Abstract
We demonstrate tracking of protein structural changes with time-resolved wide-angle X-ray scattering (TR-WAXS) with nanosecond time resolution. We investigated the tertiary and quaternary conformational changes of human hemoglobin under nearly physiological conditions triggered by laser-induced ligand photolysis. We also report data on optically induced tertiary relaxations of myoglobin and refolding of cytochrome c to illustrate the wide applicability of the technique. By providing insights into the structural dynamics of proteins functioning in their natural environment, TR-WAXS complements and extends results obtained with time-resolved optical spectroscopy and X-ray crystallography.
The dynamics and function of proteins are intimately connected to their three-dimensional structure, which is characterized by fluctuations in atomic positions and conformational transitions covering a broad range of time scales (from sub-picoseconds to seconds) and amplitudes of motion (from sub-angstroms to tens of angstroms). Protein structural changes in solution have so far been characterized mainly by time-resolved optical spectroscopic methods, which give signals that are only indirectly related to three-dimensional structures. For protein crystals, a combination of high time resolution and structural sensitivity became available with the advent of time-resolved Laue crystallography1–3, but its applicability has been limited to a few model systems because of the stringent prerequisites such as the need for highly ordered and radiation-resistant single crystals. More notably, crystal packing constraints might hinder biologically relevant motions. To obtain information about protein motions in a more natural environment, nuclear magnetic resonance (NMR) and X-ray scattering methods have been used as direct structural probes of protein structure in solution4–6, but these methods also have limitations. Small-angle X-ray scattering probes the overall size and shape of the protein whereas wide-angle X-ray scattering (WAXS) gives more detailed information such as the fold of helices and sheets7; however, the time resolution has so far been limited to 160 µs at best8,9. NMR is a powerful technique for structure determination in solution, but it works best for small proteins, needs properly labeled samples10, and the time resolution of protein NMR is inherently limited to milliseconds.
Here we demonstrate that time-resolved wide-angle X-ray scattering (TR-WAXS) using synchrotron radiation can be used to accurately probe structural changes of proteins in solution with nanosecond time resolution. TR-WAXS combines the high time resolution already proven to be important for studies on biological samples11,12 with the high structural sensitivity demonstrated for WAXS studies13,14. TR-WAXS is complementary to time-resolved optical spectroscopy as it allows for tracking of tertiary and quaternary structural changes of a protein with global sensitivity; it is sensitive to changes in the position of all the atoms in the protein rather than to modifications around a given spectroscopic marker. TR-WAXS is applicable to many biologically relevant systems in solution, thus allowing a wide range of experimental parameters such as pH or salt and protein concentrations to be varied.
We report results of TR-WAXS experiments performed mainly on human hemoglobin (Hb), a tetrameric protein made of two identical αβ dimers that is known to adopt at least two different quaternary structures in solution: a ‘relaxed’ (R) structure stabilized by the presence of ligands like CO and O2, and a ‘tense’ (T) structure that is stable when the protein is unligated15,16. The ligated-to-unligated transition in Hb involves both conformational changes within the subunits (tertiary structure transition) and changes in the relative disposition of the subunits (quaternary structure transition). The R-T transition has been studied over the last decades and is often used as a paradigm of cooperativity in molecular biology. Our data explain the pathway followed by Hb molecules while switching from the ligated to the unligated state. We obtained information on both the kinetics of the R-T transition and the sequence of structural changes taking place during the transition. In addition, we report preliminary experiments on sperm whale myoglobin (Mb) and horse heart cytochrome c (Cyt-c); the Mb data show the sensitivity of TR-WAXS to local tertiary conformational changes, and the Cyt-c data demonstrate the applicability of the technique to analyze protein folding.
