Abstract
Pneumocystis jirovecii has been detected in lung tissue from patients with chronic obstructive pulmonary disease (COPD) and is associated with disease severity. The regional distribution of the organism in lungs is unknown, but differences in distribution of Pneumocystis could affect estimates of colonization prevalence. We examined the distribution of Pneumocystis in the lungs of 19 non-HIV-infected patients with COPD who were undergoing lung transplantation. DNA was extracted from explanted lungs. We found Pneumocystis colonization in lung tissue of 42.1% of patients with advanced COPD; however, there was significant regional variation in colonization between lung segments of individual patients. Colonization was detected more commonly in the lower and middle lobes than the upper lobes. These findings suggest that single samples from an individual may underestimate the prevalence of Pneumocystis colonization and future studies may obtain a higher yield of Pneumocystis colonization detection when sampling the lower lobes.
Keywords: chronic obstructive pulmonary disease, Pneumocystis jirovecii, lung
Introduction
The presence of Pneumocystis jirovecii (Pc) in respiratory specimens in the absence of clinical infection has been defined as colonization [Morris et al., 2010]. Polymerase chain reaction (PCR) assays are very sensitive, enabling detection of limited numbers of organisms, even in cases where routine histochemical staining methods are negative [Wakefield et al., 1990]. PCR can detect colonization in diverse respiratory samples (e.g., sputum, bronchalveolar lavage, and lung tissue), but the operating characteristics of these assays may be influenced by the type and location of the respiratory sample, as well as the number and volume of samples. A recent study found the sensitivity of PCR for detection of Pc colonization in the lungs of normal subjects was increased by analyzing a large volume of lung tissue obtained from the right upper lobe [Ponce et al., 2010]. Combined analysis of both an oropharyngeal wash and a nasal swab has also been reported to improve detection of Pc in a population of older, healthy adults [Vargas et al., 2010]. The distribution of Pc colonization within the lung, however, has not been well-studied, and it is unknown whether sampling of upper lobe versus lower lobe, or apical versus basal lung regions within or among lobes affects Pc detection.
Sample site may be particularly important in lung diseases such as chronic obstructive pulmonary disease (COPD) that may have differential expression throughout the lung. Pc colonization has been associated with development of COPD-like changes in non-human primates and mouse models and with increased severity of COPD in humans [Morris et al., 2004, Calderón et al., 2007, Christensen et al., 2008, Shipley et al., 2010], although not all studies have corroborated this association [Maskell et al., 2003]. This discrepancy may be due in part to differences in study populations, but might also result from regional lung variation in colonization. In addition, there is growing interest in the lung microbiome in an effort to understand the relationship of microbes to the lung in health and disease [Friaza et al., 2010]. Microbial flora has been shown to vary within specific locations of the skin, the gastrointestinal tract, and the mouth [Cowan et al., 2008]. It is not known if the same regional variation occurs in the lung, and it remains unclear if detection of colonization in a single lung sample accurately reflects the entire lung. Regional differences in microbial detection throughout the lung would have important implications for the design and interpretation of studies of the lung microbiome or of specific organisms. We performed a cross-sectional study to determine differential anatomic distribution of Pc as a representative organism in the lungs of patients with COPD.
Materials and methods
Subjects were undergoing lung transplantation for end-stage COPD at the University of Pittsburgh Medical Center (Pittsburgh, PA) between March 2008 and August 2009. Clinical data were collected prospectively, prior to transplantation, and included demographic information, history of previous pneumonia, medications and chemical exposures, as well as completion of St. George’s Respiratory Questionnaires, pulmonary function tests, and chest radiographs. Chest computed tomography (CT) scans were visually scored for emphysema presence and severity based on the National Emphysema Treatment Trial [Fishman et al., 2003]. CT scans were rated as normal, trace (1–10%), mild (11–25%), moderate (26–49%), marked (50–74%) and severe (>75%) emphysema [NETT website]. The University of Pittsburgh Institutional Review Board approved this study, and all participants provided informed consent.
Tissue was obtained from surgical explants of the native lungs. Samples were randomly acquired from subpleural apical and basal regions in each lobe of the explanted lung(s). Two 200mg pieces of tissue were obtained from the upper, middle and lower lobes in explanted right lungs and/or upper and lower lobes of the explanted left lung. Individual sterile equipment was used to obtain samples within the same lung, and lungs were processed individually on separate days. DNA was extracted from approximately 200mg of tissue from each site. Tissue was homogenized with a Bullet BlenderTM (Next Advance, Averill Park, NY, USA) using extraction buffer (10× PCR buffer, 50μm MgCl2 and distilled water) and stainless steel beads for 2 minutes. New beads and tubes were used for each sample. DNA extraction was performed as previously described [Morris et al., 2004].
