Abstract
Thymic stromal lymphopoietin (TSLP) triggers dendritic cell–mediated T helper (Th) 2 inflammatory responses. A single-nucleotide polymorphism (SNP), rs3806933, in the promoter region of the TSLP gene creates a binding site for the transcription factor activating protein (AP)–1. The variant enhances AP-1 binding to the regulatory element, and increases the promoter–reporter activity of TSLP in response to polyinosinic-polycytidylic acid (poly[I:C]) stimulation in normal human bronchial epithelium (NHBE). We investigated whether polymorphisms including the SNP rs3806933 could affect the susceptibility to and clinical phenotypes of bronchial asthma. We selected three representative (i.e., Tag) SNPs and conducted association studies of the TSLP gene, using two independent populations (639 patients with childhood atopic asthma and 838 control subjects, and 641 patients with adult asthma and 376 control subjects, respectively). We further examined the effects of corticosteroids and a long-acting β2-agonist (salmeterol) on the expression levels of the TSLP gene in response to poly(I:C) in NHBE. We found that the promoter polymorphisms rs3806933 and rs2289276 were significantly associated with disease susceptibility in both childhood atopic and adult asthma. The functional SNP rs3806933 was associated with asthma (meta-analysis, P = 0.000056; odds ratio, 1.29; 95% confidence interval, 1.14–1.47). A genotype of rs2289278 was correlated with pulmonary function. Moreover, the induction of TSLP mRNA and protein expression induced by poly(I:C) in NHBE was synergistically impaired by a corticosteroid and salmeterol. TSLP variants are significantly associated with bronchial asthma and pulmonary function. Thus, TSLP may serve as a therapeutic target molecule for combination therapy.
Keywords: asthma, TSLP, bronchial epithelial cells, combination therapy, genetic polymorphisms
CLINICAL RELEVANCE.
Our findings suggest that thymic stromal lymphoprotein (TSLP) polymorphisms functionally contribute to the disease susceptibility to asthma. The susceptible functional allele may contribute to the T helper 2–polarized immunity in asthma during viral respiratory infections. We also found the synergistic suppression of polyinosinic-polycytidylic acid–induced TSLP production by dexamethasone and salmeterol.
Thymic stromal lymphoprotein (TSLP) is an epithelial cell–derived cytokine that triggers dendritic cell–mediated T helper (Th) 2 inflammatory responses, and plays an important role in the initiation and maintenance of the allergic immune response (1–6). A recent study showed that TSLP is released by human epithelial cells in response to microbes, trauma, or inflammation, and potently activates mast cells (7). In humans, TSLP is highly expressed by airway epithelial cells during allergic inflammation (2), and the expression of the TSLP gene in asthmatic airways is correlated with both the expression of Th2-attracting chemokines and disease severity (3).
Large numbers of association studies on asthma and asthma-related phenotypes using genetic polymorphisms were performed in different populations (8). Recent studies showed roles of human genetic polymorphisms of the TSLP gene. A variant in TSLP was associated with reductions in IgE in response to cockroaches and total IgE in a sex-stratified analysis (9). A functional single-nucleotide polymorphism (SNP), rs3806933, was identified in the regulatory element of TSLP (10). The variant creates a binding site for activating protein–1 (AP-1), and affects the transcriptional efficiency of the long form of TSLP induced by stimulation with polyinosinic-polycytidylic acid (poly[I:C]) in bronchial epithelial cells (10).
The majority of exacerbations of asthma coincide with respiratory viral infections, most commonly by rhinoviruses (11). Double stranded RNA (dsRNA), a Toll-like receptor (TLR)–3 ligand, is a potent stimulus for innate antiviral immune responses, and poly(I:C) is thought to mimic the effects of dsRNA (12). The inflammatory mediators IL-1β and TNF-α regulate human TSLP gene expression, and human TSLP mRNA concentrations increase after exposure to TLR2, TLR3, TLR8, and TLR9 ligands in airway bronchial epithelial cells (13, 14). A suppressive effect of glucocorticoid on the expression of TSLP induced in airway epithelial cells by stimulation with the TLR3 ligand and Th2 cytokines was reported (14). In addition, several clinical studies showed that the combination of an inhaled corticosteroid and long-acting β-adrenergic agonist (LABA) is more efficacious in asthma than either alone, and reduces exacerbations (15–18). We investigated the effects of dexamethasone (DEX) and salmeterol (SAL) on the induction of TSLP by poly(I:C).
In this study, we found that the promoter polymorphisms reference SNPs (rs)3806933 and rs2289276 were significantly associated with susceptibility to both childhood atopic and adult asthma, using a case-control association study. We also found a significant correlation between lung function and the genotype of rs2289278. Functional analyses of the related variant rs2289276 were performed. We also found that corticosteroids and LABA (SAL) synergistically suppressed the expression levels of TSLP mRNA and TSLP protein production induced by dsRNA in bronchial epithelial cells. Some results of this studies were previously reported in the form of an abstract (19).
MATERIALS AND METHODS
Additional details on methods are provided in the online supplement.
Study Subjects and Genotyping
All subjects with asthma were diagnosed according to the criteria of the National Institutes of Health (National Heart, Lung, and Blood Institute), as described elsewhere (20–22). Data for forced expiratory volume in 1 second (FEV1), forced vital capacity (FVC), and FEV1/FVC were available for subjects with adult asthma. The clinical parameters of participants are summarized in Table 1. Genomic DNA was prepared in accordance with standard protocols. Genotyping was performed using the TaqMan allele-specific amplification method (TaqMan-ASA; Applied Biosystems, Foster City, CA). All recruited individuals gave written, informed consent to participate in the study, in accordance with the rules of the Process Committee at the Center for Genomic Medicine, Institute of Physical and Chemical Research (Kanagawa, Japan).
