Summary
Somatic cell nuclear transfer, cell fusion, or expression of lineage-specific factors have been shown to induce cell-fate changes in diverse somatic cell types1–12. We recently observed that forced expression of a combination of three transcription factors, Brn2 (also known as Pou3f2), Ascl1, and Myt1l can efficiently convert mouse fibroblasts into functional induced neuronal (iN) cells13. Here, we show that the same three factors can generate functional neurons from human pluripotent stem cells as early as 6 days after transgene activation. When combined with the basic helix-loop-helix transcription factor NeuroD1, these factors could also convert fetal and postnatal human fibroblasts into iN cells displaying typical neuronal morphologies and expressing multiple neuronal markers, even after downregulation of the exogenous transcription factors. Importantly, the vast majority of human iN cells were able to generate action potentials and many matured to receive synaptic contacts when co-cultured with primary mouse cortical neurons. Our data demonstrate that non-neural human somatic cells, as well as pluripotent stem cells, can be directly converted into neurons by lineage-determining transcription factors. These methods may facilitate robust generation of patient-specific human neurons for in vitro disease modeling or future applications in regenerative medicine.
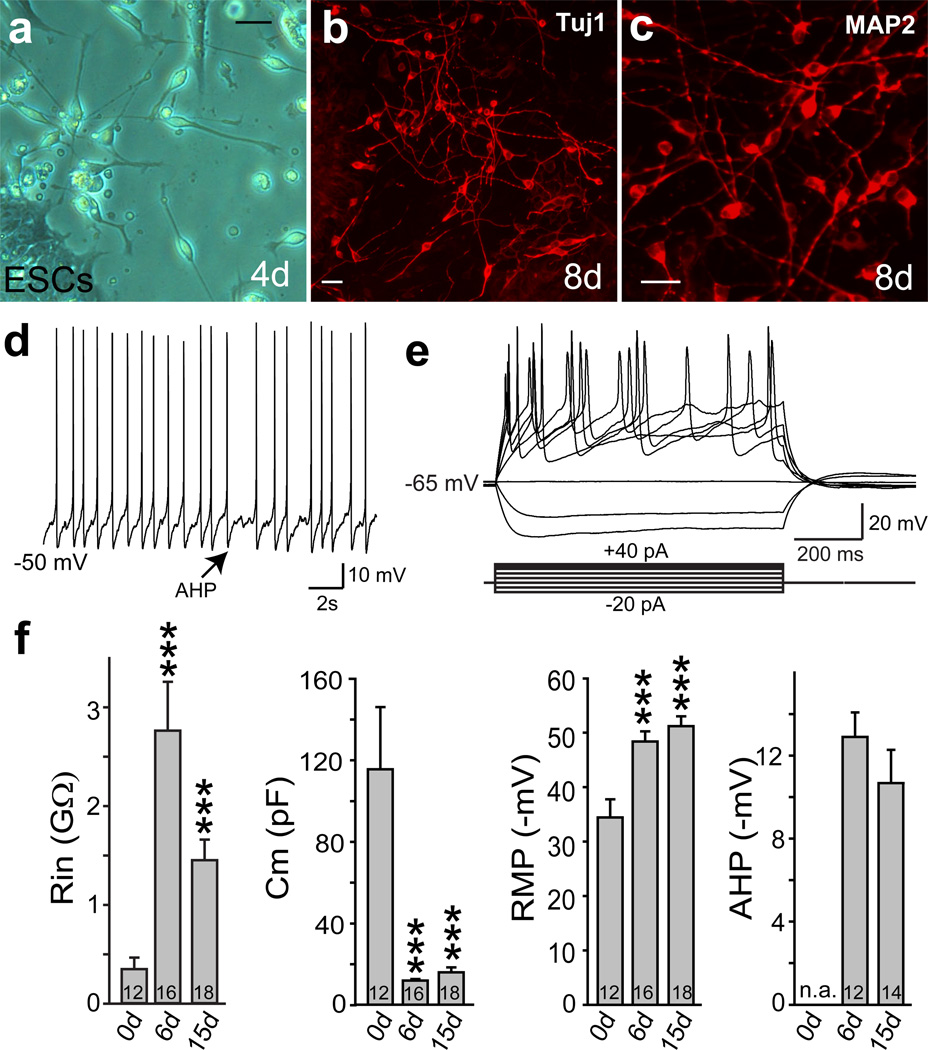
Encouraged by our recent findings in mouse cells13, we explored in this study whether also human fibroblasts could be directly induced to become functional neurons. This was unclear given the differences in the gene regulatory networks governing human and rodent neural development14–16. First, we sought to determine whether forced expression of transcription factors could induce a neuronal fate in human pluripotent cells. To that end, we infected undifferentiated human ES cells in chemically defined N3 media 17 with Brn2, Ascl1, and Myt1l (BAM) using doxycycline (dox)-inducible lentiviral vectors together with an EGFP virus. The majority of ES cells were EGFP-positive 24 hours after addition of doxycycline (Supplementary Fig. 1). Strikingly, as early as 3 days after dox treatment, we observed bipolar neuron-like cells surrounding nearly all ES cell colonies (Fig. 1a; Supplementary Fig. 1). By day 8, cells with more mature neuronal morphologies that expressed both β-III-tubulin (Tuj1) and MAP2 had migrated away from ES cell colonies and were present throughout the plate (Fig. 1b,c). In contrast, after infection with EGFP virus alone, no neuronal cells were generated during the same timeframe, and nearly all ES cells had died due to the Ara-C treatment. We then determined the relative contribution of the three factors and found that Ascl1 alone was sufficient to induce MAP2-positive cells (Supplementary Fig. 2). The addition of Brn2 or Myt1l or both did not increase the efficiency of neuronal differentiation but induced more complex morphologies. Cells infected with all three factors together displayed the most mature neuronal morphologies (Supplementary Fig. 2). Electrophysiological analysis surprisingly revealed that as early as 6 days after induction all recorded cells (n=16) generated action potentials (Fig. 1d,e). At day 15 after dox, the average resting membrane potential of neuronal cells was −51±1.8 mV (mean±SEM, n=18) (Fig. 1f, Supplementary Table 1). These ES-iN cells exhibited prominent after-hyperpolarization potentials (AHPs) following action potentials (Fig. 1d and f). Similar findings could be observed when human iPS cells were infected (Supplementary Fig. 3). Thus, the BAM factors rapidly induce neuronal differentiation of human pluripotent stem cells.