Nested PCR was performed at the Pneumocystis mitochondrial large subunit (mtLSU) rRNA gene using first round primers primers PAZ 102-E and PAZ102-H and second round primers PAZ 292-R and PAZ 102-X as previously reported [Morris et al., 2004]. Negative and positive samples (DNA from lung tissue known to contain human P. jirovecii) were included in all reactions. Positive results were determined by visual inspection and confirmed by sequencing. All PCR was performed by personnel blinded to subject identities and were performed in duplicate. DNA extraction and PCR were carried out in separate rooms, and all PCR reactions were performed in an ultraviolet box. The presence of adequate DNA and lack of PCR inhibitors in each sample was confirmed by performing PCR for human beta-globin [Spencer et al., 2008].
Subjects were considered Pc-colonized if at least one lung sample had P. jirovecii identified by sequencing. Genecodes Sequencher 4.9 sequence analysis software was used to identify the mtLSU rRNA genotypes based on polymorphisms at nucleotide positions 85 and 248 [Board et al., 2003].
Stata (StataCorp, College Station, Texas, USA, version 8.2) was used for analyses, and statistical significance was defined as p<0.05. Clinical, physiologic and radiographic data were compared between Pc-colonized and Pc-negative subjects using Wilcoxon rank sum tests for continuous or ordered variables or Fisher’s exact and chi-square tests for dichotomous variables.
Results
Demographic and clinical parameters of the 19 lung transplant recipients from whom lung specimens were obtained are detailed in Table 1. Subjects received either single or double lung transplants. All subjects quit smoking at least 1 year prior to transplantation. While all transplant recipients had severe COPD by Global Initiative for Chronic Obstructive Lung Disease (GOLD) stage, they had regional variability in their emphysema severity score [Pauwels et al., 2001]. The median emphysema severity score was severe (>75%, range 50–100%) in the upper lobe segment and marked (50–74%, range 26–100%) in the non-upper lobe segments; however, this difference was not significant. No subject had clinically evident pneumonia immediately prior to transplant, and none had Pneumocystis detected on routine clinical histology of the explanted lung.
Table 1.
Characteristics of subjects according to Pneumocystis colonization status
| Subject Characteristics | Pc+ (n=8) | Pc− (n=11) |
|---|---|---|
| Male, n (%) * | 8 (61.5) | 5 (38.5) |
| Caucasian, n (%) | 8 (100) | 11 (100) |
| Age, median (range) | 65 (52–74) | 61 (55–69) |
| Pack years, median (range) | 42 (30–80) | 60 (25–106) |
| Inhaled corticosteroids, n (%) | 6 (46.1) | 7 (53.5) |
| Systemic steroid use, n (%) | 3 (37.5) | 5 (62.5) |
| Current macrolide use, n (%) | 0 | 2 (18.2) |
| Prior pneumonia, n (%) | 6 (75.0) | 11 (100) |
| Environmental exposures, n (%) ** | 7 (87.5) | 10 (90.9) |
| Post bronchodilator FEV1, median (range) | 0.68 (0.58–2.07) | 0.67 (0.41–1.41) |
| FEV1 percent predicted, median (range) | 19.9 (13.8–63.4) | 22.7 (14.4–35.7) |
| DLco percent predicted, median (range) | 27.1 (16.5–34.3) | 31.5 (17.3–45) |
p=0.02
asbestos, berrylium, coal dust, cadmium, cobalt, diesel engine exhaust fumes, silica
Abbreviations: DLco, diffusing capacity for carbon monoxide; FEV1, forced expiratory volume in one second; Pc, Pneumocystis
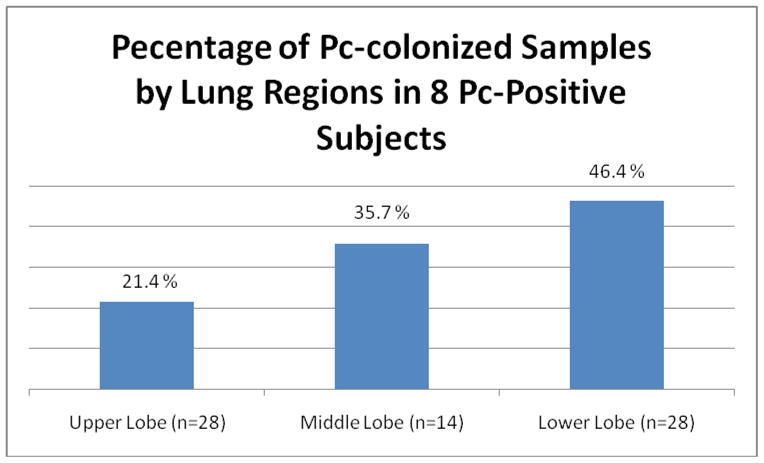
A median of 10 (range 4–10) lung segments per subject were analyzed. Three and sixteen subjects underwent single and double lung transplantation respectively. Pc was detected in at least one location in eight of 19 subjects (42.1%). Three of the eight colonized subjects (37.5%) had one positive sample and another 3 subjects (37.5%) had two samples with Pc colonization. The remaining two subjects (25.0%) had more than five positive samples. Of the six subjects with only one or two positive samples, all had colonization of the lower lobes with three (50.0%) having colonization also detected in the middle lobe. Overall, the middle and lower lobes were more likely to be colonized than the upper