Abstract
HIV-1 is relatively resistant to antibody-mediated neutralization; however, rare antibodies to the exterior envelope glycoprotein, gp120, and the transmembrane glycoprotein, gp41, can neutralize a broad array of isolates. Two antibodies, 2F5 and 4E10, are directed against the gp41 membrane proximal external region (MPER); however, the kinetic neutralization signature of these antibodies remains unresolved. Previously, we reported that the fully cleaved, cell surface envelope glycoproteins (Env) derived from the primary isolate, JR-FL, are well recognized exclusively by gp120-directed neutralizing ligands and not by nonneutralizing gp120 antibodies. However, the gp120 nonneutralizing antibodies can recognize HIV spikes that are rendered fully cleavage defective by site-directed mutagenesis. Here, we extended such analysis to gp41 neutralizing and nonneutralizing antibodies and, relative to the rules of gp120-specific antibody recognition, we observed marked contrasts. Similar to gp120 recognition, the nonneutralizing gp41 cluster 1 or cluster 2 antibodies bound much more efficiently to cleavage-defective spikes when compared to their recognition of cleaved spikes. In contrast to gp120 neutralizing antibody recognition, the broadly neutralizing gp41 antibodies 2F5 and 4E10, like the nonneutralizing gp41 antibodies, did not efficiently recognize the predominantly cleaved, primary isolate JR-FL spikes. However, if the spikes were rendered cleavage defective, recognition by both the neutralizing and nonneutralizing ligand markedly increased. CD4 interaction with the cleaved spikes markedly increased recognition by most nonneutralizing gp41 antibodies, whereas such treatment had a minimal increase of 2F5 and 4E10 recognition. These data indicate again the profound influence that cleavage imposes on the quaternary packing of primary isolate spikes and have important implications for soluble trimer candidate immunogens.
Introduction
The HIV-1 exterior envelope glycoprotein, gp120, and the transmembrane glycoprotein, gp41, are derived from the cleavage of gp160 precursor protein and are the only virally encoded proteins on the surface of the virus. These noncovalently associated glycoproteins form the trimeric functional spike on the virus surface and mediate vial entry. The gp120 subunit binds the primary receptor, CD4, and following gp120 association with the coreceptor, usually CCR5, the gp41 subunit then participates in accomplishing virus-to-cell membrane fusion and entry of viral genomic information into the target cell.1–7
Viral entry into cells can be blocked by elicited antibodies that can efficiently recognize the native functional spike. Historically, only four human monoclonal antibodies, derived from HIV-1-infected individuals, were identified that can neutralize a broad spectrum of primary isolates in vitro.8–13 Of these four antibodies, two are directed against epitopes that reside in gp120 and are known as 2G12 and b12. The other two antibodies, called 2F5 and 4E10, recognize contiguous epitopes within the gp41 membrane proximal external region (MPER). Over the past decade, it was shown that all four of these broadly neutralizing antibodies can protect against several routes of virus challenge if they are passively administered at high systemic concentrations before, or immediately following, exposure to virus (SHIV).14–23
Due to the limited numbers of broadly neutralizing antibodies, considerable interest has recently increased in defining the breadth of neutralization elicited in HIV-1-infected individuals. Approximately 10–20% of chronically infected individuals display the property of “neutralization breadth.” Sometimes the broad neutralizing activity elicited by natural HIV-1 infection can be mapped to subregions of Env. And in these cases, the activity often maps to the gp120 subunit of the HIV Env.24–27 Recently, two new broadly neutralizing antibodies were isolated from an HIV-1-infected individual and are known as PG9 and PG16.27 And although these related antibodies map to the gp120 Env subunit, they recognize gp120 in the context of a trimer and display remarkable breadth and potency of neutralization.27 In addition, very recently an isolated broad and potent CD4 binding site (CD4-bs) antibody, VRC01, was cloned from an HIV-1-infected individual who possessed neutralization breadth previously mapped to the CD4 interactive region.26,28 Another CD4-bs antibody, VRC03, was also isolated from this same patient and another relatively broad CD4-bs antibody, HJ16, was also recently isolated.29 In other broadly neutralizing patient sera, in rare instances, the specificity of the broad neutralizing activity can be mapped to the gp41 MPER region. In these sera, the neutralizing activity appears similar to the specificity displayed by 4E1030–32 and in one report, the activity could be mapped to the 2F5 epitope region.33
Previously, it was shown that following transient transfection of plasmid DNA, primary isolate Env glycoproteins expressed on cell surfaces are predominantly trimeric.34 However, only for selected Envs efficient cleavage of the gp160 precursor protein occurs, presumably to form functional Env spikes on the cell surface of the transfected cells. In our previous report, we document the efficient and, within the limits of detection, complete gp160 precursor cleavage when the cell surface Env is derived from the primary isolate, JR-FL.35 Other Env derived from isolates, such as YU2,35 and several others examined do not exhibit the nearly complete precursor cleavage as displayed by JR-FL-derived Env (data not shown). In our previous work, using fluorescence-activated cell sorting (FACS)-based cell surface staining and gp120-directed neutralizing and nonneutralizing antibodies, we demonstrate that there is a direct association between increased antibody binding to efficiently cleaved, functional JR-FL spikes and ligand neutralization capacity.35 Other studies have reported data consistent with this association.36–39 The availability of a JR-FL transient transfection system, which displays a high degree of cleaved spikes, and can be engineered to then display fully noncleaved oligomeric spikes by elimination of the natural gp120–gp41 furin cleavage site,34,35 allows us now to extend our analysis of functional spike recognition by antibody to the gp41 subunit of Env.
In the study presented here, we sought to determine if the HIV-1 gp41 neutralizing antibodies 2F5 or 4E10 efficiently recognize the fully cleaved, functional primary isolate JR-FL static spikes. The general question regarding 2F5 and 4E10 spike recognition has been explored previously in the literature, but to date remains not fully resolved,40,41 and several of these studies predate the appreciation of differences between the Envs on laboratory-adapted compared to primary isolates.42–51 The JR-FL cleaved/uncleaved cell surface spike system previously described,35 and now utilized in this study, allowed us to address further the impact of cleavage on Env architecture, specifically in the gp41 MPER and associated regions. Here, we have demonstrated that, in contrast to the efficient recognition of cleaved spikes by the broadly neutralizing gp120-directed ligands,35 the broadly neutralizing 2F5 and 4E10 antibodies poorly recognized the cleaved, functional JR-FL oligomers expressed on the cell surface. The nonneutralizing gp41 cluster 1 and cluster 2 antibodies also bound poorly to these highly cleaved spikes, similar to our previous report for the nonneutralizing, gp120-directed antibodies. If the JR-FL oligomers were intentionally rendered cleavage defective, then recognition by both the neutralizing and nonneutralizing gp41 increased dramatically, again enforcing the notion that the rules of recognition for the gp41 neutralizing determinants differ from those in gp120. Incubation of cleaved spikes with soluble CD4 caused gp120 shedding as previously reported52 and increased recognition of the gp120-deficient spikes by some nonneutralizing cluster 1 and 2 antibodies. Recognition of the CD4-treated spikes caused a moderate, but not substantial, increase in 2F5 or 4E10 recognition in the JR-FL-based system. These data have important implications for the structure or exposure of discreet gp41 epitopes in the context of the static, functional primary isolate HIV-1 spike.
