Abstract
Selenium (Se), in the form of selenoproteins, imparts many health benefits with antiinflammatory properties. Previous studies have shown that Se supplementation of macrophages negatively regulates the LPS-dependent production of inducible NO synthase (iNOS), a proinflammatory gene. Therefore, we hypothesized that l-arginine, a substrate for iNOS, is acted upon by arginase-I (Arg-I), contributing to the resolution of inflammation. We investigated the antiinflammatory activity of Se using LPS and IL-4–treated C57BL/6 murine bone marrow-derived macrophages (BMDM) from mice fed Se-deficient and Se-adequate diets. Supplementation with Se (100 nmol/L) of IL-4–treated macrophages significantly increased the expression of alternatively activated macrophage (M2) markers, Arg-I, Fizz1, and Mrc-1. Se treatment also increased the enzymatic activity of Arg-I and surface expression of Mrc-1. Conversely, expression of classically activated macrophage (M1) markers, TNFα, and IL-1β, was significantly decreased in LPS-treated macrophages that were cultured in Se and IL-4, suggesting a synergistic effect between Se and IL-4. Additionally, Arg-I activity was decreased in BMDM harvested from glutathione peroxidase (GPX) knockout mice compared to GPX wild-type mice, further establishing an important role for selenoproteins. Furthermore, BMDM treated with inhibitors of PPARγ and STAT6, pivotal transcription factors that mediate the activity of Se and IL-4, respectively, showed complete ablation of Se-dependent expression of M2 markers. In summary, these studies suggest that Se supplementation of macrophages produces endogenous activators to mediate the PPARγ-dependent switch from M1 to M2 phenotype in the presence of IL-4, possibly affecting pathways of wound healing and inflammation resolution.
Introduction
Macrophages are key components of the innate immune system that play a pivotal role in pathogen clearance and resolution of inflammation (1). It is well established that macrophages are activated along 2 distinct pathways: the classical (M1) pathway and the alternative (M2) pathway (2). Known to initiate an inflammatory response, M1 macrophages possess an enhanced phagocytic and antimicrobial phenotype (3–5). Moreover, M1 macrophages become primed to release increased amounts of reactive oxygen nitrogen species. NO, a known reactive oxygen nitrogen species, is produced by the enzyme iNOS6 from l-arginine (l-Arg), an available substrate found in macrophage cells (6).
Alternatively activated macrophages are stimulated by the T helper 2 cytokines, IL-13 and IL-4 (2, 7–9). M2 macrophages function in resolving inflammation while promoting cell proliferation and wound healing (3, 5). The stimulation of M2 macrophages by IL-4 leads to the production of arginase-I (Arg-I) (3, 6). Arg-I acts on l-Arg, the same substrate that is acted upon by iNOS, to produce l-ornithine (l-Orn) and urea, precursors of polyamines and collagen (5, 10). This competition for substrate acts as a way to control the production of NO (10, 11). In addition to Arg-I, IL-4 stimulates the expression of other M2 markers, such as Fizz-1 (found in inflammatory zone-1), and Mrc-1 (mannose receptor 1) (9, 12, 13), while also playing a role in the activation of STAT6 and PPARγ (4, 5, 7). PPARγ, peroxisome proliferator-activated receptor, a fatty acid sensor that plays a critical role in atherosclerosis and glucose metabolism (14), can be activated by both exogenous and endogenous ligands (14). Cyclopentenone prostaglandins (e.g. 15d-PGJ2) are cyclooxygenase- and H-PGDS–catalyzed products of arachidonic acid that are known to function as endogenous ligands of PPARγ (14, 15). In addition, the transcription factor STAT6 has been implicated as an integral participant of many cellular functions, including Arg-I expression (5, 16). IL-4 causes dimerization of STAT6 followed by translocation into the nucleus to modulate the expression of antiinflammatory markers by upregulating the expression of PGC-1β, a PPARγ coactivator (5, 16). Thus, in the presence of IL-4 and 15d-PGJ2, both the pathways synergize to drive the expression of M2 pathway markers. Previous studies in our laboratory demonstrated enhanced activation of PPARγ via the increased production of 15d-PGJ2 in macrophages that were supplemented with selenium (Se).
Se, an essential micronutrient, has both antiinflammatory and cancer chemopreventative properties (17–20) and is found in the body in the form of selenoproteins; proteins that contain an Sec group covalently bound to Se (17, 19, 21). Sec, also known as the 21st amino acid, is coded for by the UGA codon and is recognized by a specific tRNA, Sec tRNA (17, 20). Among the identified selenoproteins, GPX and thioredoxin reductase are the 2 most well characterized and abundantly expressed (15, 17). Recent research has shown an inverse causal relationship between Se deficiency and many diseases and disorders (17, 21). The Se found in foods is in its organic form, with the most common form being Se-Met (22). A commonly used inorganic form is sodium selenite (22). Both forms of Se are metabolized via hydrogen selenide (H2Se) for incorporation into selenoproteins (22). Due to the lack of lyase expression in cell culture models, Se in most organic forms is not readily bioavailable. To circumvent this major metabolic limitation, MSA, a readily available organo-Se source, has been used in most cancer prevention studies (23).
Previously, we demonstrated the ability of Se to downregulate the LPS-induced expression of iNOS (24) and other proinflammatory genes (15) in macrophages. Here, we demonstrate the ability of Se, in the form of selenoproteins, to effectively switch macrophage activation from M1 toward a M2 phenotype, upregulating the expression of M2 markers while decreasing the expression of M1 markers. Using murine macrophages, we examined the effect of Se on Arg-I at the transcriptional, protein expression, and enzymatic activity levels, as well as other M2 markers, following stimulation with LPS or IL-4. We describe specific pathways that are important in mediating the effects of Se. In conclusion, our data show that optimal Se status is critical for alternative macrophage activation, leading to attenuated expression of proinflammatory mediators.
Materials and Methods
Cell culture.
