Abstract
Asthma is the leading serious pediatric chronic illness in the United States, affecting 7.1 million children. The prevalence of asthma in children under 4 years of age has increased dramatically in the last 2 decades. Existing evidence suggests that this increase in prevalence derives from early environmental exposures acting on a pre-existing asthma-susceptible genotype. We studied the origins of asthma susceptibility in developing lung in rat strains that model the distinct phenotypes of airway hyperresponsiveness (Fisher rats) and atopy (brown Norway [BN] rats). Postnatal BN rat lungs showed increased epithelial proliferation and tracheal goblet cell hyperplasia. Fisher pups showed increased lung resistance at age 2 weeks, with elevated neutrophils throughout the postnatal period. Diverse transcriptomic signatures characterized the distinct respiratory phenotypes of developing lung in both rat models. Linear regression across age and strain identified developmental variation in expression of 1,376 genes, and confirmed both strain and temporal regulation of lung gene expression. Biological processes that were heavily represented included growth and development (including the T Box 1 transcription factor [Tbx5], the epidermal growth factor receptor [Egfr], the transforming growth factor beta-1-induced transcript 1 [Tgfbr1i1]), extracellular matrix and cell adhesion (including collagen and integrin genes), and immune function (including lymphocyte antigen 6 (Ly6) subunits, IL-17b, Toll-interacting protein, and Ficolin B). Genes validated by quantitative RT-PCR and protein analysis included collagen III alpha 1 Col3a1, Ly6b, glucocorticoid receptor and Importin-13 (specific to the BN rat lung), and Serpina1 and Ficolin B (specific to the Fisher lung). Innate differences in patterns of gene expression in developing lung that contribute to individual variation in respiratory phenotype are likely to contribute to the pathogenesis of asthma.
Keywords: asthma susceptibility, lung development, developmental gene expression
Asthma, the leading serious chronic illness of children in the United States, affects an estimated 7.1 million children under age 18 years (1). The prevalence of asthma in children under 4 years of age has increased by 74% in the last 2 decades (2, 3). Abnormalities of lung structure and function already exist in infants who will subsequently develop asthma. A growing body of evidence points to genes important in lung development as determinants of asthma susceptibility.
A total of 90% of all asthma originates in childhood. A number of observations suggest that early influences on lung development contribute to asthma susceptibility: in utero exposure to allergen increases risk of atopy in children (4, 5); mechanical ventilation, chronic lung disease, and early glucocorticoid (GC) therapy increase the risk for pediatric asthma (2); boys (in whom lung development lags behind girls) have a higher incidence of and more severe asthma; airway biopsies in children show tissue restructuring up to 4 years before the onset of asthmatic symptoms (6); and infants of smoking mothers have reduced lung function (compliance/resistance of the respiratory system) that tracks into later life (7, 8). Collectively, these findings suggest that intrinsic and induced changes in the lung in infancy (or earlier) are programmed into the long-term respiratory phenotype, conferring susceptibility to asthma.
Lung development is determined by complex interactions between epithelial and mesenchymal cells, mediated by multiple growth and differentiation factors (9, 10). Beginning in late gestation (36 wk) and continuing through early postnatal life (2–3 yr), immature lung saccules are gradually remodeled into mature alveoli. This “remodeling” process involves reduction of the extracellular matrix (ECM), thinning of alveolar walls, and expansion of capillary networks. In the asthmatic lung, repair of damaged epithelium is associated with reactivation of many of these same growth factors, which can, in turn, induce ECM deposition (11), subepithelial fibroblast proliferation, myofibroblast activation, and, eventually, airway remodeling (12–14). It has been suggested that variation in the expression of factors associated with subclinical changes in normal developmental remodeling could confer increased risk for remodeling of damaged airways in asthma (6, 15–19).
Three commonly studied rat strains (Lewis, brown Norway [BN], and Fisher) have long been used to model asthma (20). These strains have distinct sets of characteristics comparable to asthma in humans, each providing insight into distinct phenotypes associated with human asthma and airway hyperresponsiveness (AHR). The normoresponsive Lewis rat serves as a control (21). Although appearing normal at baseline, the defining features of asthma, including airway constriction, eosinophilic inflammation, and AHR, can be reproduced in the Lewis rat after allergen sensitization and challenge (22). In contrast, the BN rat is naturally atopic, and has long been known to produce very high levels of IgE in response to allergen (23, 24). In common with human allergic asthma, BN rats have early and late allergic responses and T cell–mediated eosinophilic airway inflammation (22). With repeated allergen challenge, they develop AHR to methacholine (MCh) (25), and have structural changes in the airways, including increased airway smooth muscle (ASM) mass (26–28). The Fisher rat is innately hyperresponsive, and has increased ASM mass (21), though it is not atopic.
We hypothesized that innate differences in patterns of gene expression in developing lung confer individual variation in asthma susceptibility. We show that distinct developmental respiratory phenotypes of rat animal models of atopy and innate AHR are associated with highly diverse transcriptome signatures.
MATERIALS AND METHODS
Animals
Lewis, Fisher, and BN male and female rats (Charles River Laboratories, Saint Constant, PQ, Canada) were housed in the Montreal Children's Hospital Animal Facility. All procedures involving animals were conducted according to the criteria established by the Canadian Council for Animal Care, and approved by the Animal Care Committee of the McGill University Health Centre (Montreal, PQ, Canada).
Airway Responsiveness
At 14 days of age airway responsiveness was measured with a computer-controlled small animal ventilator (flexiVent; SCIREQ, Montreal, PQ, Canada). Rats were deeply anesthetized by an intraperitoneal injection of xylaine (8 mg/kg) intraperitoneally and pentobarbital (70 mg/kg), tracheotomized and ventilated quasisinusoidally at a frequency of 150 breaths/min, and tidal volume of 10 ml/kg at positive end-expiratory pressure level of 3 cm H2O. Subsequently, they were paralyzed by an intraperitoneal injection of pancuronium bromide (0.8 mg/kg). Baseline respiratory system resistance was measured at 150 breaths/min. Maximal resistance was recorded before and after increasing doses of aerosolized MCh (6.25, 12.5, 25, and 50 mg/ml; n ≥ 5 for each rat strain).
Bronchoalveolar Lavage
Bronchoalveolar lavage (BAL) was performed by instilling the lungs four times with 0.4, 0.7, 0.7, and 1.0 ml of cold PBS for Postnatal Day (PN) 1, 4, 7, and 14, respectively, through a tracheal cannula. Lavage fluid was centrifuged and pellets were resuspended in 0.5 ml cold saline. Animals used for BAL collection were not subjected to flexiVent analysis. Total cell numbers were counted with a hemacytometer. Cytospin slides (Cytospin 4; Shandon, Pittsburgh, PA), prepared for differential cells counts, were stained with Diff-Quick (Hema3 stainset; Fisher Scientific, Middletown, VA) and counted (>200 cells/slide) to determine the percentage of each cell type (n ≥ 5 for each rat strain).
Rat Lung Primary Cell Culture
Isolation and primary culture of Lewis, Fisher, and BN rat lung cells was performed at PN1, 4, 7, and 14, as we previously described (29). Rats were killed with pentobarbital, lungs were excised, and fibroblast and epithelial cells were isolated. Briefly, lung tissue was digested with 2.5% trypsin and filtered repeatedly through a membrane. After incubating with 1% collagenase, cells were plated in 60 × 15-mm-well plates, and, after 1 hour, nonadherent cells were removed and replated to isolate epithelial cells from fibroblasts. Cells were grown in Eagle's minimum essential medium plus 10% FBS and counted with a hemocytometer at 2, 4, and 6 days after initial plating by removal with 0.25% trypsin plus EDTA (n = 4 cultures, all groups). Each culture represented pooled lungs from at least four animals. Viability and purity of the cultures were comparable to previously published data (29). Epithelial cells express phenotypic features of type II cells (30). Lung cell cultures are routinely tested for purity, as assessed by positive staining for cytokeratin (mouse monoclonal antibody; Santa Cruz Biotechnology, Santa Cruz, CA), and negative staining for vimentin (monoclonal antibody; Sigma-Aldrich, Oakville, ON, Canada).