Figure 1. Rapid generation of functional neurons from human ES cells.
a, Four days after induction, ES-iN cells exhibited bipolar neuronal morphologies. b–c, Eight days after induction, ES-iN cells expressed Tuj1 (b) and MAP2 (c). d, Spontaneous action potentials presumably caused by membrane potential fluctuations recorded from an ES-iN cell 6 days after induction. Arrow: pronounced AHP. e, Representative traces of action potentials in response to step current injections 15 days after induction. Membrane potential was maintained at ~ –63mV. f, Quantification of intrinsic membrane properties in control ES cells (0 day) before and after viral transduction. membrane input resistance (Rin), resting membrane potential (RMP), capacitance (Cm), after hyperpolarization potentials (AHP). Scale bars: 10µm (a,b,c). Numbers of cells recorded are labeled in the bars. Note the heterogeneity of the parameters (see also Suppl. Fig. 1). Data are presented with mean±SEM. * p<0.05.
Next, we asked whether also human fibroblasts could be directly converted into neurons. To this end, we derived three independent primary human fetal fibroblast lines (HFFs) (see methods) and performed an extensive characterization of these cultures in various growth conditions to confirm that they lack spontaneous neuronal differentiation potential and do not contain detectable amounts of neural crest stem cells (see Supplementary Fig. 4). Strikingly, 7–10 days after infection with the BAM factors we could detect cells with immature neuronal morphologies. These cells expressed Tuj1 (Supplementary Fig. 5a), but remained functionally immature as revealed by their inability to generate action potentials 20 days after dox treatment (Supplementary Fig. 5b). Thus, the BAM factors appeared to induce neuronal features but were insufficient to generate functional neurons from human fetal fibroblasts under these conditions.
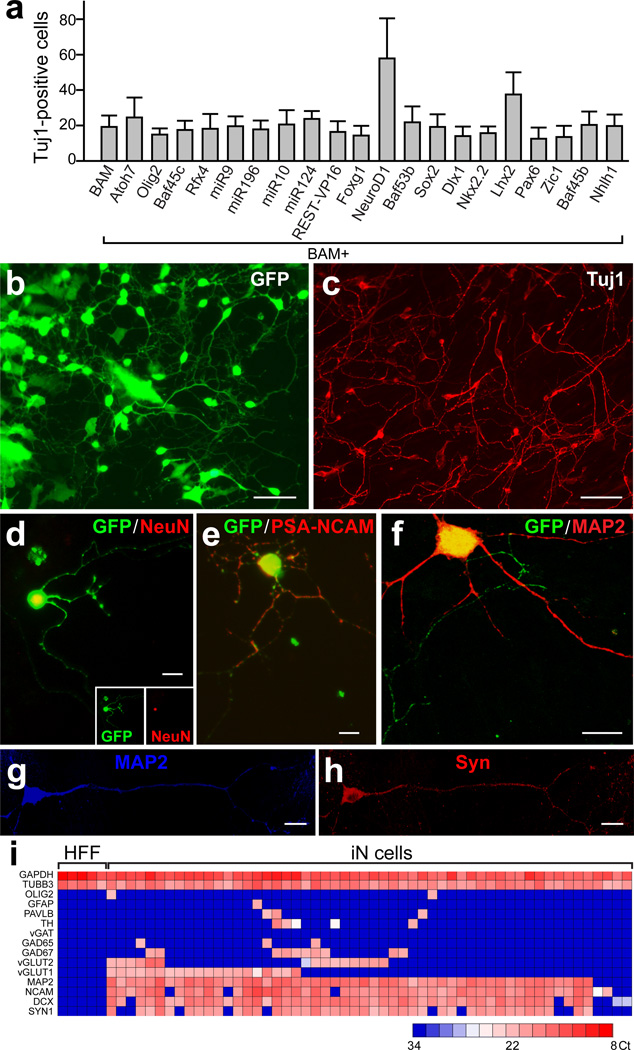
Therefore, we screened 20 additional factors that could improve the generation of neuronal cells in combination with the BAM pool. We observed that NeuroD1, another basic helix-loop-helix transcription factor, improved the efficiency of generating Tuj1-positive neuronal cells 2–3 fold after 3 weeks (Fig. 2a). To determine the relative contribution of NeuroD1, we tested various combinations of these four factors. NeuroD1 alone had no effect, but surprisingly in combination with Brn2 it was sufficient to generate a similar number of Tuj1-positive neuronal cells compared to the BAN, BMN and BAMN pools (Supplementary Fig. 6a). However, further morphological and functional characterization showed that the BAMN combination generated the most mature neuronal cells (Supplementary Fig. 6b). We therefore decided to focus the further analysis on BAMN-iN cells.