BMDM were prepared from 3-mo-old C57Bl/6 mice maintained on Se-deficient (SE-D; 0.01 mg/kg), Se-adequate (Se-A; 0.1 mg/kg), and Se-supplemented (Se-S; 0.4 mg/kg) diets (Harlan Teklab custom diets). All mice, purchased from Charles River Laboratories, were 3 wk old upon arrival to the animal facilities. In accordance with Penn State University IACUC guidelines, all mice were appropriately maintained and ethically treated. Mice were killed using a CO2 chamber. BMDM cells were collected and cultured as previously described (15). Previous studies have been done to establish the Se status of each group of mice. Total blood and erythrocyte levels were used as part of a standard procedure (15). Additionally, BMDM were prepared from GPX KO mice. The specific procedure used to create the GPX KO mice has been previously described (25). All mice used to collect BMDM were age and sex matched. RAW 264.7 macrophage cells were obtained from ATCC and maintained as previously described (15). Specific concentrations of exogenous Se were as follows: 0 nmol/L in Se-D cells and 100 nmol/L in Se-A cells.
Custom diet compositions.
All custom diets were purchased from Harlan Teklab. The 3 diets, Se-D, Se-A, and Se-S were all composed of the same base materials: torula yeast, 300 g/kg; dl-methionine, 3 g/kg; sucrose, 590 g/kg; corn oil, 50 g/kg; mineral mix (Harlan Teklab product no. 80313) (26), 35 g/kg; calcium carbonate, 11 g/kg; and vitamin mix (Harlan Teklab product no. 40060) (26), 10 g/kg. The sodium selenite concentrations within each diet were as follows: Se-D diets, 0 g/kg; Se-A diets, 0.178 g/kg; and Se-S diets, 0.89 g/kg.
Treatments.
Upon reaching 80% confluency, macrophages were pretreated for 2–3 h with synthetic compounds in various combinations: rosiglitazone (PPARγ agonist; 2 μmol/L), GW9662 (PPARγ antagonist; 1 μmol/L), leflunomide (STAT6 inhibitor; 100 μmol/L), and HQL-79 (H-PGDS inhibitor; 25 μmol/L). Rosiglitazone, GW9662, and leflunomide were purchased from Sigma-Aldrich and HQL-79 was purchased from Cayman Chemicals. Following pretreatment, the cells were stimulated with 5 or 10 μg/L of recombinant mouse IL-4 (R&D Systems) for 20 h or 0.1–1 mg/L LPS (Sigma-Aldrich) for 2–12 h. Rosiglitazone, GW9662, and leflunomide were dissolved in DMSO, and HQL-79 was dissolved in 0.1 mol/L citric acid. Cells were stimulated with DMSO (0.1%, Sigma-Aldrich) and citric acid (0.1 mol/L, Sigma-Aldrich) for 20–23 h as vehicle controls. In addition, organo-Se compounds were added to cells. BMDM’s and RAW 264.7 cells were also supplemented with either SeMet (100 nmol/L, Sigma-Aldrich) or MSA (100 nmol/L, Sigma-Aldrich).
Immunoblotting.
Whole cell lysates from BMDM’s and RAW 264.7 cells were prepared as previously described (15). The following primary antibodies were used to probe the membranes: purified anti-mouse Arg-I (BD Transduction Laboratories), anti-rabbit polyclonal GPX1 (Abcam), and anti-mouse monoclonal GAPDH (Fitzgerald). Near equal loading of protein was confirmed using GAPDH as the control. Chemiluminescent detection by autoradiography was used to visualize bands followed by densitometric evaluation using Image J program (NIH).
Arg assay.
Arg activity, assessed by a colorimetric assay that detects urea production generated by Arg hydrolysis of l-Arg, has been previously described (27). Se-D and Se-A BMDM and RAW264.7 macrophages assayed, were cultured as described earlier (15). OD at 560 nm was recorded on a Packard plate reader. We used a urea standard calibration curve (0–1 μmol; y = 9 × 10−5x + 0.0007; R2 = 0.99) to calculate the Arg-I activity. Enzyme activity is expressed as μmol of urea produced/mg of protein (27).
Real-time PCR.
Total RNA from BMDM and RAW 264.7 cells was extracted using Isol-RNA lysis reagent (5 Prime). RNA purity and concentrations were determined by agarose gel electrophoresis and UV spectroscopy, respectively. Precisely 1 μg total RNA was reverse transcribed into cDNA using a High Capacity cDNA Reverse Transcriptase kit per the manufacturer’s instructions (Applied Biosystems). For the analysis of M1 and M2 markers, cDNA was analyzed using TaqMan probes for Arg-I, Fizz1, Ym1, IL-12, iNOS, MSR, IL-β, TNFα, and Mrc-1. A GAPDH probe was used as an internal control to normalize the data. Amplifications were performed using PerfeCTa qPCR SuperMix Master mix (Quanta Biosciences) in a 7300 real-time PCR system (Applied Biosystems). ΔCt (Ct Gene– Ct GAPDH) was calculated for each sample and used for analysis of transcript abundance with respect to the untreated negative control as described (28).
Flow cytometry.
RAW 264.7 cells were cultured in their appropriate media and prepared for FACS analysis. Cells were washed with ice-cold 2% FBS in PBS. Samples were blocked with purified rat anti-mouse CD16/CD32 Fc Block (BD Pharmingen) for 10 min on ice. Without removing the Fc block, samples were treated with anti-mouse Mrc-1 antibody (Abcam) for 40 min on ice, followed by anti-mouse IgG1 FITC-labeled secondary antibody for 40 min on ice in the dark. Samples were centrifuged, washed twice in 2% FBS, and resuspended in 500 μL of 2% FBS in PBS. Samples were analyzed on the FC500 Benchtop Cytometer using CXP software (Beckman Coulter).
Statistical analysis.
Results are presented as mean ± SEM. Significant differences between 2 groups were analyzed by Student’s t test using GraphPad Prism. Significant differences comparing more than 2 groups were analyzed by ANOVA, with appropriate post hoc testing, using GraphPad Prism. Results were considered significant at P < 0.05. Three-way ANOVA with appropriate testing was used to analyze the interaction among diets, treatment groups, and stimulation (diet × treatment × stimulation). All experiments were performed in triplicate, where triplicate indicates BMDM came from 3 separate mice. RAW 264.7 cells were cultured in triplicate. The sum of means of treatments (Se and IL-4) was used to determine additive compared to synergistic relationship.