Isolation of Total Lung and Lung Primary Cell RNA
RNA from whole lung tissue and primary lung cells was isolated with Trizol reagent (Invitrogen, Burlington, ON, Canada) according to the manufacturer's instructions. RNA was resuspended in 1× RNASecure (Ambion, Austin, TX) and treated with the Turbo DNase-free kit (Ambion) to remove any traces of DNA.
Quantitative Real-Time RT-PCR
Quantitative real-time RT-PCR (qRT-PCR) was performed on the Mx4000 QPCR system (Stratagene, La Jolla, CA) with the QuantiTect One-Step Probe RT-PCR Kit or the QuantiTect SYBR green RT-PCR kit (Qiagen, Mississauga, ON, Canada). Gene-specific primers and 6-carboxy-fluorescein (FAM)-labeled probes for rat collagen III alpha 1 (Col3a1), glucocorticoid receptor (GR), Importin (IPO)-13, proliferating cell nuclear antigen (PCNA), and 18 s were created with Qiagen's QuantiProbe Custom Design Software. Gene-specific primers for SYBR green detection of Serpina1, Ficolin B (Fcnb), lymphocyte antigen 6-B (Ly6b), and glyceraldehyde 3-phosphate dehydrogenase were predesigned (Qiagen QuantiTect primer assays). One-step real-time RT-PCR reactions were performed in 25 μl volumes for 40 cycles, with 10 ng of total or cellular RNA for the genes analyzed, and 50 pg for 18 seconds, which was used to normalize for the input of RNA. Synthesis of cDNA for the SYBR green RT-PCR protocol was prepared from an initial 250 ng/μl of RNA. RNA was incubated at 65°C for 5 minutes with 1 μg/μl of random primers and 10 mM deoxynucleotide triphosphates (dNTPs). To this, 5× first-strand buffer, 1 mM DTT, 1 μl of RNase OUT (Invitrogen) and 1 μl of Superscript II (Invitrogen) was added and incubated at 42°C for 1 hour and at 70°C for 15 minutes. RT-PCR was performed in 25 μl reactions for 40 cycles with 1 μl of cDNA. Relative mRNA expression levels for Col3a1, GR, IPO13, and PCNA were analyzed according to the standard curve method. Typically, our experiments yield RNA amounts in the range of 5–50 ng. Results for Serpina1, Fcnb, and Ly6b, which were optimized for the SYBR green detection system, were analyzed using the delta-delta cycle threshold method (n ≥ 4 for all assays).
Antibodies
Antibodies used for these experiments were as follows: Fcnb, 1:200 (Santa Cruz Biotechnology); α1-antitrypsin (α1-AT; Serpina1), 1:200 (Abbiotec, San Diego, CA); collagen III, 1:200 (Abcam, Cambridge, MA); IPO13, 1:100; GR, 1:50 (Santa Cruz Biotechnology); β-actin, 1:5000 (Sigma-Aldrich); anti-rabbit IgG–horseradish peroxidase (HRP) conjugated, 1:10000 (Amersham, Little Chalfont, Buckinhamshire, UK); bovine anti-goat IgG-HRP conjugated, 1:10,000 (Santa Cruz Biotechnology); goat anti-rabbit biotinylated, 1:300 (Sigma-Aldrich); monoclonal vimentin, 1:200 (Sigma-Aldrich); and monoclonal cytokeratin, 1:200 (Santa Cruz Biotechnology).
Western Blotting
Total protein (15 μg) isolated from rat lung with KPO4 buffer was boiled for 5 minutes in SDS loading buffer before being electrophoresed on 10% SDS-polyacrylamide gels and transferred to polyvinylidene fluoride membranes (Bio-Rad Laboratories, Hercules, CA). Membranes were blocked overnight at 4°C with 10% milk/PBS-Tween and incubated with primary antibody for 1 hour. The blots were washed in PBS-Tween buffer, incubated with HRP-conjugated secondary antibodies, developed with the ECL Plus Western Blotting Detection System (GE Healthcare, Little Chalfont, Buckinghamshire, UK), and exposed to Bioflex film (Clonex Corp., Markham, Ontario, Canada).
Lung Fixation
Lungs were inflated in 4% paraformaldehyde at a pressure of 20 cm H2O and fixed overnight. Samples were dehydrated through a series of ethanol washes before being embedded in paraffin. Slices (5 μm thick) of whole lung were sectioned onto slides.
Immunohistochemistry
Slides were rehydrated through decreasing ethanol washes, rinsed with PBS–0.03% Triton, boiled in 10 mM sodium citrate for antigen retrieval, and then incubated in H2O2 with methanol for 20 minutes to block endogenous peroxidase activity. Slides were blocked with PBS–0.03% Triton containing 5% normal goat serum and 1% BSA. Primary antibodies were incubated overnight at 4°C, and secondary biotinylated antibodies were incubated for 2 hours at room temperature. Slides were washed with PBS–0.03% Triton and then stained according to the avidin–biotin peroxide complex method with the Vecstatin kit (Vector Laboratories, Burlingame, CA). Finally, they were exposed to 3,3′-diaminobenzidine (Sigma-Aldrich), and then counterstained with hematoxylin. Immunohistochemistry (IHC) for PCNA, which was used as a marker of cell proliferation, was performed with the Zymed PCNA staining kit (Zymed Laboratories, San Diego, CA). Slide preparation was performed as outlined previously here, and the staining and visualization were followed according to the manufacturer's specifications (n ≥ 4 for all IHC).
Goblet Cells
Slides were rehydrated through a series of decreasing ethanol washes and stained with the periodic acid–Schiff kit (Sigma-Aldrich) to visualize goblet cells. Images in figures are representative of sections from at least four animals.
Illumina Microarrays
Global gene expression analysis was performed at the McGill University and Genome Quebec Innovation Centre with the Illumina RatRef-12 array. The raw microarray data were standard background corrected and quantile normalized with the Bioconductor package, lumi (http://www.bioconductor.org/packages/2.0/bioc/html/lumi.html). The data set contains 22,523 features (probes for gene expression). These microarray data are available at the National Center for Biotechnology Information Gene Expression Omnibus (http://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc+GSE19666).
Statistical and Bioinformatics Analyses
All results for respiratory phenotypes are presented as means (±SEM). Statistical significance of differences between group averages was determined by one-way or two-way ANOVA, except where noted. Pair-wise comparisons for respiratory phenotypes were assessed by the two-tailed Student's t test, with significance defined as a P value of 0.05 or less. Initial analysis of microaarray data was performed with FlexArray, a software package that was designed at the McGill University and Genome Quebec Innovation Centre (Montreal, PQ, Canada; http://genomequebec.mcgill.ca/FlexArray/). To find probes (genes) that were significantly differentially expressed between any two sample groups in the microarray dataset, we used the randomized variance model (RVM) t test in FlexArray with a fold magnitude cutoff of two and a P value of 0.05 or less (31). RVM is a linear model for microarray-reported gene expression that takes into account the different sample-wise variance of residuals of individual genes. These individual gene variances are modeled as random selections from an inverse gamma distribution, the parameters of which are estimated with the residual sum of squares per gene. The difference between two sample means is then assessed with the standard t statistic with its modified variance term from RVM. Principal component analysis (PCA), a statistical tool for feature reduction, was used to investigate the dissimilarities between global gene expression (transcriptome) sample profiles. PCA assesses the relative impact of factors such as an experimental treatment, biological variation between individual animals, date of microarray hybridization, et cetera, to sample variation in an unsupervised fashion. Each point in a PCA plot corresponds to a sample transcriptome profile. After PCA analysis, we considered the effects of strain and age, as well as the interaction of age and strain, by performing linear regression analysis with the Bioconductor package, limma (32).