Figure 2. NeuroD1 increases reprogramming efficiency in primary human fetal fibroblasts.
a, Quantification of Tuj1-positive BAM-iN cells with indicated factors, 3 weeks after dox. b–c, Three weeks after dox BAM+NeuroD1 iN cells exhibited neuronal morphologies (b) and expressed Tuj1 (c) . d–f, iN cells expressed NeuN (d,) PSA-NCAM (e), and MAP2 (f) 2 weeks after dox. g–h, An iN cell expressing MAP2 (g) and synapsin (h) 4 weeks after dox and co-cultured with primary astrocytes. i, Single cell gene expression profiling using Fluidigm dynamic arrays. Rows represent the evaluated genes and columns represent individual cells. Heatmap (blue to red) represents the threshold Ct values as indicated. Data in (a) are presented as mean±SD. Scale bars: 100 µm (b, c), 10 µm (d–h).
Two weeks after induction, BAMN-iN cells exhibited neuronal morphologies and were labeled with pan-neuronal antibodies such as Tuj1, NeuN, PSA-NCAM and MAP2 (Fig. 2b–f). After extended culture periods of 4–5 weeks, we could detect cells expressing neurofilaments (Supplementary Fig. 7a), and rare neuronal processes decorated with punctate staining of synapsin and synaptotagmin, two synaptic vesicle proteins (Fig. 2g,h, Supplementary Fig. 7b). To ensure the co-expression of pan-neuronal and subtype specific markers, we performed single-cell gene-expression profiling of iN cells using Fluidigm™ dynamic RT-PCR arrays18. We analyzed 54 single HFF-iN cells 34 days after dox from two independently infected cultures (Supplementary Fig. 8). These data revealed robust co-expression of multiple pan-neuronal and synaptic markers in 50/54 HFF-iN cells (β-III-tubulin, DCX, MAP2, NCAM, synapsin). Over half (29/54) of the iN cells analyzed expressed mRNAs typical for glutamatergic neurons, such as vGLUT1, vGLUT2 or both (Fig. 2i). Only 2 iN cells expressed GAD67 in the absence of vGLUT1 or 2, and no iN cell expressed the inhibitory marker gene vGAT. Five cells expressed the catecholaminergic neuron marker tyrosine hydroxylase. Immunofluorescence analysis revealed that 5 weeks after infection 17±8% of iN cells expressed the forebrain marker Tbr1, 21±9 % expressed the marker of peripheral neurons Peripherin, while En1, a marker of midbrain neurons, serotonin and Cholinacetyltransferase were not detectable (Supplementary Fig. 9).
To assess whether the iN cell state was stable without continued transgene expression, we monitored mRNA expression levels of endogenous and exogenous BAMN genes before and after dox induction and after dox withdrawal. While the exogenous transgenes were clearly dox dependent, the 4 corresponding endogenous genes were rapidly induced and exhibited increasing expression levels over time even after dox withdrawal (Supplementary Fig. 10). Similarly, HFF-iN cells could be maintained in the absence of dox for 3 weeks (Supplementary Fig. 11).