Results
Effect on GPX1 expression in macrophages supplemented with Se.
To determine the most effective Se concentrations to use in our experiments, we used BMDM from mice fed Se-D (0.01 mg/kg) and Se-A (0.1 mg/kg) diets and RAW264.7 macrophages cultured in media containing 0 nmol/L or 100 nmol/L Se. Using cytosolic GPX1 as a marker of Se status, we examined the expression of GPX1 in BMDM cell lysates treated with LPS or IL-4 in the presence or absence of Se. There was a clear distinction in GPX1 expression between Se-D and Se-A cell lysates, indicating an increased expression of GPX1 in BMDM from mice fed Se-A diets (Supplemental Fig. 1A). Similarly, RAW264.7 cells treated with LPS or IL-4 increased in GPX1 expression in those cultured in the presence of 100 nmol/L Se but not in those cultured in 0 nmol/L Se (Supplemental Fig. 1B). Furthermore, the presence of IL-4 or LPS did not affect the expression of GPX1 in the Se-D or Se-A BMDM or RAW264.7 cells (P > 0.05).
Se supplementation of macrophages increases Arg-I expression and activity.
Given that the BMDM responded to exogenous Se by increasing the GPX1 expression levels, we examined if supraphysiological (250 nmol/L) levels of exogenous Se would further increase the activity of Arg-I. Arg-I activity in BMDM from Se-A mice treated with IL-4 (20 h) was greater than the activity in BMDM from Se-D mice (P < 0.05) (Fig. 1A). However, there was no significant difference between BMDM from Se-A mice and Se-S mice (data not shown). Thus, given that supraphysiological levels of Se do not necessarily increase the Arg-I activity, we examined the modulation of Arg-I within mice fed Se-D and Se-A diets. Arg-I activity in IL-4–stimulated BMDM was greater than LPS-stimulated and unstimulated BMDM from Se-A mice (Fig. 1B). IL-4–stimulated BMDM increased in activity in Se-A–fed compared to Se-D–fed mice (P < 0.001) (Fig. 1B). Similar results were obtained in experiments performed with Se-D (0 nmol/L Se) and Se-A (100 nmol/L Se) RAW 264.7 macrophages (P < 0.001) (Fig. 1C). Furthermore, RT-PCR analysis of Arg-I in BMDM indicated a similar pattern; Arg-I mRNA expression in BMDM treated with IL-4 from Se-A mice was significantly greater than in Se-D BMDM with IL-4 treatment (Fig. 1D). Western-blot analysis of BMDM extracts showed a greater expression of Arg-I in IL-4–stimulated BMDM from Se-A mice compared to BMDM from Se-D mice (Supplemental Fig. 2, compare lanes 3 and 6). Surprisingly, Arg-I protein expression was greater in Se-A BMDM than in Se-D BMDM prior to treatment with LPS or IL-4 (Supplemental Fig. 2, compare lanes 1 and 4) (P < 0.05). Taken together, these data clearly indicate that Se status plays an important role in the expression of Arg-I in macrophages.
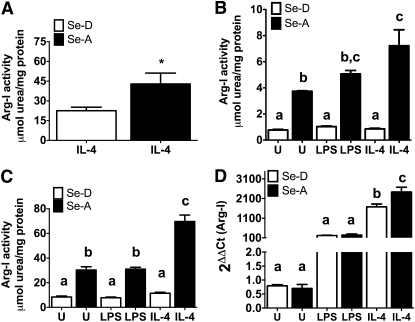
FIGURE 1.
Effect of Se on the expression and activity of Arg-I in bone marrow-derived macrophages (BMDM) and RAW264.7 macrophages. (A) Arg-1 activity measured in BMDM from Se-deficient diet (Se-D) and Se-adequate diet (Se-A) fed mice, stimulated with 5 mg/L IL-4. (B) BMDM from Se-D and Se-A mice and (C) RAW 264.7 cells, stimulated with IL-4 (10 mg/L, 20 h) and LPS (1 mg/L, 12 h). (D) Arg-I mRNA expression determined in BMDM from Se-D and Se-A mice by real-time RT-PCR. Values are means ± SEM, n = 3. (A) *Different from Se-D, P < 0.05. (B–D) Means without a common letter differ, P < 0.01.
Se supplementation of macrophages leads to differential modulation of M1 and M2 markers.
Having established that Se supplementation significantly increases Arg-I expression, we examined the modulation of 3 other M2 macrophage markers: Mrc-1, Fizz1, and Ym-1 (29, 30). RAW 264.7 macrophages and BMDM were treated with IL-4 (20 h) and LPS (4 h). IL-4 treatment only, but not LPS, increased the expression of Mrc-1 in Se-A RAW 264.7 cells compared to Se-D cells (P < 0.001) (Fig. 2A). Expression of Fizz-1 and Ym-1 in Se-A BMDM treated with IL-4 was greater than that in IL-4–treated Se-D BMDM (P < 0.01) (Fig. 2B,C). Furthermore, flow cytometric analysis of IL-4–treated RAW 264.7 macrophages had a greater surface expression of Mrc-1 in Se-A cells than in Se-D cells (P < 0.05) (Fig. 2D). These data complement the real-time PCR results described above.
FIGURE 2.
Modulation of expression of M1 and M2 macrophage markers by Se in RAW264.7 macrophage cells and bone marrow-derived macrophages. (A) RAW 264.7 macrophages and (B–I) Bone marrow-derived macrophages were cultured with (Se-A) and without (Se-D) Se and stimulated with IL-4 (5 mg/L, 20 h) or LPS (0.1 mg/L, 4 h). Real-time RT-PCR expression of Mrc-1 (A), Fizz1 (B), Ym1 (C), TNFα (E), IL-1β (F), MSR (G), IL-12 (H), and iNOS (I). (D) RAW 264.7 macrophage cells were cultured with and without Se, followed by stimulation with IL-4 (5 mg/L, 20 h) or LPS (0.1 mg/L, 4 h). Flow cytometry was used to analyze the expression of Mrc-1. Values are means ± SEM, n = 3. Within each graph, means without a common letter differ, P < 0.01.