TABLE 1.
BASELINE CHARACTERISTICS OF PARTICIPANTS
| Characteristic | Childhood Asthma | Control Group 1 | Adult Asthma | Control Group 2 |
|---|---|---|---|---|
| Total n | 639 | 838 | 641 | 376 |
| Male (%) | 59.4 | 73.4 | 42.8 | 46.8 |
| Age (years) | ||||
| Mean | 9.3 | 49.8 | 51.8 | 50.7 |
| Range | 4–15 | 20–75 | 20–75 | 29–72 |
| Atopy (n/n tested) (%) | 639/639 | 531/596 | ||
| Serum IgE (IU) | 1,101.5 ± 94.9 | 621.9 ± 62.6 | 93.9 ± 6.9 | |
| Eosinophil count (n/μl) | 518.1 ± 15.0 | 392.4 ± 16.0 | ||
| FVC (percent predicted value) | 84.0 ± 0.84 | |||
| FEV1 (percent predicted value) | 71.1 ± 0.88 | |||
| FEV1/FVC (%) | 66.8 ± 0.78 |
Definition of abbreviations: FEV1, forced expiratory volume in 1 second; FVC, forced vital capacity.
Values with ± signs represent means ± standard error.
Statistical Analysis
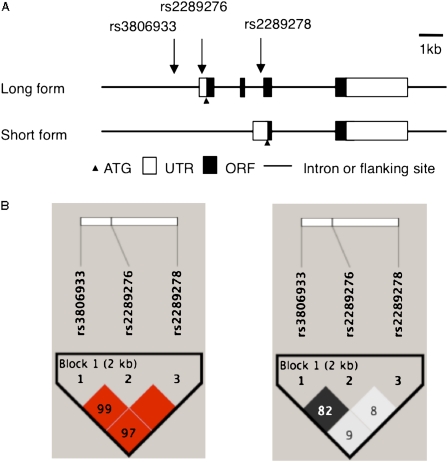
In total, 23 polymorphisms in the TSLP gene were identified in the Japanese population (10). Pairwise linkage disequilibrium (LD) was calculated as D′/LOD and r2, and three Tag SNPs (i.e., representative SNP in a genomic region with high LD), rs3806933, rs2289276, and rs2289278, were selected among seven SNPs with a frequency of greater than 10% by using the Haploview 4.1 program (http://www.broad.mit.edu/mpg/haploview/) (10) (Figure 1A). The functional promoter SNP rs3806933 had an allele-specific effect on expression through altering the affinity for AP-1, and the SNP was in strong LD with rs2289276 (r2 = 0.82) (Figure 1B). To test the association between TSLP variants and bronchial asthma, we performed a contingency χ2 test. We applied Bonferroni correction. As the number of Tag SNPs were three,we performed multiplication of P value by three. In the association study, corrected P values of less than 0.05 were judged to be significant. The Mantel-Haenszel method was used to combine allele frequency datasets. Odds ratios (ORs) with 95% confidence intervals (CIs) were also calculated. Haplotype frequencies for three loci were estimated, and haplotype association tests were performed using Haploview 4.1.
Figure 1.
Single-nucleotide polymorphisms (SNPs) and pairwise linkage disequilibrium (LD) map of the thymic stromal lymphoprotein (TSLP) gene. (A) Graphic overview of polymorphisms identified in relation to the exon/intron structure of the human TSLP gene. Three polymorphisms were genotyped in this study. The ATG (initiation codon), untranslated region (UTR), and the open reading frame (ORF) are indicated by solid triangles, open boxes, and solid boxes, respectively. (B) Pairwise D′/LOD (left) and r2 (right) for all combinations of single-nucleotide polymorphism (SNP) pairs are shown.
We further investigated associations between asthma-related phenotypes. We examined associations between asthma-related phenotypes (eosinophil count, serum total IgE, lung functions, and disease severity) and variants within patients with asthma, as described previously (22, 23). Comparisons of mRNA expression analysis and protein expression analysis were performed with the Student t test. Statistically significant differences in the luciferase assay were assessed with the Bonferroni-Dunn test with two-factor factorial ANOVAs. Statistical significance was defined at the standard 5% level.
Computational Analysis of Transcription Factor–Binding Sites and Biotinylated Oligonucleotide Precipitation Assay
TRANSFAC Professional 10.3 (http://www.biobase.de/pages/) was used to predict putative transcription factor–binding sites under the minimizing condition of the sum of false positives and false negatives.
A biotinylated oligonucleotide precipitation assay was conducted as described elsewhere (10). The oligonucleotides for the precipitation assay included, for−82C oligo, 5′-TGGCCCCTAAGGCAGGCCTTACAG-3′; and for −82T oligo, 5′-TGGCCTCTAAGGCAGGCCTTACAG -3′. AP-2α was detected by immunoblotting with an anti–AP-2α antibody (C-18; Santa Cruz Biotechnology, Inc., Santa Cruz, CA).
Quantitative Real-Time RT-PCR, ELISA, and Luciferase Assay
The expression of TSLP was determined by real-time quantitative RT-PCR, using SYBR Premix Ex Taq (Takara, Shiga, Japan). TSLP in culture supernatants was measured using ELISA kits (R&D Systems, Inc., Minneapolis, MN), and the reporter luciferase assays were conducted as described previously (10).
