Abstract
Most membrane enveloped viruses bud from infected cells by hijacking the host ESCRT machinery. The ESCRTs are recruited to bud sites by viral proteins that contain short proline-rich motifs (PRMs) known as late domains. The late domains probably evolved by co-opting host PRMs involved in the normal functions of ESCRTs in endosomal sorting and cytokinesis. The solution and crystal structures of PRMs bound to their interaction partners explain the conserved roles of Pro and other residues that predominate in these sequences. PRMs are often grouped together in much larger proline-rich regions (PRRs) of as many as 150 residues. The PRR of the ESCRT-associated protein ALIX autoregulates its conformation and activity. The robustness of different viral budding and host pathways to impairments in proline-based interactions varies considerably. The known biology of proline rich motif recognition in the ESCRT pathway seems, in principle, compatible with antiviral development, given our increasingly nuanced understanding of the relative weakness and robustness of the host and viral processes.
Keywords: protein structure, virus budding, ALG-2, endosome, cytokinesis, ALIX, TSG101, UEV domain, WW domain, CEP55
Introduction
Many membrane-enveloped viruses hijack the host ESCRT machinery in order to escape from cells (1–6). The ESCRTs are responsible for severing the narrow membrane neck connecting the nascent virion to the plasma membrane (7). ESCRT-dependent viruses recruit the ESCRTs principally via short peptide motifs called “late domains” (8). The three classes of late domain involved in ESCRT recruitment are all proline-rich motifs (PRMs), and have the form PPXY, P(S/T)AP, or LYPXnL. Viral late domain PRMs evolved to mimic the normal functions of PRM in host physiology. Viruses have had a lot to teach cell biologists, the ESCRT pathway being an excellent case in point. Indeed, the discovery of the viral PRM-ESCRT interactions led to the characterization of the first example of a host-encoded ESCRT-binding PRM. Within cellular regulatory and trafficking proteins, PRMs very often occur within the context of larger proline-rich regions (PRRs). One of the questions addressed in this review is whether PRRs have functions beyond being a collection of contiguous PRMs.
Here, we consider PRRs and PRMs as a central theme in the normal physiology and structural organization of the ESCRT pathway and as potential targets for antivirals. In addition to the long PRRs within ESCRT proteins themselves, we also consider short PRMs. We define a PRM here to be a short segment that has an identified interaction and function and at least one conserved and functionally required Pro. PRMs are common in the sequences of ESCRT proteins and in other cell and viral proteins that bind to ESCRTs, and occur both within larger PRRs and in isolation. In this review, we summarize the accumulated knowledge about what these regions do in the ESCRT pathway, what their structures look like, and prospects for their targetability in antiviral drug design.
Conformational properties of Pro and Pro-rich sequences
Before delving into the biology of PRRs and PRMs in the ESCRT pathway, we will briefly review why Pro is such a special amino acid. Pro is unique among the amino acid constituents of proteins because the Cδ of its side-chain is cyclized to its backbone amide nitrogen. This has several consequences for the structural and binding properties of Pro (9). The backbone amide of Pro cannot donate a hydrogen bond. Pro is thus a disruptor of hydrogen bonding in both α-helices and β-sheets, even though individual Pro residues can still adopt these conformations. On the other hand, Pro is strongly favored at certain positions of type I and type II β-turns. The φ conformational torsion angle of Pro is restricted by the cyclization. The bulk of the cyclized N-Cδ linkage constrains the conformational space of the residue preceding the Pro, disfavoring the α-helical conformation. When multiple Pro residues occur consecutively, their conformations are so restricted that they become essentially locked into the type II polyproline (PPII) helical conformation. Uninterrupted polyPro tracts are thus far more rigid than the other sequences. Finally, Pro is the only residue for which the cis peptide conformation occurs with any frequency. Thus PRRs have unique conformational constraints that govern the structures of proteins that contain them, but since these segments are typically removed from constructs used in crystallography and NMR, their structural roles are underappreciated.
In addition to these unique conformational restrictions, Pro has a hydrophobic ring which can form complementary packing interactions with the flat rings of the aromatic side-chains Phe, Tyr, and Trp, the flat guanidino group of Arg, and of course, other Pro side-chains. The rigidity of PRMs has important consequences for binding. Because the unbound PRM has fewer conformational degrees of freedom, these sequences lose less conformational entropy upon complex formation. Because the entropic free energy penalty for binding is smaller, fewer molecular contacts are needed to yield the same binding affinity, as compared to a non-Pro based sequence. Unlike the sometimes bewildering and uncooperative full-length PRRs, the PRMs, once whittled down to short peptides, readily give their secrets up to structural and molecular biologists.
PRRs of the ESCRTs
ALIX
ALIX has a key role in ESCRT-mediated membrane abscission in cytokinesis, and mediates the LYPXnL motif-driven budding of viruses by the ESCRTs. ALIX also seems to have roles in apoptosis and endocytosis, and it remains unclear whether and how these latter functions are related to the ESCRT-associated roles of ALIX (10). ALIX contains a N-terminal Bro1 domain that recruits and/or regulates the membrane scission-promoting ESCRT-III subunit CHMP4. This is followed by a central V domain that is the locus for binding to the LYPXnL motif, discussed below as one of the ESCRT-interacting PRMs. Most important for purposes of this review is the C-terminal 150 amino acid PRR (Fig. 1). The ALIX PRR has a 33 % Pro content and is also enriched in Gly, Ala, Ser, Thr, Tyr, Gln, and Asn, all typical of low complexity sequences.
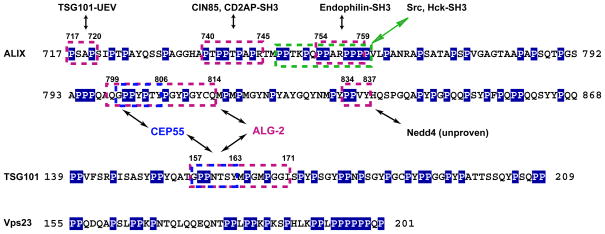
Figure 1. PRRs of the ESCRTs.
The Proline-rich regions of human ALIX, human TSG101, and Saccharomyces cerevisiae Vps23. Pro residues are highlighted in blue background. PRMs with identified functions are boxed. The arrows points to their binding partners as annotated on the figure.