Given that Se status of macrophages determined whether M1 or M2 pathway was activated, we also examined Se’s modulation of an array of M1 markers; TNFα, IL-1β, IL-12, iNOS, and MSR. Stimulation of Se-A BMDM with LPS alone showed significantly decreased marker expression compared to their Se-D counterparts ( ). Treatment with IL-4 did not induce the expression of the M1 markers compared to their Se-D counterparts (P > 0.05). Upon further examination, Mrc-1 and Ym-1 showed a >2-fold change in transcript expression in Se-A cells treated with IL-4 compared to Se-D cells similarly treated (Fig. 2A,C). IL-12 showed similar changes in Se-A BMDM treated with LPS compared to Se-D BMDM (Fig. 2G). Conversely, Fizz-1, TNFα, IL-1β, iNOS, and MSR showed additive changes in transcript expression in Se-A cells, treated with either IL-4 or LPS, compared to their Se-D counterparts (Fig. 2B,E,F,H,I). These results indicate that Se synergizes with IL-4 to upregulate the expression of M2 markers, while downregulating the expression of LPS induced M1 markers, facilitating the switch toward an alternative pathway of macrophage activation.
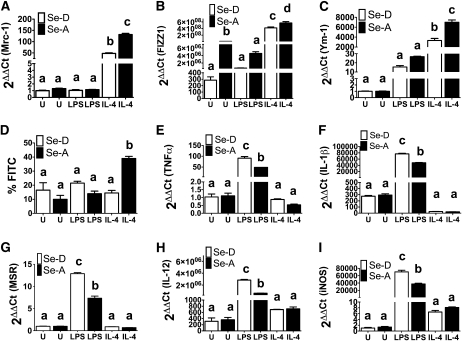
Selenoproteins are required for Arg-I expression.
Because Se is incorporated into proteins via a cotranslational mechanism as Sec, we examined if Se in the form of selenoproteins were required for the expression of Arg-I. RAW 264.7 macrophage cells were supplemented with 2 different organic forms of Se: Se-Met and MSA. The difference between Se-Met and MSA is that MSA can form selenoproteins, whereas Se-Met is unable to release Se without γ-lyase, particularly in macrophages. Much like sodium selenite, MSA also increased Arg-I activity compared to Se-D cells (P < 0.001) (Fig. 3A). Interestingly, RAW 264.7 macrophages supplemented with Se-Met did not show any increase in Arg-I activity compared to Se- and MSA-supplemented RAW 264.7 cells (P > 0.05) (Fig. 3A). Furthermore, IL-4 treatment did not increase the Arg-I activity in Se-Met–supplemented RAW cells (P > 0.05) (Fig. 3A). Western-blot analysis of GPX1 mimicked these results, exhibiting a clear expression in MSA and sodium selenite-supplemented cells, whereas Se-Met cells showed no GPX1 expression (Supplemental Fig. 3).
FIGURE 3.
Se in the form of selenoproteins is essential for Arg-I expression in RAW 264.7 macrophage cells and bone marrow-derived macrophage cells. (A) RAW 264.7 cells were cultured with 100 nmol/L methylseleninic acid (MSA) or 100 nmol/L selenomethionine (Se-Met) for 4 d and stimulated with IL-4 (5 mg/L, 20 h). Values are means ± SEM, n = 3. **P < 0.01, ***P < 0.001. All means were compared to one control (Se-deficient diet RAW 264.7 cells) and analyzed using ANOVA with Dunnett’s post hoc testing. (B) Bone marrow-derived macrophage cells isolated from glutathione peroxidase-1 (GPX1) knockout (KO) and glutathione peroxidase-1 wild-type (WT) C57bl/6 mice were cultured with 100 nmol/L sodium selenite for 3 d and stimulated with IL-4 (5 mg/L, 20 h) and LPS (1 mg/L, 12 h). Values are means ± SEM, n = 3. Within each graph, means without a common letter differ, P < 0.01. Se-A, selenium-adequate; Se-D, selenium-deficient.
To further confirm the effects of selenoproteins on Arg-I expression, we utilized a GPX1 KO mouse model. BMDM macrophages from WT and GPX1 KO mice were cultured and treated with LPS (12 h) and IL-4 (20 h). Arg-I activity was higher in the WT BMDM treated with IL-4 compared to the GPX1 KO cells treated with IL-4 (P < 0.001) (Fig. 3B). Interestingly, untreated and LPS-treated BMDM from GPX1 KO, compared to GPX 1 WT mice, did not show an increase in Arg-I activity (Fig. 3B). Both the ex vivo and in vivo results clearly demonstrate a requirement for Se in the form of selenoproteins to increase Arg-I activity.
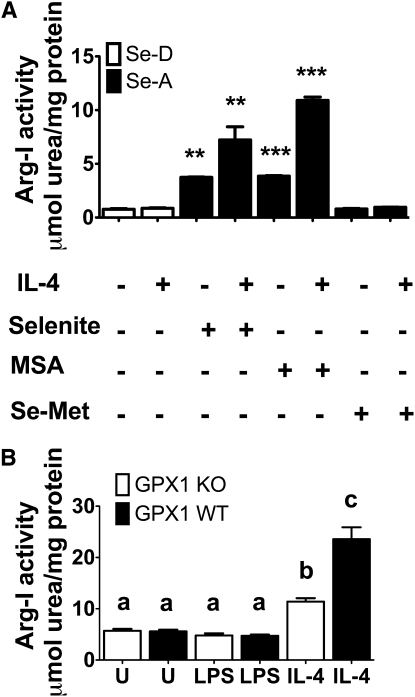
PPAR#x03B3 and STAT6 are both essential for the Se-dependent upregulation of Arg-I.