RESULTS
Identification of TSLP Polymorphisms and Haplotypes Associated with Asthma Susceptibility
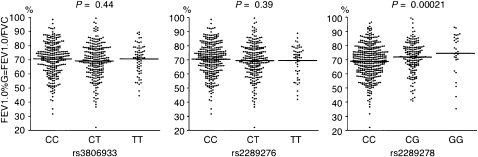
All genotype and allele frequencies are shown in Table 2, and those for the control and asthma groups were in Hardy-Weinberg equilibrium. We found that the functional SNP rs3806933 was associated with childhood atopic asthma (P = 0.0063; OR, 1.25; 95% CI, 1.07–1.47) and adult asthma (P = 0.0023; OR, 1.37; 95% CI, 1.12–1.67). Another SNP, rs2289276, was also significantly associated with childhood atopic asthma (P = 0.00066; OR, 1.33; 95% CI, 1.13–1.57) under our allelic model (Table 2). The directions of associations of the two SNPs were similar in both populations. We combined the results using a Mantel-Haenszel meta-analysis, and observed the most significant association at rs3806933 (meta-analysis, P = 0.000056; OR, 1.29; 95% CI, 1.14–1.47) (Table 2). We also found a significant association under the dominant model at rs3806933 (meta-analysis, P = 0.00013; OR, 1.37; 95% CI, 1.16–1.62) and at rs2289276 (meta-analysis, P = 0.00019; OR, 1.36; 95% CI, 1.16–1.61) (Table 2). We further conducted an association study, using adult asthma cases that were diagnosed in childhood (Table E1 in the online supplement). We confirmed a significant association between rs3806933 and adult asthma diagnosed in childhood with a similar direction of association. A recent study showed a female-specific association between a variant of the TSLP gene and total serum IgE and IgE to cockroach (9). We performed a further sex-stratified analysis, but could not determine the female-specific effect on the associations. Although we surveyed associations between the three SNPs and patients with asthma who had high eosinophil counts, high serum IgE concentrations, and disease severity, we could not find any association. However, the rs2289278 C allele was significantly correlated with decreased FEV1/FVC in adult asthma (P = 0.00021 according to the Jonckheere-Terpstra test) (Figure 2). We next constructed the haplotypes of the three SNPs and estimated the frequency of each haplotype in control subjects and patients with bronchial asthma (Table E2). We identified three common haplotypes in the population. Haplotype T-T-C of TSLP was significantly associated with childhood atopic asthma and adult asthma. We obtained P values of 0.00070 and 0.032, respectively, by using the Haploview 4.1 program.
TABLE 2.
GENOTYPE COUNTS, FREQUENCIES, AND CASE-CONTROL ASSOCIATION TEST RESULTS
| db Single-Nucleotide Polymorphism Identification | Case |
Control |
Minor Allele Frequencies |
P Values and Odds Ratios (with 95% Confidence Intervals) |
|||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Allele1/2 | 1/1 | 1/2 | 2/2 | Sum | 1/1 | 1/2 | 2/2 | Sum | Case | Control | Allelic | Dominant | Recessive |
| Childhood atopic asthma | Control 1 | ||||||||||||
| rs3806933 | 294 | 269 | 68 | 631 | 446 | 311 | 72 | 829 | 0.32 | 0.27 | 0.0063 | 0.0064 | 0.18 |
| −847C/T | 0.47 | 0.43 | 0.11 | 0.54 | 0.38 | 0.09 | 1.25 (1.07–1.47) | 1.34 (1.09–1.64) | 1.27 (0.90–1.80) | ||||
| rs2289276 | 322 | 256 | 60 | 638 | 493 | 288 | 56 | 837 | 0.29 | 0.24 | 0.00066 | 0.0013 | 0.055 |
| −82C/T | 0.5 | 0.4 | 0.09 | 0.59 | 0.34 | 0.07 | 1.33 (1.13–1.57) | 1.41 (1.14–1.73) | 1.45 (0.99–2.12) | ||||
| rs2289278 | 418 | 195 | 25 | 638 | 537 | 261 | 38 | 836 | 0.19 | 0.20 | 0.52 | 0.61 | 0.56 |
| 1560C/G | 0.66 | 0.31 | 0.04 | 0.64 | 0.31 | 0.05 | 0.94 (0.78–1.13) | 0.95 (0.76–1.17) | 0.86 (0.51–1.43) | ||||
| Adult asthma | Control 2 | ||||||||||||
| rs3806933 | 289 | 274 | 71 | 634 | 204 | 143 | 27 | 374 | 0.33 | 0.26 | 0.0023 | 0.0060 | 0.039 |
| −847C/T | 0.46 | 0.43 | 0.11 | 0.55 | 0.38 | 0.07 | 1.37 (1.12–1.67) | 1.43 (1.11–1.85) | 1.62 (1.02–2.58) | ||||
| rs2289276 | 322 | 264 | 53 | 639 | 213 | 138 | 23 | 374 | 0.29 | 0.25 | 0.034 | 0.044 | 0.21 |
| −82C/T | 0.5 | 0.41 | 0.08 | 0.57 | 0.37 | 0.06 | 1.25 (1.02–1.53) | 1.30 (1.01–1.68) | 1.38 (0.83–2.29) | ||||
| rs2289278 | 415 | 187 | 29 | 631 | 232 | 127 | 17 | 376 | 0.19 | 0.21 | 0.28 | 0.19 | 0.96 |
| 1560C/G | 0.66 | 0.30 | 0.05 | 0.62 | 0.34 | 0.05 | 0.88 (0.71–1.11) | 0.84 (0.64–1.09) | 1.02 (0.55–1.88) | ||||
| Combined (Mantel-Haenszel) | |||||||||||||
| rs3806933 | 583 | 543 | 139 | 1265 | 650 | 454 | 99 | 1203 | 0.32 | 0.27 | 0.000056 | 0.00013 | 0.022 |
| −847C/T | 0.46 | 0.43 | 0.11 | 0.54 | 0.38 | 0.08 | 1.29 (1.14–1.47) | 1.37 (1.16–1.62) | 1.39 (1.05–1.85) | ||||
| rs2289276 | 644 | 520 | 113 | 1277 | 706 | 426 | 79 | 1211 | 0.29 | 0.24 | 0.000076 | 0.00019 | 0.026 |
| −82C/T | 0.50 | 0.41 | 0.09 | 0.58 | 0.35 | 0.07 | 1.3 (1.14–1.48) | 1.36 (1.16–1.61) | 1.42 (1.04–1.96) | ||||
| rs2289278 | 833 | 382 | 54 | 1269 | 769 | 388 | 55 | 1212 | 0.19 | 0.21 | 0.25 | 0.23 | 0.69 |
| 1560C/G | 0.66 | 0.30 | 0.04 | 0.63 | 0.32 | 0.05 | 0.92 (0.79–1.06) | 0.90 (0.76–1.07) | 0.92 (0.61–1.39) | ||||
P values of the two populations represent the χ2 test for case-control comparisons under each model. db, the Single Nucleotide Polymorphism Database (dbSNP).