The ALIX PRR contains five PRMs of known function. The 717PSAP720 sequence binds to the UEV domain of the ESCRT-I subunit TSG101 (11–13). The 740PTPAPR745 PRM binds to SH3 domains of the endocytic adaptor proteins CIN85 (14) and CD2AP (15). The SH3 domain is a ubiquitous PRM binding domain, and indeed was the first one to be described. The ALIX PTPAPR sequence belongs to an unusual subset of two-faced SH3 binding-PRMs, which bind to two SH3 domains at once, so dimerizing the SH3 domain-containing proteins (16). The 748PPTKPQPPARPPP760 region contains the binding site for the conventional SH3 domains of the Src (17, 18) and Hck kinases (19) and the endocytic protein endophilin (20). The multi-SH3 domain ubiquitin ligase POSH binds to the ALIX PRR (21), probably through one of these SH3 binding PRMs. Thus far, none of the three PRMs described above have been shown to be essential for any of the ESCRT-related functions of ALIX. The midbody protein CEP55 binds to the 800GPPYPTY806 sequence in ALIX, and this interaction is required for ESCRT-mediated membrane abscission in cytokinesis (22–25). The CEP55 binding site overlaps with the 799QGPPYPTYPGYPGYCQ814 sequence that binds to the Ca2+-binding apoptosis regulator ALG-2 (26). The ALIX PRR contains one PPXY motif, 834PPVY837, which could potentially serve as the binding site for the WW domain-containing ubiquitin ligase Nedd4 (27, 28). Taking the above described PRMs together, they account for 47 of the 150 residues of the PRR.
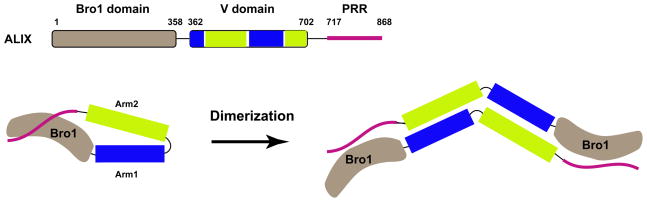
The ALIX PRR has been implicated in both the dimerization and autoinhibition of ALIX. ALIX dimerizes both through its V domain (29, 30) and the 852PSYP855 sequence close to its C-terminus (31) (Fig. 2). The ability to dimerize via the PRR appears to be important for ALIX-mediated HIV-1 budding, although this requirement can be overridden in the case of another virus, EIAV, that seem to interact more strongly with ALIX than HIV-1 (31). There is clear evidence that the PRR autoinhibits both the CHMP4 and HIV-1 LYPXnL motif binding activities of ALIX (32–34). The autoinhibition seems to involve a contact between the N-terminus of the PRR and a Src-phosphorylation site (17) on a tip of the Bro1 domain (32) known as “patch 2” (35). This contact is not compatible with the conformation of the ALIX Bro1-V construct (ALIX-ΔPRR) in the crystallized monomer (36) or the solution dimer (30). These ALIX-ΔPRR structures likely represent an active conformation of ALIX. The current autoinhibition model thus suggests that the conformation of full-length autoinhibited ALIX will be different from any state observed so far (Fig. 2). In summary, current data suggest a dual role for the ALIX PRR as a locus for multiple PRMs and as a regulator of ALIX conformation and activity.
Figure 2. Autoregulation of ALIX by its PRR.
The domain organization of ALIX is depicted schematically and colored separately: Bro1 domain (light brown), V domain (blue (Arm1), green (Arm2)) and PRR region (purple). The PRR tail autoinhibits ALIX, shown in a putative closed conformation (left panel) (32, 33). ALIX appears to function as a dimer (31), hence the active form is shown as a dimer on the basis of the solution structure of dimeric ALIX-ΔPRR (30).
TSG101/Vps23
TSG101 is one of four subunits of the ESCRT-I complex. Its N-terminal UEV domain binds both to the ubiquitin tag on ESCRT cargo (37) and to a PRM of the form P(S/T)AP (38–41). The UEV domain is connected to the heterotetrameric core of ESCRT-I (42) by a 70-residue PRR with a 30 % Pro content. The only PRM of known function within this region is the CEP55 binding sequence 157GPPNTSY163 (22, 23, 43) (Fig. 1). As described below, the TSG101 PRR also binds ALG-2, but the precise motif has not been mapped. Apart from providing a tether connecting the UEV domain to the core, it is unclear why this region has evolved to be as big or as Pro-rich as it is. The yeast ortholog of TSG101, Vps23, contains a 47 residue PRR with a 45 % Pro content within its UEV-core linker (Fig. 1). The Vps23 PRR contains no identified PRMs. Indeed, none of the human PRM binding protein mentioned above are conserved in yeast, except for Bro1 (ALIX) and Vps23 (TSG101) themselves. The Vps23 PRR does contain one contiguous tract of six consecutive Pro residues. The Pro6 tract seems at least as likely to contribute to the mechanical rigidity of the Vps23 PRR as it does to molecular recognition, and one wonders if the most ancient role for the PRR might not be a mechanical and conformational one.
PRMs and ESCRTs
P(S/T)AP and SDP motifs
The P(S/T)AP motif is best known as the major late domain of HIV-1, HTLV-1, Ebola, and a half-dozen or so other characterized viruses (3) (Table 1). In human cells, the best characterized P(S/T)AP motif is that of the Hrs subunit of ESCRT-0 (44–46). The P(S/T)AP sequence itself is abundant in the human proteome, but many P(S/T)AP sequences are buried within folded domains and so not functionally available for ESCRT interactions. TSG101 itself contains an internal PTAP motif, but this sequence is predicted to be buried within the stably folded headpiece domain based on the conformation of its ortholog in the crystallized yeast ESCRT-I core complex (42). Indeed, short peptides spanning internal P(S/T)AP sequences in both TSG101 and another human ESCRT-I subunit, VPS37B, bind to the UEV domain in vitro (47, 48), but they do not seem to be functional in the context of the complete ESCRT-I complex in cells (48).
Table 1.
ESCRT-binding and ESCRT-associated PRMs
| Motif1 [preferred motif] | Interacting domain | Inactivating mutant in domain (human sequence numbering except where noted otherwise) | Viral protein with functional motif2 | Cellular ESCRT-interacting protein with physiologically functional motif | Cellular ESCRT-interacting protein with motif whose physiological function is unconfirmed |
| (P/A/S)(S/T)AP [P(S/T)APP] | TSG101 UEV |
Y63A, N69P, M95A | HIV-1 p6, FIV p2, Mason-Pfizer monkey Gag, HTLV-1 Gag, PFV Gag, VSV M, Ebola VP40, Japanese encephalitis NS3, Lassa Z, Bluetongue NS3 | Hrs, Tal, Tom1L1, GGA3, ALIX | Sec16A, Sec24B, Caprin-1, eIF-4B, PABP1, EWS, CPSF7, NFκ-b2, TFG, ATXL2, BAT3, TB182, TRI47, VASP, WDR26 |
| SDP | Vps23 UEV |
W16A3 | Vps27, Rim8 | ||
| LYPX1/3L | ALIX/Bro1 V | F676D | HIV-1 p6, EIAV p9, PFV Gag, Sendai C, Vaccinia F13L, RSV Gag | Aspergillus PacC | |
| PPXY [PPXYESψΦ] | Nedd4/Rsp5, WWP1, WWP2, and Itch WW | H470A4 | Marburg VP40, RSV Gag, Mason-Pfizer monkey Gag; murine leukemia Gag, HTLV-1 Gag, VSV M, Rabies M, Ebola VP40, LMCV Z, Lassa Z, Bluetongue NS3 | (selected) ENaCβand γ subunits, ARRDC1-4, TXNIP, yeast Art1-9 | ALIX |
| GPPX3Y | CEP55 EABR |
R191A, W184A | TSG101, ALIX, TEX14 | ||
| PPYPXYPXYP | ALG-2 | Y124A, R125A | ALIX | TSG101 |
ψ and Φ refer to aromatic and hydrophobic residues, respectively.