Materials and Methods
Transient transfection of envelope glycoprotein expression plasmids
One day prior to transfection, 10 × 106 293T cells in DMEM and containing 10% heat-inactivated fetal calf serum (HIFBS), 1% penicillin-streptomycin were seeded in a 150-mm tissue culture dish. The cells were transfected with the pSVIII expressor plasmids encoding JR-FL cleavage-competent wild-type and cleavage-defective Env, along with cotransfection of the tat expression plasmid, pctat using Fugene6 (Roche) at a ratio DNA/Fugene6 of 1:3 and 5 μg total DNA per 1 × 106 cells. As previously described, Quick Change mutagenesis (Stratagene) was used to introduce mutations at the natural gp120–gp41 cleavage site boundary that changes the amino acid sequence from REKR to SEKS (residues 508–511, HXBc2 numbering53). These subtle and conservative changes confirmed by sequencing render the cleavage-competent Env cleavage defective.
FACS staining of cell surface HIV-1 Env
FACS staining was performed as previously described.54 Forty-eight hours following transfection, the cells were harvested and washed in FACS buffer [phosphate-buffered saline (PBS), 5% HIFBS, 0.02% azide] and stained with a panel of monoclonal antibodies that were also used in viral neutralization assays. The monoclonal antibody–cell mixture was washed extensively in FACS buffer and antihuman phycoerythrin (PE) (Sigma) at a 1:200 dilution was added for 1 h, followed by extensive washing to remove unbound secondary antibody. To study the effect of CD4 on the binding of selected antibodies, 50 μg/ml of sCD4 (Progenics) was added to the transfected cells and incubated for 1 h on ice with occasional shaking. The mixture was washed with FACS buffer and incubated with the antibodies for 1 h either at room temperature (RT) or on ice with intermittent shaking. The stained cells were analyzed by FACS on a Beckman Coulter Caliber Instrument or occasionally on a BD LSR-II (see Supplemental Data).
Purification of JR-FL gp140-foldon trimeric Env and PAGE
The envelope glycoproteins (gp140-FT-His) were expressed by transfecting the 293F cell line (Invitrogen, Carlsbad, CA) and incubating for 96 h in shaking suspension culture in serum-free media following the manufacturer's directions (Invitrogen) at 37°C in 5% CO2. Prior to transfection, the cells were grown to high density (i.e., 2.4 × 106 cells/ml) and immediately before transfection the cells were diluted with at least 50% fresh medium to a density of 1.2 × 106 cells/ml. The cells were transfected with 250 μg of the plasmid, pcDNA3.1 (−) expressing selected Env sequences and incubated in shake flasks. Cell-free supernatants were collected by centrifugation at 3500 × g for 20 min at 4°C to remove the cells. Prior to purification, supernatants were filtered through sterile 0.2 μm filters and stored at 4°C in the presence of protease inhibitors. Proteins were purified by lentil lectin affinity chromatography followed by chelating chromatography over a Ni2+ column (GE Health Care, Piscataway, NJ). For sodium dodecyl sulfate polyacrylamide gel electrophoresis (SDS-PAGE), equal quantities (10 μg) of each protein were resolved on an SDS (4 to 14%) polyacrylamide gel (NuPAGE, Invitrogen) and detected by staining with 0.2% Coomassie blue.
Immunoprecipitation and Western blot analysis
The shed gp120 was detected by immunoprecipitation (IP) of cell-free supernatants from JR-FL Env expressing 293 cells. Briefly, supernatants (1 ml) of 293 cells treated with or without CD4 were analyzed by immunoprecipitation with 1 μg of the anti-V3 antibody, 39F, to detect soluble gp120 in the media. Immune complexes were separated by boiling in sample buffer containing SDS and resolved on 4 to 15% SDS gels and detected by immunoblotting using rabbit anti-gp120 polyclonal antibody.
JR-FL HIV-1 Env gp140-foldon enzyme-linked immunosorbent assay
The antigenicity of the purified Env probes was determined by assessing Env recognition by a panel of monoclonal antibodies using an enzyme-linked immunosorbent assay (ELISA) as described elsewhere.55 In brief, each well of a Maxisorp high binding plate (Nunc) was coated with 100 μl (200 ng) of protein in PBS, pH 7.4, overnight at 4°C. Starting from a concentration of 2 μg/ml, the antibodies were 10-fold serially diluted: (1) the CD4 binding site-directed antibodies, b12 and F105 (kind gifts of D. Burton and M. Posner); (2) the glycan-directed antibody, 2G12 (H. Katinger); (3) the MPER-directed antibodies, 2F5 and 4E10 (H. Katinger); and (4) the cluster 1 and 2 gp41-directed antibodies, 7B2 and 22B (J. Robinson), 50-69, 98-6, 126-6, and 240-D (a kind gift from Susan Zolla-Pazner and Mirek Gorny at NYU) were used for binding the Env coated on the plate. Horseradish peroxidase (HRP)-conjugated antihuman or antimouse IgG were used as secondary antibodies and the TMB (3,3,5,5′-tetramethylbenzidine) peroxidase immunoassay substrate (Bio-Rad) was used as the colorimetric reporter reagent. The optical density (OD) was determined at 450 nm on a microplate reader (Molecular Devices).
Results
FACS of gp41 antibodies with the JR-FL envelope glycoproteins highlights the impact of precursor cleavage on structure
To perform cell surface staining analysis of HIV-1 Env trimers, the cytoplasmic tails of the Env were genetically deleted to enhance cell surface expression, increasing the sensitivity of the analysis as previously described.35,56–58 The 293T cells transfected with the plasmids encoding both cleavage-competent and genetically engineered cleavage-defective JR-FL gp160 glycoproteins were first incubated with a panel of monoclonal antibodies and then analyzed by FACS as described previously.35,54
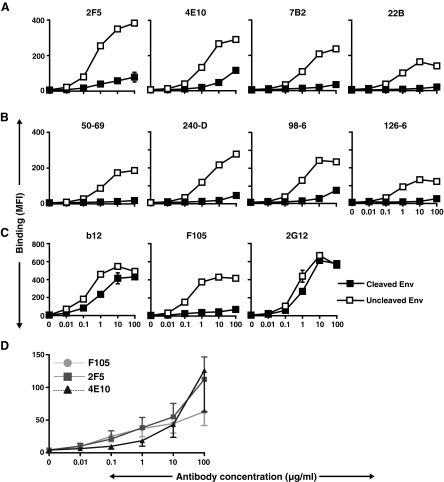
Previously, we reported that there were clear differences in the recognition of gp120-directed neutralizing ligands (b12, 2G12, sCD4, HIVIG) compared to the nonneutralizing ligands (F105, 17b, IgGb6, 39F, 211c, C11) to the cleavage-competent JR-FL cell surface Env.35 Therefore, in this study we made use of these previous observations as controls for the system to assess if there was any differential effect of Env cleavage on binding by either neutralizing (2F5 and 4E10) or nonneutralizing (7B2, 22B, 50-69, 240-D, 98-6, and 126-6) gp41-directed antibodies. The mean fluorescence intensities (MFI) of different concentrations of ligands specifically interacting with cell surface Env were derived from the FACS histograms. These data were then used to generate the binding curves shown in Fig. 1. From the FACS-based binding curves shown in Fig. 1A and B, it was clear that both gp41 neutralizing antibodies, 2F5 and 4E10, and the set of gp41 nonneutralizing antibodies, bound more efficiently to the engineered uncleaved JR-FL Env spikes than to the predominantly cleaved Env spikes. There was detectable binding of 2F5 and 4E10 at the highest concentrations tested; however, this was clearly at much lower levels than that achieved in the cleavage-defective state and was observed for the nonneutralizing antibody, 98-6, as well. These data again demonstrated the substantial effects that cleavage, or the lack thereof, imparts on exposure of many elements of the functional Env spike. That the gp41-directed nonneutralizing cluster 1 antibodies efficiently recognized noncleaved cell surface Env is a bit surprising as the cluster 1 antibodies are thought to recognize predominantly postfusion gp41 conformations of Env.59
FIG. 1.