RESULTS
Structural sensitivity and signal-to-noise ratio of TR-WAXS
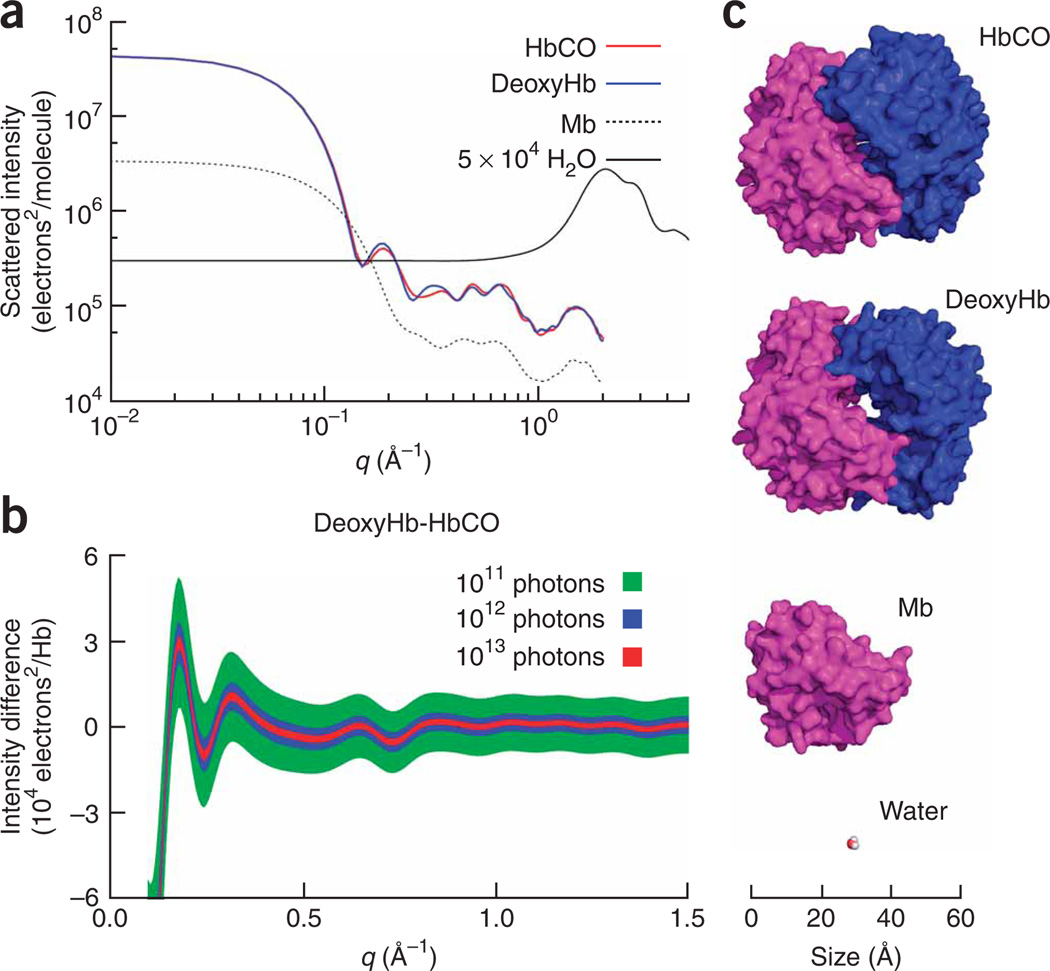
One of the advantages of scattering as compared to spectroscopic techniques is that once a collection of atomic positions is given, it is straightforward to calculate the scattering pattern for an isotropic ensemble of molecules (Fig. 1). Recently developed codes17 allow calculation of the scattering intensity starting from a given protein structure deposited in the protein data bank (PDB). The small-angle X-ray scattering range (q = 4π sin(θ/2)λ−1 < 0.2 Å−1), where q is the magnitude of the scattering vector, θ is the scattering angle and λ is the X-ray wavelength, has been greatly exploited18 as biologically relevant phenomena sizably affect the scattering in this q region. At small angles all the electrons in a protein scatter in phase, thus substantially boosting the protein contribution to the scattered intensity against the water background (Fig. 1a). The protein scattering pattern is more or less featureless for q < 0.1 Å−1. At higher q values, a more structured pattern appears mainly due to correlations among the protein subunits (0.15 < q < 0.25 Å−1), helices or sheets (0.25 < q < 0.6 Å−1) and between the atoms forming the secondary structures (q > 0.6 Å−1)7,14. Differences in the scattering patterns of carbonmonoxy-Hb (HbCO) and deoxygenated Hb (deoxyHb) are evident for q > 0.1 Å−1 (Fig. 1a,b). The difference between the two patterns reaches a maximum in the 0.1–0.5 Å−1 region as expected; indeed, the two Hb structures differ mainly in their tertiary and quaternary structure (Fig. 1c). The dominating water scattering makes it more difficult to measure scattering differences at q > 1 Å−1. The calculated deoxyHb-HbCO scattering difference reported in Figure 1b is an estimation of the effect of the Hb structural transition and highlights the advantage of using scattering differences for monitoring protein structural changes.
