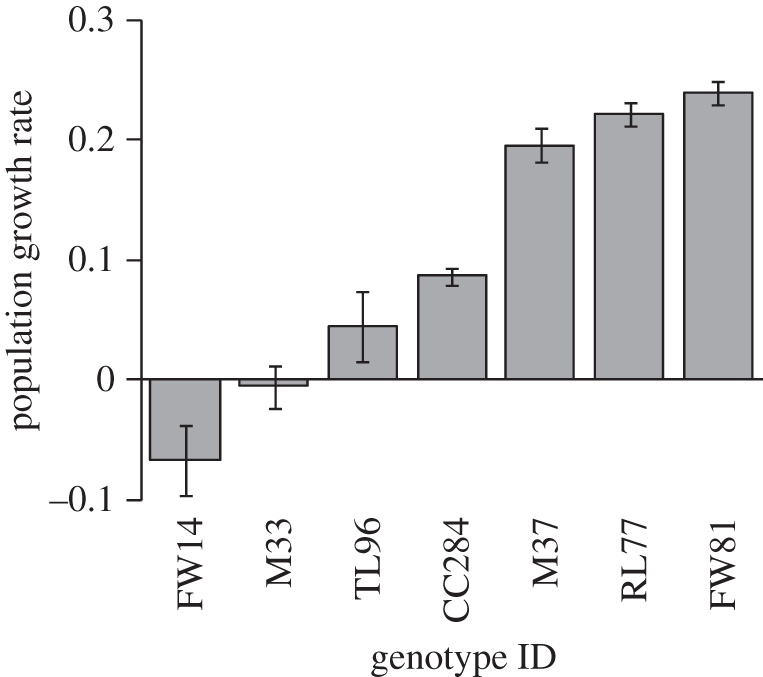
Population growth rate of the aphid on each genotype. Population growth rates of aphids in 10 days, starting from 10 adults. Error bars indicate s.e.
Figure 3.
Interactions between genotypes of the tall goldenrod S. altissima and the aphid U. nigrotuberculatum. (a) Effect of plant genotypes on aphid density. (b) Residual number of aphids from the regression against plant height. Plant order is based on the population growth rates of aphids in figure 2. Error bars indicate s.e.
In the second experiment, there was a significant genotypic difference in plant height (F9,90 = 10.25, p < 0.0001), and aphid density significantly increased with plant height (r2 = 0.27, p < 0.0001; see the electronic supplementary material, figure S2). However, residuals differed significantly among genotypes (F9,90 = 12.39, p < 0.0001; figure 3b), indicating that variation in quality among genotypes made a strong contribution to the variation in aphid density. Residual values of genotypes tended to increase as aphid density increased from negative to positive, indicating that genotypic variation in plant resistance to aphids made a large contribution to the variation in aphid density among plants. In the rest of this report, we will use the aphid densities as a measure of plant genotype ‘resistance’: the lower the aphid density, the more ‘resistant’ the genotype.
(b). Effect of genotype mixture on aphid population size
The treatments in the third experiment had a strong impact on the plot-level aphid densities (figure 4a; treatment: F2,27 = 2.35, p = 0.11; time: F1,27 = 47.94, p < 0.0001; interaction: F2,27 = 14.48, p < 0.0001). There was no significant change in the number of aphids per plot in the single-genotype treatment during the experiment (paired t-test: t = 0.86, d.f. = 9, p = 0.41), but there were significant increases in aphid density in the mixed genotype (t = 3.86, d.f. = 9, p < 0.01) and the mixed-genotype-with-cage treatments (t = 5.71, d.f. = 9, p < 0.001). The final aphid density in the mixed-genotype treatment was significantly greater than the expected density calculated from the single-genotype treatment (t = 2.56, d.f. = 9, p = 0.03; figure 4b), indicating significant non-additive, synergistic effects of plant genotypic diversity on population size of the aphid.
Figure 4.