RESULTS
Fisher Rat Pups Display Impaired Lung Function
In rodents, alveolarization is exclusively postnatal. We initially asked whether naive rat models of atopy and innate hyperresponsiveness would exhibit differences in lung function in the newborn period. Airway responsiveness was measured in BN, Fisher, and Lewis rat pups at PN14 (the earliest time at which lung function can be reliably assessed; n ≥ 5 all groups) with a small animal ventilator. Fisher rat pups displayed significantly elevated baseline lung resistance when compared with Lewis pups. Administration of increasing doses of MCh, a smooth muscle agonist, dramatically enhanced this effect (Figure 1, P < 0.05). BN rats had a limited increase in baseline lung resistance when compared with Lewis rats, which was not affected by increasing doses of MCh (Figure 1, P < 0.05). These findings confirm that changes in lung function that associate with asthma susceptibility and AHR in these models are present very early in life.
Figure 1.
Fisher rat pups display increased lung resistance at age 2 weeks. Airway responsiveness was measured at Postnatal Day (PN) 14 with a small animal ventilator. Fisher rat pups displayed significantly elevated baseline lung resistance, an effect that was enhanced by methacholine (MCh; n ≥ 5; P < 0.05). Brown Norway (BN) rats had a limited increase in baseline lung resistance.
BN Rats Display Increased Cell Proliferation
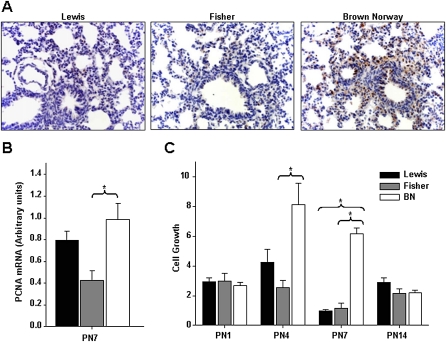
We next assessed whether the differences in lung function observed during the newborn period would reflect changes in lung growth and structure. Lung and body weight and lung to body weight ratios were normal. No obvious interstrain differences in gross morphology of the lung were seen during the first 2 weeks of life. Restructuring of asthmatic lungs is associated with epithelial proliferation. We asked whether developmental patterns of respiratory epithelial proliferation distinguished the three rat strains. PCNA expression in whole lung tissue (n ≥ 4, all groups) was assessed by immunohistochemistry (Figure 2A) and qRT-PCR (Figure 2B). At PN7, BN rat lungs displayed increased epithelial proliferation when compared with Lewis and Fisher rat lungs, as assessed by PCNA immunostaining (Figure 2A). This was associated with elevated expression of PCNA mRNA (Figure 2B, P > 0.05).
Figure 2.
BN rats display increased cell proliferation. (A) Immunostaining for proliferating cell nuclear antigen (PCNA) shows an increase in proliferation in BN rat lungs when compared with Lewis and Fisher rat lungs at PN7 (n ≥ 4, all groups). (B) PCNA expression in whole lung tissue was assessed by quantitative real-time PCR. At PN7, BN rat lungs had significantly elevated PCNA levels (n ≥ 4; P < 0.05). (C) Primary cultures of lung epithelial cells were isolated from rat pups at PN1, 4, 7, and 14. At PN4 and 7, the BN rat lung shows significantly elevated cell proliferation (n ≥ 4; P < 0.05).
As an additional assessment of cellular proliferation, we cultured primary airway epithelial cells isolated from rat lungs at PN1, 4, 7, and 14. Cultures were grown for 2, 4, and 6 days, after which the cells were collected and counted to determine the relative rate of cell proliferation. At PN4 and 7, when the normal rat airway epithelium undergoes a shift from an essentially proliferating to a differentiating state, the BN rat lung epithelial cells continued to show significantly elevated cell proliferation (n ≥ 4 cultures each pooled from ≥ 4 lungs; P < 0.05; Figure 2C).
Neutrophils Are Elevated in BAL from Fisher Pups
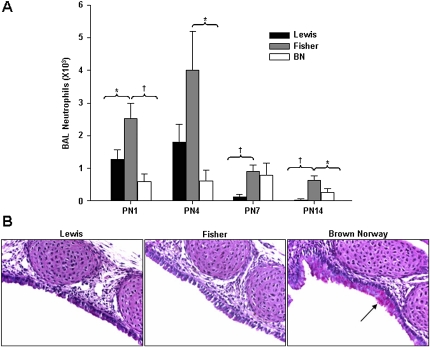
Inflammation in asthma is associated with the recruitment of inflammatory cells, mainly eosinophils, activated mast cells, and T helper type 2 cells into the airways. We assessed inflammatory cell infiltrates in BAL fluid isolated from rat pups at PN1, 4, 7, and 14 (n = 9, PN1, 7, and 14; n = 6, PN4). There were no marked differences in total cell counts between the strains. Fisher pups had significantly elevated neutrophils when compared with Lewis pups at PN1, 7, and 14. Compared with BN pups, neutrophils in Fisher pups were significantly elevated at PN1, 4, and 14 (Figure 3A, P < 0.05).
Figure 3.
(A) Postnatal Fisher pups show increased neutrophils in bronchoalveolar lavage (BAL) infiltrates. Inflammatory cell infiltrates were assessed in BAL fluid isolated from rat pups at PN1, 4, 7, and 14. Fisher pups had significantly elevated neutrophils when compared with Lewis pups at PN1, 7, and 14. Compared with BN pups, neutrophils in Fisher pups were significantly elevated at PN1, 4, and 14 (n = 9, PN1, 7, and 14; n = 6, PN4; *P < 0.05, †P < 0.01). (B) Newborn BN pups display goblet cell hyperplasia. Periodic acid–Schiff staining was used to assess goblet cells in trachea of BN, Fisher, and Lewis rats. Few goblet cells were present in normal lung between PN1 and 14. By contrast, trachea of naive BN rats had significantly elevated goblet cell numbers at PN1. By contrast, trachea of naive BN rats displayed both accumulation and hypertrophy of goblet cells at PN1.
Newborn BN Pups Display Goblet Cell Hyperplasia
Substantial increases in the number of goblet cells lining the airway epithelium is a characteristic feature of inflammation in asthmatic airways and neonatal lung injury. We used periodic acid–Schiff staining to assess goblet cells in trachea and bronchi of developing BN, Fisher, and Lewis rats (n = 4 all groups). Few goblet cells were present in postnatal lung of Lewis and Fisher rat between PN1 and 14. By contrast, trachea of naive BN rats displayed both accumulation and hypertrophy of goblet cells at PN1 (Figure 3B).
Transcriptome Profiles of Developing Lungs in BN, Fisher, and Lewis Rats Are Highly Distinct
Having established developmental differences in the respiratory phenotype of these models, we next explored associated changes in gene expression. Illumina genome-wide profiling was performed with the RatRef12 array on RNA isolated at PN1, 7, and 14 from the three rat strains (n = 4, all strain–time combinations, except for BN [n = 5] and Lewis [n = 3] at PN1). PCA was used to assess patterns of global gene expression in all strains over time. All three strains showed clustering of global patterns of gene expression. The PCA plot at PN7 is illustrated in Figure 4A. Considerably more overlap was noted between the Fisher and Lewis strains (for example, similar values along the Principal Component 1 axis in Figure 4A) than between either strain compared with BN rats, suggesting greater genetic differences between the naturally atopic BN and the Lewis and Fisher strains.