See ref. (3) for the original citations for viral proteins, which are not included here for reasons of space.
Numbering is for S. cerevisiae.
Numbering is for the Nedd4 sequence from rat; this mutation inactivates only the 3rd WW domain of Nedd4. Abbreviations: TSG101, tumor suppressor gene 101; UEV, ubiquitin E2 variant; HIV, human immunodeficiency virus; FIV, feline immunodeficiency virus; HTLV, human T cell leukemia virus; PFV, prototypic foamy virus; VSV, vesicular stomatitis virus; EIAV, equine infectious anemia virus; RSV, Rous sarcoma virus; LCMV, lymphocytic choriomeningitis virus; Hrs, HGF receptor substrate; Tal, TSG101-associated ligase; Tom1L1, target of myb1-like 1; GGA3, Golgi-associated, gamma adaptin ear containing, ARF binding protein 3; ALIX, ALG-2 interacting protein X; PABP1, Poly(A)-binding protein 1; CPSF7, Cleavage and polyadenylation specificity factor subunit 7; EWS, RNA-binding protein; NFκ-b2, Nuclear factor NF-kappa-B p100 subunit; ATXL2, Ataxin-2-like protein; TB182, 182 kDa tankyrase-1-binding protein; TRI47, Tripartite motif-containing protein 47; VASP, Vasodilator-stimulated phosphoprotein; WDR26, WD repeat-containing protein 26; Nedd4, Neuronal precursor cell expressed developmentally downregulated 4; ENaC, epithelial sodium channel; ARRDC, arrestin domain-containing protein; TXNIP, thioredoxin interacting protein; ART, arrestin-related trafficking; CEP55, centrosomal protein of 55 kDa; EABR, ESCRT and ALIX binding region; TEX14, testis expressed 14; ALG-2, apoptosis linked gene-2.
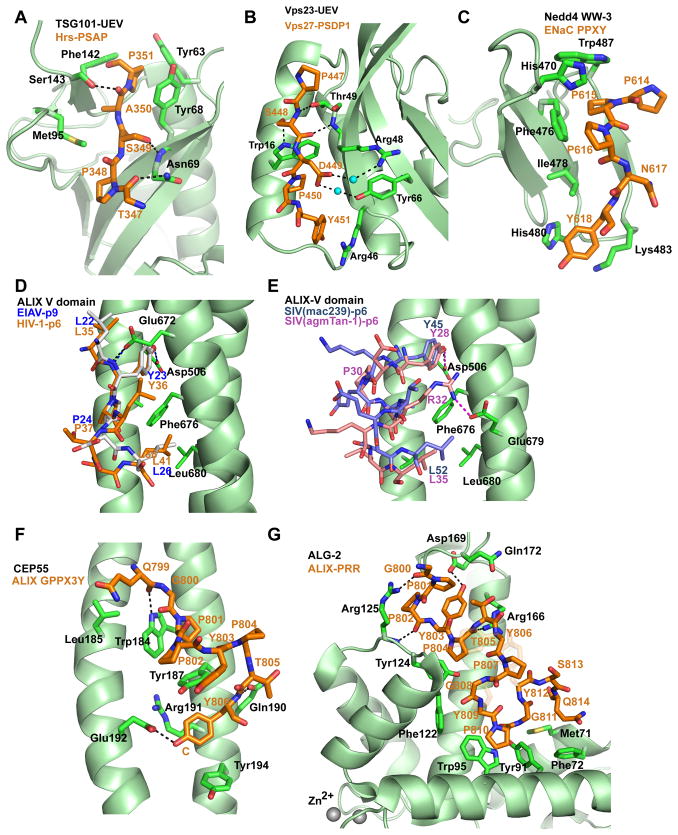
Evidence for a physiological ESCRT-binding role for the P(S/T)AP is available for a handful of proteins besides Hrs. These include Tom1L1 (49), GGA3 (50), and the ubiquitin ligase Tal (51). However, at least fifteen additional P(S/T)AP-containing proteins bind to TSG101 in cells, including proteins of RNA processing and silencing, vesicular trafficking from the endoplasmic reticulum, and transcriptional regulation (48). The only known ligand for the P(S/T)AP motif is the UEV domain of the TSG101 subunit of ESCRT-I. The affinity of the shortest P(S/T)AP peptides tested is modest, in the range of 50–300 μM (52, 53), with slightly higher affinities seen for longer HIV-1 Gag constructs (54). P(S/T)AP peptides bind in an extended (β-strand) conformation to a site involving the C-terminus of the UEV domain (53, 55) (Fig. 3A). The Ser/Thr of the motif is required because the side-chain hydroxyl makes a hydrogen bond with the main-chain backbone nitrogen of Asn69. The Ala is required because it packs in a pocket that is too small to allow any other residue. The final Pro of the motif is also tightly packed into a pocket that fits its side-chain uniquely. On the other hand, the first Pro of the motif is not strictly required (48). Indeed, some synthetic peptides with hydrophobic substituents added to the first Pro actually have a higher affinity for the UEV domain (52, 56). The yeast ortholog of Hrs is Vps27, and also uses a PRM to bind to the UEV domain of Vps23. The relevant PRM in yeast has the form SDP (57), and is important for the co-localization of ESCRT-0-mediated cargo clusters with ESCRT-I-mediated membrane buds (58). The yeast SDP motif binds to a completely different site on the UEV domain as compared to the P(S/T)AP motif of multicellular eukaryotes (59) (Fig. 3B). SDP motifs seem to be important for the recruitment of the ESCRTs in other contexts in yeast, for example, one of the yeast arrestin-related trafficking proteins (ARTs) is apparently recruited by this mechanism (60). The P(S/T)AP and SDP sequences are not functionally interchangeable. The larger theme of PRM-UEV interactions is preserved across the eukarya, but the structural details have almost nothing in common between yeast and multicellular eukaryotes.
Figure 3. PRM recognition.