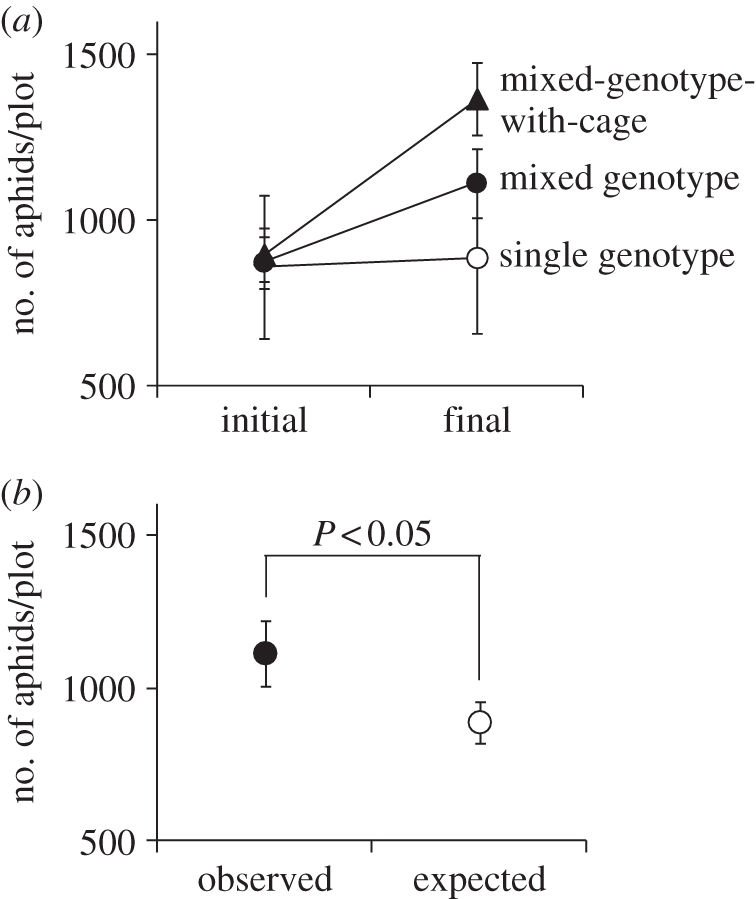
The effect of the single genotype, mixed genotype, and mixed-genotype-with-cage treatment on population sizes of the aphids. (a) Changes in aphid density in the single genotype, mixed genotype and mixed-genotype-with-cage treatments. (b) Expected density from additive effect of genotypes alone versus observed density in mixture. Error bars indicate s.e.
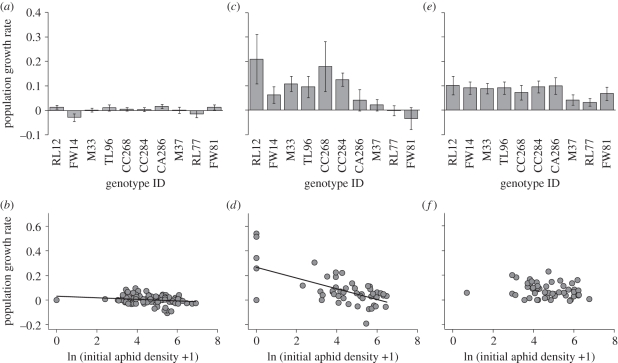
Population growth rates on plant genotypes and the relationship between population growth rates and initial aphid densities differed among treatments (figure 5 and table 1). In the single-genotype treatment, the differences among plant genotypes in aphid population growth rate were not significant (figure 5a). There was a significant negative relationship between the initial aphid density and population growth rate (figure 5b). In contrast, plant genotype had a highly significant effect on aphid population growth rate in the mixed-genotype treatment, in which population growth rates were significantly greater on the more resistant plants in the mixed-genotype treatment (figure 5c). The regression showed a significant negative association between initial density and population growth rate in the mixed-genotype treatment (figure 5d). Aphid movement among plants may have played an important role in determining the final densities, as indicated by the presence of wingless aphids on plants where their initial densities were zero (see figure 5d). In contrast to the mixed-genotype treatment, we found no significant effects of genotype or initial density on population growth rates in the mixed-genotype-with-cage treatment (figure 5e,f). This indicates that preventing aphid interplant movement and/or natural enemy attack preserved the equivalence of population growth rates as were found on these genotypes in the single-genotype treatment, and this resulted in a lack of density-dependent population growth based on the initial density. Furthermore, aphid growth rates were significantly higher on more resistant genotypes in the mixed-genotype plots (resistance level: F1,30 = 6.79, p = 0.014; slope = 0.017), while genotype combination did not significantly affect this pattern (plot: F9,30 = 0.98, p = 0.47; interaction: F9,30 = 1.09, p = 0.39).