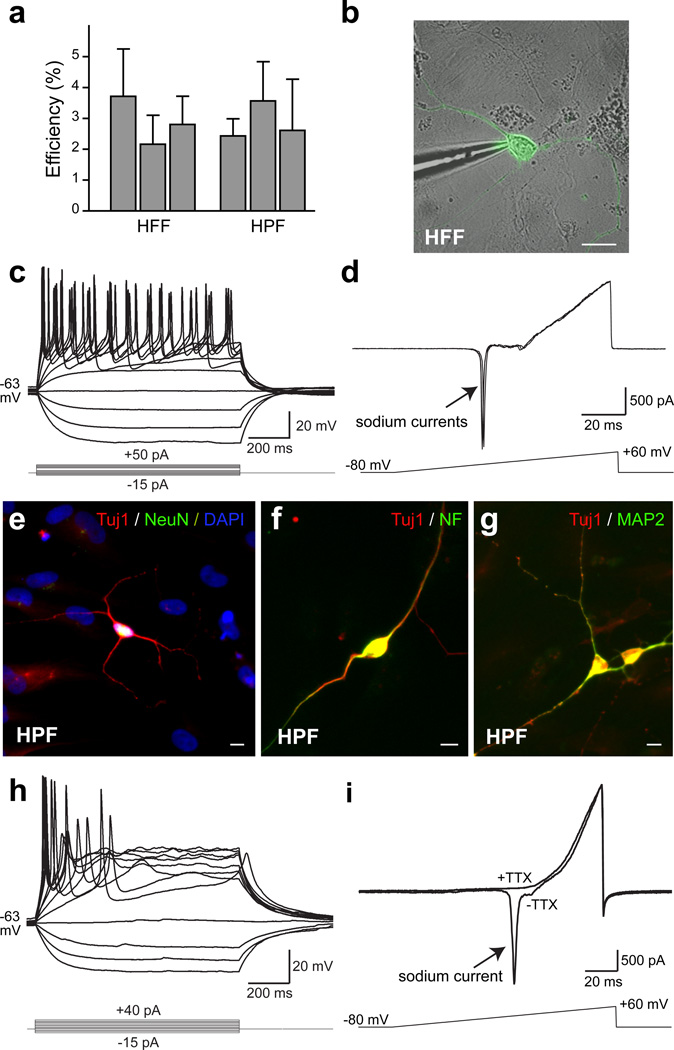
We next asked whether iN cells generated from HFFs exhibited active membrane properties. iN cells were identified by EGFP fluorescence (Fig. 3b) and whole-cell recordings were performed 14–35 days after dox. 14–25 days after addition of dox the average resting membrane potential of HFF-iN cells was −52.2 ± 2.2 mV (mean±SEM, n=41). When HFF-iN cells were step depolarized, action potentials could be detected in many iN cells at 14–25 days, and in all recorded iN cells at days 34–35 (Fig. 3c, Supplementary Figs. 7 & 12 and Table 1). Fast-activating and inactivating inward Na+ currents as well as outward K+ currents were also observed (Fig. 3d).
Figure 3. Membrane properties of fibroblast iN cells.
a, Quantification of Tuj1-positive neuronal cells from HFFs (line HFF-A) 3 weeks after dox or HPFs (line HPF-B) 4 weeks after dox. N=3 independent experiments. b, Patch clamp recording was conducted on HFF-iN cells identified by EGFP fluorescence and DIC microcopy. c, Representative traces of membrane potentials in response to step current injections (lower panel) from an HFF-iN cell 19 days after dox. Membrane potential was maintained at ~ −63 mV. d, Representative traces of membrane currents recorded with a ramp protocol (lower panel). Fast activating and inactivating Na+ currents were prominent. Three traces are shown superimposed. e–g, HPF-iN cells express Tuj1 (red) and NeuN (green) (e), Neurofilament (green) (f) and MAP2 (green) (g). h, Representative traces of membrane potentials in response to step current injections in HPF-iN cells. Action potentials were generated in cultures without glia. i, Representative traces of membrane currents recorded following a ramp protocol (lower panel) in HPF-iN cells. The Na+ currents could be blocked by TTX. Data in (a) are presented as mean±SD. Scale bars: 10 µm (a, e–g).
To determine whether the BAMN factors were also capable of converting more mature human fibroblasts into iN cells, we derived primary human postnatal fibroblasts (HPFs) from 3 different perinatal foreskin resections. In all three HPF lines, expression of BAMN factors reproducibly generated neuron-like cells with co-expression of multiple pan-neuronal markers (Fig. 3e–g). Intriguingly, the efficiencies of iN cell generation from fetal and postnatal fibroblasts were similar (2–4% of cells plated; Fig. 3a). Single-cell gene-expression profiling of iN cells revealed that 46/51 HPF iN cells co-expressed pan-neuronal and synaptic markers 42 days after infection; the majority of the HPF iN cells (37/51) appear to be glutamatergic neurons. Immunofluorescence analysis revealed that 6 weeks after infection, 81±17% of iN cells expressed Tbr1 above the levels of fibroblasts, 15.2±6.6% were peripherin-positive (Supplementary Fig. 13). Unlike fetal fibroblasts, most postnatal fibroblasts showed weak but specific Tbr1 staining (Supplementary Fig. 13a–d). Electrophysiological recordings demonstrated the presence of regenerative action potentials as well as voltage-dependent channel activities in the majority of cells analyzed from 2 lines (e.g. 17/18 cells from line HPF-B, Fig. 3h–i, Supplementary Table 1). Furthermore, iN cells exhibiting active membrane properties could be generated from dermal fibroblasts derived from an 11-year-old human subject (Supplementary Fig. 14).