The structural basis of PRM recognition by ESCRTs and ESCRT-associated proteins. All models are in a ball-and-stick representation, with the PRM peptide colored in brown and protein domains colored in green (except for C and D, where multiple peptides are shown). Key hydrogen bonds between protein domain and peptide are shown as black dashed lines. (A) P(S/T)AP recognition. Structure of human TSG101 UEV domain bound to the human Hrs PSAP peptide (PDB id: 3OBQ). (B) SDP recognition by the yeast Vps23 UEV domain. Structure of the Vps23 UEV domain bound to a yeast SDP peptide (PDB id: 3R42). Water molecules are shown as spheres. (C) The PPXY motif. The structure of human Nedd4 WW-3 domain in complex with ENaC PPNY peptide (PDB id: 1I5H). (D, E) Structural comparison of the ALIX V domain in complex with late-domain peptides of viruses. (D) HIV-1 p6 Gag (PDB id: 2R02, brown) and EIAV p9-Gag (PDB id: 2R03, gray) are overlaid together binding to ALIX V domain. The hydrogen bonds between ALIX and HIV-1 p6 are shown in black dashed lines; The H-bonds between ALIX and EIAV p9 are shown in blue dashed lines. (E) SIV(mac239) p6-Gag peptide (PDB id: 2XS1, blue) and SIV(agmTan-1) p6-Gag (PDB id: 2XS8, light purple) are overlaid together in complex with ALIX V domain. The hydrogen bond between ALIX Asp506 and SIVmac239 Y45 is labeled using black dashed line; The hydrogen bonds between ALIX and SIV(agmTan-1) are shown in magenta dashed line. (F) ALIX GPPX3Y peptide bound to CEP55 (PDB id: 3E1R). (G) ALIX PPR peptide interacts with ALG-2 (PDB id: 2ZNE). Protein chain A and peptide chain C are shown here, Zn2+ ions are shown as spheres. The sticks of Y806, Y809 and Y812 in ALIX PRR peptide are shown in a transparent model in order to highlight the backbone conformation of the ALIX PRR peptide.
The PTAP-UEV domain interaction is essential for HIV-1 budding. Current AIDS antivirals target the three enzymes encoded by the viral genome. Because of the prospect of drug resistance, there is considerable interest in targeting host-virus interactions such as that of the Gag PTAP and TSG101 UEV domain. The obvious concern in targeting host proteins is toxicity from interference with vital cellular functions. Fortunately, mutational disruption of the P(S/T)AP binding site on the TSG101 UEV domain does not seem to impair receptor downregulation by the ESCRTs, at least not for the degradation of endogeneous EGF receptors (53). On the other hand, mutational disruption of the site does lead to a partial defect in ESCRT-mediated cytokinesis (22). Thus a spectrum of phenotypes is seen following interference with P(S/T)AP recognition: strong (HIV-1 release), intermediate (cytokinesis), or none (downregulation of endogenous EGF receptor).
PPXY motifs
The main function of PPXY motifs is to target the proteins containing them to interact with WW domains (so named for their two signature Trp residues). The ESCRT complexes per se do not contain WW domains, but the WW domain-containing Nedd4 family ubiquitin ligases (Rsp5 in yeast) are intimately connected with the ESCRTs. PPXY late domains of many viruses, including Rous sarcoma, Marburg, Ebola, and Rabies viruses (3) (Table 1), recruit the Nedd4 family ligases WWP1, WWP2, and Itch to the site of virus budding (61). The link from these ligases to the ESCRTs probably involves more than simply the ubiquitination of viral proteins. The arrestin-related trafficking (ART) proteins (62) contain PPXY motifs of their own, bind directly to ESCRTs, and appear likely to play a role as bridges connected the ligases to the ESCRTs in PPXY-dependent budding (63). Some cargo, such as the epithelial sodium channel ENaC, contain PPXY motifs and appear to be shunted into the ESCRT pathway through a direct interaction with WW domains of Nedd4 family ligases. WW domains fall into classes with additional specificity determinants. The third WW domain of Nedd4 belongs to class I, and its most preferred motif is PPXYESψΦ, where ψ and Φ refer to aromatic and hydrophobic residues, respectively (64). The ENaC β subunit has a PPXY peptide conforming to the preferred motif, and it binds to the third WW domain of Nedd4 with Kd = 20 μM (65). The structure of the ENaC PPXY motif with this WW domain showed that the first three residues of the PPXY motif are in a PPII conformation and directly contact one of the signature Trp residues (65) (Fig. 3C). The final Tyr residue of the motif contacts the ring of an essential His side-chain of the WW domain and makes other hydrophobic contacts. Most of the Nedd4-family substrates that go on to become ESCRT cargo do not contain PPXY motifs. Of the various mechanisms for the ubiquitination and entry of these cargo into the ESCRT pathway, it currently appears that ART proteins may in many cases be the direct cargo selectors (62). The ARTs then contribute their own PPXY motifs to recruit the Nedd4 family ligases, leading to cargo ubiquitination and finally internalization into multivesicular bodies (MVBs) by the ESCRTs.
LYPXnL motifs
The LYPXnL motif, where n=1 or 3, is the third example of a Pro-containing ESCRT-interacting sequence first discovered as a viral late domain. These motifs bind to the V domain of ALIX with low micromolar affinity (29, 36, 66, 67). The N-terminal LYP sequence and the C-terminal Leu bind in the same pockets on arm 2 of the V domain (67) regardless of whether they are separated by one or three residues. In the case of the motif from HIV-1 Gag p6, where n = 3, the intervening residues form one turn of an α-helix (67). In EIAV p9, n = 1, and the single intervening residue is in an extended conformation (67) (Fig. 3D). ALIX and the Gag LYPXnL motif sustain a significant level of residual HIV-1 budding in the absence of the TSG101-PTAP interaction. Indeed, in Jurkat cells, > 20 % particle release is seen even when both the PTAP and LYPXnL motifs are crippled (68). Any concerted therapeutic program to shut down ESCRT-mediated HIV budding would at a minimum need to target the ALIX V domain in addition to the TSG101 UEV domain, so it is fortunate that all of these structures are now in hand.
The concept of the LYPXnL motif and the ALIX V domain as canonical motif-domain interaction pair is elegant, but we now know it is too simple. Certain SIV Gag proteins lack LYPXnL motifs, yet still bind ALIX with affinities in the tens of micromolar (69) (Fig. 3E). These motifs all bind to the same site on the V domain, and all have a Tyr residue that interacts with the same pocket on the V domain. Otherwise, these sequences have almost nothing in common; one of the sequences does not even have a Pro among its binding determinants. So far, the LYPXnL motif has been a less productive guide to normal ESCRT physiology than the other viral PRMs. In Aspergillus, the transcription factor PacC contains such a motif, and interacts with an ALIX homolog in the course of its pH-dependent cleavage and activation (70). In the canonical ESCRT pathway, clear cut functions for V domain interactors have yet to be established. The absence of known functions for this domain in human physiology is at least encouraging with respect to its targetability for antivirals. Perhaps the ALIX V domain will, in the course of its normal function in the ESCRT pathway, turn out to use nonstandard interactions such as with the SIV Gag sequences.