FACS-based binding curves derived by antibody binding to cell surface JR-FL Env. (A, B) Mean fluorescent intensity (MFI) values of both neutralizing (2F5 and 4E10) and nonneutralizing gp41-directed antibodies (7B2, 22B, 50-69, 240-D, 98-6, and 126-6) are shown to cleavage-competent (▪) and cleavage-defective (□) Env. (C) As controls, neutralizing (b12 and 2G12) and nonneutralizing (F105) gp120-directed antibody-binding profiles are shown. The data shown were derived from the same representative experiment performed in duplicate. The standard errors between duplicates were small and are not always visible since the error bars are often obscured by the symbols; data generated from as many as five other independent experiments displayed similar trends. (D) Comparative binding analysis of 2F5, 4E10, and the nonneutralizing antibody F105 to cleavage-competent JR-FL Env. Note that here the MFI data are presented on a different scale. The curves were generated by averaging the MFI values at each antibody concentration and were derived from five independent experiments; error bars are shown.
In contrast, and as expected, the gp120-directed neutralizing antibody, b12, efficiently recognized (Fig. 1C) the cleavage-competent Env, consistent with the finding from our previous study.35 Also as expected, the neutralizing gp120-directed antibody, 2G12, bound to cleavage-competent Env. Since this antibody also binds to cleavage-defective Env, we used 2G12 to confirm the equivalent expression of the JR-FL trimeric Env on the cell surface (Fig. 1C). Finally, the nonneutralizing gp120-directed antibody, F105, was used as a negative control and, as expected, could not efficiently recognize functional JR-FL Env spikes on cell surfaces (Fig. 1C). Note that F105, as well as the 98-6 antibody, neither of which can neutralize the JR-FL virus, bound slightly to the cleavage-competent spikes at high antibody concentrations. At these high antibody concentrations, 2F5 and 4E10 also recognized slightly the cleavage-competent spikes. To determine if this low level recognition was similar to that displayed by the nonneutralizing antibodies, we compared multiple curves generated by F105, 2F5 and 4E10 binding to Env. (Fig. 1D and Supplementary Fig. S1; Supplementary Data are available online at www.liebertonline.com/aid). Because the low levels of recognition by both the nonneutralizing and neutralizing antibodies are overlapping, and because it is thought that nonneutralizing antibodies cannot efficiently recognize the functional spike, we conclude that this binding is likely due to a small, but previously nondetected fraction of uncleaved, or defective, JR-FL spikes expressed on the cell surface. However, this binding could reflect that very inefficient recognition of the cleaved spikes may be accomplished by 2F5 or 4E10 at high antibody concentrations. We did confirm that this low level recognition was specific as antibody recognition was completely abrogated when cell surface Env harboring point mutations in the 2F5 and 4E10 core epitopes were utilized as targets (Fig. S2). Finally, to ensure that the low level 2F5 (4E10) binding was not due to 2F5 (4E10)-induced shedding of gp120 (similar to CD4-induced shedding), followed by a conformational change in gp41 that then caused 2F5 (or 4E10) to dissociate and result in the low level of cleaved Env recognition by 2F5 (4E10) as reported here, we did an experiment in which we incubated the JR-FL spikes with 2F5, then followed the incubation with 2G12 binding analysis. The preincubation of 2F5 with the spikes did not decrease subsequent spike recognition by 2G12 in any way, suggesting that 2F5 does not appreciably shed gp120 from the JR-FL cell surface spikes. These data are presented in Supplementary Fig. S3. Note that the b12:F105 ratio was very high, indicating that efficient cleavage of the spikes had occurred.
The tail-truncated JR-FL Env trimers behave similarly to wild-type spikes on virus
To confirm that the tail-truncated JR-FL Env used for the binding analysis did not differ substantially from the full length JR-FL envelope, gp160, we made pseudoviruses using either the tail-truncated or full-length Env gp160. Accordingly, the neutralization profiles were analyzed with the gp120- and gp41-directed antibodies (Table 1). The data presented in Table 1 demonstrate that there are only minor differences in the amount of antibodies needed for 80% neutralization of each virus type and moderate differences needed for 50% neutralization of virus. Importantly, the antibodies that did not neutralize the wild-type virus also failed to neutralize the tail-deleted mutant virus. That both types of pseudo-virus showed similar sensitivity against the neutralizing and non-neutralizing antibodies suggested that the tail-truncated JR-FL Env maintained a topology similar to that of the full-length spikes on the viral surface.
Table 1.
Inhibitory Concentrations of Antibodies for JR-FL and JR-FL ΔCT Virus (μg/ml)
| |
gp41-directed antibodies |
|||||||
|---|---|---|---|---|---|---|---|---|
| |
Neutralizing |
Nonneutralizing |
||||||
| Virus | 2F5 | 4E10 | 7B2 | 22B | 240-D | 50-69 | 126-6 | 98-6 |
| JR-FL Full length | ||||||||
| IC50 | 7.42 | 2.75 | >50 | >50 | >50 | >50 | >50 | >50 |
| IC80 | >50 | 20.9 | >50 | >50 | >50 | >50 | >50 | >50 |
| IC90 | >50 | >50 | >50 | >50 | >50 | >50 | >50 | >50 |
| JR-FL ΔCT (+) | ||||||||
| IC50 | 0.88002 | 0.13537 | >50 | >50 | >50 | >50 | >50 | >50 |
| IC80 | 11.7 | 6.11 | >50 | >50 | >50 | >50 | >50 | >50 |
| IC90 | >50 | >50 | >50 | >50 | >50 | >50 | >50 | >50 |
| |
gp120-directed antibodies |
||||
|---|---|---|---|---|---|
| |
Neutralizing |
Nonneutralizing |
|||
| Virus | b12 | 2G12 | 447 | 17b | F105 |
| JR-FL Full length | |||||
| IC50 | 0.00339 | 0.4791 | 9.26 | >50 | >50 |
| IC80 | 0.02241 | 2.51 | >50 | >50 | >50 |
| IC90 | 0.06767 | >5 | >50 | >50 | >50 |
| JR-FL ΔCT (+) | |||||
| IC50 | 0.00055 | 0.12546 | 1.3 | >50 | >50 |
| IC80 | 0.00865 | 3.21 | >50 | >50 | >50 |
| IC90 | 0.0433 | >5 | >50 | >50 | >50 |
Impact of CD4 on neutralizing and nonneutralizing gp41-directed antibody recognition
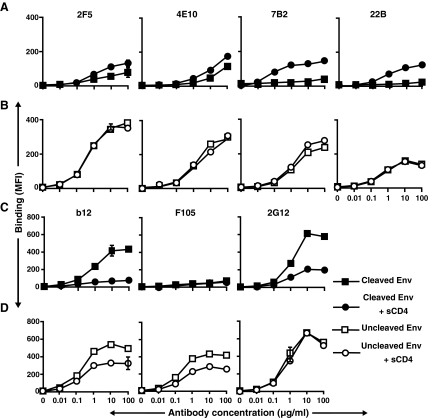
As shown in Fig. 1A, the 2F5 and 4E10 antibodies differed considerably in the level of binding to cleaved compared to noncleaved JR-FL Env. Neither of these MPER-directed antibodies efficiently recognized the cleavage-competent primary isolate JR-FL Env. This suggested that consistent with several other studies, these antibodies required receptor triggering for efficient recognition of the HIV-1 Env functional spike.60 Therefore, we asked if a soluble form of the primary receptor for virus (sCD4) could permit binding, presumably by inducing a substantial conformational change in Env following interaction with gp120. When cells expressing cleaved Env were incubated with sCD4 prior to exposure to the antibodies, the binding of both MPER-directed antibodies 2F5 and 4E10 revealed only slight increases in recognition of the cleavage-competent JR-FL spikes (Fig. 2A). In contrast, and as expected, most of the nonneutralizing cluster 1 and 2 antibodies bound better to the cleavage-competent Env preincubated (or coincubated) with sCD4, compared to the low level of binding to the untreated JR-FL “control” spikes (Fig. 2A). In fact, sCD4 treatment had a much greater effect on induction of the epitopes for several of the nonneutralizing cluster 1 and 2 antibodies, when compared to induction of 2F5 and 4E10 recognition (Fig. 2A and Fig. S4). For both neutralizing and nonneutralizing gp41 antibodies, when sCD4 was added to cells expressing cleavage-defective Env prior to the antibodies, recognition remained essentially unaltered (Fig. 2B).