Figure 5.
Population growth rates in the mixture experiment. Mean population growth rates on genotypes are presented in (a) the single-genotype treatment, (c) mixed-genotype treatment and (e) mixed-genotype-with-cage treatment. Plants are ordered the same as figure 2. Error bars indicate s.e. On (b,d,f), relationships between initial densities and population growth rates in the single genotype, mixed genotype, and mixed-genotype-with-cage treatment are presented, respectively. Each point indicates the value on each plant individual. Solid lines indicate significant regression (b: r2 = 0.035, y =−0.006x + 0.030, p = 0.039; d: r2 = 0.36, y =−0.044x + 0.266, p < 0.0001). In the mixed-genotype-with-cage treatment, there was no significant relationship (e: r2 = 0.04, p = 0.17).
Table 1.
Summary of the ANOVA results demonstrating the effects of plant genotype, initial aphid density, and their interaction on population growth rates of the aphid on individual plants in the single genotype, mixed genotype and mixed-genotype-with-cage treatment.
| treatment | factor | d.f. | F | p |
|---|---|---|---|---|
| single genotype | genotype (G) | 9,74 | 1.08 | 0.38 |
| initial density (I) | 1,74 | 23.34 | <0.0001 | |
| G × I | 9,74 | 6.88 | <0.001 | |
| mixed genotype | genotype (G) | 9,30 | 4.49 | <0.001 |
| initial density (I) | 1,30 | 31.27 | <0.0001 | |
| G × I | 9,30 | 3.59 | 0.003 | |
| mixed-genotype- | genotype (G) | 9,30 | 0.79 | 0.63 |
| with-cage | initial density (I) | 1,30 | 0 | 0.99 |
| G × I | 9,30 | 0.95 | 0.50 |
4. Discussion
The present study demonstrated that the population size of the aphid U. nigrotuberculatum could be affected by the genotypic variation in plant resistance in the plot. Non-additive effects were also revealed by the higher aphid population size in genotypically diverse plots of Solidago plants than that expected from additive effects alone.
(a). Genotypic variation in plant resistance
Our study supported the hypothesis that intraspecific genotypic variation in plant resistance can influence aphid density. It has previously been demonstrated that there is variation in resistance among S. altissima genotypes to a wide range of herbivorous insects (e.g. [21,22]). A wide variety of plant traits could contribute to genotypic variation in resistance to aphids, including the chemical composition of the phloem sap, amino acid and soluble nitrogen concentration, and/or stem trichome density [6,23,24]. In addition, genotypic variation in plant traits may also indirectly affect aphid abundance through herbivore-induced trait modification [25,26]. For example, Pilson [25] observed that S. altissima genotype was a significant predictor of the distribution of the aphid Uroleucon tissoti among plants only when a suite of branch-inducing herbivores was present. Also, the density of gall-making insects is influenced by genetic variation in S. altissima, and gallers can have large plant-mediated indirect effects on other herbivorous insects [10,27]. However, these indirect effects could not have influenced our results because we used herbivore-free plants in the first experiment, and did not use galled plants in the second and third experiments.
Plant genotype, environment, and genotype × environment all have the potential to determine the population dynamics and community structure of arthropods associated with a plant [8,28]. Although a few studies on these effects have suggested that local adaptation by plants or arthropods can weaken the effects of plant genotype on associated communities at large regional scales [8,28], we found that the relative resistance of S. altissima genotypes was similar in Japan and Minnesota, USA, despite the potential for differences in the aphid populations, and in the abiotic environments such as the length of the growing season, temperature and humidity, between the two countries that could influence plant genotype × environment interactions and the aphid. The lack of differences in the response by the aphid to S. altissima genotypes in the two countries might be owing to the recent introduction of the plant and aphid to Japan. Solidago altissima was introduced to Japan from North America 100 years ago and the aphid invasion occurred only in the early 1990s [29]. There may not yet have been enough time for the aphid to adapt to local environments in Japan.