Figure 2.
Relationship of TSLP rs3806933, rs2289276, and rs2289278 genotype with lung function in adult patients with asthma. The ratio of forced expiratory volume in 1 second (FEV1) to forced vital capacity (FVC) is plotted, and horizontal bars represent the mean for each genotype group.
To test the generalizability of our findings in other ethnic populations, we compared our results with two recent genome-wide association studies (GWASs) of asthma (24, 25). Markers rs3806932, rs2289276, and rs11466741 were included in the Illumina panels used in the studies. Marker rs3806933 was in complete LD with rs3806932 (D′ = 1.00 and r2 = 1.00), and rs2289276 was in complete LD with rs11466741 (D′ = 1.00 and r2 = 1.00) in a Japanese population (10). However, we could not replicate the association between these TSLP variants and asthma in the samples with African ancestry and European ancestry (Table E3).
Transcription Factor Binding to the rs2289276 SNP
We predicted a potential allelic difference in cis-acting regulatory function in transcription via a bioinformatics approach, and rs2289276 (−82C/T) was potentially found to alter the affinity of a transcription factor, AP-2α, between two alleles (Figure 3A). The sequence containing the −82C SNP on the protective allele corresponded to the putative binding site to AP-2α, a possible transcription suppression factor (26). We next examined the binding of AP-2α protein to the sequences containing the −82C/T SNP, and binding was clearly detectable in both oligonucleotides without stimulation (Figure 3B). The binding ability of −82T was diminished compared with that of −82C, regardless of poly(I:C) stimulation, suggesting that the higher AP-2α binding to −82C (on the protective allele) may have reduced its transcriptional activity through repressive effects on transcription.
Figure 3.
(A) DNA sequences of transcription factor–binding motifs around rs2289276 SNP. The positions of potential AP-2α binding sites are shown in the open box. *SNPs. (B) Binding affinity of transcription factors to oligonucleotides (oligo) in vitro. Normal human bronchial epithelia (NHBEs) were stimulated with or without 10 μg/ml polyinosinic-polycytidylic acid (poly[I:C]) for 1 hour. Proteins interacting with the double-stranded oligonucleotides were precipitated and analyzed by immunoblotting with the indicated antibodies. Three independent experiments were performed with similar results. AP, activating protein. w.b., Western blotting.
The Induction of TSLP mRNA and Protein Expression Induced by Poly(I:C) in Normal Human Bronchial Epithelial Cells Is Synergistically Impaired by a Corticosteroid and SAL
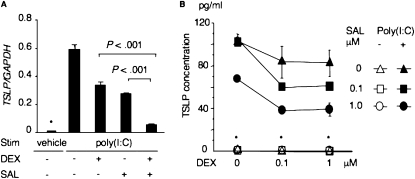
We investigated the effects of DEX and SAL on the induction of TSLP by poly(I:C). We first confirmed, by ELISA, the capability of DEX to suppress poly(I:C)-induced TSLP protein production in normal human bronchial epithelial (NHBE) cells. The addition of DEX 0.5 hour before or simultaneously with poly(I:C) stimulation dramatically reduced TSLP production in a dose-dependent manner (Figure E1). Even if DEX was added to the NHBE medium at 1 hour after poly(I:C) stimulation, TSLP production was also significantly impaired (more than 50% suppression) (Figure E1). Next we investigated the effects of SAL on the expression of the TSLP gene and TSLP protein production. NHBE cells were stimulated with or without poly(I:C) for 4 hours. DEX (1 μM) or SAL (1 μM) was added to medium 0.5 hour before stimulation with poly(I:C). The relative expression of TSLP in NHBE was normalized with glyceraldehyde-3-phosphate dehydrogenase (GAPDH) expression. The expression of TSLP mRNA was decreased by DEX and SAL. The greatest suppressive effect was evident after concurrent exposure of the cells to DEX and SAL (Figure 4A). Data regarding expression are representative of three independent experiments, and similar results were obtained using NHBE cells from three individuals.
Figure 4.
Suppression of TSLP production by dexamethasone (DEX) and salmeterol (SAL) in NHBE. (A) Quantitative RT-PCR assay of TSLP. P < 0.001, according to Student t test. (B) Effects of DEX and SAL on TSLP protein production in NHBE stimulated (Stim) by poly(I:C). Concentrations of TSLP in supernatants were measured by ELISA. *Not detectable.