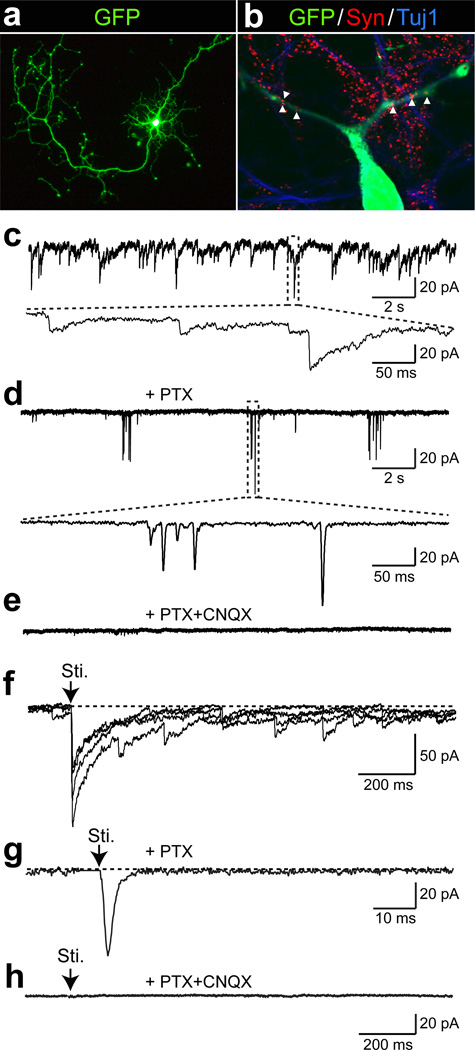
Finally, we determined whether human iN cells can express functional neurotransmitter receptors and form functional synapses. Application of either GABA or l-glutamate to HFF-iN cells induced current responses that could be blocked by picrotoxin and CNQX, respectively (Supplementary Fig. 12e, f). We then dissociated HFFs 4–7 days after infection with the BAMN factors and EGFP and plated them onto previously established mouse cortical neuronal cultures. These co-cultures were maintained for up to 5 weeks thereafter. HFF-iN cells were identified by the EGFP expression (Fig. 4a). Whole-cell recordings after 2 to 3 weeks of co-culture showed no synaptic activity (n=20) but after 4–5 weeks approximately half of human iN cells recorded exhibited spontaneous postsynaptic currents (PSCs) with variable kinetics (n=21, Fig. 4c, Supplementary Table 1). Immunostaining with synapsin antibodies confirmed the presence of scattered synaptic puncta on the dendrites of EGFP-positive cells (Fig. 4b). When the GABAA receptor inhibitor picrotoxin was applied, the majority of the spontaneous PSCs were blocked, demonstrating that they were inhibitory (IPSCs) (Fig. 4d). In the presence of picrotoxin, bursting of spontaneous excitatory postsynaptic currents (EPSCs) was revealed and could be blocked by the AMPA receptor blocker CNQX (Fig. 4e). Focal stimulation evoked both IPSCs and EPSCs that could be blocked by picrotoxin and CNQX (Fig. 4f–h). Importantly, PSCs could also be recorded from HPF-iN cells co-cultured with mouse cortical neurons 4 weeks after infection (Supplementary Fig. 15). These results demonstrate that fetal and postnatal fibroblast-derived iN cells could form functional synapses and integrated into pre-existing neuronal networks.
Figure 4. Synaptic responses of HFF-iN cells.
a, An HFF-iN cell expressing EGFP co-cultured with mouse cortical neurons at day 35 after dox. b, Synapsin positive puncta co-localize with neurites extending from HFF-iN cells (arrow heads). c, Thirty-five days after dox, spontaneous PSCs were recorded in HFF-iN cells. d, The slow responses could be blocked by PTX. Bursting events of EPSCs were recorded in the presence of PTX. The insert shows the fast kinetics of the responses. e, In the presence of PTX and CNQX (50 µM), no spontaneous activities were observed. f, Evoked postsynaptic responses. Four traces were super imposed. Sti. = stimulation. g, In the presence of PTX, electric stimulation evoked fast-kinetic excitatory PSCs (EPSCs). h, No evoked synaptic responses were observed in the presence of PTX and CNQX. Scale bars: 100 µm (a); 10µm (b).
In this report, we have identified a combination of transcription factors that are capable of directly converting human fibroblasts into neurons. Like mouse iN cells13, ES cell19–21 - and iPS cell-derived neurons22,23, the human iN cells appear relatively immature, as indicated by their slightly depolarized membrane potentials and the relatively low-amplitude synaptic responses. Compared to mouse iN cells, human iN cells required longer culture periods to develop synaptic activity. Future studies will be necessary to thoroughly optimize conditions for human iN cell generation and maturation, which would facilitate applications of this method for the study of human neuronal development and disease.
Methods Summary
Cell culture
H9 human ES cells (WiCell Research Resources) and iPS cells were expanded in mTeSR1 (Stem Cell Technologies) and passaged as clumps or single cells24. Primary human fetal fibroblasts (HFFs) were isolated from the distal half of the limbs of GW8-10 fetuses obtained from Advanced Bioscience Resources Inc. Primary human postnatal fibroblasts (HPFs) were established from foreskin. Primary mouse cortical cultures and glial monolayer cultures were established as described previously13.
Lentiviral Infections
Lentiviral production and fibroblast infections were performed as described previously13. Primary fibroblasts and pluripotent stem cells were infected with concentrated lentivirus and treated with doxycycline (dox, 2 µg/µL) 16–24 hours later.
Electrophysiology and expression analysis
Cells were analyzed by immunofluorescence and electrophysiology as described elsewhere13,25. Single-cell gene expression profiling was performed utilizing the Fluidigm Biomark dynamic array18,26 according to the manufacturer’s instructions.