GPPX3Y motifs
Only three functional occurrences of this specialized motif have been reported to date. These are within the PRRs of ALIX and TSG101, described above, and that of the germ cell cytokinesis inhibitor TEX14 (71). The function of the GPPX3Y motif is to target the protein containing it to the midbody connecting a pair of dividing cells. There, the ESCRTs are involved in the process of constricting and finally severing the narrow membrane neck connecting the two daughter cells (22–25). Peptides derived from the GPPX3Y sequences of ALIX and TSG101 bind with Kd ~ 1 μM affinity to the coiled-coil midbody protein CEP55 (43). One peptide binds to one CEP55 dimer. Despite its conservation between ALIX and TSG101, the Gly of the motif is not particularly important for binding. The Pro residues are much more important, and the Tyr is critical (43). Some questions remain, for example, it is not clear how essential is the spacing of three residues between the diPro and the Tyr. A more complete understanding of the binding determinants for this site, such as from a peptide library study, would be helpful and might perhaps identify other protein ligands for CEP55. In contrast to other PRMs, which are bound by discrete, specialized protein domains, the GPPX3Y motifs bind to a region within the much larger CEP55 coiled coil. The ESCRT- and ALIX binding region (EABR) within CEP55 is differentiated from the rest of the coiled coil because it possesses unusual bulky and charged residues at the a and d positions. These unusual features force the two coils apart, creating a single binding site for the GPPX3Y motif. In spite of the presence of a diPro sequence, the motif does not have a PPII conformation. The structure of the peptide is best described as being in a β-conformation over the GPP residues, followed by the X3Y residues in a β-turn. The GPPX3Y motif on CEP55 consists of two subsites, one that binds to the GPP portion of the motif, and a second that surrounds the Tyr (Fig. 3F). The site seems to be optimized primarily to bind the surfaces of the Pro and Tyr rings as opposed to selecting a special conformation.
ALIX and ALG-2
ALG-2 (apoptosis-linked gene 2) is a dimeric Ca2+-binding EF hand protein that binds to the PRRs of both ALIX (20) and TSG101 (72). The binding is Ca2+-dependent, and the ALG-2 dimer appears to be capable of acting as a Ca2+-dependent bridge to recruit ALIX and TSG101 to one another in cells (73). The physiological ramifications of Ca2+ signaling into the ESCRT system have yet to be deeply explored, but we now understand a great deal about this interaction at least at the structural level. The binding site for ALG-2 on ALIX has been mapped to a Pro, Gly, and Tyr rich section that overlaps with all of the CEP55 binding sequence described above, and extends beyond it. Clearly, ALG-2 and CEP55 cannot bind to ALIX simultaneously. The corresponding sequence in TSG101 has not been mapped in detail, but by analogy to ALIX probably corresponds more or less to 158PPNTSYMPGMPGGI171. The ALIX peptide was co-crystallized with ALG-2 (26), and the segment 801PPYPTYPGYPGY812 interacts extensively with ALG-2 (Fig. 3G). The ALG-2 pocket that enfolds the N-terminal PPYP residues only forms the binding-competent conformation in the presence of Ca2+ (or Zn2+ as a surrogate), explaining the Ca2+-dependence of binding. Despite the high density of Pro residues in this motif, there are apparently enough non-Pro residues that the type II polyproline helix is not the preferred conformation. Instead, the peptide manifests a series of 2–3 residue stretches that are in a β conformation. These are connected by a series of ~90 degree bends at Gly and Pro residues (and in one case, Thr805). The turn-favoring propensity of Gly and Pro probably facilitates these bends and may explain the conserved role of so many Gly and Pro residues in this PRM. Two Tyr residues (803 and 809) are deeply buried in hydrophobic pockets, while another two interact more superficially. Three of the Pro residues (802, 804, and 810) form extensive hydrophobic interactions. On the other hand, the surface-exposed Pro807 seems to have a purely conformational role.
Concluding remarks
The past few years have shown us just how complex the rules can be when it comes to PRMs in ESCRT function. The P(S/T)XP:UEV domain case showed how two seemingly similar motifs, differing only by an Ala versus an Asp at the X position, can turn out to interact at completely different binding sites on one conserved domain in different species. Just as two apparently similar motifs can interact with very different binding sites, a single binding site can recruit very different motifs. The clearest case amongst the ESCRT PRMs are the unusual ALIX V domain binding sequences in certain SIV strains that, beyond a single Tyr residue, have no obvious common features with the canonical motif. In an exciting development, the combination of bioinformatics and proteomics is beginning to provide us with a more reliable picture of the PRM interactome (48). With respect to the long PRRs of ESCRT proteins such as ALIX and TSG101, two-thirds or more of the residues within the PRR cannot be accounted for in terms of known interactions. One wonders if it is that the majority of binding partners have yet to be discovered, or is it the unique conformational properties of Pro in rigidifying and extending the reach of the PRR in three dimensions make these regions so important. With respect to targeting PRM-ESCRT interactions with antivirals, the accumulated data reveal significant differences in the relative robustness of viral and host pathway dependencies to interference with ESCRT-PRM interactions. In principle, this should be favorable to their targeting by antivirals. The potential existence of currently uncharacterized physiological roles of ESCRT PRMs in areas such as RNA processing could introduce complications, however. The biggest challenge in targeting this interaction may be at the level of chemistry rather than biology, given the issues with the screenability and drugability of peptide-protein interactions. Nevertheless, vigorous efforts are being devoted to overcoming these obstacles (52, 56, 74).
Acknowledgments
This work was supported by the Intramural Program of the NIH, NIDDK and the Intramural AIDS Targeted Anti-viral Program of the Office of the Director, NIH (J. H. H.), and an Intramural AIDS Research Fellowship to X. R.