Figure 1.
Plausibility of TR-WAXS based on photon statistics considerations. (a) Calculated X-ray patterns for the relevant protein structures together with the water pattern scaled for a 1 mM protein solution (~5 × 104 H2O molecules for each Hb molecule). The scattered intensity is expressed in units of electrons2/protein molecule. The HbCO, deoxyHb and Mb scattering patterns were calculated using the PDB files 1BBB, 2HHB and 1DWR, respectively. (b) Calculated deoxyHb-HbCO difference pattern for three photon fluxes. The line thickness represents one s.d. of the signal that had been calculated under the assumption of shot noise–limited data. At least 1012 photons had to be accumulated to resolve the characteristic oscillations of the difference patterns. (c) Snapshots of the molecular structures used in the calculations of scattering patterns generated using PyMOL (DeLano, W.L. The PyMOL User’s Manual, DeLano Scientific, 2002). In the case of Hb, the αβ dimers involved in the quaternary structure transition are colored differently.
Pump-probe experiments are typically ‘X-ray photon–starved’, and a critical component in the signal-to-noise ratio of difference patterns is photon statistics. Under the assumption of photon counting limited data, we estimated that ~1012 photons impinging on the sample are needed to resolve the typical oscillations of difference patterns (Fig. 1b and Supplementary Methods online).
TR-WAXS methodology and data processing
In a TR-WAXS experiment (Fig. 2), a laser pulse is used to trigger the protein structural change, and transient structures are then followed by delayed X-ray pulses from an undulator in a straight section of the synchrotron. Structural changes occurring in the sample (Fig. 2d) leave their ‘fingerprints’ in the differences between the signals measured before and after the laser initiates the reaction (Figs. 2b,c) and can be monitored as a function of time. In the case of hemoglobin (Fig. 2d), a laser pulse in the green light wavelength (527 nm in our experiment) can break (photolyze) the Hb-CO bonds. Immediately after photolysis, the iron-heme displacement triggers a tertiary relaxation that is dominated by local structure changes. Strain at the Hb subunit interfaces drives a large-amplitude quaternary structural transition leading to T-state Hb. Finally, CO recombines bimolecularly with Tstate Hb to regenerate the R state.
Figure 2.
TR-WAXS methodology and data processing. (a) Schematic representation of the experimental setup. Transient structures were generated by a laser pulse (green beam) that triggered the conformational change (through photolysis of the Hb-CO bond), which was followed by delayed quasi-monochromatic X-ray pulses (100 ps long) extracted from the synchrotron (blue circle). Scattered X-rays were recorded in the forward direction by a sensitive CCD camera. The red arrow indicates the direction of electron flow inside the synchotron. (b) Each image was azimuthally averaged and converted into a one-dimensional curve. I, intensity. (c) After normalization, a reference scattering pattern, which probed the unexcited sample, was subtracted from the scattering pattern at a given time delay. These difference patterns are ’fingerprints’ of the transient protein structural changes. (d) Low-resolution snapshots depict the expected time-dependent structural changes. Regions of the protein involved in structural rearrangements are colored red. As the laser pulse (green arrow) photolyzes Hb-CO bonds, the protein undergoes tertiary structural changes (in less than 150 ns) followed by the R to T transition (3 µs time scale). Full rebinding occurs at slower time scales (1–10 ms).