(b). Non-additive effects of plant genotypic diversity
In the third common garden experiment, plant genotypic diversity caused non-additive effects on aphid population size. In the single-genotype treatment, the number of aphids on individual plants did not significantly change during the experiment, probably because aphid abundance on each plant had reached an equilibrium density prior to the initiation of the treatments. Aphid densities peaked early in the common garden in August 2008 and remained relatively constant thereafter (electronic supplementary material, figure S3), in agreement with the pattern found by Cappuccino [30]. The significant genotype-by-density effect indicated that density-dependent per capita population growth rate differed among genotypes. However, the slope of the density dependence was near zero in the single-genotype plots (figure 5b). These results suggest that aphid densities were near equilibrium on the experimental plants during the third experiment. Hence, no significant differences in population growth rates were found among the genotypes in the single-genotype treatment where populations were high at the initiation of the experiment (179.0 ± 181.5 (mean ± s.d.) aphids/plant), whereas we found a significant difference among genotypes in the first experiment, which was initiated with 10 aphids per plant. In the mixed-genotype-with-cage treatment, aphid densities were higher compared with the single-genotype treatment because the equilibrium population size had increased owing to the removal of natural enemies.
In the mixed-genotype treatment, we detected significant effects of genotypes and genotype-by-density on the population growth rates. Because there were no genotype effects on the population growth rates in other treatments, the difference in population growth rates among genotypes in the mixed-genotype treatment did not result from changes in plant quality and quantity. Hence, our findings suggest that plant genotypic diversity could have produced non-additive effects on the plot-level population size by influencing herbivore and/or natural enemy movement among plants via the following processes.
First, we consider the hypothesis that herbivore movement produced the non-additive effects of genotypic diversity on aphid population size [7,20]. Theory [20] indicates that non-additive increase in herbivore population size owing to source–sink dynamics requires a positive correlation across plant types between r and K of herbivore populations. Using the population growth rates in the first experiment as r and the final densities on the caged plants in the third experiment as K, a significant positive correlation between r and K was found (rPearson = 0.86, p = 0.013). This suggests that the source–sink dynamics would increase overall population size compared with that expected from the average genotype quality [31]. The source–sink mechanism also requires density-dependent population growth [20]. This was indicated by the significant density effects found in the single-genotype plots, which suggested a density-dependent aphid population growth.
We argue that plant genotypes with low resistance produced high aphid densities (high K plants), and that they served as a ‘source’ of aphids that immigrated to more resistant genotypes (low K plants), where aphid reproductive success was low and served as a ‘sink’. In this model, when aphids were above the carrying capacity on the preferred low resistance genotypes and were suffering density-dependent processes, they were induced to move to the higher resistance plants, which were less-crowded in the genotypically diverse plots [32]. This aphid movement would have resulted in significantly higher population growth rates on lower K plants within plots in the mixed-genotype treatment that was conducted when the preferred plants were near their carrying capacity. This reverses the trend of aphid population growth rates seen in the first experiment (rPearson = −0.75, p = 0.05).
Evidence of the movement of aphids among plants was also provided by the presence of aphids on plants where they were absent in the initial census in the mixed-genotype treatment (figure 5d). Furthermore, most of these newly established aphids were wingless morphs. Wingless aphids often walk across bare the soil surface from one plant to another neighbouring one, which is stimulated by various cues including density, host plant quality and physical disturbance [33,34]. Thus, we suggest that a non-additive increase in population size in the mixed-genotype plots would be caused primarily by short-distance movement of wingless aphids (among genotypes within a plot) rather than long-distance movement of alate aphids (among plots or from sources outside the experiment). If long-distance dispersal occurred more frequently than short-distance movement, immigration from other plots or from outside of the common garden should have resulted in an increase in population size in the single-genotype treatment, as well as the mixed-genotype treatment. Therefore, we conclude that source–sink dynamics via short-distance movement within a plot are likely to have caused the non-additive increase in aphid population size in response to genotypic diversity of host plants [7].