FIG. 2.
Effects of sCD4 on the binding of antibodies to cell-surface JR-FL Env. FACS-based cell-surface staining curves indicating the binding of gp41-directed antibodies to JR-FL (A) cleavage-competent without sCD4 (▪) and with sCD4 (•) and (B) cleavage-defective envelope without sCD4 (□) and with sCD4 (o). Both neutralizing (2F5 and 4E10) and nonneutralizing antibodies (7B2, 22B) directed to gp41 Env are shown in (A) and (B). (C) The bindings of both neutralizing (b12 and 2G12) and nonneutralizing (F105) antibodies directed toward gp120 in the absence and presence of sCD4 are shown as controls in (C) and (D). The data shown are from a single representative experiment, performed in duplicate, and the error bars are not often visible. The data generated from five other independent experiments showed similar trends.
As previously reported, the gp120-directed, nonneutralizing CD4bs antibody, F105, bound much better to the cleavage-defective Env than to the isogenic cleaved version (Fig. 1C). The low level recognition of the cleaved spikes by F105 remained unaltered in the presence of sCD4, while its binding with the cleavage-defective Env decreased approximately 50% (Fig. 2D), suggesting a substantial degree of sCD4 competition in the cleavage-defective context. In the presence of sCD4, IgGb12 binding was substantially reduced to the cleavage-competent spike, and, to a lesser extent, the cleavage-defective spike (Fig. 2C). On the cleaved spike, reduced b12 recognition may be due to either CD4-induced shedding of gp120 or direct cross-competition. In the presence of sCD4, the recognition of cleaved spikes by 2G12 was greatly reduced, while there was no reduction in binding to cleavage-defective Env (Fig. 2C and D). These results suggest that the reduction of IgGb12 and 2G12 binding is due to CD4-induced shedding of gp120 from the cell surface.
Induction of gp41 antibodies following sCD4 addition is due to gp120 shedding
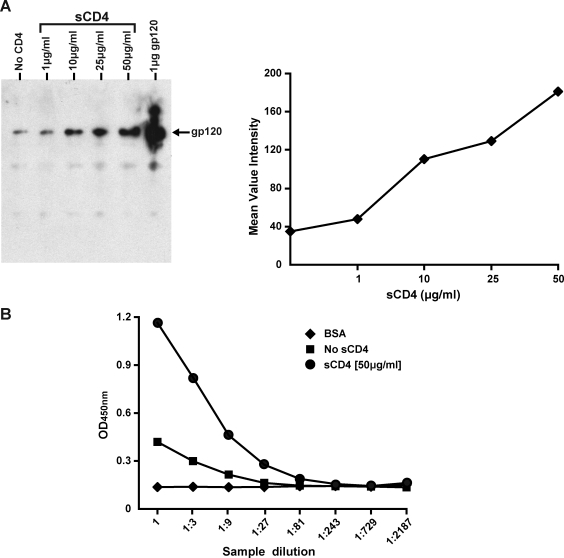
Although with much precedent,61 we sought to confirm that following incubation of the Env cleavage-competent expressing cells with sCD4, both the induction of binding by the cluster 1 and 2 antibodies and the reduced binding of IgGb12 and 2G12 were unequivocally due to shedding of gp120 from cleaved Env. Therefore, Env-expressing cells were treated with increasing concentrations of sCD4. In parallel, an equivalent number of cells was treated similarly, but without sCD4, to monitor gp120 “spontaneous shedding.” The cell-free supernatants from both sCD4-treated and -untreated Env-expressing cells were immunoprecipitated with V3 antibody, 39F. The intensity of the bands (Fig. 3A, left panel) in the Western blot increased as increasing amounts of sCD4 were added to the cells, indicating CD4-induced shedding of gp120 from the cleaved Env expressed on the cell surface as also observed by others.52 Figure 3A (right panel) shows the graphic representation of a gradual increase in shedding of gp120 when the cells were incubated with increasing amounts of sCD4. Further confirmation of the shedding was obtained by detecting the shed gp120 in the supernatant using a lectin-capture ELISA.55 The sCD4-treated samples contained higher amounts of gp120 in the supernatant compared to the untreated control (Fig. 3B). In addition, preincubation of the Env-expressing cells with b12, but not 2G12, could abrogate sCD4-induced shedding of gp120 (not shown). As expected, we observed reduced levels of cell-surface binding with IgGb12 and 2G12 to the cells in this experiment when the spikes were incubated with sCD4, compared to the untreated control (data not shown).
FIG. 3.
CD4-induced shedding of gp120 from JR-FL cell surface Env spikes. (A) The gp120 present in the cell supernatants (100 μl) derived from the JR-FL Env-expressing cells treated with increasing concentration of sCD4 (1–50 μg/ml) was immunoprecipitated by the anti-V3 monoclonal, 39F. The gp120 glycoproteins were resolved by sodium dodecyl sulfate polyacrylamide gel electrophoresis (SDS–PAGE) and the Western blot was developed by polyclonal rabbit anti-gp120 primary antibody followed by horseradish peroxidase (HRP)-conjugated goat antirabbit IgG secondary antibody. Untreated cells were used to evaluate the spontaneous shedding of gp120 from the cell surface spikes. Purified gp120 was used as a positive control (far right lane). The bands on the film were quantitated using the Quantity one software (Bio-Rad) and are plotted in the right panel. (B) The same set of cell supernatants was analyzed by ELISA to detect the presence of shed gp120 as shown. The gp120 present in the samples was captured on the ELISA plate precoated with lectin. Lectin-captured gp120 was then detected using the 39F monoclonal antibody.
