Abstract
MicroRNAs (miRNAs) are important negative regulators of gene expression in plant and animals, which are endogenously produced from their own genes. Computational comparative approach based on evolutionary conservation of mature miRNAs has revealed a number of orthologs of known miRNAs in different plant species. The homology-based plant miRNA discovery, followed by target prediction, comprises several steps, which have been done so far manually. Here, we present the bioinformatics pipeline miRTour which automates all the steps of miRNA similarity search, miRNA precursor selection, target prediction and annotation, each of them performed with the same set of input sequences.
Availability
The database is available for free at http://bio2server.bioinfo.uni-plovdiv.bg/miRTour/
Keywords: microRNAs, microRNAs targets, plant miRNA homologs
Background
MicroRNAs (miRNAs) are important components of the regulatory networks in plant and animals. Plant miRNAs have been well characterized in several model systems for plant genomics such as Arabidopsis, Brachypodium, Oryza and Populus [1–6]. They are single-stranded 20–22 nt small RNAs that are endogenously produced from their own genes [2]. MiRNA genes in plants are systematically grouped into families that yield almost identical miRNAs. Currently, 2952 miRNA genes across 43 plant species were identified and annotated in miRBase v16 [7]. Here, we present the computational approach miRTour for homology-based discovery of plant miRNA and their targets from sequencing datasets (EST, GSS, SRA, etc.). The program automates all the steps of miRNA similarity search, miRNA precursor selection, target prediction and annotation, each of them performed with the same set of input sequences. The miRTour software has a user friendly web-interface which provides a comprehensive pipeline for identification of plant miRNA homologs.
Implementation
We developed a web-based application for identification of new homologs of plant miRNAs in EST/GSS/SRA or assembled contig dataset. The program uses a local database that consists of all plant mature miRNA sequences from miRBase v16 (http://www.mirbase.org). The first step in our algorithm is mapping the plant mature miRNA sequences onto the ESTs/GSS dataset provided by the user. For the short sequence mapping, we employ the Gassst (Global Alignment Short Sequence Search Tool) v1.27, with no more than 4 mismatches. [8]. In the next step, the miRTour filters out the protein-coding ESTs/GSSs with miRNA hits applying BLASTX with E value 10-6 with Arabidopsis and Orysa protein sequences (A local copy of the BLAST tool was obtained from the NCBI FTP server, ftp://ftp.ncbi. nih.gov/blast). Next, putative miRNA foldback precursors are extracted from the non-coding ESTs/GSSs that contain matched known miRNAs. For selection of new premiRNA and miRNA candidates, the following criteria are taken into consideration: (1) MFEI (minimum free energy index) for the predicted premiRNA secondary structure to be greater than 0.7; (2) the number of the mismatches in miRNA/miRNA* must be less than 5; (3) the number of consecutive mismatches in miR/miR* must be less than 4. The secondary structure of the putative precursor sequence is predicted and visualized with the RNAfold algorithm from Vienna RNA package, on which bases the described criteria are evaluated [9]. Using ClustalW, the miRTour creates a colour alignment of the predicted precursor sequence with sequences of known plant mature miRNAs. To find the protein-coding targets of newly identified miRNAs in the same initial EST/GSS dataset, we have wrapped the software for target searching TargetFinder v1.6 in our script [10]. The annotation of the protein-coding target sequences is performed against all known Arabidopsis and Orysa protein sequences by BLASTX.
Software Input
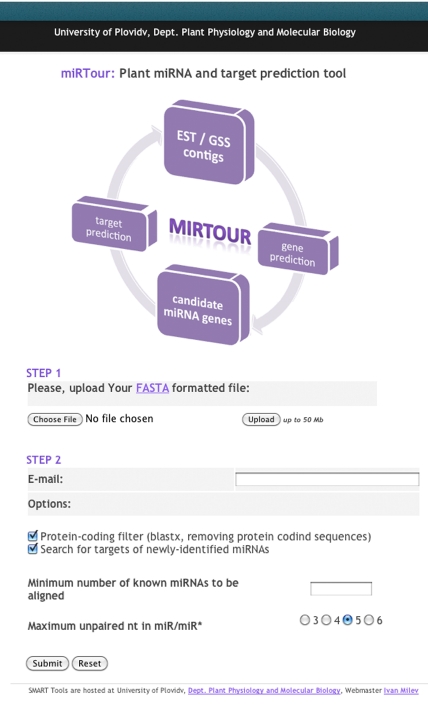
The miRTour is freely available at http://bio2server.bioinfo.uniplovdiv.bg/miRTour/. It has a simple and intuitive web-based user interface (Figure 1). In order to run a job on the miRTour, the following data are required as an input: Step 1: First, the user has to upload a file with EST/GSS/contig sequences (up to 50MB) in multi FASTA format. Step 2: The user has to input an e-mail address. This step is obligatory since the batch process of a large dataset requires more server-side time. The user can turn on/off both, the BLASTX filter and the target search module, if needed. Other options that the user should specify are the minimum number of plant homologs that the miRTour must align and the maximum number of unpaired bases in miR/miR* (the default parameter is 5).
Figure 1.
The miRTour software web-interface.
Software Output
The output results are sent automatically by e-mail in HTML and PDF formats. The result data will be kept on the server for 5 days from the moment of submission. For each query dataset submitted to the miRTour, the output consists of sections with putative pre-miRNA and mature miRNA sequences; homology alignment of the newly-identified miRNA precursors to known plant miRNAs; details for secondary structure of the precursor - MFEI, CG% and number of mismatches in miR/miR*; predicted target sequences for the new mature miRNAs.
Conclusion
High-throughput parallel sequencing technologies have made possible the production and accumulation of large genome and transcriptome datasets (EST/GSS contigs). Homology-based discovery of miRNAs in plant species, along with target prediction step, were a comprehensive assignment, which have been done so far manually. The developed miRTour automates those tasks and provides a pipeline for identification of new miRNAs and prediction of their targets from a single dataset of sequences.
Caveat and future development
Due to server limitation, large datasets (more than 50MB) cannot be submitted for analysis by the miRTour. As the future development of our tool, we plan to extend our server capacity in a way that will allow larger file uploading. Another stage of development is to upgrade the miRTour to work with animal miRNAs, but it will take a longer time since the prediction of animal miRNA targets is more complex task compared to plant miRNA target prediction.
Footnotes
Citation:Milev et al, Bioinformation 6(6): 248-249 (2011)
References
- 1.V Baev, et al. Genomics. 2011;97:282. [Google Scholar]
- 2.DP Bartel, et al. Cell. 2004;116:281. [Google Scholar]
- 3.D Klevebring, et al. BMC Genomics. 2009;10:620. doi: 10.1186/1471-2164-10-620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.R Sunkar, et al. Plant Cell. 2005;17:1397. [Google Scholar]
- 5.R Sunkar, G Jagadeeswaran. BMC Plant Biol. 2008;8:37. doi: 10.1186/1471-2229-8-37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.R Sunkar, et al. BMC Plant Biol. 2008;8:25. doi: 10.1186/1471-2229-8-25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Griffiths-Jones S, et al. Nucleic Acids Res. 2008;36:D154. doi: 10.1093/nar/gkm952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.G Rizk, D Lavenier. Bioinformatics. 2010;26:2534. doi: 10.1093/bioinformatics/btq485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.IL Hofacker, et al. Nucleic Acids Res. 2003;31:3429. doi: 10.1093/nar/gkg599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.N Fahlgren, et al. PLoS One. 2007;2:e219. [Google Scholar]