lobes (p=0.05, odds ratio 2.86, 95% confidence interval 1.01–8.33) (Figure 1). All subjects with more than five Pc-positive samples had upper, middle and lower lobe colonization, but subjects with only one or two colonized samples had no upper lobe colonization. There were no differences in the likelihood of detecting Pc within the apical or basal regions with 13 of 24 (54.2%) and 11 of 24 (45.8%) Pc–positive samples respectively (p=0.68). The concentration of DNA obtained between lobes was not significantly different with upper and lower lobe medians of 0.2 μg/μL and 0.1 μg/μL respectively (p=0.74). Pc-positive and Pc-negative samples also had no significant difference in DNA concentration with medians of 1.0 μg/μL and 1.1 μg/μL respectively (p=0.221).
Figure 1.
Medication use, prior pneumonia, and physiologic and radiographic variables were not significantly different in Pc-positive and Pc-negative subjects (Table 1); however, Pc-positive subjects were more likely to be male. Of five subjects with Pc in multiple locations, two were colonized with more than one Pc genotype within the same lung, but not within the same lobe (Table 2).
Table 2.
Genotype of 8 Pc-positive subjects
| Genotype | Subjects (n=8) |
|---|---|
| c/c | 1 |
| a/c | 1 |
| t/c | 2 |
| c/t | 2 |
| mixed | 2 |
Discussion
Approximately 40% of subjects with severe COPD were found to be Pc-colonized using sensitive PCR detection assays and samples from multiple lung regions. There was a regional distribution of colonization with more colonization in the lower lobes, but there did not appear to be a difference in colonization frequency according to apical or basal regions within each lobe. The majority of multiply-colonized subjects had the same genotype throughout the lung, but a small percentage displayed more than one genotype. There was no dominant genotype in our study, and no subjects had Pc detected by routine clinical staining.
Our results demonstrate that the selection of lung region for sampling significantly affects determination of colonization prevalence in those with COPD. For example, if upper lobe samples had been used exclusively, only 10.5% of subjects would have been considered colonized. This observation could partly explain the differences in colonization detection in the COPD literature which varies between 7–41% [Calderón et al., 1996, Morris et al., 2004]. While differences in respiratory sample used (i.e. oral wash, sputum, or bronchoalveolar lavage), use of detection methods with differing sensitivities, or severity of COPD in study cohorts likely account for some of the disparities seen in the various studies, our results suggest that lung region sampled also influences colonization detection.
There are several possible explanations of the apparent regional differences of Pc colonization. Pc detection could be less sensitive in the upper lobes of COPD patients because the disease has already become end-stage in these areas or these areas have lower tissue content. Although emphysema was more pronounced in the upper lobe samples, disease was still extensive in the lower lobes, and DNA content was not significantly different in upper versus lower lobe samples. The relative propensity for lower lobe colonization could also be a result of greater lower lung ventilation compared to the upper regions of the lung [Glenny, 2009]. Other factors related to the particular growth requirements of Pc might also result in a propensity of the organism for the lower lobes.
In our study, males were more likely to be Pc-positive perhaps reflecting greater environmental exposure. There was no significant difference in exposure between genders to various environmental agents including asbestos, silica, coal dust, diesel engine exhaust, beryllium or cadmium. Other environmental factors could be involved or the discrepancy could be related to the sample size. We also found no predominant Pc genotype, possibly due to the small sample size or reflecting the different geographic areas where subjects lived as the lung transplant population originates from various parts of the state and country.