Recognition of the soluble gp140 protein as stable foldon trimers by selected antibodies
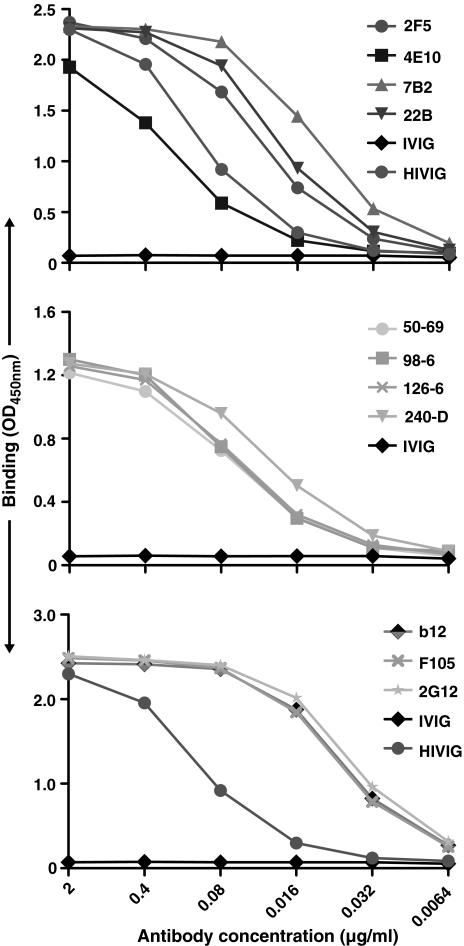
To assess the specificity and recognition pattern of the gp41 antibodies for a relatively well-characterized form of soluble Env, we expressed the soluble JR-FL gp140 stabilized by the heterologous trimerization motif, foldon.62 Following purification, we tested recognition of the soluble JR-FL trimers by the antibodies used in this study, with a focus on the gp41 antibodies. Not surprisingly, both the neutralizing MPER antibodies 2F5 and 4E10 and the nonneutralizing gp41 antibodies recognized the soluble, cleavage-defective JR-FL trimers (Fig. 4).
FIG. 4.
ELISA binding of selected monoclonal antibodies to soluble JR-FL gp140-foldon glycoprotein. (Top) ELISA binding of gp41-directed antibodies to purified JR-FLgp140-foldon trimers coated on the ELISA plate. (Middle) ELISA binding of the cluster 1 and cluster 2 gp41 antibodies to JR-FL gp140-foldon trimers. (Bottom) Selected control gp120-directed antibodies binding to the JR-FL gp140-foldon trimers.
Discussion
In this study, we again demonstrate that precursor cleavage has a substantial impact on overall HIV-1 Env spike architecture as assessed by differential antibody recognition. We utilized selected antibodies to the gp41 MPER to probe cleavage-competent versus cleavage-defective JR-FL cell surface functional spikes. We extended our previous observations of gp120-directed ligand recognition of cell surface, JR-FL spikes and observed that there are markedly different rules of engagement for gp41 neutralizing and nonneutralizing antibodies compared to their gp120 counterparts. We showed that although the cleavage-defective spike conformation permited recognition by both gp41 neutralizing and nonneutralizing antibodies (similar to gp120), for gp41, not even the broadly neutralizing antibodies 2F5 and 4E10 could efficiently recognize the fully cleaved, primary isolate JR-FL, tail-truncated spikes. These data are consistent with a recent report in which an oligomeric MPER-containing construct could adsorb 2F5 and 4E10 activity after incubation of the antibodies with primary isolate virus.40 The data are not entirely consistent with the report that 4E10, and especially 2F5, bind to JR-FL SOS trimeric functional spikes in a gel shift assay.63 Perhaps this type of cysteine-stabilized SOS functional spike is captured in a transitional state between native folding and fusion, consistent with reports of 17b interaction with these glycoproteins.60 Here, at the highest antibody concentrations tested, we did observe a very low level binding of 2F5 to the tail-truncated JR-FL spikes and this may reflect the slightly increased sensitivity of the tail-truncated Envs to 2F5-mediated neutralization. The difference in rules of engagement for gp120 compared to gp41 antibodies may be related to the more profound conformational changes that gp41 undergoes following receptor–coreceptor engagement compared to postreceptor-induced gp120 conformational changes.
These data also have important implications on HIV-1 Env-based trimer design. The goal of the trimeric Env immunogens is to mimic, in a soluble form, the functional Env trimeric spike present on the virions. However, due to the labile nature of the gp120-gp41, and with the exception of one design strategy (SOS), most other approaches utilize uncleaved oligomeric forms of soluble Env antigen.55,62,64–68 In terms of immunogenicity, it is hoped that a soluble mimetic of the functional spike would elicit antibodies capable of spike recognition and mediate virus neutralization.24 In fact, there are several reports indicating that trimeric Env immunogens induce antibodies that display increased breadth of neutralization in animals.68–72 However, in light of the data presented here and previously, that uncleaved Env trimers expose the many more nonneutralizing epitopes differently than their cleaved counterparts,35,48,73 Env cleavage is a reasonable property to include in a soluble trimeric immunogen. Accordingly, attempts have been described to make trimeric envelope immunogen with efficient cleavage and test them in vivo.37,65,74 It is not yet fully clear if the soluble, cleaved forms of the Env trimer will be superior immunogens than their uncleaved trimeric relatives, but to date neither elicits optimal neutralization breadth in test subjects, when subsequently analyzed in vitro.
The results presented here support our previous observation that cleavage is an important event for maintaining the conformational integrity of the native functional Env. The consequence is to efficiently occlude or shield the epitopes recognized by many and most (nonneutralizing) antibodies. The poor recognition by gp41-directed neutralizing antibodies to cleaved spikes is possibly due to either occlusion of these sites in native cleaved envelope, or to the fact that they are exposed only during the process of receptor engagement, or during formation of a fusion-competent transitional intermediate as previously suggested. Further assessment of the precise kinetics of MPER exposure following receptor engagement in a variety of HIV-1 Envs may be warranted and may shed some light on the differential resistance/sensitivity of an array of laboratory-adapted and primary isolates to 2F5 and 4E10 neutralization.
The binding of some cluster 1 and 2 antibodies was enhanced after treatment of sCD4 to the cells expressing cleaved Env. At the same time, we have demonstrated that CD4 induces the shedding of gp120. It is likely that the enhanced binding of these gp41-directed nonneutralizing antibodies is due to either exposure of the epitope after the shedding of gp120 from the surface or due to conformational changes in gp41 after CD4 treatment. The improvement of exposure of a number of gp41 epitopes upon shedding induced by soluble CD4 was reported previously by others.52 The exposure of the nonneutralizing epitopes on purified gp140 was found to be dependent on the cleavage of the envelope.71 The inability to neutralize the JR-FL virus by various cluster antibodies provides evidence of the hidden epitopes in the functional spikes that evade binding to those antibodies. Further studies will be needed to clarify the molecular mechanisms by which 2F5 and 4E10 antibodies neutralize the virus even though they bind poorly to the static primary isolate spike. These data have implications for a better understanding of functional spike architecture on the free virion and better informed Env trimer-based immunogen design.
Supplementary Material
Acknowledgments
We thank Brenda Hartman for help with the figures and Dennis Burton, Hermann Katinger, and Marshal Posner for the monoclonal antibodies b12, 2G12, 2F5, 4E10, and F105. We thank Mirek Gorny and Susan Zolla-Pazner for the antibodies 50-69, 98-6D, 126-D, and 240-D provided by funding from the NYU CFAR (AI27742). This study was funded by the NIH intramural research program and funding was also provided by the International AIDS Vaccine Initiative and the Bill and Melinda Gates Foundation.
