Abstract
Hepatitis delta virus (HDV) and cytoplasmic polyadenylation element-binding protein 3 (CPEB3) ribozymes form a family of self-cleaving RNAs characterized by a conserved nested double-pseudoknot and minimal sequence conservation. Secondary structure-based searches were used to identify sequences capable of forming this fold, and their self-cleavage activity was confirmed in vitro. Active sequences were uncovered in several marine organisms, two nematodes, an arthropod, a bacterium, and an insect virus, often in multiple sequence families and copies. Sequence searches based on identified ribozymes showed that plants, fungi, and a unicellular eukaryote also harbor the ribozymes. In Anopheles gambiae, the ribozymes were found differentially expressed and self-cleaved at basic developmental stages. Our results indicate that HDV-like ribozymes are abundant in nature and suggest that self-cleaving RNAs may play a variety of biological roles.
Self-cleaving ribozymes are autocatalytic RNAs that include the hammerhead, hepatitis delta virus (HDV), hairpin, the Neurospora Varkud satellite, and the bacterial GlmS motifs (1). Recently, an HDV ribozyme-like fold was found in the human CPEB3 gene, but sequence-based searches were unable to identify this class of RNAs outside of mammals (2). Because in HDVand CPEB3 ribozymes only six nucleotides (nt) are invariant, whereas about 60 nt are required to form the minimal fold, the conserved nested double-pseudoknot secondary structure (Fig. 1A and fig. S1) was used to search genomic databases for RNAs capable of magnesium-dependent self-cleavage (3–5).
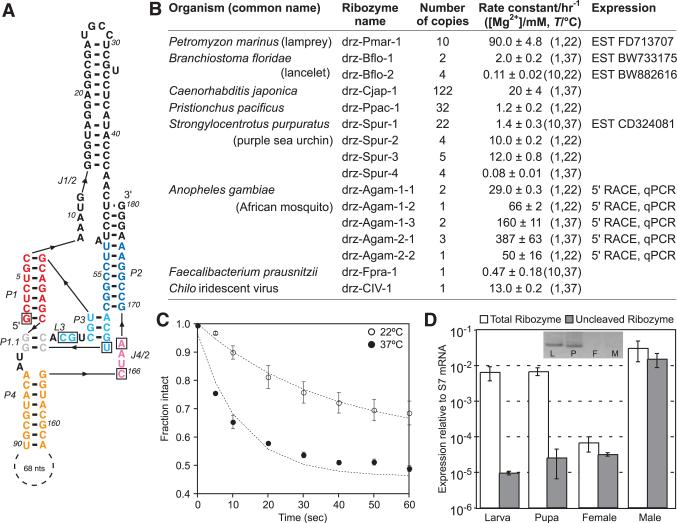
Fig. 1.
A. gambiae drz-Agam-2-1 ribozyme. (A) Core elements are colored by region corresponding to the HDV ribozyme (1). Boxed letters are the six invariant positions used in secondary structure searches that identified the ribozymes listed in (B). (C) Graph of in vitro self-cleavage in 1 mM MgCl2 and (D) RT-qPCR of drz-Agam-2-1 isolated from indicated developmental stages. (Inset) 5′ RACE of the ribozyme. All data are average values ± average deviations.
In vitro self-scission was initially confirmed in six eukaryotes, one bacterium, and one insect virus (Fig. 1B, table S1, and figs. S2 to S18). All eukaryotes harbor several HDV-like ribozymes, and some contain multiple sequence families (figs. S2 to S11). These sequences were used to search the GenBank Expressed Sequence Tags database (dbEST), where additional ribozymes, often in apparently self-cleaved form, were identified in plants, fungi, fish, insects, a tapeworm, and a unicellular ciliate (table S2).