Although we examined colonization with a single organism thought to be important in one type of lung disease, these results have implications for the use of molecular methods to detect low levels of other microbes in the lung in both health and disease. There has been growing interest in the examination of the human microbiome. In sites such as the skin, the gastrointestinal tract, and the mouth, microbial flora has been shown to vary with specific locations within each of these structures [Cowan et al., 2008]. It is not known if the same regional variation occurs in the lung. Our results suggest that organism detection varies in different lung regions. We cannot determine whether the variation seen is specific only to Pc in COPD, but the findings raise the question of appropriate sampling methods for future microbiome studies.
There are several limitations to this study. First, because of the nature of the study population, we had a relatively small sample size. An increased sample size would likely have increased the prevalence of colonization detected, but the results support a greater tendency than chance to find colonization in certain areas and highlight the need to interpret studies of colonization using a single sample with caution. It also would have been of interest to compare bronchoalveolar lavage and noninvasive respiratory samples to lung tissue, but these samples were not available. We might also have found a higher prevalence of Pc colonization if we used a greater mass of lung tissue. A recent study found a high prevalence of Pc colonization in normal subjects when analyzing large volumes of lung tissue, but such volumes would not routinely be available for research purposes [Ponce et al., 2010]. In contrast to the current study, Ponce also found high prevalence of colonization in the upper lobes, but did not sample the lower lobes, so that we cannot determine if the variation in prevalence by location was similar to ours [Ponce et al., 2010]. Although we performed PCR in duplicates in addition to sequencing, additional testing of another DNA locus was not performed. Finally, we also lacked a healthy control group to determine if a similar distribution of colonization is seen in healthy subjects, but these types of studies would be difficult to perform in the normal population [Lieberman et al., 2006, Vargas et al., 2010].
In summary, the lung region studied alters the estimate of Pneumocystis colonization in persons with COPD. Our results suggest that selection of lung region to sample could significantly alter estimation of Pc colonization prevalence and supports use of lower lobe samples to improve diagnostic yield in subjects with COPD.
Acknowledgments
This work was supported by NIH HL095370, HL083461 (AM) and NIH HL084948 (FCS, SRD) We thank Chad Karoleski, Joseph Latoche and Joseph Pilewski for technical assistance and graciously allowing access to lung tissue.
Footnotes
Parts of this manuscript have been presented as a poster discussion at the 2010 American Thoracic Society conference in New Orleans, USA.
The authors have no conflict of interest.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Board KF, Patil S, Lebedeva I, Capuano S, Trichel AM, Murphey-Corb M, Rajakumar PA, Flynn JL, Haidaris CG, Norris KA. Experimental Pneumocystis carinii pneumonia in simian immunodeficiency virus infected rhesus macaques. J Infect Dis. 2003;187:576–588. doi: 10.1086/373997. [DOI] [PubMed] [Google Scholar]
- 2.Calderón EJ, Regordan C, Medrano FJ, Ollero M, Varela JM. Pneumocystis carinii infection in patients with chronic bronchial disease. Lancet. 1996;347(9006):977. doi: 10.1016/s0140-6736(96)91468-3. [DOI] [PubMed] [Google Scholar]
- 3.Calderón EJ, Rivero L, Respaldiza N, Morilla R, Montes-Cano MA, Friaza V, Muñoz-Lobato F, Varela JM, Medrano FJ, Horra CL. Systemic inflammation in patients with chronic obstructive pulmonary disease who are colonized with Pneumocystis jiroveci. Clin Infect Dis. 2007;45(2):e17–9. doi: 10.1086/518989. [DOI] [PubMed] [Google Scholar]