Methods
Cell culture
H9 human ES cells (WiCell Research Resources) were expanded in mTeSR1 (Stem Cell Technologies). The generation and characterization of the iPS cells used in this study will be published elsewhere (Sebastiano & Wernig). The day before infection, cells were treated with accutase and seeded as single cells in 3.5 cm tissue culture dishes on matrigel in mTeSR1 containing 2 µM Thiazovivin (Bio Vision)24. To inhibit the growth of uninfected ES cells and select for post-mitotic neurons, we added 4 µM Cytosine β-d-arabinofuranoside (Ara-C) to the media 48 hours after dox addition. Primary human fetal fibroblasts were isolated from the distal half of the limbs of 8–10 week old fetuses (Advanced Bioscience Resources Inc.). The tissue was dissociated after trypsin digestion and plated in MEF media (DMEM high glucose, calf serum, sodium pyruvate, non-essential amino acids, penicillin/streptomycin and β-mercaptoethanol). Primary human postnatal fibroblasts (HPFs) were established from dissociated foreskin tissue derived from 1–3 day-old newborns. Before being used for experiments, primary fibroblast cells were passaged for at least 3 times. Primary mouse cortical cultures and glial monolayer cultures were established as previously described13. To maintain the iN cell culture, cells were either cultured in N3 media media (DMEM/F2 (Invitrogen), apotransferrin (100 µg/ml), insulin (5 µg/ml), sodium selenite (30 nM), progesterone (20 nM), putrescine (100 nM), penicillin/streptomycin) supplemented with neurotrophic factors including brain derived neurotrophic factor, glial cell-derived neurotrophic factor, neurotrophin-3 and ciliary neurotrophic factor (R&D systems) or dissociated using papain and replated onto previously established monolayer culture of primary mouse glia or primary neurons from mouse cortex in neuronal growth media (MEM (Gibco) supplemented with B27 (Gibco), glucose (5mg/ml), transferring (10 µg/ml), 5% fetal bovine serum and Ara-C (2µM, Sigma)13,25.
Virus infections
Lentiviral production and fibroblast infections were performed as described previously13. Briefly, primary human fetal or postnatal fibroblasts were plated and infected with concentrated lentiviral particles and polybrene (8 µg/µL) in fresh MEF media. Viral media was removed after 16–24 hours and replaced with MEF media containing doxycycline (dox, 2 µg/µL). After 24–48 hours, media was changed to N3 media containing dox (2µg/µL). For human ES cell infections, H9 human embryonic stem cells were switched into N3 media containing polybrene (2 µg/µL) 24 hours after re-plating, and concentrated lentiviral particles were added. After 16–24 hours, cultures were switched into N3 media containing dox (2 µg/µL) and changed daily before dissociation. Forty-eight hours after the initial addition of dox, Ara-C (4 µg/µL) was added to the media to inhibit proliferation of uninfected ES cells until analysis 6 days after the addition of dox. All chemicals were purchased from Sigma-Aldrich™ if not otherwise specified.
Single cell gene expression analysis (FluidigmTM dynamic array)
Single-cell gene expression profiling was performed utilizing the Fluidigm Biomark dynamic array according to the manufacturer’s protocol18,26. Briefly, single cells growing on culture dishes after 5 or 6 weeks of transduction were collected by aspiration into patch electrodes and ejected into 2× cells direct buffer (Invitrogen), flash frozen and kept at −80°C until further processing. Thawed cells were subject to target-specific reverse-transcription and 18 cycles of PCR pre-amplification with a mix of primers specific to the target genes (STA). STA products were then processed for real-time PCR analysis on Biomark 48:48 Dynamic Array integrated fluidic circuits (Fluidigm). To ensure the specificity of the amplification, titrations of total human brain RNA were included in each experiment, and only primers that demonstrated a linear amplification were analyzed. Furthermore, melting curves of the PCR products were compared between the single cells and the control RNA to ensure the specificity of the PCR products.
Electrophysiology
Action potentials were recorded with current-clamp whole-cell configuration. The pipette solution for current-clamp experiments contained (in mM): 123 K-gluconate, 10 KCl, 1 MgCl2, 10 HEPES, 1 EGTA, 0.1 CaCl2, 1 K2ATP, 0.2 Na4GTP and 4 glucose, pH adjusted to 7.2 with KOH. Membrane potentials were kept around −65 to −70 mV, and step currents were injected to elicit action potentials. For whole-cell voltage dependent current recordings, the same internal solution as aforementioned was used. For synaptic functional evaluation, the internal solution contained (in mM): CsCl 135, HEPES 10, EGTA 1, Mg-ATP 4, Na4GTP 0.4, and QX-314 10, pH7.4. The bath solution contained (in mM): NaCl 140, KCl 5, CaCl2 2, MgCl2 2, HEPES 10, and glucose 10, pH7.4. Synaptic responses were measured as described previously13,25. Stimulus artifacts for evoked synaptic responses were removed for graphic representation. Electrophysiological data are presented as mean ± SEM. All statistical comparisons were made using Student’s t-test.