A second hypothesis explaining the non-additive effects on population size would be the effects of natural enemies. A significant impact of natural enemies on aphid population density was indicated by the increased population growth rate on each genotype when natural enemy attack was prevented. We suggest that the following two mechanisms of predators could increase the population size and produce variation in aphid population growth rates in the mixed-genotype treatment. First, colonization by natural enemies into the mixed-genotype plots may be reduced due to olfactory interference [15,16]. Because plants often have genotypic variation in herbivore-induced volatile emission (e.g. [35,36]), searching efficiency of natural enemies may be reduced in the mixed-genotype treatment. Movement by natural enemies within plots is a second mechanism by which predators could have influenced the differential aphid population growth on genotypes in the mixed-genotype treatment. Natural enemies may track the abundance of prey populations at a small spatial scale (e.g. within plots) more effectively than larger scales (e.g. among plots) [37,38]. In this case, natural enemies that had colonized plants previous to the start of the experiment would cause higher mortality on the less-resistant genotypes where aphid densities were high than on more resistant genotypes where aphid densities were low. In addition, it should be noted that non-consumptive effects of natural enemies on the aphids, such as behavioural changes in response to the presence of predation risk may have enhanced the source–sink dynamics of the aphid. The presence of natural enemies has been demonstrated to increase aphid movement [33,39]. Aphids may have moved from high-density to low-density plants both to avoid natural enemies and to avoid negative density-dependent effects as proposed earlier. The aphid movement and natural enemy hypotheses are not mutually exclusive.
(c). Associational susceptibility
The synergistic effects of plant genotypes can increase the herbivore loads of individual plants in genotypically diverse plots, resulting in associational susceptibility [40,41]. This contrasts with the prediction of the resource concentration hypothesis [14], which proposes that herbivore density will be higher in monocultures when compared with polycultures owing to increased immigration to, decreased emigration from, or increased reproduction within a monoculture. This hypothesis has been more frequently supported in specialist herbivores than generalist herbivores, which have a wide range of adaptations for responding to plant species diversity [16,41]. Generalist herbivores can gain resources by feeding on a range of plant species and generalists' abundance can increase with plant species diversity [42]. Because specialist herbivores can move among and feed on diverse genotypes of a host species, we argue that specialist herbivores feeding on a diverse range of genotypes of a host species have similarities to generalist herbivores feeding on a diverse range of species. Our results show that aphid population density increases in patches with a diversity of genotypes, just as generalist herbivore density increases in a patch with a diversity of species.
The present study shows that genotypic diversity of host plants can non-additively increase the population size of an herbivorous insect. This is the first empirical support for the hypothesis that the genotypic diversity of host plants would produce source–sink dynamics in herbivore populations resulting in increased herbivore population size. Future studies should focus on the spatial context, in particular the movement of plant-associated arthropods, to better understand effects of plant genetic diversity on population dynamics of plant-associated arthropods.
Acknowledgements
We thank J. K. Itami for extensive assistance in the field and for editorial comments on this manuscript, and also thank J. Bailey, M. Johnson, E. Nakajima and anonymous reviewers for their valuable comments on an earlier draft. This study was supported by the Ministry of Education, Culture, Sports, Science and Technology under the JSPS Core-to-core programme (20004), Global COE programme (A06) of Kyoto University, and Grant-in Aids for Scientific Research to T.O. (B-20370010) and for JSPS Research Fellowship to S.U. (22-9260), and by an NSF Grant (DEB-0949280) to T.P.C.