To connect the increased activation of PPARγ to Arg-I expression in the context of Se status, we used rosiglitazone, a synthetic PPARγ agonist, and GW9662, a PPARγ antagonist (31). RAW 264.7 and BMDM were pretreated with either rosiglitazone or GW9662 for 2 h. Following pretreatment, cells were stimulated with IL-4 as described earlier. Both rosiglitazone and GW9662 remained on the cells for a total of 22 h. Pretreatment of cells with rosiglitazone at 1 μmol/L significantly increased the activity of Arg-I in IL-4–treated Se-A RAW264.7 macrophages compared to Se-D RAW 264.7 macrophages treated with IL-4 (P < 0.001) (Fig. 4A). Moreover, rosiglitazone pretreatment increased Arg-I activity in IL-4–treated Se-A RAW 264.7 macrophages compared to vehicle control IL-4–treated Se-A RAW 264.7 macrophages (P < 0.001) (Fig. 4A). Pretreatment of BMDM from Se-A mice with GW9662 at 1 μmol/L completely blocked the effect of Se while greatly inhibiting activity in IL-4–treated cells, such that the Arg-I activity in Se-A BMDM did not differ from that in IL-4–treated Se-D BMDM (Fig. 4B). In contrast to the rosiglitazone treatment, GW9662 treatment decreased Arg-I activity in IL-4–treated Se-A BMDM compared to vehicle control IL-4–treated Se-A BMDM (P < 0.01) (Fig. 4B). Furthermore, GW9662-treated Se-A RAW264.7 cells inhibited the IL-4–dependent increase in Mrc-1 expression compared to the vehicle control, suggesting a critical role for PPARγ in the regulation of M2 markers by Se (P < 0.001) (Fig 4C).
FIGURE 4.
Se acts through PPARγ to upregulate the expression of Arg-I and Mrc-1. (A) RAW 264.7 cells were prestimulated with 1 μmol/L rosiglitazone for 2 h, followed by IL-4 (5 mg/L, 20 h) and LPS (1 mg/L, 12 h). (B) Bone marrow-derived macrophages isolated from Se-deficient (Se-D) diet and Se-adequate (Se-A) diet-fed mice were prestimulated with 1 μmol/L GW9662 for 2 h followed by IL-4 (5 mg/L, 20 h) and LPS (1 mg/L, 12 h). (C) RAW 264.7 macrophage cells were cultured with and without Se followed by pretreatment with 1 μmol/L GW9662 for 2 h prior to stimulation with IL-4 (5 mg/L, 20 h) and LPS (0.1 mg/L, 4 h). Values are means ± SEM, n = 3. Within each graph, means without a common letter differ, P < 0.01.
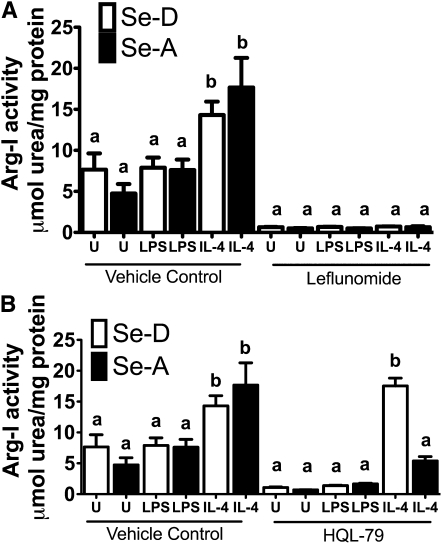
IL-4 mediates PPARγ-dependent gene expression through a STAT6-dependent mechanism involving enhanced recruitment of PGC-1β, a well-known PPARγ coactivator (5). To examine the role of STAT6, we utilized leflunomide, which inhibits the phosphorylation of STAT6, preventing the nuclear translocation and subsequent binding to cognate sites on the DNA (32). BMDM were pretreated with 100 μmol/L leflunomide for 2 h, followed by IL-4 treatment as described earlier. Consistent with the PPARγ antagonist results, leflunomide treatment inhibited Arg-I activity in IL-4–treated Se-A BMDM (compare vehicle control Se-A BMDM to leflunomide–treated Se-A BMDM) (P < 0.001) (Fig. 5A). These data suggest Se upregulates Arg-I expression through a STAT6-dependent pathway.
FIGURE 5.
STAT6 pathway and hematopoietic PG D2 synthase inhibitor treatment plays a critical role in the expression of Arg-1 in Se-adequate (Se-A) macrophages. (A) Bone marrow-derived macrophages isolated from Se-deficient (Se-D) and Se-A mice were pretreated with 100 μmol/L leflunomide for 2 h, followed by IL-4 (5 mg/L, 20 h) and LPS (1 mg/L, 12 h). (B) Bone marrow-derived macrophages isolated from Se-deficient and Se-A mice were pretreated with 25 μmol/L HQL-79 for 2 h followed by stimulation with IL-4 (5 mg/L, 20 h) and LPS (1 mg/L, 12 h). Values are means ± SEM, n = 3. Within each graph, means without a common letter differ, P < 0.01.
Se-dependent upregulation of Arg-I expression is mediated via arachidonic acid metabolism by H-PGDS.
To provide a mechanistic explanation of the effect of Se on the PPARγ-dependent expression of Arg-I, we examined the role of endogenous ligands of PPARγ, particularly 15d-PGJ2 formed through an H-PGDS pathway. Se-A BMDM were pretreated with 25 μmol/L HQL-79 for 2 h, followed by IL-4 and LPS treatments as described earlier. HQL-79 inhibits H-PGDS, a cytosolic enzyme responsible for the synthesis of PGD2 from PGH2, an arachidonic acid-derived cyclooxygenase metabolite (33, 34). Treatment of Se-A BMDM with HQL-79 and IL-4 decreased the Se-dependent expression of Arg-I (compare vehicle control Se-A BMDM to HQL-79–treated Se-A BMDM) (P < 0.01) (Fig. 5B). Though IL-4–treated Se-A BMDM had higher activity than unstimulated and LPS-treated samples, they were still lower than vehicle control-treated Se-A BMDM. Taken together, these results suggest that the arachidonic acid pathway plays a critical role in mediating the effect of Se.