- 4.Christensen PJ, Preston AM, Ling T, Du M, Fields WB, Curtis JL, Beck JM. Pneumocystis murina infection and cigarette smoke exposure interact to cause increased organism burden, development of airspace enlargement, and pulmonary inflammation in mice. Infect Immun. 2008;76(8):3481–90. doi: 10.1128/IAI.00165-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cowan MK, Talaro KP. Microbiology: An Organ Systems Approach. Chapter 13 USA: McGraw Hill Higher Education; 2008. Microbe-Human Interactions: Infection and Disease. [Google Scholar]
- 6.Fishman A, Martinez F, Naunheim K, Piantadosi S, Wise R, Ries A, Weinmann G, Wood DE National Emphysema Treatment Trial Research Group. A randomized trial comparing lung-volume-reduction surgery with medical therapy for severe emphysema. N Engl J Med. 2003;348(21):2059–73. doi: 10.1056/NEJMoa030287. [DOI] [PubMed] [Google Scholar]
- 7.Friaza V, la Horra C, Rodríguez-Domínguez MJ, Martín-Juan J, Cantón R, Calderón EJ, Del Campo R. Metagenomic analysis of bronchoalveolar lavage samples from patients with idiopathic interstitial pneumonia and its antagonic relation with Pneumocystis jirovecii colonization. J Microbiol Methods. 2010;82(1):98–101. doi: 10.1016/j.mimet.2010.03.026. [DOI] [PubMed] [Google Scholar]
- 8.Glenny RW. Determinants of regional ventilation and blood flow in the lung. Intensive Care Med. 2009;35:1833–1842. doi: 10.1007/s00134-009-1649-3. [DOI] [PubMed] [Google Scholar]
- Lieberman D, Shleyfer E, Castel H, Terry A, Harman-Boehm I, Delgado J, Peled N. Nasopharyngeal versus oropharyngeal sampling for isolation of potential respiratory pathogens in adults. J Clin Microbiol. 2006;44:525–8. doi: 10.1128/JCM.44.2.525-528.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maskell NA, Waine DJ, Lindley A, Pepperell JC, Wakefield AE, Miller RF, Davies RJ. Asymptomatic carriage of Pneumocystis jiroveci in subjects undergoing bronchoscopy: a prospective study. Thorax. 2003;58:594–7. doi: 10.1136/thorax.58.7.594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Morris A, Sciurba FC, Lebedeva IP, Githaiga A, Elliott WM, Hogg JC, Huang L, Norris KA. Association of chronic obstructive pulmonary disease severity and Pneumocystis colonization. Am J Respir Crit Care Med. 2004;170:408–13. doi: 10.1164/rccm.200401-094OC. [DOI] [PubMed] [Google Scholar]
- 10.National Emphysema Treatment Trial. Available from: www.nhlbi.nih.gov/health/prof/lung/nett/lvrsweb.htm.
- 11.Pauwels RA, Buist AS, Calverley PM, Jenkins CR, Hurd SS GOLD Scientific Committee . Global strategy for the diagnosis, management, and prevention of chronic obstructive pulmonary disease. NHLBI/WHO Global Initiative for Chronic Obstructive Lung Disease (GOLD) Workshop summary. Am J Respir Crit Care Med. 2001;163:1256–76. doi: 10.1164/ajrccm.163.5.2101039. [DOI] [PubMed] [Google Scholar]
- 12.Ponce CA, Gallo M, Bustamante R, Vargas SL. Pneumocystis colonization is highly prevalent in the autopsied lungs of the general population. Clin J Infect Dis. 2010;50:347–53. doi: 10.1086/649868. [DOI] [PubMed] [Google Scholar]
- 13.Shipley TW, Kling HM, Morris A, Patil S, Kristoff J, Guyach S, Murphy JE, Shao X, Sciurba FC, Rogers RM, Richards T, Thompson P, Montelaro RC, Coxson HO, Hogg JC, Norris KA. Persistent Pneumocystis colonization leads to the development of chronic obstructive pulmonary disease in a nonhuman primate model of AIDS. J Infect Dis. 2010;202(2):302–12. doi: 10.1086/653485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Spencer L, Ukwu M, Alexander T, Valadez K, Liu L, Frederick T, Kovacs A, Morris A. Epidemiology of Pneumocystis colonization in families. Clin Infect Dis. 2008;46:1237–1240. doi: 10.1086/533449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vargas SL, Pizarro P, López-Vieyra M, Neira-Avilés P, Bustamante R, Ponce CA. Pneumocystis colonization in older adults and diagnostic yield of single versus paired noninvasive respiratory sampling. Clin Infect Dis. 2010;50:e19–21. doi: 10.1086/649869. [DOI] [PubMed] [Google Scholar]
- 15.Wakefield AE, Pixley FJ, Banerji S, Sinclair K, Miller RF, Moxon ER, Hopkin JM. Detection of Pneumocystis carinii with DNA amplification. Lancet. 1990;336:451–453. doi: 10.1016/0140-6736(90)92008-6. [DOI] [PubMed] [Google Scholar]