References
- 1.Whitham T. G., et al. 2003. Community and ecosystem genetics: a consequence of the extended phenotype. Ecology 84, 559–573 10.1890/0012-9658(2003)084[0559:CAEGAC]2.0.CO;2 (doi:10.1890/0012-9658(2003)084[0559:CAEGAC]2.0.CO;2) [DOI] [Google Scholar]
- 2.Whitham T. G., et al. 2006. A framework for community and ecosystem genetics: from genes to ecosystems. Nat. Rev. Genet. 7, 510–523 10.1038/nrg1877 (doi:10.1038/nrg1877) [DOI] [PubMed] [Google Scholar]
- 3.Wimp G. M., Young W. P., Woolbright S. A., Martinsen G. D., Keim P., Witham T. G. 2004. Conserving plant genetic diversity for dependent animal communities. Ecol. Lett. 7, 776–780 10.1111/j.1461-0248.2004.00635.x (doi:10.1111/j.1461-0248.2004.00635.x) [DOI] [Google Scholar]
- 4.Crutsinger G. M., Collins M. D., Fordyce J. A., Gompert Z., Nice C. C., Sanders N. J. 2006. Plant genotypic diversity predicts community structure and governs an ecosystem process. Science 313, 966–968 10.1126/science.1128326 (doi:10.1126/science.1128326) [DOI] [PubMed] [Google Scholar]
- 5.Johnson M. T. J., Lajeunesse M. J., Agrawal A. A. 2006. Additive and interactive effects of plant genotypic diversity on arthropod communities and plant fitness. Ecol. Lett. 9, 24–34 10.1111/j.1461-0248.2005.00833.x (doi:10.1111/j.1461-0248.2005.00833.x) [DOI] [PubMed] [Google Scholar]
- 6.Johnson M. T. J. 2008. Bottom-up effects of plant genotype on aphids, ants, and predators. Ecology 89, 145–154 10.1890/07-0395.1 (doi:10.1890/07-0395.1) [DOI] [PubMed] [Google Scholar]
- 7.Underwood N. 2009. Effect of genetic variance in plant quality on the population dynamics of a herbivorous insect. J. Anim. Ecol. 78, 839–847 10.1111/j.1365-2656.2009.01540.x (doi:10.1111/j.1365-2656.2009.01540.x) [DOI] [PubMed] [Google Scholar]
- 8.Bangert R. K., Allan G. J., Turek R. J., Wimp G. M., Meneses N., Martinsen G. D., Keim P., Whitham T. G. 2006. From genes to geography: a genetic similarity rule for arthropod community structure at multiple geographic scales. Mol. Ecol. 15, 4215–4228 10.1111/j.1365-294X.2006.03092.x (doi:10.1111/j.1365-294X.2006.03092.x) [DOI] [PubMed] [Google Scholar]
- 9.Crutsinger G. M., Reynolds W. N., Classen A. T., Sanders N. J. 2008. Disparate effects of plant genotypic diversity on foliage and litter arthropod communities. Oecologia 158, 65–75 10.1007/s00442-008-1130-y (doi:10.1007/s00442-008-1130-y) [DOI] [PubMed] [Google Scholar]
- 10.Crawford K. M., Crutsinger G. M., Sanders N. J. 2007. Host-plant genotypic diversity mediates the distribution of an ecosystem engineer. Ecology 88, 2114–2120 10.1890/06-1441.1 (doi:10.1890/06-1441.1) [DOI] [PubMed] [Google Scholar]
- 11.Loreau M., Hector A. 2001. Partitioning selection and complementarity in biodiversity experiments. Nature 412, 72–76 10.1038/35083573 (doi:10.1038/35083573) [DOI] [PubMed] [Google Scholar]
- 12.Hooper D. U., et al. 2005. Effects of biodiversity on ecosystem functioning: a consensus of current knowledge. Ecol. Monogr. 75, 3–35 10.1890/04-0922 (doi:10.1890/04-0922) [DOI] [Google Scholar]
- 13.Hughes A. R., Inouye B. D., Johnson M. T. J., Underwood N., Vellend M. 2008. Ecological consequences of genetic diversity. Ecol. Lett. 11, 609–623 10.1111/j.1461-0248.2008.01179.x (doi:10.1111/j.1461-0248.2008.01179.x) [DOI] [PubMed] [Google Scholar]