References
- 1.Morita E, Sundquist WI. Retrovirus budding. Annu Rev Cell Dev Biol. 2004;20:395–425. doi: 10.1146/annurev.cellbio.20.010403.102350. [DOI] [PubMed] [Google Scholar]
- 2.Fujii K, Hurley JH, Freed EO. Beyond Tsg101: the role of Alix in ‘ESCRTing’ HIV-1. Nat Rev Microbiol. 2007;5:912–916. doi: 10.1038/nrmicro1790. [DOI] [PubMed] [Google Scholar]
- 3.Chen BJ, Lamb RA. Mechanisms for enveloped virus budding: Can some viruses do without an ESCRT? Virology. 2008;372:221–232. doi: 10.1016/j.virol.2007.11.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Usami Y, Popov S, Popova E, Inoue M, Weissenhorn W, Gottlinger HG. The ESCRT pathway and HIV-1 budding. Biochem Soc Trans. 2009;37:181–184. doi: 10.1042/BST0370181. [DOI] [PubMed] [Google Scholar]
- 5.Bieniasz PD. The Cell Biology of HIV-1 Virion Genesis. Cell Host Microbe. 2009;5:550–558. doi: 10.1016/j.chom.2009.05.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Carlton JG, Martin-Serrano J. The ESCRT machinery: new functions in viral and cellular biology. Biochem Soc Trans. 2009;37:195–199. doi: 10.1042/BST0370195. [DOI] [PubMed] [Google Scholar]
- 7.Hurley JH, Hanson PI. Membrane budding and scission by the ESCRT complexes: It’s all in the neck. Nat Rev Mol Cell Biol. 2010;11:556–566. doi: 10.1038/nrm2937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Freed EO. Viral late domains. J Virol. 2002;76:4679–4687. doi: 10.1128/JVI.76.10.4679-4687.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Williamson MP. The structure and function of proline-rich regions in proteins. Biochem J. 1994;297:249–260. doi: 10.1042/bj2970249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Odorizzi G. The multiple personalities of Alix. J Cell Sci. 2006;119:3025–3032. doi: 10.1242/jcs.03072. [DOI] [PubMed] [Google Scholar]
- 11.Strack B, Calistri A, Craig S, Popova E, Gottlinger HG. AIP1/ALIX is a binding partner for HIV-1 p6 and EIAV p9 functioning in virus budding. Cell. 2003;114:689–699. doi: 10.1016/s0092-8674(03)00653-6. [DOI] [PubMed] [Google Scholar]
- 12.von Schwedler UK, Stuchell M, Muller B, Ward DM, Chung HY, Morita E, Wang HE, Davis T, He GP, Cimbora DM, Scott A, Krausslich HG, Kaplan J, Morham SG, Sundquist WI. The protein network of HIV budding. Cell. 2003;114:701–713. doi: 10.1016/s0092-8674(03)00714-1. [DOI] [PubMed] [Google Scholar]
- 13.Martin-Serrano J, Yaravoy A, Perez-Caballero D, Bieniasz PD. Divergent retroviral late-budding domains recruit vacuolar protein sorting factors by using alternative adaptor proteins. Proc Natl Acad Sci U S A. 2003;100:12414–12419. doi: 10.1073/pnas.2133846100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chen B, Borinstein SC, Gillis J, Sykes VW, Bogler O. The glioma-associated protein SETA interacts with AIP1/Alix and AZIG-2 and modulates apoptosis in astrocytes. J Biol Chem. 2000;275:19275–19281. doi: 10.1074/jbc.M908994199. [DOI] [PubMed] [Google Scholar]
- 15.Usami Y, Popov S, Gottlinger HG. Potent rescue of human immunodeficiency virus type 1 late domain mutants by ALIX/AIP1 depends on its CHMP4 binding site. J Virol. 2007;81:6614–6622. doi: 10.1128/JVI.00314-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Jozic D, Cardenes N, Deribe YL, Moncalian G, Hoeller D, Groemping Y, Dikic I, Rittinger K, Bravo J. Cbl promotes clustering of endocytic adaptor proteins. Nat Struct Mol Biol. 2005;12:972–979. doi: 10.1038/nsmb1000. [DOI] [PubMed] [Google Scholar]
- 17.Schmidt MHH, Dikic I, Bogler O. Src phosphorylation of Alix/AIP1 modulates its interaction with binding partners and antagonizes its activities. J Biol Chem. 2005;280:3414–3425. doi: 10.1074/jbc.M409839200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Dejournett RE, Kobayashi R, Pan S, Wu C, Etkin LD, Clark RB, Bogler O, Kuang J. Phosphorylation of the proline-rich domain of Xp95 modulates Xp95 interaction with partner proteins. Biochem J. 2007;401:521–531. doi: 10.1042/BJ20061287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Shi XL, Opi S, Lugari A, Restouin A, Coursindel T, Parrot I, Perez J, Madore E, Zimmermann P, Corbeil J, Huang MD, Arold ST, Collette Y, Morelli X. Identification and biophysical assessment of the molecular recognition mechanisms between the human haemopoietic cell kinase Src homology domain 3 and ALG-2-interacting protein X. Biochem J. 2010;431:93–102. doi: 10.1042/BJ20100314. [DOI] [PubMed] [Google Scholar]
- 20.Chatellard-Causse C, Blot B, Cristina N, Torch S, Missotten M, Sadoul R. Alix (ALG-2-interacting protein X), a protein involved in apoptosis, binds to endophilins and induces cytoplasmic vacuolization. J Biol Chem. 2002;277:29108–29115. doi: 10.1074/jbc.M204019200. [DOI] [PubMed] [Google Scholar]
- 21.Tsuda M, Seong KH, Aigaki T. POSH, a scaffold protein for JNK signaling, binds to ALG-2 and ALIX in Drosophila. FEBS Lett. 2006;580:3296–3300. doi: 10.1016/j.febslet.2006.05.005. [DOI] [PubMed] [Google Scholar]
- 22.Carlton JG, Martin-Serrano J. Parallels between cytokinesis and retroviral budding: a role for the ESCRT machinery. Science. 2007;316:1908–1912. doi: 10.1126/science.1143422. [DOI] [PubMed] [Google Scholar]
- 23.Morita E, Sandrin V, Chung HY, Morham SG, Gygi S, Rodesch CK, Sundquist WI. Human ESCRT and ALIX proteins interact with proteins of the midbody and function in cytokinesis. EMBO J. 2007;26:4215–4227. doi: 10.1038/sj.emboj.7601850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Guizetti J, Schermelleh L, Mäntler J, Maar S, Poser I, Leonhardt H, Müller-Reichert T, Gerlich DW. Cortical constriction during abscission involves helices of ESCRT-III-dependent filaments. Science. 2011;331:1616–1620. doi: 10.1126/science.1201847. [DOI] [PubMed] [Google Scholar]
- 25.Elia N, Sougrat R, Spurlin T, Hurley JH, Lippincott-Schwartz J. Dynamics of ESCRT machinery during cytokinesis and its role in abscission. Proc Natl Acad Sci U S A. 2011;108:4846–4851. doi: 10.1073/pnas.1102714108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Suzuki H, Kawasaki M, Inuzuka T, Okumura M, Kakiuchi T, Shibata H, Wakatsuki S, Maki M. Structural Basis for Ca2+-Dependent Formation of ALG-2/Alix Peptide Complex: Ca2+/EF3-Driven Arginine Switch Mechanism. Structure. 2008;16:1562–1573. doi: 10.1016/j.str.2008.07.012. [DOI] [PubMed] [Google Scholar]