Discussion
Macrophages are well-known effectors that have a significant bearing on the duration, magnitude, and quality of the immune response. Mounting evidence describes a more complex model that involves multiple macrophage phenotypes, which influences immunity not only via their ability to downregulate the production of proinflammatory mediators but also to facilitate pathways of resolution and wound healing. The latter property arises from a subset of macrophages (M2a-c) that express a battery of cytokines and cell surface receptors to promote catabasis responses leading to tissue repair and angiogenesis (2, 35). We present here novel findings that Se status of macrophages is critical to promote the expression of an alternatively activated phenotype that is linked to wound healing and collagen synthesis. Such a switch from the M1 toward the M2 pathway is further complemented by a substantial decrease in the expression of NF-κB–dependent proinflammatory genes, such as TNFα, IL-1β, and iNOS, which are markers of the classically activated macrophages (15, 24). Our studies have shown that macrophages cultured with Se produce an endogenous lipid mediator, 15d-PGJ2, which activates PPARγ-dependent pathways of anti-inflammatory gene expression (15), while repressing expression of proinflammatory genes. In addition, we demonstrate the ability of Se to synergize with IL-4–dependent activation of STAT6 to activate PPARγ.
Previously, it was shown that IL-4 can act as a stimulus to drive the expression of many M2 markers, such as Arg-I, Mrc-1, and Fizz-1 (9, 29, 30). In Se-A cells, IL-4 treatment significantly increased Arg-I at the activity, protein, and transcript levels. Arg-I activity increased in both the Se-D and Se-A macrophages. However, the greatest increase in activity levels was in the Se-A macrophages treated with IL-4 compared to their Se-D counterpart, suggesting a synergistic effect between IL-4 and Se. Further assessment revealed there was no interaction. Additionally, Arg-I activity levels were assessed using IL-13, another T helper 2 cytokine shown to act as a stimulus of M2 marker expression (9, 29). Although IL-13 treatment of Se-A macrophages increased the activity of Arg-I, the magnitude of the increase was much lower than with IL-4 (data not shown). Seen even in a proinflammatory state, Se shifts the l-Arg metabolism from LPS-induced production of NO via iNOS toward production of l-Orn and polyamines that are important for wound healing. Arg-I is a key determinant of M2 macrophage activation that is widely studied as a marker of M2 macrophages in murine macrophages; however, Arg-I is not expressed in human macrophages (2). Given that Fizz-1 and Ym-1 are also restricted to the murine system, we examined Mrc-1, which is expressed in both murine and human systems (2, 30). As reported earlier, IL-4 stimulation greatly enhanced the expression of Mrc-1 (9, 30). Upon Se supplementation and IL-4 treatment, our data showed a significant increase in Mrc-1 transcript levels compared to the cells cultured in Se-D media with IL-4. Intriguingly, exogenous addition of Se showed no increase in the levels of Mrc-1 expression with LPS compared to the Se-D cells cultured in the presence of LPS. A similar pattern was also observed with Fizz1 expression. Although expression of Ym-1 was significantly increased by LPS treatment of Se-A macrophages compared to the Se-D cells, its levels were far below those with IL-4 treatment. Although the repression of IL-4–dependent M2 genes by LPS is thought to be an event required for the polarization of macrophages toward the M1 pathway, Se supplementation appears to favor the polarization toward a resolution response, which might be essential to prevent the exacerbated activation of proinflammatory genes leading to tissue destruction and inflammation. On the other hand, IL-12, iNOS, IL-1β, and TNFα, all M1 markers, showed the opposite trend upon stimulation with LPS. LPS increased IL-1β and IL-12 transcript expression, whereas IL-4 had very little to no effect. Interestingly, IL-4 significantly increased transcript expression of TNFα. However, their expression levels were far below those seen with LPS. Indeed, expressions of all markers were significantly decreased with Se supplementation. Although there were differences between BMDM and RAW264.7 cells, which represent primary and immortalized cultures, respectively, the trend toward increased M2 markers in Se-A cells with IL-4 was near identical. Such a concordance in results further lends credence to the idea that Se status indeed does play a pivotal role in the expression of some of the M2 markers.
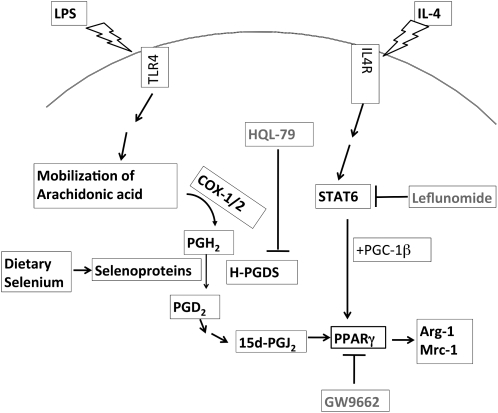
The mechanisms for the upregulation of Arg-I by Se are still not well understood. Our earlier work conclusively showed that Se-supplemented macrophages produced high amounts of the endogenous PPARγ agonist, 15d-PGJ2, to accompany the activation of PPARγ (15). Such an activation of PPARγ was inhibited by the antagonist, GW9662. Given that PPARγ activation upregulated Arg-I expression (5) and our results showed that the use of HQL-79 (H-PGDS inhibitor) completely blocked the effect of the Se-dependent increase in Arg-I, these results further confirm the ability of Se to enhance the production of an endogenous ligand that plays a major role in the expression of Arg-I. In addition to PPARγ, the use of leflunomide also indicates a potential cross-talk between IL-4 activation of STAT6 and PPARγ, where Se plays a key role as a positive modulator, perhaps by recruiting PGC-1β, a PPARγ coactivator (Fig. 6). Intriguingly, the ability of Se to increase Arg-I activity was only seen with those selenocompounds that led to the cellular synthesis of selenoproteins. Given that GPX1 is an abundant selenoprotein whose expression is increased by bioavailable Se, the use of a genetic GPX1 KO mouse model was pertinent in confirming this observation. Our results conclusively show that the absence of GPX1 significantly decreases Arg-I activity compared to WT, confirming the need for Se in the form of selenoproteins to effectively modulate the redox status of cells. Furthermore, the addition of IL-4 confirmed the synergistic relationship observed with Se. Further examination of the Arg-I promoter for the binding of transcription factors and accessory proteins as a function of Se status is awaited to obtain a complete picture of the underlying basis of the molecular transition of M1 to M2 phenotypes by specific selenoproteins.