Author Disclosure Statement
No competing financial interests exist.
References
- 1.Center RJ. Leapman RD. Lebowitz J. Arthur LO. Earl PL. Moss B. Oligomeric structure of the human immunodeficiency virus type 1 envelope protein on the virion surface. J Virol. 2002;76(15):7863–7867. doi: 10.1128/JVI.76.15.7863-7867.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Choe H. Farzan M. Sun Y, et al. The beta-chemokine receptors CCR3 and CCR5 facilitate infection by primary HIV-1 isolates. Cell. 1996;85(7):1135–1148. doi: 10.1016/s0092-8674(00)81313-6. [DOI] [PubMed] [Google Scholar]
- 3.Dalgleish AG. Beverley PC. Clapham PR. Crawford DH. Greaves MF. Weiss RA. The CD4 (T4) antigen is an essential component of the receptor for the AIDS retrovirus. Nature. 1984;312(5996):763–767. doi: 10.1038/312763a0. [DOI] [PubMed] [Google Scholar]
- 4.Hallenberger S. Bosch V. Angliker H. Shaw E. Klenk HD. Garten W. Inhibition of furin-mediated cleavage activation of HIV-1 glycoprotein gp160. Nature. 1992;360(6402):358–361. doi: 10.1038/360358a0. [DOI] [PubMed] [Google Scholar]
- 5.Weissenhorn W. Dessen A. Harrison SC. Skehel JJ. Wiley DC. Atomic structure of the ectodomain from HIV-1 gp41. Nature. 1997;387(6631):426–430. doi: 10.1038/387426a0. [DOI] [PubMed] [Google Scholar]
- 6.Willey RL. Bonifacino JS. Potts BJ. Martin MA. Klausner RD. Biosynthesis, cleavage, and degradation of the human immunodeficiency virus 1 envelope glycoprotein gp160. Proc Natl Acad Sci USA. 1988;85(24):9580–9584. doi: 10.1073/pnas.85.24.9580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wu L. Gerard NP. Wyatt R, et al. CD4-induced interaction of primary HIV-1 gp120 glycoproteins with the chemokine receptor CCR-5. Nature. 1996;384(6605):179–183. doi: 10.1038/384179a0. [DOI] [PubMed] [Google Scholar]
- 8.Bolognesi DP. Matthews TJ. HIV vaccines. Viral envelope fails to deliver? Nature. 1998;391(6668):638–639. doi: 10.1038/35504. [DOI] [PubMed] [Google Scholar]
- 9.Burton DR. Montefiori DC. The antibody response in HIV-1 infection. AIDS. 1997;11(Suppl A):S87–98. [PubMed] [Google Scholar]
- 10.Chan DC. Fass D. Berger JM. Kim PS. Core structure of gp41 from the HIV envelope glycoprotein. Cell. 1997;89(2):263–273. doi: 10.1016/s0092-8674(00)80205-6. [DOI] [PubMed] [Google Scholar]
- 11.Eckert DM. Malashkevich VN. Hong LH. Carr PA. Kim PS. Inhibiting HIV-1 entry: Discovery of D-peptide inhibitors that target the gp41 coiled-coil pocket. Cell. 1999;99(1):103–115. doi: 10.1016/s0092-8674(00)80066-5. [DOI] [PubMed] [Google Scholar]
- 12.Musey L. Hughes J. Schacker T. Shea T. Corey L. McElrath MJ. Cytotoxic-T-cell responses, viral load, and disease progression in early human immunodeficiency virus type 1 infection. N Engl J Med. 1997;337(18):1267–1274. doi: 10.1056/NEJM199710303371803. [DOI] [PubMed] [Google Scholar]
- 13.Scheffold A. Lohning M. Richter A, et al. Analysis and sorting of T cells according to cytokine expression. Eur Cytokine Netw. 1998;9(3 Suppl):5–11. [PubMed] [Google Scholar]
- 14.Andre S. Seed B. Eberle J. Schraut W. Bultmann A. Haas J. Increased immune response elicited by DNA vaccination with a synthetic gp120 sequence with optimized codon usage. J Virol. 1998;72(2):1497–1503. doi: 10.1128/jvi.72.2.1497-1503.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bures R. Gaitan A. Zhu T, et al. Immunization with recombinant canarypox vectors expressing membrane-anchored glycoprotein 120 followed by glycoprotein 160 boosting fails to generate antibodies that neutralize R5 primary isolates of human immunodeficiency virus type 1. AIDS Res Hum Retroviruses. 2000;16(18):2019–2035. doi: 10.1089/088922200750054756. [DOI] [PubMed] [Google Scholar]
- 16.Hessell AJ. Poignard P. Hunter M, et al. Effective, low-titer antibody protection against low-dose repeated mucosal SHIV challenge in macaques. Nat Med. 2009;15(8):951–954. doi: 10.1038/nm.1974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hessell AJ. Rakasz EG. Poignard P, et al. Broadly neutralizing human anti-HIV antibody 2G12 is effective in protection against mucosal SHIV challenge even at low serum neutralizing titers. PLoS Pathog. 2009;5(5):e1000433. doi: 10.1371/journal.ppat.1000433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hessell AJ. Rakasz EG. Tehrani DM, et al. Broadly neutralizing monoclonal antibodies 2F5 and 4E10, directed against the human immunodeficiency virus type 1 (HIV-1) gp41 membrane proximal external region (MPER), protect against SHIVBa-L mucosal challenge. J Virol. 2010;84(3):1302–1313. doi: 10.1128/JVI.01272-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Manz R. Assenmacher M. Pfluger E. Miltenyi S. Radbruch A. Analysis and sorting of live cells according to secreted molecules, relocated to a cell-surface affinity matrix. Proc Natl Acad Sci USA. 1995;92(6):1921–1925. doi: 10.1073/pnas.92.6.1921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Montefiori DC. Robinson WE., Jr. Schuffman SS. Mitchell WM. Evaluation of antiviral drugs and neutralizing antibodies to human immunodeficiency virus by a rapid and sensitive microtiter infection assay. J Clin Microbiol. 1988;26(2):231–235. doi: 10.1128/jcm.26.2.231-235.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Muster T. Steindl F. Purtscher M, et al. A conserved neutralizing epitope on gp41 of human immunodeficiency virus type 1. J Virol. 1993;67(11):6642–6647. doi: 10.1128/jvi.67.11.6642-6647.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ogg GS. Jin X. Bonhoeffer S, et al. Quantitation of HIV-1-specific cytotoxic T lymphocytes and plasma load of viral RNA. Science. 1998;279(5359):2103–2106. doi: 10.1126/science.279.5359.2103. [DOI] [PubMed] [Google Scholar]
- 23.Robey WG. Arthur LO. Matthews TJ, et al. Prospect for prevention of human immunodeficiency virus infection: Purified 120-kDa envelope glycoprotein induces neutralizing antibody. Proc Natl Acad Sci USA. 1986;83(18):7023–7027. doi: 10.1073/pnas.83.18.7023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Burton DR. Desrosiers RC. Doms RW, et al. HIV vaccine design and the neutralizing antibody problem. Nat Immunol. 2004;5(3):233–236. doi: 10.1038/ni0304-233. [DOI] [PubMed] [Google Scholar]
- 25.Haynes BF. Montefiori DC. Aiming to induce broadly reactive neutralizing antibody responses with HIV-1 vaccine candidates. Expert Rev Vaccines. 2006;5(4):579–595. doi: 10.1586/14760584.5.4.579. [DOI] [PubMed] [Google Scholar]