- 27.Nikko E, Andre B. Split-ubiquitin two-hybrid assay to analyze protein-protein interactions at the endosome: Application to Saccharomyces cerevisiae Bro1 interacting with ESCRT complexes, the Doa4 ubiquitin hydrolyase, and the Rsp5 ubiquitin ligase. Eukaryot Cell. 2007;6:1266–1277. doi: 10.1128/EC.00024-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sette P, Jadwin JA, Dussupt V, Bello NF, Bouamr F. The ESCRT-associated protein Alix recruits the ubiquitin liages Nedd4–1 to facilitate HIV-1 release through the LYPXnL L domain motif. J Virol. 2010;84:8181–8192. doi: 10.1128/JVI.00634-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Munshi UM, Kim J, Nagashima K, Hurley JH, Freed EO. An Alix fragment potently inhibits HIV-1 budding - Characterization of binding to retroviral YPXL late domains. J Biol Chem. 2007;282:3847–3855. doi: 10.1074/jbc.M607489200. [DOI] [PubMed] [Google Scholar]
- 30.Pires R, Hartlieb B, Signor L, Schoehn G, Lata S, Roessle M, Moriscot C, Popov S, Hinz A, Jamin M, Boyer V, Sadoul R, Forest E, Svergun DI, Gottlinger HG, et al. A Crescent-Shaped ALIX Dimer Targets ESCRT-III CHMP4 Filaments. Structure. 2009;17:843–856. doi: 10.1016/j.str.2009.04.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Carlton JG, Agromayor M, Martin-Serrano J. Differential requirements for Alix and ESCRT-III in cytokinesis and HIV-1 release. Proc Natl Acad Sci U S A. 2008;105:10541–10546. doi: 10.1073/pnas.0802008105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zhou X, Si J, Corvera J, Gallick GE, Kuang J. Decoding the intrinsic mechanism that prohibits ALIX interaction with ESCRT and viral proteins. Biochem J. 2010;432:525–534. doi: 10.1042/BJ20100862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zhou X, Pan S, Sun L, Corvera J, Lin SH, Kuang J. The HIV-1 p6/EIAV p9 docking site in ALIX is autoinhibited as revealed by a conformation-sensitive anti-ALIX monoclonal antibody. Biochem J. 2008;414:215–220. doi: 10.1042/BJ20080642. [DOI] [PubMed] [Google Scholar]
- 34.Pan S, Wang R, Zhou X, He G, Koomen J, Kobayashi R, Sun L, Corvera J, Gallick GE, Kuang J. Involvement of the adaptor protein alix in actin cytoskeleton assembly. J Biol Chem. 2006;281:34640–34650. doi: 10.1074/jbc.M602263200. [DOI] [PubMed] [Google Scholar]
- 35.Kim J, Sitaraman S, Hierro A, Beach BM, Odorizzi G, Hurley JH. Structural basis for endosomal targeting by the Bro1 domain. Dev Cell. 2005;8:937–947. doi: 10.1016/j.devcel.2005.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Fisher RD, Chung HY, Zhai Q, Robinson H, Sundquist WI, Hill CP. Structural and biochemical studies of ALIX/AIP1 and its role in retrovirus budding. Cell. 2007;128:841–852. doi: 10.1016/j.cell.2007.01.035. [DOI] [PubMed] [Google Scholar]
- 37.Katzmann DJ, Babst M, Emr SD. Ubiquitin-dependent sorting into the multivesicular body pathway requires the function of a conserved endosomal protein sorting complex, ESCRT-I. Cell. 2001;106:145–155. doi: 10.1016/s0092-8674(01)00434-2. [DOI] [PubMed] [Google Scholar]
- 38.Garrus JE, von Schwedler UK, Pornillos OW, Morham SG, Zavitz KH, Wang HE, Wettstein DA, Stray KM, Cote M, Rich RL, Myszka DG, Sundquist WI. Tsg101 and the vacuolar protein sorting pathway are essential for HIV-1 budding. Cell. 2001;107:55–65. doi: 10.1016/s0092-8674(01)00506-2. [DOI] [PubMed] [Google Scholar]
- 39.VerPlank L, Bouamr F, LaGrassa TJ, Agresta B, Kikonyogo A, Leis J, Carter CA. Tsg101, a homologue of ubiquitin-conjugating (E2) enzymes, binds the L domain in HIV type 1 Pr55(Gag) Proc Natl Acad Sci U S A. 2001;98:7724–7729. doi: 10.1073/pnas.131059198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Demirov DG, Ono A, Orenstein JM, Freed EO. Overexpression of the N-terminal domain of TSG101 inhibits HIV-1 budding by blocking late domain function. Proc Natl Acad Sci U S A. 2002;99:955–960. doi: 10.1073/pnas.032511899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Martin-Serrano J, Zang T, Bieniasz PD. HIV-I and Ebola virus encode small peptide motifs that recruit Tsg101 to sites of particle assembly to facilitate egress. Nat Med. 2001;7:1313–1319. doi: 10.1038/nm1201-1313. [DOI] [PubMed] [Google Scholar]
- 42.Kostelansky MS, Schluter C, Tam YYC, Lee S, Ghirlando R, Beach B, Conibear E, Hurley JH. Molecular architecture and functional model of the complete yeast ESCRT-I heterotetramer. Cell. 2007;129:485–498. doi: 10.1016/j.cell.2007.03.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Lee HH, Elia N, Ghirlando R, Lippincott-Schwartz J, Hurley JH. Midbody targeting of the ESCRT machinery by a noncanonical coiled coil in CEP55. Science. 2008;322:576–580. doi: 10.1126/science.1162042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Bache KG, Brech A, Mehlum A, Stenmark H. Hrs regulates multivesicular body formation via ESCRT recruitment to endosomes. J Cell Biol. 2003;162:435–442. doi: 10.1083/jcb.200302131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Pornillos O, Higginson DS, Stray KM, Fisher RD, Garrus JE, Payne M, He GP, Wang HE, Morham SG, Sundquist WI. HIV Gag mimics the Tsg101-recruiting activity of the human Hrs protein. J Cell Biol. 2003;162:425–434. doi: 10.1083/jcb.200302138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Lu Q, Hope LWQ, Brasch M, Reinhard C, Cohen SN. TSG101 interaction with HRS mediates endosomal trafficking and receptor down-regulation. Proc Natl Acad Sci U S A. 2003;100:7626–7631. doi: 10.1073/pnas.0932599100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Stuchell MD, Garrus JE, Muller B, Stray KM, Ghaffarian S, McKinnon R, Krausslich HG, Morham SG, Sundquist WI. The human endosomal sorting complex required for transport (ESCRT-I) and its role in HIV-1 budding. J Biol Chem. 2004;279:36059–36071. doi: 10.1074/jbc.M405226200. [DOI] [PubMed] [Google Scholar]
- 48.Schlundt A, Sticht J, Piotukh K, Kosslick D, Jahnke N, Keller S, Schuemann M, Krause E, Freund C. Proline-rich Sequence Recognition - II. PROTEOMICS ANALYSIS OF Tsg101 UBIQUITIN-E2-LIKE VARIANT (UEV) INTERACTIONS. Mol Cell Proteomics. 2009;8:2474–2486. doi: 10.1074/mcp.M800337-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Puertollano R. Interactions of Tom1L1 with the multivesicular body sorting machinery. J Biol Chem. 2005;280:9258–9264. doi: 10.1074/jbc.M412481200. [DOI] [PubMed] [Google Scholar]
- 50.Puertollano R, Bonifacino JS. Interactions of GGA3 with the ubiquitin sorting machinery. Nat Cell Biol. 2004;6:244–251. doi: 10.1038/ncb1106. [DOI] [PubMed] [Google Scholar]