We also measured protein concentrations of TSLP under the same conditions. NHBE cells were stimulated with or without poly(I:C), and concentrations of TSLP in supernatants 24 hours after stimulation were measured by ELISA. DEX and SAL were added to the medium 0.5 hour before the stimulation with poly(I:C) at the indicated doses. The synergistic suppression of TSLP protein production was also evident after concurrent exposure to DEX and SAL, and DEX or SAL caused a dose-dependent decrease in TSLP protein production (Figure 4B). The data in Figure 4B represent the mean ± SD, and are representative of two independent experiments performed using NHBE cells from two individuals. Similar results were obtained, implying that DEX and SAL may synergistically suppress TSLP production in human airway epithelial cells during viral respiratory infections.
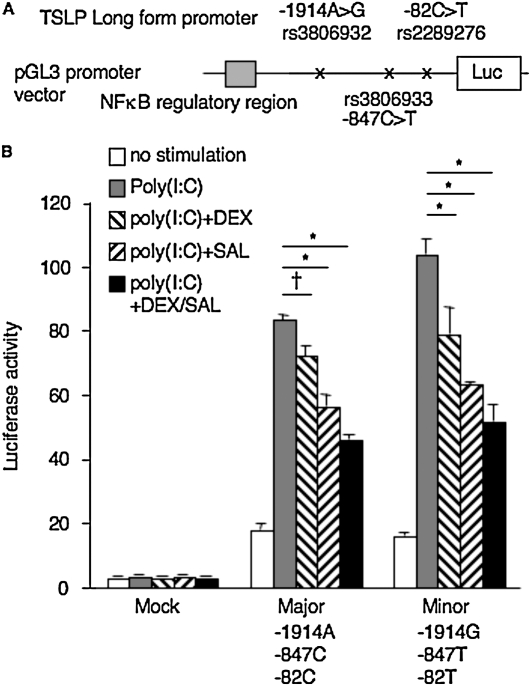
Effects of Dexamethasone and Salmeterol on Luciferase Transcription
Rs3806932 (−1914T/G) was in complete LD with rs3806933 (−847C/T) (D′ = 1.0,0 and r2 = 1.00) (10). The promoter activity of reporter constructs containing the −1914G-847T-82T (minor) haplotype showed significantly greater activity than the other haplotype, −1914A-847C-82C (major), in response to poly(I:C), as described previously (10). Here we examined the effects of DEX and SAL on the promoter activities of these reporter constructs (Figure 5A). DEX and SAL suppressed reporter activities, and their synergistic suppression of luciferase activities was also evident after concurrent exposure to DEX and SAL in both haplotypes (Figure 5B). However, no difference was evident in the activities of luciferase between the major and minor haplotypes after concurrent exposure to DEX and SAL (P = 0.096 by the Bonferroni-Dunn test, with two-factor ANOVA).
Figure 5.
(A) Luciferase (Luc) constructs. Gray box indicates NF-κB regulatory region. (B) Effects of DEX and SAL on luciferase transcriptional activities of haplotypes of the long form of TSLP in NHBE. Data represent mean ± SD and are from three experiments in triplicate. *P < 0.0001 and †P = 0.0021, according to the Bonferroni-Dunn test with two-factor ANOVA.
DISCUSSION
We report that a functional promoter SNP (rs3806933) of TSLP that creates a binding site for the transcription factor AP-1 enhances AP-1 binding to the regulatory element (10). The functional variant also increases the promoter–reporter activity of long-form TSLP in response to poly(I:C) stimulation in NHBE (10). In this study, we found that the TSLP functional polymorphism was associated with both childhood atopic and adult asthma in a Japanese population. A synergistic relationship is evident between viral infection and allergen sensitization and exposure in provoking exacerbations, and the major trigger for exacerbations in both children and adults is viral infection (27–29). Clinical studies suggest that patients with asthma are more susceptible to rhinovirus infections than normal individuals, and sustain a longer duration of lower respiratory tract symptoms when infected with rhinoviruses (30–32). TSLP appears to be involved in the development of bronchial asthma through functional genetic polymorphisms that may contribute to Th2-polarized immunity through higher TSLP production by bronchial epithelial cells in response to viral respiratory infections.
A recent study showed that the expression of TSLP is increased in asthmatic airways, and correlates with lung function (3). The numbers of both epithelial and submucosal cells expressing TSLP mRNA correlated inversely with FEV1 (3). We found a significant correlation between the rs2289278 genotype and lung function (FEV1/FVC). The rs2289278 variant is located in intron 2 of long-form TSLP and in the 5′ untranslated region of short-form TSLP. However, the genetic influences of the polymorphism on the function of the TSLP gene in asthma are unclear. Further investigation of the functional role of the variant needs to be conducted.
A recent study showed that an SNP in TSLP is associated with cockroach-allergy IgE in Costa Rican girls and with total IgE in girls from two populations (9). That study showed significant evidence of linkage to IgE produced in response to cockroach allergy on chromosome 5q23 in female subjects (9). TSLP is located near the linkage peak, and exerts female-specific effects on lung disease in mice (33). Moreover, rs2289276 is reportedly associated with reductions in IgE in the response to cockroach and total IgE (9). In our Japanese population, we could not find an association between total IgE and the rs2289276 SNP among patients with asthma in this study and a female-specific effect on the associations. Although inconsistent associations may reflect sample size, phenotype heterogeneity, or gene–environment interactions in the population, further replication studies of these findings are needed.
We could not replicate our findings in the populations that were examined in two recent GWASs (24, 25). Differences in minor allele frequencies (MAFs) of the variants were evident among the control populations. The MAFs of the three variants, rs3806932, rs2289276, and rs11466741, in the Japanese population were 0.29, 0.26, and 0.26, respectively (10). To test for replication at the level of the gene rather than the SNP, further fine mapping around the TSLP gene and association studies would be needed.