Figure 4.
Transcriptome profiles of developing lungs in BN, Fisher, and Lewis rats are highly distinct. (A) Principal component analysis plot of lung global gene expression profile at PN7 illustrating the highly distinct transcriptome signatures of the three rat models. Note that the expression profiles of the BN rat lung are more different from the Lewis rat lung than the Fisher rat lung (n = 4, all strain–time combinations, except for BN rat at PN1 [n = 5] and Lewis at PN1 [n = 3]). (B) The Venn diagrams show overlapping patterns of gene expression at PN1, 7, and 14, and illustrate strain and temporal regulation of lung gene expression in the neonatal period. Fold change limits set at greater than +2 or less than −2 (P < 0.05, all comparisons). (C) Biological processes heavily represented among variably expressed genes (from a linear regression analysis accounting for strain and age) are illustrated in the heat maps of samples at PN7. The processes highlighted are growth and development, extracellular matrix (ECM) and cell adhesion, and immune function. Novel genes of interest identified here, including members of gene families previously implicated in asthma, are indicated by blue arrows. PC, principal component.
PCA analysis provided evidence of distinct transcriptome profiles across strains. The Venn diagrams (fold change > +2 or < −2; P < 0.05, all comparisons) in Figure 4B show overlapping patterns of gene expression at PN1, 7, and 14, and illustrate strain and temporal regulation of lung gene expression in the neonatal period. For additional information on temporal overlap of gene expression, see the Venn diagrams provided in Figure E1 in the online supplement. Finally, we used linear regression analysis to identify specific genes that are variably expressed during postnatal lung development in a strain-dependent fashion. Linear regression accounting for age and strain effects identified developmental variation in expression of 1,376 such genes. Biological processes heavily represented among these variably expressed genes included growth and development (including the T Box 1 transcription factor 5 [Tbx5], the epidermal growth factor receptor [EGFR], and the transforming growth factor β-1-induced transcript 1 [Tgfbr1l1], ECM and cell adhesion (including multiple collagen and integrin genes), and immune function (including Ly6 subunits, IL-17b, and the Toll-interacting protein) (Figure 4C). A subset of nine genes with unique expression profiles in the lungs of BN or Fisher rats were chosen for validation by qRT-PCR and protein analysis. Five were clearly validated: Col3a1, Egfr, Fcnb, Ly6b Serpina1. Among these, validation of four is described subsequently here. The role of Egfr, which is up-regulated in BN rat lungs and down-regulated in Fisher rat lungs, is currently the subject of a larger study.
Increased Expression of Collagen III a1, a Key Structural Component of the Lung, Is Up-Regulated in Postnatal Lungs of BN Rats
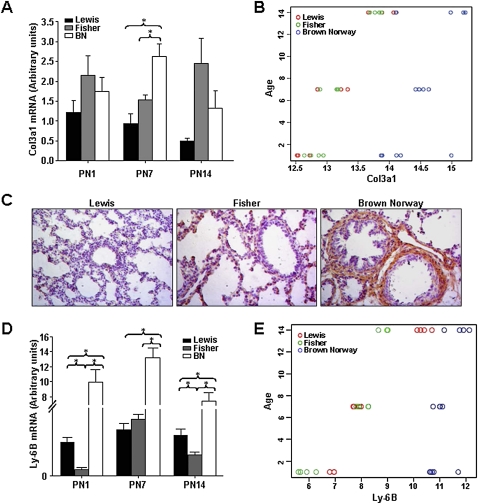
Collagen and elastin are key components of the architectural framework of alveoli that are essential to the establishment of normal mechanical properties of lung parenchyma. Increased collagen deposition in the lung is a marker for airway remodeling in asthma (33). Col3a1 encodes the α1 chain of collagen III. Given the mesenchymal expression of Col3a1, we assessed Col3a1 mRNA by qRT-PCR in primary cultures of postnatal lung fibroblasts. We observed a significant increase in Col3a1 in BN rat as compared with Lewis and Fisher rat at PN7, but not at PN1 or 14 (Figure 5A, n = 4, P < 0.05). Figure 5B illustrates the interaction of age (PN1–PN14) and strain for Col3a1 mRNA, as assessed by regression analysis. These data, demonstrating strain- and time-specific variance in gene expression, suggest that Col3a1 expression increases from PN1 to 14 in all three strains. Moreover, at all time points, Col3a1 levels were significantly higher in BN rat lungs. Representative images of Collagen III α1 immunostaining in lungs of BN rats at PN7 (Figure 5C, n = 4) demonstrate an associated increase in protein expression.
Figure 5.
Expression of Col3a1 and the Ly6b antigen are up-regulated in postnatal lungs of BN rat. (A) Col3a1 mRNA expression in whole lung tissue was assessed by quantitative real-time PCR at PN1, 7, and 14. A significant increase in Col3a1 mRNA in isolated BN rat lung fibroblasts was observed at PN7 (n = 4; P < 0.05). (B) Linear regression analysis shows that Col3a1 expression increases from PN1 to PN14 in all three strains. At all time points, Col3a1 levels are significantly higher in BN rat than in Lewis and Fisher rat lungs. (C) Immunostaining showing increased collagen III a1 peptide in lungs of BN rat at PN7. (D) Ly6b mRNA expression in whole lung tissue was assessed by quantitative real-time PCR at PN1, 7, and 14. A very significant increase in Ly6b mRNA is observed in BN rat lungs at all three time points (n = 4; P < 0.005). (E) Linear regression analysis shows that Ly6b expression increased from PN1 to PN14 in lungs of all three rat strains. At all time points, Ly6b mRNA levels were highest in BN rat lungs.
The Ly6b Antigen Is Dramatically Elevated in Postnatal Lungs of BN Rat
The Ly6 genetic locus controls expression of antigenic specificities on multiple cell types, including lung, trachea, and immune cells (34, 35). Expression of the secreted Ly6/PLAUR (plasminogen activator urokinase receptor) domain containing protein 1 (Slurp1) of the Ly6 family is regulated by epidermal growth factor(EGF), retinoic acid, and IFN-γ. Ly6 antigens have been implicated in structural integrity of the skin and in lymphocyte activation (34–36). Illumina array analysis identified the Ly6b, and Ly6 g6e antigens (see heatmap, Figure 4C, “Immune Function”) as being elevated in BN rat lung at PN7. A highly significant increase in Ly6b mRNA in lungs of BN rats was validated by qRT-PCR at PN1, 7, and 14 (n = 4, P < 0.005 [Figure 5D]). Figure 5E illustrates the interaction of age (PN1–PN14) and strain for Ly6b mRNA, as assessed by regression analysis. Ly6b expression showed a temporal increase in expression from PN1 to 14 in the lungs of all three rat strains. At all time points, Ly6b mRNA levels were highest in BN rat lungs. No antibody is currently available for rat Ly6b.