- 51.Amit I, Yakir L, Katz M, Zwang Y, Marmor MD, Citri A, Shtiegman K, Alroy I, Tuvia S, Reiss Y, Roubini E, Cohen M, Wides R, Bacharach E, Schubert U, et al. Tal, a Tsg101-specific E3 ubiquitin ligase, regulates receptor endocytosis and retrovirus budding. Genes Dev. 2004;18:1737–1752. doi: 10.1101/gad.294904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Liu F, Stephen AG, Waheed AA, Aman MJ, Freed EO, Fisher RJ, Burke TR. SAR by oxime-containing peptide libraries: Application to Tsg101 ligand optimization. ChemBioChem. 2008;9:2000–2004. doi: 10.1002/cbic.200800281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Im YJ, Kuo L, Ren X, Burgos PV, Zhao XZ, Lui F, Burke TR, Bonifacino JS, Freed EO, Hurley JH. Crystallographic and functional analysis of the ESCRT-1/HIV-1 Gag PTAP interaction. Structure. 2010;18:1536–1547. doi: 10.1016/j.str.2010.08.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Pornillos O, Alam SL, Rich RL, Myszka DG, Davis DR, Sundquist WI. Structure and functional interactions of the Tsg101 UEV domain. EMBO J. 2002;21:2397–2406. doi: 10.1093/emboj/21.10.2397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Pornillos O, Alam SL, Davis DR, Sundquist WI. Structure of the Tsg101 UEV domain in complex with the PTAP motif of the HIV-1 p6 protein. Nat Struct Biol. 2002;9:812–817. doi: 10.1038/nsb856. [DOI] [PubMed] [Google Scholar]
- 56.Kim SE, Liu F, Im YJ, Stephen AG, Waheed AA, Freed EO, Fisher RJ, Hurley JH, Burke TR. Elucidation of new binding interactions with the tumor suppressor gene 101 (tsg101) protein using modified HIV-1 Gag-p6 derived peptide ligands. ACS Med Chem Lett. 2011 doi: 10.1021/ml1002579. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Bilodeau PS, Winistorfer SC, Kearney WR, Robertson AD, Piper RC. Vps27-Hse1 and ESCRT-I complexes cooperate to increase efficiency of sorting ubiquitinated proteins at the endosome. J Cell Biol. 2003;163:237–243. doi: 10.1083/jcb.200305007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Wollert T, Hurley JH. Molecular mechanism of multivesicular body biogenesis by ESCRT complexes. Nature. 2010;464:864–873. doi: 10.1038/nature08849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Ren X, Hurley JH. Structural basis for endosomal recruitment of ESCRT-I by ESCRT-0 in yeast. EMBO J. 2011 doi: 10.1038/emboj.2011.122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Herrador A, Herranz S, Lara D, Vincent O. Recruitment of the ESCRT Machinery to a Putative Seven-Transmembrane-Domain Receptor Is Mediated by an Arrestin-Related Protein. Mol Cell Biol. 2010;30:897–907. doi: 10.1128/MCB.00132-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Martin-Serrano J, Eastman SW, Chung W, Bieniasz PD. HECT ubiquitin ligases link viral and cellular PPXY motifs to the vacuolar protein-sorting pathway. J Cell Biol. 2005;168:89–101. doi: 10.1083/jcb.200408155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Lin CH, MacGum JA, Chu T, Stefan CJ, Emr SD. Arrestin-Related Ubiquitin-Ligase Adaptors Regulate Endocytosis and Protein Turnover at the Cell Surface. Cell. 2008;135:714–725. doi: 10.1016/j.cell.2008.09.025. [DOI] [PubMed] [Google Scholar]
- 63.Rauch S, Martin-Serrano J. Multiple interactions between the ESCRT machinery and arrestin-related proteins: implications in PPXY-dependent budding. J Virol. 2011;85:3546–3556. doi: 10.1128/JVI.02045-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Kasanov J, Pirozzi G, Uvegas AJ, Kay BK. Characterizing class I WW domains defines key specificity determinants and generates mutant domains with novel specificities. Chem Biol. 2001;8:231–241. doi: 10.1016/s1074-5521(01)00005-9. [DOI] [PubMed] [Google Scholar]
- 65.Kanelis V, Rotin D, Forman-Kay JD. Solution structure of a Nedd4 WW domain-ENaC peptide complex. Nat Struct Biol. 2001;8:407–412. doi: 10.1038/87562. [DOI] [PubMed] [Google Scholar]
- 66.Lee S, Joshi A, Nagashima K, Freed EO, Hurley JH. Structural basis for viral late-domain binding to Alix. Nat Struct Mol Biol. 2007;14:194–199. doi: 10.1038/nsmb1203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Zhai Q, Fisher RD, Chung HY, Myszka DG, Sundquist WI, Hill CP. Structural and functional studies of ALIX interactions with YPXnL late domains of HIV-1 and EIAV. Nat Struct Mol Biol. 2008;15:43–49. doi: 10.1038/nsmb1319. [DOI] [PubMed] [Google Scholar]
- 68.Fujii K, Munshi UM, Ablan SD, Demirov DG, Soheilian F, Nagashima K, Stephen AG, Fisher RJ, Freed EO. Functional role of Alix in HIV-1 replication. Virology. 2009;391:284–292. doi: 10.1016/j.virol.2009.06.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Zhai Q, Landesman MB, Robinson H, Sundquist WI, Hill CP. Identification and structural characterization of the ALIX-binding late domains of simian immunodeficiency virus SIVmac239 and SIVagmTan-1. J Virol. 2011;85:632–637. doi: 10.1128/JVI.01683-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Vincent O, Rainbow L, Tilburn J, Arst HN, Penalva MA. YPXL/I is a protein interaction motif recognized by Aspergillus PalA and its human homologue, AIP1/Alix. Mol Cell Biol. 2003;23:1647–1655. doi: 10.1128/MCB.23.5.1647-1655.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Iwamori T, Iwamori N, Ma L, Edson MA, Greenbaum MP, Matzuk MM. TEX14 Interacts with CEP55 To Block Cell Abscission. Mol Cell Biol. 2010;30:2280–2292. doi: 10.1128/MCB.01392-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Katoh K, Suzuki H, Terasawa Y, Mizuno T, Yasuda J, Shibata H, Maki M. The penta-EF-hand protein ALG-2 interacts directly with the ESCRT-I component TSG101, and Ca2+-dependently co-localizes to aberrant endosomes with dominant-negative AAA ATPase SKD1/Vps4B. Biochem J. 2005;391:677–685. doi: 10.1042/BJ20050398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Okumura M, Ichioka F, Kobayashi R, Suzuki H, Yoshida H, Shibata H, Maki M. Penta-EF-hand protein ALG-2 functions as a Ca2+-dependent adaptor that bridges Alix and TSG101. Biochem Biophys Res Comm. 2009;386:237–241. doi: 10.1016/j.bbrc.2009.06.015. [DOI] [PubMed] [Google Scholar]
- 74.Tavassoli A, Lu Q, Gam J, Pan H, Benkovic SJ, Cohen SN. Inhibition of HIV Budding by a Genetically Selected Cyclic Peptide Targeting the Gag-TSG101 Interaction. ACS Chem Biol. 2008;3:757–764. doi: 10.1021/cb800193n. [DOI] [PubMed] [Google Scholar]