- 26.Li Y. Migueles SA. Welcher B, et al. Broad HIV-1 neutralization mediated by CD4-binding site antibodies. Nat Med. 2007;13(9):1032–1034. doi: 10.1038/nm1624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Walker LM. Phogat SK. Chan-Hui PY, et al. Broad and potent neutralizing antibodies from an African donor reveal a new HIV-1 vaccine target. Science. 2009;326(5950):285–289. doi: 10.1126/science.1178746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wu X. Yang ZY. Li Y, et al. Rational design of envelope identifies broadly neutralizing human monoclonal antibodies to HIV-1. Science. 2010;329(5993):856–861. doi: 10.1126/science.1187659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Corti D. Langedijk JP. Hinz A, et al. Analysis of memory B cell responses and isolation of novel monoclonal antibodies with neutralizing breadth from HIV-1-infected individuals. PLoS One. 2010;5(1):e8805. doi: 10.1371/journal.pone.0008805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Gray ES. Moore PL. Bibollet-Ruche F, et al. 4E10-resistant variants in a human immunodeficiency virus type 1 subtype C-infected individual with an anti-membrane-proximal external region-neutralizing antibody response. J Virol. 2008;82(5):2367–2375. doi: 10.1128/JVI.02161-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Li M. Salazar-Gonzalez JF. Derdeyn CA, et al. Genetic and neutralization properties of subtype C human immunodeficiency virus type 1 molecular env clones from acute and early heterosexually acquired infections in Southern Africa. J Virol. 2006;80(23):11776–11790. doi: 10.1128/JVI.01730-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sather DN. Armann J. Ching LK, et al. Factors associated with the development of cross-reactive neutralizing antibodies during human immunodeficiency virus type 1 infection. J Virol. 2009;83(2):757–769. doi: 10.1128/JVI.02036-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Tomaras GD. Yates NL. Liu P, et al. Initial B-cell responses to transmitted human immunodeficiency virus type 1: Virion-binding immunoglobulin M (IgM) and IgG antibodies followed by plasma anti-gp41 antibodies with ineffective control of initial viremia. J Virol. 2008;82(24):12449–12463. doi: 10.1128/JVI.01708-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Grundner C. Mirzabekov T. Sodroski J. Wyatt R. Solid-phase proteoliposomes containing human immunodeficiency virus envelope glycoproteins. J Virol. 2002;76(7):3511–3521. doi: 10.1128/JVI.76.7.3511-3521.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Pancera M. Wyatt R. Selective recognition of oligomeric HIV-1 primary isolate envelope glycoproteins by potently neutralizing ligands requires efficient precursor cleavage. Virology. 2005;332(1):145–156. doi: 10.1016/j.virol.2004.10.042. [DOI] [PubMed] [Google Scholar]
- 36.Sattentau QJ. Moore JP. Conformational changes induced in the human immunodeficiency virus envelope glycoprotein by soluble CD4 binding. J Exp Med. 1991;174(2):407–415. doi: 10.1084/jem.174.2.407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Binley JM. Sanders RW. Clas B, et al. A recombinant human immunodeficiency virus type 1 envelope glycoprotein complex stabilized by an intermolecular disulfide bond between the gp120 and gp41 subunits is an antigenic mimic of the trimeric virion-associated structure. J Virol. 2000;74(2):627–643. doi: 10.1128/jvi.74.2.627-643.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Dey AK. David KB. Lu M. Moore JP. Biochemical and biophysical comparison of cleaved and uncleaved soluble, trimeric HIV-1 envelope glycoproteins. Virology. 2009;385(1):275–281. doi: 10.1016/j.virol.2008.12.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Herrera C. Klasse PJ. Michael E, et al. The impact of envelope glycoprotein cleavage on the antigenicity, infectivity, and neutralization sensitivity of Env-pseudotyped human immunodeficiency virus type 1 particles. Virology. 2005;338(1):154–172. doi: 10.1016/j.virol.2005.05.002. [DOI] [PubMed] [Google Scholar]
- 40.Alam SM. Morelli M. Dennison SM, et al. Role of HIV membrane in neutralization by two broadly neutralizing antibodies. Proc Natl Acad Sci USA. 2009;106(48):20234–20239. doi: 10.1073/pnas.0908713106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Shen X. Dennison SM. Liu P, et al. Prolonged exposure of the HIV-1 gp41 membrane proximal region with L669S substitution. Proc Natl Acad Sci USA. 2009;107(13):5972–5977. doi: 10.1073/pnas.0912381107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Daar ES. Li XL. Moudgil T. Ho DD. High concentrations of recombinant soluble CD4 are required to neutralize primary human immunodeficiency virus type 1 isolates. Proc Natl Acad Sci USA. 1990;87(17):6574–6578. doi: 10.1073/pnas.87.17.6574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Hwang SS. Boyle TJ. Lyerly HK. Cullen BR. Identification of envelope V3 loop as the major determinant of CD4 neutralization sensitivity of HIV-1. Science. 1992;257(5069):535–537. doi: 10.1126/science.1636088. [DOI] [PubMed] [Google Scholar]
- 44.Koito A. Harrowe G. Levy JA. Cheng-Mayer C. Functional role of the V1/V2 region of human immunodeficiency virus type 1 envelope glycoprotein gp120 in infection of primary macrophages and soluble CD4 neutralization. J Virol. 1994;68(4):2253–2259. doi: 10.1128/jvi.68.4.2253-2259.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.O'Brien WA. Chen IS. Ho DD. Daar ES. Mapping genetic determinants for human immunodeficiency virus type 1 resistance to soluble CD4. J Virol. 1992;66(5):3125–3130. doi: 10.1128/jvi.66.5.3125-3130.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Orloff SL. Kennedy MS. Belperron AA. Maddon PJ. McDougal JS. Two mechanisms of soluble CD4 (sCD4)-mediated inhibition of human immunodeficiency virus type 1 (HIV-1) infectivity and their relation to primary HIV-1 isolates with reduced sensitivity to sCD4. J Virol. 1993;67(3):1461–1471. doi: 10.1128/jvi.67.3.1461-1471.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Parren PW. Wang M. Trkola A, et al. Antibody neutralization-resistant primary isolates of human immunodeficiency virus type 1. J Virol. 1998;72(12):10270–10274. doi: 10.1128/jvi.72.12.10270-10274.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Si Z. Phan N. Kiprilov E. Sodroski J. Effects of HIV type 1 envelope glycoprotein proteolytic processing on antigenicity. AIDS Res Hum Retroviruses. 2003;19(3):217–226. doi: 10.1089/088922203763315722. [DOI] [PubMed] [Google Scholar]
- 49.Sullivan N. Sun Y. Li J. Hofmann W. Sodroski J. Replicative function and neutralization sensitivity of envelope glycoproteins from primary and T-cell line-passaged human immunodeficiency virus type 1 isolates. J Virol. 1995;69(7):4413–4422. doi: 10.1128/jvi.69.7.4413-4422.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Sullivan N. Sun Y. Sattentau Q, et al. CD4-Induced conformational changes in the human immunodeficiency virus type 1 gp120 glycoprotein: Consequences for virus entry and neutralization. J Virol. 1998;72(6):4694–4703. doi: 10.1128/jvi.72.6.4694-4703.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Zhang YJ. Fredriksson R. McKeating JA. Fenyo EM. Passage of HIV-1 molecular clones into different cell lines confers differential sensitivity to neutralization. Virology. 1997;238(2):254–264. doi: 10.1006/viro.1997.8812. [DOI] [PubMed] [Google Scholar]