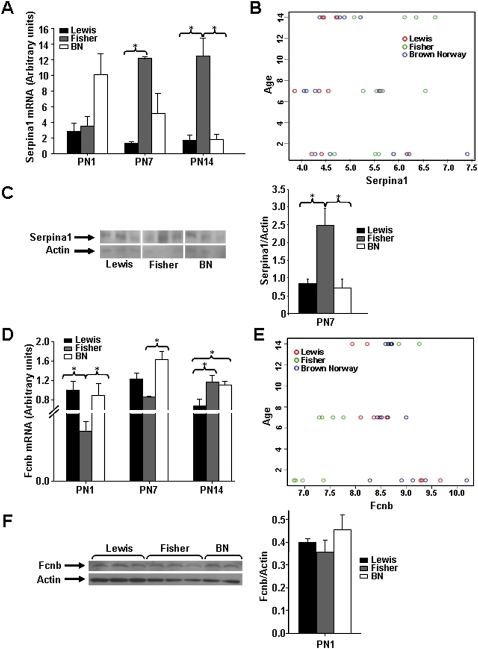
α1-AT, the Major Inhibitor of Neutrophil Elastase, Is Up-Regulated in Postnatal Lungs of Fisher Rats
The serine protease inhibitor, α1-AT, encoded by the Serpina1 gene, inhibits the activity of neutrophil elastase, a protease secreted by neutrophils during inflammation to protect the host from bacteria. Circulating α1-AT is taken up by the lung, where it acts in tissue defense against elastolysis. Serum α1-AT levels rise in situations of inflammation. Illumina array analysis identified an increase in Serpina1 mRNA in lungs of Fisher rats compared with Lewis and BN rats at PN7 (see heatmap, Figure 4C, “Growth and Development”). An increase in Serpina1 mRNA in lungs of Fisher rats at PN7 and 14 was validated by qRT-PCR (n = 4, P < 0.006 [Figure 6A]). Figure 6B illustrates the interaction of age (PN1–PN14) and strain for Serpina1 mRNA, as assessed by regression analysis. Consistent with array and qRT-PCR results, temporal changes in Serpina1 expression were limited, and elevated levels of Serpina1 in lungs of Fisher rats were observed at PN7 and 14 only. Figure 6C illustrates a representative Western blot (n = 4) showing elevated α1-AT levels in the lung of Fisher rats at PN7.
Figure 6.
Expression of Serpina1 is up-regulated, and Ficolin B (Fcnb) expression is suppressed, in lungs of postnatal Fisher rat. (A) Serpina1 mRNA expression in whole lung tissue was assessed by quantitative real-time PCR at PN1, 7, and 14. A significant increase in Serpina1 mRNA in lungs of Fisher compared with Lewis and BN rats was observed at PN7 and 14 (n = 4; P < 0.006). (B) Linear regression analysis shows that temporal changes in Serpina1 expression were limited. Elevated levels of Serpina1 in lungs of Fisher rats are apparent at PN7 and 14 only. (C) Western blot showing increased expression of Serpina1 (α1-antitrypsin) in lungs of Fisher rats compared with Lewis and BN rats (n = 3; P < 0.05). (D) Fcnb mRNA expression in whole lung tissue was assessed by quantitative real-time PCR at PN1, 7, and 14. A significant decrease in Fcnb mRNA in lungs of Fisher compared with Lewis and BN rats was observed at PN1 and 7 (n = 4; P < 0.02). (E) Linear regression analysis shows that Fcnb expression in Fisher rat lung is below that of Lewis and BN rat lung at PN1 and 7. At PN14, the reverse is true. (F) Western blot showing a trend toward decreased expression of Fcnb in lungs of Fisher rats compared with Lewis and BN rats at PN1.
Fcnb, a Pattern-Recognition Regulator of Innate Immunity, Is Suppressed in Postnatal Lungs of Fisher Rats
Ficolins are a family of pattern-recognition proteins believed to have a role in innate immune response (37). Fcnb is absent in plasma, and localizes to macrophages and neutrophils. Illumina array analysis showed a highly significant reduction in Fcnb in lungs of neonatal Fisher rats at PN1 (not shown in heatmap, log2FC = −2.43; P = 4.6 × 10−8), which was confirmed by qRT-PCR (n = 4, P ≤ 0.02 [Figure 6D]). Figure 6E illustrates the interaction of age and strain for Fcnb, as assessed by regression analysis. Note that, although Fcnb expression in Fisher rat lung was below that of Lewis and BN rat lung at PN1 and 7, the reverse is true at PN14. Figure 6F illustrates a representative Western blot (n = 3) of Fcnb expression in the lungs of Fisher rats at PN1. A trend toward reduced Fcnb did not reach significance, likely due to the limited sample size.
Temporal Up-regulation of GR and Down-regulation of its Nuclear Transport Carrier IPO13 in Postnatal Lungs of BN Rats
The GR is a critical mediator of GC effects during lung development. GR also mediates effects of exogenous GC in the control of asthma exacerbations. We did not detect significant differences in GR expression in the lungs of rat models by Illumina array analysis. Nevertheless, based on the importance of steroids in lung development and asthma management, and taking into account the evidence that children with asthma express greater quantities of GR mRNA than do healthy children, we elected to investigate expression of GR in the lungs of the three rat models. Of particular interest was whether the atopic BN rats would show increased GR expression in the lung during respiratory development.
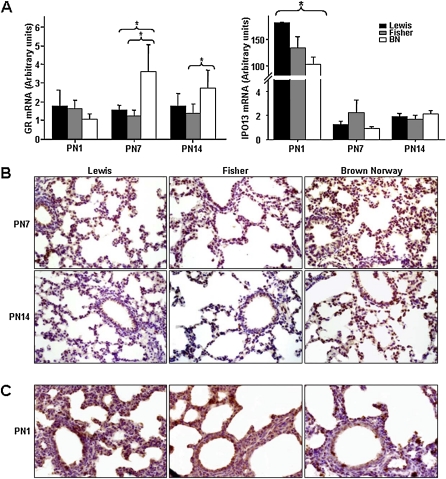
We assessed expression of GR mRNA and protein by qRT-PCR and immunohistochemistry, as illustrated in Figure 7. A significant increase in GR mRNA (Figure 7A, P < 0.006, n = 4) was observed at PN7 and 14. Representative images of GR immunostaining (Figure 6B, n = 4) show increased nuclear GR in the lungs of BN rats at PN7 and 14, consistent with mRNA findings. The developmentally regulated nuclear transport receptor, IPO13 (previously referred to as Lgl2), mediates nuclear import of GR in airway epithelial cells (38). IPO13 genetic variation is associated with improved airway responsiveness in childhood asthma (39). A significant reduction in IPO13 mRNA was observed in the lungs of BN compared with Lewis rats at PN1 (n = 4, P < 0.05 [Figure 7A]). Note also the dramatic drop in IPO13 expression in lungs of all rat models by PN7, consistent with the reported developmental regulation of this transport receptor (40). Representative images of IPO13 immunostaining (n = 4 [Figure 7C]) also show reduced nuclear IPO13 in the lungs of BN rats at PN1.
Figure 7.
Expression of the glucocorticoid receptor (GR) and importin 13 (IPO13, mediates nuclear import of GR) are temporarily up-regulated in postnatal lungs of the BN rat. (A) GR and IPO13 expression in whole lung tissue was assessed by quantitative real-time PCR. At PN7 and 14, a significant increase in GR mRNA was seen in BN rat (n = 4; P < 0.006). At PN1 only, a significant decrease in IPO13 mRNA was seen in BN rat (n = 4; P < 0.05). (B) Representative images of GR immunostaining showing increased nuclear GR in the lungs of BN rats at PN7 and 14. (C) Representative images of IPO13 immunostaining show decreased nuclear IPO13 in the lungs of BN rats at PN1.
DISCUSSION
Although a growing body of epidemiologic evidence points to aberrant lung development as a significant contributing factor to chronic respiratory disease, including asthma, little information is available on the molecular determinants of respiratory susceptibility established during pulmonary maturation. One factor that may contribute to asthma susceptibility is the long maturation period of the lung that occurs postnatally, thus limiting human studies. To our knowledge, this is the first study reporting specific developmental differences in lung growth, respiratory function, and gene expression associated with atopic and AHR-specific phenotypes in the rat in the absence of provoked allergen exposures. We report on the interaction of temporal and strain-specific variance in expression of genes important in respiratory growth and development, ECM remodeling, and immune function. Our studies allow us to begin to assemble a genotype–phenotype correlation linking the program of postnatal lung development with specific asthma susceptibility traits.