- 14.Root R. B. 1973. Organization of a plant–arthropod association in simple and diverse habitats: the fauna of collards (Brassica oleracea). Ecol. Monogr. 43, 95–124 10.2307/1942161 (doi:10.2307/1942161) [DOI] [Google Scholar]
- 15.Tahvanainen J. O., Root R. B. 1972. The influence of vegetational diversity on the population ecology of a specialized herbivore, Phyllotreta cruciferae (Coleoptera: Chrysomelidae). Oecologia 10, 321–346 10.1007/BF00345736 (doi:10.1007/BF00345736) [DOI] [PubMed] [Google Scholar]
- 16.Andow D. A. 1991. Vegetational diversity and arthropod population response. Annu. Rev. Entomol. 36, 561–586 10.1146/annurev.en.36.010191.003021 (doi:10.1146/annurev.en.36.010191.003021) [DOI] [Google Scholar]
- 17.Siemann E., Tilman D., Haarstad J., Ritchie M. 1998. Experimental tests of the dependence of arthropod diversity on plant diversity. Am. Nat. 152, 738–750 10.1086/286204 (doi:10.1086/286204) [DOI] [PubMed] [Google Scholar]
- 18.Holt R. D. 1985. Population dynamics in two-patch environments: some anomalous consequences of an optimal habitat distribution. Theor. Popul. Biol 28, 181–208 10.1016/0040-5809(85)90027-9 (doi:10.1016/0040-5809(85)90027-9) [DOI] [Google Scholar]
- 19.Pulliam H. R. 1988. Sources, sinks, and population regulation. Am. Nat. 132, 652–661 10.1086/284880 (doi:10.1086/284880) [DOI] [Google Scholar]
- 20.Underwood N. 2004. Variance and skew of the distribution of plant quality influence herbivore population dynamics. Ecology 85, 686–693 10.1890/03-0030 (doi:10.1890/03-0030) [DOI] [Google Scholar]
- 21.Maddox G. D., Root R. B. 1987. Resistance to 16 diverse species of herbivorous insects within a population of goldenrod, Solidago altissima: genetic variation and heritability. Oecologia 72, 8–14 10.1007/BF00385037 (doi:10.1007/BF00385037) [DOI] [PubMed] [Google Scholar]
- 22.Craig T. P., Itami J. K. 2008. Evolution of preference and performance relationships. In Specialization, speciation, and radiation: the evolutionary biology of herbivorous insects (ed. Tilmon K. J.), pp. 20–28 Berkeley, CA: University of California Press [Google Scholar]
- 23.Agrawal A. A. 2004. Plant defense and density dependence in the population growth of herbivores. Am. Nat. 164, 113–120 10.1086/420980 (doi:10.1086/420980) [DOI] [PubMed] [Google Scholar]
- 24.Wimp G. M., Whitham T. G. 2007. Host plants mediate ant–aphid mutualism and their effects on community structure and diversity. In Ecological communities: plant mediation in indirect interaction webs (eds Ohgushi T., Craig T. P., Price P. W.), pp. 275–305 Cambridge: Cambridge University Press [Google Scholar]
- 25.Pilson D. 1992. Aphid distribution and the evolution of goldenrod resistance. Evolution 46, 1358–1372 10.2307/2409942 (doi:10.2307/2409942) [DOI] [PubMed] [Google Scholar]
- 26.Ohgushi T. 2005. Indirect interaction webs: herbivore-induced effects through trait change in plants. Annu. Rev. Ecol. Evol. Syst. 36, 81–105 10.1146/annurev.ecolsys.36.091704.175523 (doi:10.1146/annurev.ecolsys.36.091704.175523) [DOI] [Google Scholar]
- 27.Craig T. P., Itami J. K., Craig J. V. 2007. Host plant genotype influences survival of hybrids between Eurosta solidaginis host races. Evolution 61, 2607–2613 10.1111/j.1558-5646.2007.00209.x (doi:10.1111/j.1558-5646.2007.00209.x) [DOI] [PubMed] [Google Scholar]
- 28.Johnson M. T. J., Agrawal A. A. 2005. Plant genotype and environment interact to shape a diverse arthropod community on evening primrose (Oenothera biennis). Ecology 86, 874–885 10.1890/04-1068 (doi:10.1890/04-1068) [DOI] [Google Scholar]