- 52.Sattentau QJ. Zolla-Pazner S. Poignard P. Epitope exposure on functional, oligomeric HIV-1 gp41 molecules. Virology1995. 206(1):713–717. doi: 10.1016/s0042-6822(95)80094-8. [DOI] [PubMed] [Google Scholar]
- 53.Korber BT. Foley F. Kuiken C. Pillai S. Sodroski J. Numbering positions in HIV relative to HXBc2. In: Sodroski J, editor. Human Retroviruses and AIDS. Los Alamos National Laboratory; Los Alamos, NM: 1998. pp. iii-102–iii-103. [Google Scholar]
- 54.Koch M. Pancera M. Kwong PD, et al. Structure-based, targeted deglycosylation of HIV-1 gp120 and effects on neutralization sensitivity and antibody recognition. Virology. 2003;313(2):387–400. doi: 10.1016/s0042-6822(03)00294-0. [DOI] [PubMed] [Google Scholar]
- 55.Chakrabarti BK. Kong WP. Wu BY, et al. Modifications of the human immunodeficiency virus envelope glycoprotein enhance immunogenicity for genetic immunization. J Virol. 2002;76(11):5357–5368. doi: 10.1128/JVI.76.11.5357-5368.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Berlioz-Torrent C. Shacklett BL. Erdtmann L, et al. Interactions of the cytoplasmic domains of human and simian retroviral transmembrane proteins with components of the clathrin adaptor complexes modulate intracellular and cell surface expression of envelope glycoproteins. J Virol. 1999;73(2):1350–1361. doi: 10.1128/jvi.73.2.1350-1361.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.LaBranche CC. Sauter MM. Haggarty BS, et al. A single amino acid change in the cytoplasmic domain of the simian immunodeficiency virus transmembrane molecule increases envelope glycoprotein expression on infected cells. J Virol. 1995;69(9):5217–5227. doi: 10.1128/jvi.69.9.5217-5227.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Si Z. Cayabyab M. Sodroski J. Envelope glycoprotein determinants of neutralization resistance in a simian-human immunodeficiency virus (SHIV-HXBc2P 3.2) derived by passage in monkeys. J Virol. 2001;75(9):4208–4218. doi: 10.1128/JVI.75.9.4208-4218.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Gorny MK. Zolla-Pazner S. Recognition by human monoclonal antibodies of free and complexed peptides representing the prefusogenic and fusogenic forms of human immunodeficiency virus type 1 gp41. J Virol. 2000;74(13):6186–6192. doi: 10.1128/jvi.74.13.6186-6192.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Binley JM. Cayanan CS. Wiley C. Schulke N. Olson WC. Burton DR. Redox-triggered infection by disulfide-shackled human immunodeficiency virus type 1 pseudovirions. J Virol. 2003;77(10):5678–5684. doi: 10.1128/JVI.77.10.5678-5684.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Thali M. Furman C. Helseth E. Repke H. Sodroski J. Lack of correlation between soluble CD4-induced shedding of the human immunodeficiency virus type 1 exterior envelope glycoprotein and subsequent membrane fusion events. J Virol. 1992;66(9):5516–5524. doi: 10.1128/jvi.66.9.5516-5524.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Yang X. Lee J. Mahony EM. Kwong PD. Wyatt R. Sodroski J. Highly stable trimers formed by human immunodeficiency virus type 1 envelope glycoproteins fused with the trimeric motif of T4 bacteriophage fibritin. J Virol. 2002;76(9):4634–4642. doi: 10.1128/JVI.76.9.4634-4642.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Crooks ET. Jiang P. Franti M, et al. Relationship of HIV-1 and SIV envelope glycoprotein trimer occupation and neutralization. Virology. 2008;377(2):364–378. doi: 10.1016/j.virol.2008.04.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Jeffs SA. Goriup S. Kebble B, et al. Expression and characterisation of recombinant oligomeric envelope glycoproteins derived from primary isolates of HIV-1. Vaccine. 2004;22(8):1032–1046. doi: 10.1016/j.vaccine.2003.08.042. [DOI] [PubMed] [Google Scholar]
- 65.Sanders RW. Vesanen M. Schuelke N, et al. Stabilization of the soluble, cleaved, trimeric form of the envelope glycoprotein complex of human immunodeficiency virus type 1. J Virol. 2002;76(17):8875–8889. doi: 10.1128/JVI.76.17.8875-8889.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Srivastava IK. Stamatatos L. Legg H, et al. Purification and characterization of oligomeric envelope glycoprotein from a primary R5 subtype B human immunodeficiency virus. J Virol. 2002;76(6):2835–2847. doi: 10.1128/JVI.76.6.2835-2847.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Yang X. Wyatt R. Sodroski J. Improved elicitation of neutralizing antibodies against primary human immunodeficiency viruses by soluble stabilized envelope glycoprotein trimers. J Virol. 2001;75(3):1165–1171. doi: 10.1128/JVI.75.3.1165-1171.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Zhang CW. Chishti Y. Hussey RE. Reinherz EL. Expression, purification, and characterization of recombinant HIV gp140. The gp41 ectodomain of HIV or simian immunodeficiency virus is sufficient to maintain the retroviral envelope glycoprotein as a trimer. J Biol Chem. 2001;276(43):39577–39585. doi: 10.1074/jbc.M107147200. [DOI] [PubMed] [Google Scholar]
- 69.Barnett SW. Lu S. Srivastava I, et al. The ability of an oligomeric human immunodeficiency virus type 1 (HIV-1) envelope antigen to elicit neutralizing antibodies against primary HIV-1 isolates is improved following partial deletion of the second hypervariable region. J Virol. 2001;75(12):5526–5540. doi: 10.1128/JVI.75.12.5526-5540.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Bower JF. Yang X. Sodroski J. Ross TM. Elicitation of neutralizing antibodies with DNA vaccines expressing soluble stabilized human immunodeficiency virus type 1 envelope glycoprotein trimers conjugated to C3d. J Virol. 2004;78(9):4710–4719. doi: 10.1128/JVI.78.9.4710-4719.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Dey B. Svehla K. Xu L, et al. Structure-based stabilization of HIV-1 gp120 enhances humoral immune responses to the induced co-receptor binding site. PLoS Pathog. 2009;5(5):e1000445. doi: 10.1371/journal.ppat.1000445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.VanCott TC. Mascola JR. Kaminski RW, et al. Antibodies with specificity to native gp120 and neutralization activity against primary human immunodeficiency virus type 1 isolates elicited by immunization with oligomeric gp160. J Virol. 1997;71(6):4319–4330. doi: 10.1128/jvi.71.6.4319-4330.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Schulke N. Vesanen MS. Sanders RW, et al. Oligomeric and conformational properties of a proteolytically mature, disulfide-stabilized human immunodeficiency virus type 1 gp140 envelope glycoprotein. J Virol. 2002;76(15):7760–7776. doi: 10.1128/JVI.76.15.7760-7776.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Binley JM. Sanders RW. Master A, et al. Enhancing the proteolytic maturation of human immunodeficiency virus type 1 envelope glycoproteins. J Virol. 2002;76(6):2606–2616. doi: 10.1128/JVI.76.6.2606-2616.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.