Mounting evidence suggests that aberrant signals in inflamed airways influence epithelial cell proliferation (17). Aberrant repair at the mucosal surface can, in turn, trigger abnormal behavior of the underlying mesenchyme, leading to ECM deposition and, eventually, airway remodeling. The developmental program of the lung in BN and Fisher rats model distinct asthma phenotypes that may antedate and relate to asthma susceptibility. Earlier findings demonstrated increased airway smooth muscle and innate AHR to MCh in adult Fisher rats (41). We now show that newborn Fisher rats also display significantly elevated baseline airway resistance in the absence of allergen challenge. Notable also in the innately hyperresponsive Fisher rat was an increase in neutrophils in BAL. These findings are suggestive of the presence of an early, pre- or perinatal inflammatory process associated with innate AHR. Whether the early inflammation occurs coincident to or precedes the increased airway responsiveness is still unclear, and will require further work. By contrast, the BN rat, with tracheal goblet cell hyperplasia immediately after birth and increased respiratory epithelial proliferation at a time when normal rat lung epithelium has switched from a “proliferation” to a “differentiation” mode, may represent another example of aberrant pulmonary development associated with a proinflammatory phenotype.
That gene expression profiles of developing lungs would help distinguish these rat models was predictable. The highly distinct patterns of gene expression showing interaction of strain with age throughout the period of maximal alveolarization are of great interest. Linear regression identified variant expression of 1,376 genes. The Venn diagrams in Figure 4 highlight the temporal regulation of this variance. Analysis of gene-by-strain interactions also allowed us to identify genes with variant expression restricted to either Fisher or BN rat lung at a given time point. An important objective was to identify genes that contribute to the unique features of the individual models of AHR and atopy, and we therefore focused our validation experiments on these genes.
Subepithelial airway fibrosis, a unique feature of asthma, is a consequence of excessive deposition of collagen I, III, and V in the lamina reticularis (42). The findings of increased Col3a1 mRNA and collagen III protein in postnatal lung tissue of BN rats is of particular interest in light of the findings of Fedorov and colleagues (43), demonstrating significant increases in immunoreactive collagen III in the lamina reticularis of bronchial biopsies of children with asthma. Moreover, consistent with a role for mesenchymal epithelial signaling in determining the “phenotype” of atopic risk, is the finding of a parallel increase in Egfr (data not shown) in BN rat lungs.
Members of the Ly6 multigene family encode cell surface antigens with multiple specificities that have not been well characterized. Secreted Ly6/PLAUR (plasminogen activator urokinase receptor) domain containing protein 1 (Slurp1), a Ly6b family member, has been implicated in physiological and structural integrity of the skin (36). Slurp1 is regulated by Egfr and retinoic acid—key modulators of lung development—as well as by IFN-γ. The dramatic increase in Ly6b mRNA in the BN rat is, to our knowledge, the first association of an Ly6 family member with the asthma-associated phenotype. We were unable to confirm an excess of Ly6b protein, as no rat antibody is currently available.
The combined findings of neutrophilia, increased α1-AT (encoded by Serpina1), and reduced Fcnb in the lungs of Fisher rats define a developmental respiratory phenotype distinct from that of the BN rat. Airway remodeling in asthma involves degradation of the ECM. Critical to the dynamic structure of the normal ECM is the equilibrium between synthesis and degradation of elastin, which is, in turn, dependent on the appropriate balance of elastase and its primary inhibitor, α1-AT. Neutrophils provide the major source of elastase in the lung. In sputum from subjects with asthma, significantly elevated levels of elastase and α1-AT correlate with the percentage of neutrophils (44). Increased levels of α1-AT appear, however, insufficient to counterbalance the increased levels of elastase in these patients. An imbalance of α1-AT/elastase in the lung has been proposed to contribute to airway inflammation, thereby increasing AHR and impaired lung function in children with asthma (45). The Fisher rat is innately hyperresponsive, and has increased ASM mass (21). We suggest that neutrophilia in this model may be associated with an imbalance of α1-AT/elastase that is likely to contribute to AHR in Fisher rats in association with pre-/perinatal inflammation of unknown origin.
This is, to our knowledge, the first report of altered Fcnb expression, a molecule associated with innate immunity, in the developing lung in an animal model of innate airway hyperresponsiveness. The ficolins are pattern-recognition molecules that trigger the innate immune system upon binding to microbial surfaces (37). Human M-ficolin, the homolog of rodent Fcnb, is exclusively expressed in phagocytic cells. M-ficolin, found in lung and spleen (46), has been localized to secretory granules in the cytoplasm of peripheral neutrophils and monocytes, and in type II alveolar epithelial cells (47). Mouse Fcnb is up-regulated in the lysozomes of activated macrophages (48). Different developmental spatial–temporal expression patterns of Ficolin A (Fcna) and Fcnb suggest that these molecules are likely to play distinct roles in the pre- and postnatal stages (46). Fcna and Fcnb also appear to have two distinct routes to eliminate the pathogens: one through the lectin pathway (human L-ficolin, Fcna), the other, a more primitive form of opsonophagocytosis performed by Fcnb (46). Our demonstration of reduced Fcnb in the postnatal lung of Fisher rats suggests that innate AHR in this model may be attributed, at least in part, to deficiency of essential pattern-recognition molecules that serve as a protective interface, protecting the developing lung from pathogenesis and local inflammation.
The GR is the critical mediator of GC effects during lung development. GR also mediates effects of exogenous GC in the control of asthma exacerbations. At PN7, the BN rat lungs had increased GR mRNA and protein. High levels of immunoreactive nuclear GR were observed at PN7 and 14 by immunohistochemistry. These findings are interesting in light of studies showing that steroid-insensitive subjects with asthma have high GR numbers (49–51), and that children with asthma express greater quantities of GR mRNA than do healthy children (52). Increased expression of GR may improve the ability to regulate cytokine responses. Augmented GR may be one mechanism to improve GC responsiveness in a lung that is more vulnerable to inflammation. Notably, we also observed up-regulation of Tgfbr1l1 in the BN rat throughout postnatal development. Tgfb1l1 (also known as hydrogen peroxide-inducible clone 5 protein [Hic-5]) is an important coactivator of several nuclear proteins, including GR in lung epithelium (53).
Our data suggest that inflammatory, immune, structural, and physiological (AHR) changes in the lung of the developing rat can be identified and differentiated by using the differing phenotypic susceptibilities of the inbred strains of rat, in conjunction with genomic approaches, in the developing animal. The very early identification of these abnormalities suggests that environmental and genetic processes, operating in utero and in early postnatal life, have a major influence on disease development later in life. Further dissection of these genomic signatures should give important clues that can then be tested in humans. For example, it would be of great interest to relate polymorphism in the genes identified in this investigation to the relevant asthma phenotypes in humans. Such work is currently underway.
We have used both direct assessment of specific genes previously implicated in asthma (GR, IPO13) and global gene expression analysis to identify genes associated with asthma susceptibility phenotypes in our rat models. Our findings affirm the importance of both approaches, and highlight both the power and the limitations of microarray analyses.
In conclusion, through the use of rat models of distinct asthma susceptibility phenotypes, we show unique respiratory gene expression profiles in the perinatal lung. The delineation of specific molecular determinants of respiratory susceptibility has implications for both pediatric and adult pulmonary health. The ultimate goal is to identify how in utero and early life exposures interact with pre-existing respiratory genotypes to initiate asthma pathogenesis to enable the development of early preventive strategies and novel therapeutic protocols targeted to effectively and safely reverse immune dysfunction.