FIGURE 6.
A proposed mechanism underlying Se-dependent upregulation of Arg-I, Mrc-1, and other M2 markers through the modulation of STAT6 and PPARγ-dependent pathways in macrophages.
In conclusion, our results clearly demonstrate that Se supplementation shunts macrophage activation from a proinflammatory M1 state toward an antiinflammatory M2 state. In doing so, we speculate that such macrophages become prone to helping cell proliferation and promoting cell growth after insult or injury. This is reminiscent of a recent report where Se status in T-cells was shown to be critical for activation, differentiation, and proliferation (36). Further work to validate these ex vivo studies is needed to examine the role of such a phenotypic switch in models that are known to trigger highly polarized immune responses associated with increased M2 signatures, such as helminth models, leading to parasite expulsion and regulation of inflammation.
Supplementary Material
Acknowledgments
We thank Dr. Pamela Hankey and Dr. Daniel Sharda (Penn State University) for their assistance with the Arg assay protocol and providing insightful guidance and Dr. Ann Skulas-Ray for assistance with statistics. S.M.N. and K.S.P. designed research; S.M.N. conducted research; K.S.P. and X.G.L. provided essential reagents and materials; S.M.N. and K.S.P. analyzed data; S.M.N. performed statistical analysis; S.M.N. and K.S.P. wrote the paper; and S.M.N. and K.S.P. had primary responsibility for final content. All authors read and approved the final manuscript.
Footnotes
Supported by grants from NIH (DK 077152; AT004350).
Supplemental Figures 1–3 are available from the “Online Supporting Material” link in the online posting of the article and from the same link in the online table of contents at jn.nutrition.org.
Abbreviations used: BMDM, bone marrow-derived macrophage; GPX, glutathione peroxidase; H-PGDS, hematopoietic PG D2 synthase; KO, knockout; MSA, methylseleninic acid; PGC1-β, PPARγ co-activator 1 beta; 15d-PGJ2, 15-deoxy-Δ12,14-PGJ2; Sec, selenocysteine; Se-A, Se-adequate diet; Se-D, Se-deficient diet; Se-S, Se-supplemented; Se-Met, selenomethionine; WT, wild type.
Literature Cited
- 1.Fairweather D, Cihakova D. Alternatively activated macrophages in infection and autoimmunity. J Autoimmun. 2009;33:222–30 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Martinez FO, Helming L, Gordon S. Alternative activation of macrophages: an immunologic functional perspective. Annu Rev Immunol. 2009;27:451–83 [DOI] [PubMed] [Google Scholar]
- 3.MacKinnon AC, Farnworth SL, Hodkinson PS, Henderson NC, Atkinson KM, Leffler H, Nilsson UJ, Haslett C, Forbes SJ, Sethi T. Regulation of alternative macrophage activation by galectin-3. J Immunol. 2008;180:2650–8 [DOI] [PubMed] [Google Scholar]
- 4.Pauleau AL, Rutschman R, Lang R, Pernis A, Watowich SS, Murray PJ. Enhancer-mediated control of macrophage-specific arginase I expression. J Immunol. 2004;172:7565–73 [DOI] [PubMed] [Google Scholar]
- 5.Vats D, Mukundan L, Odegaard JI, Zhang L, Smith KL, Morel CR, Wagner RA, Greaves DR, Murray PJ, Chawla A. Oxidative metabolism and PGC-1beta attenuate macrophage-mediated inflammation. Cell Metab. 2006;4:13–24 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wang WW, Jenkinson CP, Griscavage JM, Kern RM, Arabolos NS, Byrns RE, Cederbaum SD, Ignarro LJ. Co-induction of arginase and nitric oxide synthase in murine macrophages activated by lipopolysaccharide. Biochem Biophys Res Commun. 1995;210:1009–16 [DOI] [PubMed] [Google Scholar]
- 7.Gray MJ, Poljakovic M, Kepka-Lenhart D, Morris SM., Jr Induction of arginase I transcription by IL-4 requires a composite DNA response element for STAT6 and C/EBPbeta. Gene. 2005;353:98–106 [DOI] [PubMed] [Google Scholar]
- 8.Odegaard JI, Ricardo-Gonzalez RR, Goforth MH, Morel CR, Subramanian V, Mukundan L, Red Eagle A, Vats D, Brombacher F, Ferrante AW, et al. Macrophage-specific PPARgamma controls alternative activation and improves insulin resistance. Nature. 2007;447:1116–20 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Stein M, Keshav S, Harris N, Gordon S. Interleukin 4 potently enhances murine macrophage mannose receptor activity: a marker of alternative immunologic macrophage activation. J Exp Med. 1992;176:287–92 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Pesce JT, Ramalingam TR, Mentink-Kane MM, Wilson MS, El Kasmi KC, Smith AM, Thompson RW, Cheever AW, Murray PJ, Wynn TA. Arginase-1-expressing macrophages suppress Th2 cytokine-driven inflammation and fibrosis. PLoS Pathog. 2009;5:e1000371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Morris SM, Jr, Kepka-Lenhart D, Chen LC. Differential regulation of arginases and inducible nitric oxide synthase in murine macrophage cells. Am J Physiol. 1998;275:E740–7 [DOI] [PubMed] [Google Scholar]
- 12.Kreider T, Anthony RM, Urban JF, Jr, Gause WC. Alternatively activated macrophages in helminth infections. Curr Opin Immunol. 2007;19:448–53 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Mosser DM. The many faces of macrophage activation. J Leukoc Biol. 2003;73:209–12 [DOI] [PubMed] [Google Scholar]