We could not replicate our findings in European-ancestry and African-ancestry populations for which recent GWASs were performed (24, 25). Differences in MAFs of the variants were evident among the control populations. The MAFs of the three variants, rs3806932, rs2289276, and rs11466741, in the Japanese population were 0.29, 0.26, and 0.26, respectively (10), compared with 0.43, 0.29, and 0.29 in the Cjo;djppd Asthma Management Program (CAMP) population, 0.63, 0.18, and 0.31 in the African-American population, and 0.66, 0.17, and 0.28 in the African Caribbean population. Failure to observe SNP-for-SNP replication in ethnically diverse populations is not uncommon, and can result from variations in allele frequencies, population admixture, heterogeneity of the phenotype, and environmental factors. The fact that we could not replicate the association at the SNP level is therefore not surprising, and variants other than those tested in this study are likely to be causal. Elsewhere a gene-based rather than SNP-for-SNP approach was argued to provide evidence for genetic analysis at the functional level (25). Our findings suggest that replication occurs at the level of the gene rather than the SNP, and further fine mapping around the TSLP gene is warranted.
We previously showed that a functional rs3806933 SNP in the promoter region of long-form TSLP (−847T) creates a binding site for AP-1 and enhances AP-1 binding to the regulatory element (10). In this study, we found the binding of AP-2α protein, a possible transcription suppression factor, to the sequences containing the rs2289276 (−82C/T) SNP, and higher binding to −82C (on the protective allele). Both of the SNPs, rs3806933 (−847C/T) and rs2289276 (−82C/T), were significantly associated with susceptibility to asthma. We also identified a common disease-susceptible haplotype, T(−847)-T(−82)-C(1560), in both childhood atopic and adult asthma. The two different transcription factors on the two promoter SNPs may lead to preferential transcription from the susceptible haplotype T-T-C through their cooperative effects in bronchial epithelial cells. In this study, we did not examine the functional effects of polymorphisms rs3806932, rs2289277, rs10073816, and rs11466741, which were in strong LD with the related variants, rs3806933 and rs2289276. The functions of these linked polymorphisms remain to be elucidated.
Combination therapy with LABA and inhaled corticosteroids reduces the frequency of exacerbations in asthma, and it is also efficacious as an interventional therapy in asthma exacerbations (16–18). In this study, SAL was able to suppress the production of TSLP in NHBE cells, and we also demonstrated the synergistic suppression of poly(I:C)-induced TSLP production by DEX and SAL in NHBE cells. Respiratory viral infections are associated with the majority of exacerbations in bronchial asthma, and a recent study showed that combined corticosteroid/β2 agonists synergistically suppress rhinovirus-induced neutrophil (CXCL8) and lymphocyte (CCL5 and CXCL10) chemokine production in airway epithelial cells (34). Glucocorticosteroids inhibit both NF-κB and AP-1 through glucocorticoid-induced leucine zipper protein (35, 36), and an NF-κB binding site was identified 3.7 kb upstream from the start of TSLP transcription (13). Although further investigation of the molecular mechanisms mediating the suppressive effects of DEX and SAL on TSLP and chemokine production is needed, combination therapy may exert an anti-Th2 polarized inflammatory effect during respiratory viral infections through suppressive effects on TSLP and the production of other chemokines in airway epithelial cells.
In conclusion, we identified TSLP as a susceptibility gene for childhood atopic and adult asthma by means of a case-control study with SNPs, and demonstrated that glucocorticoid/LABA treatment synergistically suppressed poly(I:C)-induced TSLP in NHBE. Our data strongly support the important role of TSLP in bronchial asthma and the clinical benefits of combination therapy in asthma management.
Supplementary Material
Acknowledgments
We thank all the participants in this study. We also thank Makiko T. Shimizu, Hiroshi Sekiguchi, and Nami Kawaraichi for technical assistance, Chinatsu Fukushima for providing patient data, and Dr. Takashi Naito for correcting samples. The authors thank the investigators from the Genomic Research on Asthma in the African Diaspora consortium, including Terri H. Beaty, Ingo Ruczinski, Audrey V. Grant, Candelaria Vergara, Tanda Murray, Cassandra Foster, Peisong Gao, Alkis Togias, Nadia Hansel, Gregory Diette, Patrick Breysse, N. Franklin Adkinson, Mark Liu, Mezbah Faruque, Georgia Dunston, and Harold Watson. Genomic Research on Asthma in the African Diaspora was supported by National Institutes of Health grant HL087699.
This work was supported by grants from the Ministry of Education, Culture, Sports, Science, and Technology of Japan, and from the Ministry of Health, Labor, and Welfare of Japan. It was also supported by the members of the Rotary Club of Osaka–Midosuji District (2660 Rotary International) in Japan. K.C.B. was supported in part by the Mary Beryl Patch Turnbull Scholar Program.