Immunofluorescence and RT-PCR
For immunofluorescence experiments, cells were fixed in 4% paraformaldehyde in PBS for 10 min at room temperature. Following fixation, cells were incubated in 0.2% Triton X-100 in PBS for 5 min at room temperature. After washing twice with PBS, cells were blocked in a solution of PBS containing 4% BSA and 1% cosmic calf serum (CCS) for 30 minutes at room temperature. Primary and secondary antibodies were applied for 1 hour and 30 min, respectively. The following antibodies were used for our analysis: rabbit anti-Tuj1 (Covance, 1:1000), mouse anti-Tuj1 (Covance, 1:1000), mouse anti-MAP2 (Sigma, 1:500), mouse anti-NeuN (Millipore, 1:200), mouse anti-neurofilament (Developmental Studies Hybridoma Bank (DSHB), 2H3a, 1:1000), rabbit anti-synapsin (E028, 1:1000), anti-synaptotagmin (Synaptic systems, 41.1, 1:2000), guinea pig anti-vGLUT1 (Millipore, 1:2000), mouse anti-GAD6 (DSHB, 1:500), rabbit anti-Tbr1 (Abcam, 1:200), mouse anti-Peripherin (Sigma, 1:100), sheep anti-Tyrosine Hydroxylase (Pel-Freez, 1:500), rabbit anti-GFAP (DAKO, 1:4000), mouse anti-Sox2 (R&D Systems, 1:50), goat anti-Brn2 (clone C-20, Santa Cruz Biotechnology, 1:100), rabbit anti-Ascl1 (Abcam, 1:200), mouse anti-BrdU (Becton-Dickinson, 1:50), mouse anti-LU5 (Abcam, 1:200), goat anti-Sox10 (Santa Cruz Biotechnology, 1:40). FITC-, and Cy3-conjugated secondary antibodies were obtained from Jackson Immunoresearch. Alexa-488-, Alexa-546- and Alexa-633-conjugated secondary antibodies were obtained from Invitrogen. DAPI (Sigma, 1:10,000). For RT-PCR analysis, RNA was isolated using RNAqueous® Kit (Applied Biosystems) following the manufacturer’s instructions, treated with DNAse (Applied Biosystems) and reverse-transcribed with Superscript III (Invitrogen).
Efficiency calculation
The following method was used to calculate the efficiency of neuronal induction. The total number of Tuj1 positive cells with a neuronal morphology, defined as cells having a circular, three-dimensional appearance that extend a thin process at least three times longer than their cell body, were quantified at indicated time points. We determined this number in at least 15 randomly selected 20× visual fields on a Zeiss Axio Observer microscope. The average and standard deviation per field was determined and then used to extrapolate the total number of iN cells present in the entire dish based the known surface areas of a 20× visual field and the respective culture dish. We then divided this number by the number of cells plated before infection to get the percentage of the starting population of cells that adopted neuron-like characteristics. In multiple independent experiments we verified that this extrapolation method yields cell numbers very similar to those measured by a hemocytometer at the time of plating. Data are presented as Mean ± SD.
Supplementary Material
Acknowledgements
We would like to thank Yuko Kokubu for technical assistance in molecular cloning and Yingsha Zhang for assistance in iPS cell induced neuron culture. We also thank Dr. Yi Sun of UCLA for providing the microRNAs expression lentiviral vectors and Dr. Sadhan Majumder of UT MD Anderson Cancer Center for REST-VP16 construct. This work was enabled by start-up funds from the Institute for Stem Cell Biology and Regenerative Medicine at Stanford (M.W.), the Ellison Medical Foundation (M.W.), the Stinehard-Reed Foundation (M.W.), the Donald E. and Delia B. Baxter Foundation (M.W.), the NIH grants 1R01MH092931 (M.W. and T.C.S.) and RC4 NS073015 (M.W.). Z.P.P. is supported by 2008 and 2010 NARSAD Young Investigator Awards. T.V. is supported by the Ruth and Robert Halperin Stanford Graduate Fellowship. A.C. is supported by the AXA research fund and D.R.F. is supported by BioX Undergraduate Fellowship.
Footnotes
Author Contributions
Z.P.P., N.Y., T.V., A.O., T.C.S. and M.W. designed the experiments, analysed the data. D.R.F. and T.Q.Y helped with lentiviral production. A.C., V.S. and S.M. helped to provide experimental material and helped the analyses. Z.P.P., N.Y., T.V., T.C.S. and M.W. wrote the paper.