Supplementary Material
Acknowledgments
The authors gratefully acknowledge the assistance of Laura Montermini and Salomon Cornejo in the preparation of the manuscript.
This work was supported by operating grants from the Canadian Institutes of Health Research (F.K.) and from the National Institutes of Health (B.A.R.), and by graduate scholarships from the Montreal Children's Hospital Research Institute (N.C. and L.R.) and from the Respiratory Health Network of the Fonds de la recherche en santé and the Canadian Institutes of Health Research (L.R.).
This article has an online supplement, which is accessible from this issue's table of contents at www.atsjournals.org
Originally Published in Press as DOI:10.1165/rcmb.2009-0412OC on January 29, 2010
Author Disclosure: F.K. received a sponsored grant from Canadian Institute of Health Research for more than $100,001. A.T.K. received royalties from Massachusetts Institute of Technology Press for less than $1000 as an author of a book. He also received a sponsored grant from National Institutes of Health (NIH)/National Heart, Lung, and Blood Institute for $50,001–$100,000 and from NIH/National Institute of Neurological Disorders and Stroke (NINDS) for $10,001–$50,000. J.G.M. received lecture fees from Merck and Altana for less than $1,000 along with a sponsored grant from Merck for $50,001–$100,000, NIH for more than $100,001, and Canadian Institutes of Health Research for more than $100,001. B.A.R. received lecture fees from Novartis for $5,001–$10,000 and royalties of $5,001–$10,000 as a section editor. S.T.W. received consultancy fees from Genentech for $5,001–$10,000. None of the other authors has a financial relationship with a commercial entity that has an interest in the subject of this manuscript.
References
- 1.Centers for Disease Control and Prevention/National Center for Health Statistics. Analysis by the American Lung Association RaPSDuSaS: National Health Interview Survey raw data [Internet]. 2010: American Lung Association [accessed October 18, 2010]. Available from: http://www.lungusa.org/lung-disease/asthma/resources/facts-and-figures/asthma-children-fact-sheet.html
- 2.Grischkan J, Storfer-Isser A, Rosen CL, Larkin EK, Kirchner HL, South A, Wilson-Costello DC, Martin RJ, Redline S. Variation in childhood asthma among former preterm infants. J Pediatr 2004;144:321–326. [DOI] [PubMed] [Google Scholar]
- 3.Mannino DM, Homa DM, Akinbami LJ, Moorman JE, Gwynn C, Redd SC. Surveillance for asthma—United States, 1980–1999. MMWR Surv Summ 1999;51:1–13. [PubMed] [Google Scholar]
- 4.Warner JA, Jones C, Jones A, Warner J. Prenatal origins of allergic disease. J Allergy Clin Immunol 2000;105:S493–S496. [DOI] [PubMed] [Google Scholar]
- 5.Kihlstrom A, Lilja G, Pershagen G, Hedlin G. Exposure to high doses of birch pollen during pregnancy, and risk of sensitization and atopic disease in the child. Allergy 2003;58:871–877. [DOI] [PubMed] [Google Scholar]
- 6.Davies DE, Wicks J, Powell RM, Puddicombe SM, Holgate ST. Airway remodeling in asthma: new insights. J Allergy Clin Immunol 2003;111:215–225. [DOI] [PubMed] [Google Scholar]
- 7.Wang X, Wypij D, Gold DR, Speizer FE, Ware JH, Ferris BG Jr, Dockery DW. A longitudinal study of the effects of parental smoking on pulmonary function in children 6–18 years. Am J Respir Crit Care Med 1994;149:1420–1425. [DOI] [PubMed] [Google Scholar]
- 8.Hanrahan JP, Brown RW, Carey VJ, Castile RG, Speizer FE, Tager IB. Passive respiratory mechanics in healthy infants: effects of growth, gender, and smoking. Am J Respir Crit Care Med 1996;154:670–680. [DOI] [PubMed] [Google Scholar]
- 9.Kaplan F. Molecular determinants of fetal lung organogenesis. Mol Genet Metab 2000;71:321–341. [DOI] [PubMed] [Google Scholar]
- 10.Warburton DB. Growth factor signaling in lung morphogenetic centers: automaticity, stereotypy and symmetry. Respir Res 2003;4:5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Roche WR, Beasley R, Williams JH, Holgate ST. Subepithelial fibrosis in the bronchi of asthmatics. Lancet 1989;1:520–524. [DOI] [PubMed] [Google Scholar]
- 12.Zhang S, Smartt H, Holgate ST, Roche WR. Growth factors secreted by bronchial epithelial cells control myofibroblast proliferation: an in vitro co-culture model of airway remodeling in asthma. Lab Invest 1999;79:395–405. [PubMed] [Google Scholar]
- 13.Minoo P, King RJ. Epithelial–mesenchymal interactions in lung development. Annu Rev Physiol 1994;56:13–45. [DOI] [PubMed] [Google Scholar]
- 14.Jeffrey PK. The development of large and small airways. Am J Respir Crit Care Med 1998;157:S174–S180. [DOI] [PubMed] [Google Scholar]
- 15.Davies DE, Holgate ST. Asthma: the importance of epithelial mesenchymal communication in pathogenesis: inflammation and the airway epithelium in asthma. Int J Biochem Cell Biol 2002;34:1520–1526. [DOI] [PubMed] [Google Scholar]
- 16.Bousquet J, Jacot W, Yssel H, Vignola AM, Humbert M. Epigenetic inheritance of fetal genes in allergic asthma. Allergy 2004;59:138–147. [DOI] [PubMed] [Google Scholar]
- 17.Knight DA, Lane CL, Stick SM. Does aberrant activation of the epithelial–mesenchymal trophic unit play a key role in asthma or is it an unimportant sideshow? Curr Opin Pharmacol 2004;4:251–256. [DOI] [PubMed] [Google Scholar]
- 18.Holgate S, Holloway J, Wilson S, Bucchieri F, Puddicombe S, Davies D. Epithelial–mesenchymal communication in the pathogenesis of chronic asthma. Proc Am Thorac Soc 2004;1:93–98. [DOI] [PubMed] [Google Scholar]
- 19.Knight DA, Holgate ST. The airway epithelium: structural and functional properties in health and disease. Respirology 2003;8:432–446. [DOI] [PubMed] [Google Scholar]
- 20.Martin J, Tamaoka M. Rat models of asthma and chronic obstructive lung disease. Pulm Pharmacol Ther 2006;19:377–385. [DOI] [PubMed] [Google Scholar]
- 21.Eidelman DH, DiMaria GU, Bellofiore S, Wang NS, Guttmann RD, Martin JG. Strain-related differences in airway smooth muscle and airway responsiveness in the rat. Am Rev Respir Dis 1991;144:792–796. [DOI] [PubMed] [Google Scholar]
- 22.Eidelman D, Bellofiore S, Martin J. Late airway responses to antigen challenge in sensitized inbred rats. Am Rev Respir Dis 1988;137:1033–1037. [DOI] [PubMed] [Google Scholar]
- 23.Murphey S, Brown S, Miklos N, Fireman P. Reagin synthesis in inbred strains of rats. Immunology 1974;27:245–253. [PMC free article] [PubMed] [Google Scholar]
- 24.Murphey S, White C, Waters T, Fireman P. Reagin synthesis in inbred rats. J Allergy Clin Immunol 1976;58:381–386. [DOI] [PubMed] [Google Scholar]
- 25.Bellofiore S, Martin JG. Antigen challenge of sensitized rats increases airway responsiveness to methacholine. J Appl Physiol 1988;65:1642–1646. [DOI] [PubMed] [Google Scholar]
- 26.Sapienza S, Du T, Eidelman DH, Wang NS, Martin JG. Structural changes in the airways of sensitized brown norway rats after antigen challenge. Am Rev Respir Dis 1991;144:423–427. [DOI] [PubMed] [Google Scholar]
- 27.Salmon M, Walsh DA, Huang TJ, Barnes PJ, Leonard TB, Hay DW, Chung KF. Involvement of cysteinyl leukotrienes in airway smooth muscle cell DNA synthesis after repeated allergen exposure in sensitized brown Norway rats. Br J Pharmacol 1999;127:1151–1158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Xu KF, Vlahos R, Messina A, Bamford TL, Bertram JF, Stewart AG. Antigen-induced airway inflammation in the brown Norway rat results in airway smooth muscle hyperplasia. J Appl Physiol 2002;93:1833–1840. [DOI] [PubMed] [Google Scholar]
- 29.Kaplan F, Ledoux P, Kassamali FQ, Gagnon S, Post M, Koehler D, Deimling J, Sweezey NB. A novel developmentally regulated gene in lung mesenchyme: homology to a tumor-derived trypsin inhibitor. Am J Physiol Lung Cell Mol Physiol 1999;276:L1027–L1036. [DOI] [PubMed] [Google Scholar]
- 30.Caniggia I, Tseu I, Han RN, Smith BT, Tanswell K, Post M. Spatial and temporal differences in fibroblast behavior in fetal rat lung. Am J Physiol 1991;261:L424–L433. [DOI] [PubMed] [Google Scholar]
- 31.Wright G, Simon R. A random variance model for detection of differential gene expression in small microarray experiments. Bioinformatics 2003;18:2448–2455. [DOI] [PubMed] [Google Scholar]
- 32.Smyth G. Limma: linear models for microarray data. In: Gentleman R, Carey V, Dudoit S, Irizzary R, Hu T, editors. Bioinformatics and computational biology solutions using r and bioconductor. New York: Springer; 2005. pp. 397–420.