This article has an online supplement, which is accessible from this issue's table of contents at www.atsjournals.org
Originally Published in Press as DOI: 10.1165/rcmb.2009-0418OC on July 23, 2010
Author Disclosure: M.A. served on the advisory board for GlaxoSmithKline and Kyorin Co., Ltd., for $5,001–$10,000. M.A. also received lecture fees from GlaxoSmithKline, Boehringer Ingelheim, Abbott Japan Co., Ltd., and AstraZeneca for $1,001–$5,000. K.C.B. received sponsored grants from Sanofi-Aventis and the National Institutes of Health (HL087699) for more than $100,001 each. Q.L.D. received a sponsored grant from Canadian Institute of Health Research for more than $100,001. T.E. received lecture fees from Sanofi Aventis, Boehringer Ingelheim, and Kyowa Hakko Kirin Pharma for $1,001–$5,000 each, and Dainippon Sumitomo Pharma and Ono Pharmaceutical Co., Ltd. for less than $1,000 each. N.H. received a sponsored grant from the Japanese Society for the Promotion of Science for more than $100,001. T.M. received lecture fees from GlaxoSmithKline, Schering-Plough, AstraZeneca, and Dainippon Sumitomo Pharma Co., Ltd., for up to $1,000 each. H.S. received lecture fees from Banyu for up to $1,000 and GlaxoSmithKline for $1001–$5000. M.T. received lecture fees from Merck, GlaxoSmithKline, and Schering-Plough for up to $1,000. S.T.W received consultancy fees from Genentech for up to $1,000. S.F.Z. received expert witness fees from Amgen Corp. for $10,001–$50,000, and royalties from Immgenix for up to $1,000. S.F.Z. owns stock in Amgen Corp for $50,001–$100,000. S.F.Z. received sponsored grants from the National Institutes of Health and Juvenile Diabetes Research Foundation for more than $100,001, and the Food Allergy Initiative for $50,001–$100,000. None of the other authors has a financial relationship with a commercial entity that has an interest in the subject of this manuscript.
References
- 1.Reche PA, Soumelis V, Gorman DM, Clifford T, Liu MR, Travis M, Zurawski SM, Johnston J, Liu YJ, et al. Human thymic stromal lymphopoietin preferentially stimulates myeloid cells. J Immunol 2001;167:336–343. [DOI] [PubMed] [Google Scholar]
- 2.Soumelis V, Reche PA, Kanzler H, Yuan W, Edward G, Homey B, Gilliet M, Ho S, Antonenko S, Lauerma A, et al. Human epithelial cells trigger dendritic cell mediated allergic inflammation by producing TSLP. Nat Immunol 2002;3:673–680. [DOI] [PubMed] [Google Scholar]
- 3.Ying S, O'Connor B, Ratoff J, Meng Q, Mallett K, Cousins D, Robinson D, Zhang G, Zhao J, Lee TH, et al. Thymic stromal lymphopoietin expression is increased in asthmatic airways and correlates with expression of Th2-attracting chemokines and disease severity. J Immunol 2005;174:8183–8190. [DOI] [PubMed] [Google Scholar]
- 4.Ziegler SF, Liu YJ. Thymic stromal lymphopoietin in normal and pathogenic T cell development and function. Nat Immunol 2006;7:709–714. [DOI] [PubMed] [Google Scholar]
- 5.Liu YJ. Thymic stromal lymphopoietin: master switch for allergic inflammation. J Exp Med 2006;203:269–273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Liu YJ, Soumelis V, Watanabe N, Ito T, Wang YH, Malefyt Rde W, Omori M, Zhou B, Ziegler SF. TSLP: an epithelial cell cytokine that regulates T cell differentiation by conditioning dendritic cell maturation. Annu Rev Immunol 2007;25:193–219. [DOI] [PubMed] [Google Scholar]
- 7.Allakhverdi Z, Comeau MR, Jessup HK, Yoon BR, Brewer A, Chartier S, Paquette N, Ziegler SF, Sarfati M, Delespesse G. Thymic stromal lymphopoietin is released by human epithelial cells in response to microbes, trauma, or inflammation and potently activates mast cells. J Exp Med 2007;204:253–258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Vercelli D. Discovering susceptibility genes for asthma and allergy. Nat Rev Immunol 2008;8:169–182. [DOI] [PubMed] [Google Scholar]
- 9.Hunninghake GM, Lasky-Su J, Soto-Quiros ME, Avila L, Liang C, Lake SL, Hudson TJ, Spesny M, Fournier E, Sylvia JS, et al. Sex-stratified linkage analysis identifies a female-specific locus for IgE to cockroach in Costa Ricans. Am J Respir Crit Care Med 2008;177:830–836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Harada M, Hirota T, Jodo AI, Doi S, Kameda M, Fujita K, Miyatake A, Enomoto T, Noguchi E, Yoshihara S, et al. Functional analysis of the thymic stromal lymphopoietin variants in human bronchial epithelial cells. Am J Respir Cell Mol Biol 2009;40:368–374. [DOI] [PubMed] [Google Scholar]
- 11.Johnston SL, Pattemore PK, Sanderson G, Smith S, Lampe F, Josephs L, Symington P, O'Toole S, Myint SH, Tyrrell DA, et al. Community study of role of viral infections in exacerbations of asthma in 9–11 year old children. BMJ 1995;310:1225–1229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Alexopoulou L, Holt AC, Medzhitov R, Flavell RA. Recognition of double-stranded RNA and activation of NF-kB by Toll-like receptor 3. Nature 2001;413:732–738. [DOI] [PubMed] [Google Scholar]
- 13.Lee HC, Ziegler SF. Inducible expression of the proallergic cytokine thymic stromal lymphopoietin in airway epithelial cells is controlled by NF-kB. Proc Natl Acad Sci USA 2007;104:914–919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kato A, Favoreto S Jr, Avila PC, Schleimer RP. TLR3- and Th2 cytokine–dependent production of thymic stromal lymphopoietin in human airway epithelial cells. J Immunol 2007;179:1080–1087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Pauwels RA, Lofdahl CG, Postma DS, Tattersfield AE, O'Byrne P, Barnes PJ, Ullman A. Effect of inhaled formoterol and budesonide on exacerbations of asthma: Formoterol and Corticosteroids Establishing Therapy (FACET) International Study Group. N Engl J Med 1997;337:1405–1411. [DOI] [PubMed] [Google Scholar]