References
- 1.Blau HM, et al. Plasticity of the differentiated state. Science. 1985;230(4727):758–766. doi: 10.1126/science.2414846. [DOI] [PubMed] [Google Scholar]
- 2.Gurdon JB. From nuclear transfer to nuclear reprogramming: the reversal of cell differentiation. Annu Rev Cell Dev Biol. 2006;22:1–22. doi: 10.1146/annurev.cellbio.22.090805.140144. [DOI] [PubMed] [Google Scholar]
- 3.Heins N, et al. Glial cells generate neurons: the role of the transcription factor Pax6. Nat Neurosci. 2002;5(4):308–315. doi: 10.1038/nn828. [DOI] [PubMed] [Google Scholar]
- 4.Ieda M, et al. Direct reprogramming of fibroblasts into functional cardiomyocytes by defined factors. Cell. 2010;142(3):375–386. doi: 10.1016/j.cell.2010.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Shen CN, Slack JM, Tosh D. Molecular basis of transdifferentiation of pancreas to liver. Nat Cell Biol. 2000;2(12):879–887. doi: 10.1038/35046522. [DOI] [PubMed] [Google Scholar]
- 6.Tada M, Takahama Y, Abe K, Nakatsuji N, Tada T. Nuclear reprogramming of somatic cells by in vitro hybridization with ES cells. Curr Biol. 2001;11(19):1553–1558. doi: 10.1016/s0960-9822(01)00459-6. [DOI] [PubMed] [Google Scholar]
- 7.Takahashi K, et al. Induction of pluripotent stem cells from adult human fibroblasts by defined factors. Cell. 2007;131(5):861–872. doi: 10.1016/j.cell.2007.11.019. [DOI] [PubMed] [Google Scholar]
- 8.Wilmut I, Schnieke AE, McWhir J, Kind AJ, Campbell KH. Viable offspring derived from fetal and adult mammalian cells. Nature. 1997;385(6619):810–813. doi: 10.1038/385810a0. [DOI] [PubMed] [Google Scholar]
- 9.Xie H, Ye M, Feng R, Graf T. Stepwise reprogramming of B cells into macrophages. Cell. 2004;117(5):663–676. doi: 10.1016/s0092-8674(04)00419-2. [DOI] [PubMed] [Google Scholar]
- 10.Zhou Q, Brown J, Kanarek A, Rajagopal J, Melton DA. In vivo reprogramming of adult pancreatic exocrine cells to beta-cells. Nature. 2008;455(7213):627–632. doi: 10.1038/nature07314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Graf T, Enver T. Forcing cells to change lineages. Nature. 2009;462(7273):587–594. doi: 10.1038/nature08533. [DOI] [PubMed] [Google Scholar]
- 12.Zhou Q, Melton DA. Extreme makeover: converting one cell into another. Cell Stem Cell. 2008;3(4):382–388. doi: 10.1016/j.stem.2008.09.015. [DOI] [PubMed] [Google Scholar]
- 13.Vierbuchen T, et al. Direct conversion of fibroblasts to functional neurons by defined factors. Nature. 2010;463(7284):1035–1041. doi: 10.1038/nature08797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hansen DV, Lui JH, Parker PR, Kriegstein AR. Neurogenic radial glia in the outer subventricular zone of human neocortex. Nature. 2010;464(7288):554–561. doi: 10.1038/nature08845. [DOI] [PubMed] [Google Scholar]
- 15.Kriegstein A, Noctor S, Martinez-Cerdeno V. Patterns of neural stem and progenitor cell division may underlie evolutionary cortical expansion. Nat Rev Neurosci. 2006;7(11):883–890. doi: 10.1038/nrn2008. [DOI] [PubMed] [Google Scholar]
- 16.Zhang X, et al. Pax6 is a human neuroectoderm cell fate determinant. Cell Stem Cell. 2010;7(1):90–100. doi: 10.1016/j.stem.2010.04.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bottenstein JE, Sato GH. Growth of a rat neuroblastoma cell line in serum-free supplemented medium. Proc Natl Acad Sci U S A. 1979;76(1):514–517. doi: 10.1073/pnas.76.1.514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Guo G, et al. Resolution of cell fate decisions revealed by single-cell gene expression analysis from zygote to blastocyst. Dev Cell. 2010;18(4):675–685. doi: 10.1016/j.devcel.2010.02.012. [DOI] [PubMed] [Google Scholar]
- 19.Johnson MA, Weick JP, Pearce RA, Zhang SC. Functional neural development from human embryonic stem cells: accelerated synaptic activity via astrocyte coculture. J Neurosci. 2007;27(12):3069–3077. doi: 10.1523/JNEUROSCI.4562-06.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wu H, et al. Integrative genomic and functional analyses reveal neuronal subtype differentiation bias in human embryonic stem cell lines. Proc Natl Acad Sci U S A. 2007;104(34):13821–13826. doi: 10.1073/pnas.0706199104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Koch P, Opitz T, Steinbeck JA, Ladewig J, Brustle O. A rosette-type, self-renewing human ES cell-derived neural stem cell with potential for in vitro instruction and synaptic integration. Proc Natl Acad Sci U S A. 2009;106(9):3225–3230. doi: 10.1073/pnas.0808387106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Marchetto MC, et al. A model for neural development and treatment of Rett syndrome using human induced pluripotent stem cells. Cell. 2010;143(4):527–539. doi: 10.1016/j.cell.2010.10.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hu BY, et al. Neural differentiation of human induced pluripotent stem cells follows developmental principles but with variable potency. Proc Natl Acad Sci U S A. 2010;107(9):4335–4340. doi: 10.1073/pnas.0910012107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Xu Y, et al. Revealing a core signaling regulatory mechanism for pluripotent stem cell survival and self-renewal by small molecules. Proc Natl Acad Sci U S A. 2010;107(18):8129–8134. doi: 10.1073/pnas.1002024107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Maximov A, Pang ZP, Tervo DG, Sudhof TC. Monitoring synaptic transmission in primary neuronal cultures using local extracellular stimulation. J Neurosci Methods. 2007;161(1):75–87. doi: 10.1016/j.jneumeth.2006.10.009. [DOI] [PubMed] [Google Scholar]
- 26.Wong CC, et al. Non-invasive imaging of human embryos before embryonic genome activation predicts development to the blastocyst stage. Nat Biotechnol. 2010;28(10):1115–1121. doi: 10.1038/nbt.1686. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.