- 14.Chawla A, Barak Y, Nagy L, Liao D, Tontonoz P, Evans RM. PPAR-gamma dependent and independent effects on macrophage-gene expression in lipid metabolism and inflammation. Nat Med. 2001;7:48–52 [DOI] [PubMed] [Google Scholar]
- 15.Vunta H, Belda BJ, Arner RJ, Channa Reddy C, Vanden Heuvel JP, Sandeep Prabhu K. Selenium attenuates pro-inflammatory gene expression in macrophages. Mol Nutr Food Res. 2008;52:1316–23 [DOI] [PubMed] [Google Scholar]
- 16.Gao B. Cytokines, STATs and liver disease. Cell Mol Immunol. 2005;2:92–100 [PubMed] [Google Scholar]
- 17.Bellinger FP, Raman AV, Reeves MA, Berry MJ. Regulation and function of selenoproteins in human disease. Biochem J. 2009;422:11–22 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ganther HE. Selenium metabolism, selenoproteins and mechanisms of cancer prevention: complexities with thioredoxin reductase. Carcinogenesis. 1999;20:1657–66 [DOI] [PubMed] [Google Scholar]
- 19.Combs GF., Jr Chemopreventive mechanisms of selenium. Med Klin (Munich). 1999;94 Suppl 3:18–24 [DOI] [PubMed] [Google Scholar]
- 20.Papp LV, Lu J, Holmgren A, Khanna KK. From selenium to selenoproteins: synthesis, identity, and their role in human health. Antioxid Redox Signal. 2007;9:775–806 [DOI] [PubMed] [Google Scholar]
- 21.Holben DH, Smith AM. The diverse role of selenium within selenoproteins: a review. J Am Diet Assoc. 1999;99:836–43 [DOI] [PubMed] [Google Scholar]
- 22.Chen J, Berry MJ. Selenium and selenoproteins in the brain and brain diseases. J Neurochem. 2003;86:1–12 [DOI] [PubMed] [Google Scholar]
- 23.Jiang C, Wang Z, Ganther H, Lu J. Distinct effects of methylseleninic acid versus selenite on apoptosis, cell cycle, and protein kinase pathways in DU145 human prostate cancer cells. Mol Cancer Ther. 2002;1:1059–66 [PubMed] [Google Scholar]
- 24.Prabhu KS, Zamamiri-Davis F, Stewart JB, Thompson JT, Sordillo LM, Reddy CC. Selenium deficiency increases the expression of inducible nitric oxide synthase in RAW 264.7 macrophages: role of nuclear factor-kappaB in up-regulation. Biochem J. 2002;366:203–9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Cheng WH, Ho YS, Ross DA, Valentine BA, Combs GF, Lei XG. Cellular glutathione peroxidase knockout mice express normal levels of selenium-dependent plasma and phospholipid hydroperoxide glutathione peroxidases in various tissues. J Nutr. 1997;127:1445–50 [DOI] [PubMed] [Google Scholar]
- 26.Esworthy RS, Yang L, Frankel PH, Chu FF. Epithelium-specific glutathione peroxidase, Gpx2, is involved in the prevention of intestinal inflammation in selenium-deficient mice. J Nutr. 2005;135:740–5 [DOI] [PubMed] [Google Scholar]
- 27.Corraliza IM, Campo ML, Soler G, Modolell M. Determination of arginase activity in macrophages: a micromethod. J Immunol Methods. 1994;174:231–5 [DOI] [PubMed] [Google Scholar]
- 28.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-delta delta C(T)) method . Methods. 2001;25:402–8 [DOI] [PubMed] [Google Scholar]
- 29.Louis CA, Mody V, Henry WL, Jr, Reichner JS, Albina JE. Regulation of arginase isoforms I and II by IL-4 in cultured murine peritoneal macrophages. Am J Physiol. 1999;276:R237–42 [DOI] [PubMed] [Google Scholar]
- 30.Raes G, De Baetselier P, Noel W, Beschin A, Brombacher F, Hassanzadeh Gh G. Differential expression of FIZZ1 and Ym1 in alternatively versus classically activated macrophages. J Leukoc Biol. 2002;71:597–602 [PubMed] [Google Scholar]
- 31.Wu JS, Lin TN, Wu KK. Rosiglitazone and PPAR-gamma overexpression protect mitochondrial membrane potential and prevent apoptosis by upregulating anti-apoptotic Bcl-2 family proteins. J Cell Physiol. 2009;220:58–71 [DOI] [PubMed] [Google Scholar]
- 32.Siemasko K, Chong AS, Jack HM, Gong H, Williams JW, Finnegan A. Inhibition of JAK3 and STAT6 tyrosine phosphorylation by the immunosuppressive drug leflunomide leads to a block in IgG1 production. J Immunol. 1998;160:1581–8 [PubMed] [Google Scholar]
- 33.Aritake K, Kado Y, Inoue T, Miyano M, Urade Y. Structural and functional characterization of HQL-79, an orally selective inhibitor of human hematopoietic prostaglandin D synthase. J Biol Chem. 2006;281:15277–86 [DOI] [PubMed] [Google Scholar]
- 34.Matsushita N, Aritake K, Takada A, Hizue M, Hayashi K, Mitsui K, Hayashi M, Hirotsu I, Kimura Y, Tani T, et al. Pharmacological studies on the novel antiallergic drug HQL-79: II. Elucidation of mechanisms for antiallergic and antiasthmatic effects. Jpn J Pharmacol. 1998;78:11–22 [DOI] [PubMed] [Google Scholar]
- 35.Laskin DL. Macrophages and inflammatory mediators in chemical toxicity: a battle of forces. Chem Res Toxicol. 2009;22:1376–85 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hoffmann FW, Hashimoto AC, Shafer LA, Dow S, Berry MJ, Hoffmann PR. Dietary selenium modulates activation and differentiation of CD4+ T cells in mice through a mechanism involving cellular free thiols. J Nutr. 2010;140:1155–61 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.