- 33.Roche W. Inflammatory and structural changes in the small airways in bronchial asthma. Am J Respir Crit Care Med 1998;157:S191–S194. [DOI] [PubMed] [Google Scholar]
- 34.Leclair K, Rabin M, Nesbitt N, Pravtcheva D, Ruddle F, Palfree R, Bothwell A. Murine Ly-6 multigene family is located on chromosome 15. Proc Natl Acad Sci USA 1987;84:1638–1642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Moriwaki Y, Yoshikawa K, Fukuda H, Fujii YX, Misawa H, Kawashima K. Immune system expression of SLURP-1 and SLURP-2, two endogenous nicotinic acetylcholine receptor ligands. Life Sci 2007;80:2365–2368. [DOI] [PubMed] [Google Scholar]
- 36.Mastrangeli R, Donini S, Kelton CA, He C, Bressan A, Milazzo F, Ciolli V, Borrelli F, Martelli F, Biffoni M, et al. ARS component B: structural characterization, tissue expression and regulation of the gene and protein (SLURP-1) associated with Mal De Meleda. Eur J Dermatol 2003;13:560–570. [PubMed] [Google Scholar]
- 37.Runza VL, Schwaeble W, Männel D. Ficolins: novel pattern recognition molecules of the innate immune response. Immunobiology 2008;213:297–306. [DOI] [PubMed] [Google Scholar]
- 38.Tao T, Lan J, Lukacs GL, Hache RJG, Kaplan F. Importin 13 regulates nuclear import of the glucocorticoid receptor in airway epithelial cells. Am J Respir Cell Mol Biol 2006;35:668–680. [DOI] [PubMed] [Google Scholar]
- 39.Raby BA, Van SK, Lasky-Su J, Tantisira K, Kaplan F, Weiss ST, Raby BA, Van Steen K, Lasky-Su J, Tantisira K, et al. Importin-13 genetic variation is associated with improved airway responsiveness in childhood asthma. Respir Res 2009;10:67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Tao T, Lan J, Presley J, Sweezey N, Kaplan F. Nucleocytoplasmic shuttling of Lgl2 is developmentally regulated in fetal lung. Am J Respir Cell Mol Biol 2004;30:350–359. [DOI] [PubMed] [Google Scholar]
- 41.Martin J, Ramos-Barbon D. Airway smooth muscle growth from the perspective of animal models. Respir Physiol Neurobiol 2003;137:251–261. [DOI] [PubMed] [Google Scholar]
- 42.Batra V, Musani AI, Hastie AT, Khurana S, Carpenter KA, Zangrilli JG, Peters SP, Batra V, Musani AI, Hastie AT, et al. Bronchoalveolar lavage fluid concentrations of transforming growth factor (TGF)-beta1, TGF-beta2, interleukin (IL)-4 and IL-13 after segmental allergen challenge and their effects on alpha-smooth muscle actin and collagen III synthesis by primary human lung fibroblasts. Clin Exp Allergy 2004;34:437–444. [DOI] [PubMed] [Google Scholar]
- 43.Fedorov IA, Wilson SJ, Davies DE, Holgate ST. Epithelial stress and structural remodelling in childhood asthma. Thorax 2005;60:389–394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.VIignola A, Bonnano A, Mirabella A, Riccobono L, Mirabella F, Profita A, Bellia V, Bousquet J, Bonsignore G. Increased levels of elastase and alpha1-antitrypsin in sputum of asthmatic patients. Am J Respir Crit Care Med 1998;157:505–511. [DOI] [PubMed] [Google Scholar]
- 45.von Ehrenstein OS, Maier EM, Weiland SK, Carr D, Hirsch T, Nicolai T, Roscher AA, von Mutius E. {Alpha}1 antitrypsin and the prevalence and severity of asthma. Arch Dis Child 2004;89:230–231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Endo Y, Matsushita M, Fujita T. Role of Ficolin in innate immunity and its molecular basis. Immunobiology 2007;212:371–379. [DOI] [PubMed] [Google Scholar]
- 47.Liu Y, Endo Y, Homma S, Kanno K, Yaginuma H, Fujita T. Ficolin A, Ficolin B are expressed in distinct ontogenic patterns and cell types in the mouse. Mol Immunol 2005;42:1265–1273. [DOI] [PubMed] [Google Scholar]
- 48.Runza V, Hehlgans T, Echtenacher B, Zahringer U, Schwaeble W, Mannel D. Localization of the mouse defense lectin ficolin B in lysosomes of activated macrophages. J Endotoxin Res 2006;12:120–126. [DOI] [PubMed] [Google Scholar]
- 49.Bronnegard M, Werner S, Gustafsson JA. Regulation of glucocorticoid receptor expression in cultured fibroblasts from a patient with familial glucocorticoid resistance. J Steroid Biochem Mol Biol 1991;39:693–701. [DOI] [PubMed] [Google Scholar]
- 50.Werner S, Thoren M, Gustafsson JA, Bronnegard M. Glucocorticoid receptor abnormalities in fibroblasts from patients with idiopathic resistance to dexamethasone diagnosed when evaluated for adrenocortical disorders. J Clin Endocrinol Metab 1992;75:1005–1009. [DOI] [PubMed] [Google Scholar]
- 51.Mjaanes CM, Whelan GJ, Szefler SJ. Corticosteroid therapy in asthma: predictors of responsiveness. Clin Chest Med 2006;27:119–132. (vii.). [DOI] [PubMed] [Google Scholar]
- 52.Miller GE, Chen E. Life stress and diminished expression of genes encoding glucocorticoid receptor and B2-adrenergic receptor in children with asthma. Proc Natl Acad Sci USA 2006;103:5496–5501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Saelzler MP, Spackman CC, Liu Y, Martinez LC, Harris JP, Abe MK. ERK8 down-regulates transactivation of the glucocorticoid receptor through Hic-5. J Biol Chem 2006;281:16821–16832. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.