- 16.Calverley P, Pauwels R, Vestbo J, Jones P, Pride N, Gulsvik A, Anderson J, Maden C. Trial of Inhaled Steroids and Long-Acting Beta2 Agonists Study Group: combined salmeterol and fluticasone in the treatment of chronic obstructive pulmonary disease: a randomised controlled trial. Lancet 2003;361:449–456. [DOI] [PubMed] [Google Scholar]
- 17.Kankaanranta H, Lahdensuo A, Moilanen E, Barnes PJ. Add-on therapy options in asthma not adequately controlled by inhaled corticosteroids: A comprehensive review. Respir Res 2004;5:17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.O'Byrne PM, Bisgaard H, Godard PP, Pistolesi M, Palmqvist M, Zhu Y, Ekström T, Bateman ED. Budesonide/formoterol combination therapy as both maintenance and reliever medication in asthma. Am J Respir Crit Care Med 2005;171:129–136. [DOI] [PubMed] [Google Scholar]
- 19.Harada M, Hirota T, Jodo AI, Ziegler SF, Tamari M. Functional SNP in promoter of TSLP gene is associated with susceptibility to childhood atopic asthma. Am J Respir Crit Care Med 2008;177:A774. [Google Scholar]
- 20.Hirota T, Harada M, Sakashita M, Doi S, Miyatake A, Fujita K, Enomoto T, Ebisawa M, Yoshihara S, Noguchi E, et al. Genetic polymorphism regulating ORM1-like 3 (Saccharomyces cerevisiae) expression is associated with childhood atopic asthma in a Japanese population. J Allergy Clin Immunol 2008;121:769–770. [DOI] [PubMed] [Google Scholar]
- 21.Hirota T, Suzuki Y, Hasegawa K, Obara K, Matsuda A, Akahoshi M, Nakashima K, Cheng L, Takahashi N, Shimizu M, et al. Functional haplotypes of IL-12B are associated with childhood atopic asthma. J Allergy Clin Immunol 2005;116:789–795. [DOI] [PubMed] [Google Scholar]
- 22.Harada M, Nakashima K, Hirota T, Shimizu M, Doi S, Fujita K, Shirakawa T, Enomoto T, Yoshikawa M, Moriyama H, et al. Functional polymorphism in the suppressor of cytokine signaling 1 gene associated with adult asthma. Am J Respir Cell Mol Biol 2007;36:491–496. [DOI] [PubMed] [Google Scholar]
- 23.Harada M, Obara K, Hirota T, Yoshimoto T, Hitomi Y, Sakashita M, Doi S, Miyatake A, Fujita K, Enomoto T, et al. A functional polymorphism in IL-18 is associated with severity of bronchial asthma. Am J Respir Crit Care Med 2009;180:1048–1055. [DOI] [PubMed] [Google Scholar]
- 24.Himes BE, Hunninghake GM, Baurley JW, Rafaels NM, Sleiman P, Strachan DP, Wilk JB, Willis-Owen SA, Klanderman B, Lasky-Su J, et al. Genome-wide association analysis identifies PDE4D as an asthma-susceptibility gene. Am J Hum Genet 2009;84:581–593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Mathias RA, Grant AV, Rafaels N, Hand T, Gao L, Vergara C, Tsai YJ, Yang M, Campbell M, Foster C, et al. A genome-wide association study on African-ancestry populations for asthma. J Allergy Clin Immunol 2010;125:336–346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wang X, Bolotin D, Chu DH, Polak L, Williams T, Fuchs E. AP-2alpha: a regulator of EGF receptor signaling and proliferation in skin epidermis. J Cell Biol 2006;172:409–421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Murray CS, Simpson A, Custovic A. Allergens, viruses, and asthma exacerbations. Proc Am Thorac Soc 2004;1:99–104. [DOI] [PubMed] [Google Scholar]
- 28.Contoli M, Caramori G, Mallia P, Johnston S, Papi A. Mechanisms of respiratory virus-induced asthma exacerbations. Clin Exp Allergy 2005;35:137–145. [DOI] [PubMed] [Google Scholar]
- 29.Sears MR. Epidemiology of asthma exacerbations. J Allergy Clin Immunol 2008;122:662–668. [DOI] [PubMed] [Google Scholar]
- 30.Gern JE, Calhoun W, Swenson C, Shen G, Busse WW. Rhinovirus infection preferentially increases lower airway responsiveness in allergic subjects. Am J Respir Crit Care Med 1997;155:1872–1876. [DOI] [PubMed] [Google Scholar]
- 31.Papadopoulos NG, Stanciu LA, Papi A, Holgate ST, Johnston SL. A defective Type 1 response to rhinovirus in atopic asthma. Thorax 2002;57:328–332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wark PA, Johnston SL, Bucchieri F, Powell R, Puddicombe S, Laza-Stanca V, Holgate ST, Davies DE. Asthmatic bronchial epithelial cells have a deficient innate immune response to infection with rhinovirus. J Exp Med 2005;201:937–947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Taneda S, Segerer S, Hudkins KL, Cui Y, Wen M, Segerer M, Wener MH, Khairallah CG, Farr AG, Alpers CE. Cryoglobulinemic glomerulonephritis in thymic stromal lymphopoietin transgenic mice. Am J Pathol 2001;159:2355–2369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Edwards MR, Johnson MW, Johnston SL. Combination therapy: synergistic suppression of virus-induced chemokines in airway epithelial cells. Am J Respir Cell Mol Biol 2006;34:616–624. [DOI] [PubMed] [Google Scholar]
- 35.Barnes PJ. Corticosteroid effects on cell signalling. Eur Respir J 2006;27:413–426. [DOI] [PubMed] [Google Scholar]
- 36.Mittelstadt PR, Ashwell JD. Inhibition of AP-1 by the glucocorticoid-inducible protein GILZ. J Biol Chem 2001;276:29603–29610. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.