In Anopheles gambiae, representatives of two ribozyme families were tested in vitro (Fig. 1 and figs. S2 to S5). The drz-Agam-1 ribozymes were found by using a restrictive structure descriptor and closely resemble HDVand CPEB3 ribozymes, whereas the drz-Agam-2 family was found by using a descriptor that permitted extended J1/2 and P4 regions. Although previous work demonstrated that a variable P4 helix does not affect catalytic activity (6), an expanded J1/2 region has not been observed. This feature appears to stabilize the overall structure, with a fast cleavage rate constant of 1.7 ± 0.4 min–1 at 37°C (Fig. 1C). Rapid amplification of cDNA ends (5′ RACE) experiments performed on total RNA extracts from basic developmental stages of A. gambiae showed that the sequenced 5′ ends map to the in vitro verified self-cleavage sites. Reverse transcription quantitative polymerase chain reaction (RT-qPCR) revealed highly differential expression and self-cleavage of the ribozymes (Fig. 1D and figs. S2 to S5), suggesting that both processes are regulated.
Several other sequence families contain features unusual for HDVand CPEB3 ribozymes. In drz-Spur-3, the P1 helix is only six base pairs long, yet the ribozyme undergoes efficient self-cleavage (Fig. 1B and fig. S8). Drz-Spur-4, drz-Ppac-1, and drz-Dpap-1 contain only one adenosine in the J4 and J2 section (figs. S9, S14, and S19).
Although in vivo analysis of other ribozymes has not been conducted, several findings indicate potential biological activity. In nematodes, the ribozymes (drz-Ppac-1 and drz-Cjap-1; figs. S12 to S15) are intergenic and widely distributed, with drz-Cjap-1, or its fragments, composing up to 1/10,000 of the animal's genome. These copies reside between conserved downstream sequences and diverse upstream sequences and likely play a part in retrotransposition. A similar role is hypothesized for Strongylocentrotus purpuratus and A. gambiae ribozymes, which appear within or near genes coding for reverse transcriptase–like proteins. Other ribozymes likely partake in RNA processing: drz-CIV-1 resides between predicted transcription and translation start sites of an RNA polymerase large-subunit gene, drz-Fpra-1 maps between two metabolic genes, and drz-Dpap-1 bisects a transcript containing an upstream splice-leader sequence and downstream 5S ribosomal RNA (figs. S17 to 19).
Our results indicate that HDV-like ribozymes, and likely ribozymes in general, are widely distributed in nature and may play a variety of biological roles. Although alignment-based sequence searching can uncover conserved regions of these molecules, a structure-based approach has proven more effective at finding new functional RNAs.
Supplementary Material
Footnotes
Supporting Online Material
www.sciencemag.org/cgi/content/full/326/5955/953/DC1
Materials and Methods
Figs. S1 to S19
Tables S1 and S2
References
References and Notes
- 1.Fedor MJ. Annu. Rev. Biophys. 2009;38:271. doi: 10.1146/annurev.biophys.050708.133710. [DOI] [PubMed] [Google Scholar]
- 2.Salehi-Ashtiani K, Lupták A, Litovchick A, Szostak JW. Science. 2006;313:1788. doi: 10.1126/science.1129308. [DOI] [PubMed] [Google Scholar]
- 3.Ferre-D'Amare AR, Zhou K, Doudna JA. Nature. 1998;395:567. doi: 10.1038/26912. [DOI] [PubMed] [Google Scholar]
- 4.Materials and methods are available as supporting material on Science Online.
- 5.Gautheret D, Major F, Cedergren R. Comput. Appl. Biosci. 1990;6:325. doi: 10.1093/bioinformatics/6.4.325. [DOI] [PubMed] [Google Scholar]
- 6.Been MD, Perrotta AT, Rosenstein SP. Biochemistry. 1992;31:11843. doi: 10.1021/bi00162a024. [DOI] [PubMed] [Google Scholar]
- 7.We thank the Lupták laboratory, G. Weiss, and J. Kieft for comments and A. James for A. gambiae samples. This work was supported by the University of California, Irvine. A.L. is a member of the Chao Family Comprehensive Cancer Center. The GenBank accession numbers for the confirmed ribozymes are BK006878 to BK006897.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.