- 29.Ando Y., Ohgushi T. 2008. Ant- and plant-mediated indirect effects induced by aphid colonization on herbivorous insects on tall goldenrod. Popul. Ecol 50, 181–189 10.1007/s10144-007-0072-2 (doi:10.1007/s10144-007-0072-2) [DOI] [Google Scholar]
- 30.Cappuccino N. 1987. Comparative population dynamics of two goldenrod aphids: spatial patterns and temporal constancy. Ecology 68, 1634–1646 10.2307/1939856 (doi:10.2307/1939856) [DOI] [PubMed] [Google Scholar]
- 31.Underwood N. 2007. Variation in and correlation between intrinsic rate of increase and carrying capacity. Am. Nat. 169, 136–141 10.1086/509942 (doi:10.1086/509942) [DOI] [PubMed] [Google Scholar]
- 32.White J. A., Whitham T. G. 2000. Associational susceptibility of cottonwood to a box elder herbivore. Ecology 81, 1795–1803 10.1890/0012-9658(2000)081[1795:ASOCTA]2.0.CO;2 (doi:10.1890/0012-9658(2000)081[1795:ASOCTA]2.0.CO;2) [DOI] [Google Scholar]
- 33.Dixon A. F. G. 1998. Aphid Ecology: an optimization approach, 2nd edn. London: Chapman and Hall [Google Scholar]
- 34.Narayandas G. K., Alyokhin A. V. 2006. Interplant movement of potato aphid (Homoptera: Aphididae) in response to environmental stimuli. Environ. Entomol. 35, 733–739 10.1603/0046-225X-35.3.733 (doi:10.1603/0046-225X-35.3.733) [DOI] [Google Scholar]
- 35.Halitschke R., Keßler A., Lorenz J. K. A., Baldwin I. T. 2000. Ecophysiological comparison of direct and indirect defenses in Nicotiana attenuate. Oecologia 124, 408–417 10.1007/s004420000389 (doi:10.1007/s004420000389) [DOI] [PubMed] [Google Scholar]
- 36.Degen T., Dillmann C., Marion-Poll F., Turlings T. C. J. 2004. High genetic variability of herbivore-induced volatile emission within a broad range of maize inbred lines. Plant. Physiol. 135, 1928–1938 10.1104/pp.104.039891 (doi:10.1104/pp.104.039891) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Bernstein C. 1984. Prey and predator emigration responses in the acarine system Tetranychus urticae-Phytoseiulus persimilis. Oecologia 61, 134–142 10.1007/BF00379099 (doi:10.1007/BF00379099) [DOI] [PubMed] [Google Scholar]
- 38.Schellhorn N. A., Andow D. A. 2005. Response of coccinellids to their aphid prey at different spatial scales. Popul. Ecol. 47, 71–76 10.1007/s10144-004-0204-x (doi:10.1007/s10144-004-0204-x) [DOI] [Google Scholar]
- 39.Losey J. E., Denno R. F. 1998. The escape response of pea aphids to foliar-foraging predators: factors affecting dropping behaviour. Ecol. Entomol. 23, 53–61 10.1046/j.1365-2311.1998.00102.x (doi:10.1046/j.1365-2311.1998.00102.x) [DOI] [Google Scholar]
- 40.Atsatt P. R., O'Dowd D. J. 1976. Plant defense guilds. Science 193, 24–29 10.1126/science.193.4247.24 (doi:10.1126/science.193.4247.24) [DOI] [PubMed] [Google Scholar]
- 41.Agrawal A. A., Lau J. A., Hambäck P. A. 2006. Community heterogeneity and the evolution of interactions between plants and insect herbivores. Q. Rev. Biol. 81, 349–376 10.1086/511529 (doi:10.1086/511529) [DOI] [PubMed] [Google Scholar]
- 42.Unsicker S. B., Franzke A., Specht J., Köhler G., Linz J., Renker C., Stein C., Weisser W. W. 2010. Plant species richness in montane grasslands affects the fitness of a generalist grasshopper species. Ecology 91, 1083–1091 10.1890/09-0402.1 (doi:10.1890/09-0402.1) [DOI] [PubMed] [Google Scholar]