Although photodissociation triggers tertiary and quaternary structural relaxations to the unligated stable T structure, this process competes with CO rebinding. To enhance the quaternary transition yield and to obtain a more homogeneous population of photolyzed protein molecules, we used an intense (~2.5 mJ/mm2) 150-ns laser pulse. Such a long pulse can photolyze multiple times those subunits that undergo fast ‘geminate’ rebinding11,19. Under these experimental conditions nearly 100%of the Hb molecules are fully photolyzed, and ~70% of them are able to switch to the T state, this fraction being limited by the concomitant bimolecular rebinding of CO to Hb molecules in the R state (Supplementary Methods).
Laser-induced X-ray scattering differences contain not only fingerprints from protein structural changes but also solvent heating contributions arising from changes in solvent temperature, density and pressure20,21 that are induced by the energy transferred from the laser into the solvent via protein absorption. To decouple the two contributions, we subtracted from the difference signal at each time delay, the difference signal measured 32 ms after photolysis (Fig. 3a). At 32 ms, Hb recovered its equilibrium HbCO structure, but the energy deposited by the laser pulse has not yet diffused out of the probed sample volume. The difference signal at 32 ms is thus a fingerprint of the structural changes induced uniquely by solvent heating. We plotted the heat-removed 100 µs time-resolved data and the ‘static’ difference (deoxyHb minus HbCO) obtained in separate static experiments (Fig. 3b). This comparison showed that data-reduction procedures and the subtraction of the solvent response were valid and confirmed the structural sensitivity of TR-WAXS to the R-T transition. The good agreement between the static and time-resolved differences led us to the following conclusions: (i) TR-WAXS allows monitoring of protein conformational transitions with high fidelity and (ii) the difference signal at 100 µs is due to the R to T transition, meaning that Hb reached the equilibrium deoxygenated T state in less than 100 µs.
Figure 3.
TR-WAXS data on hemoglobin and the solvent heating contribution. (a) TR-WAXS data at 100 µs after photolysis compared with that at 32 ms (multiplied by 2); 32 ms after photolysis, the unbound CO molecules have recombined to Hb and the scattering signal is due essentially to a temperature rise of the solvent. The 32-ms pattern was used to subtract the solvent response at any time delay after proper scaling. (b) The solvent response–subtracted time-resolved data at 100 µs after photolysis compared with the static equilibrium scattering difference between a deoxyHb equilibrium sample and an HbCO sample. (c) Laser-induced changes of the scattering patterns for several selected time delays (black, after solvent response subtraction). Data at all time delays have been fitted as a linear combination of the difference patterns at 200 ns and 100 µs (red). The TR-WAXS pattern obtained at 10 ns from photolysis of MbCO is shown to demonstrate the sensitivity of the technique to local tertiary structural changes. (d) The population kinetics of R-like species and T-like species estimated from TR-WAXS data. (e) Schematic representation of Hb structural dynamics between about 100 ns and 100 ms from photolysis.
To follow the structural dynamics of Hb, we collected several time delays between ~200 ns and 32 ms (Fig. 3c). A clear difference signal was present 200 ns after photolysis. This signal was markedly different from that at 100 µs, indicating that X-rays had captured a new intermediate that may originate from a tertiary structural transition as already suggested by optical spectroscopy11,12. To provide a clear and independent experimental proof that TR-WAXS is sensitive to tertiary conformational changes, we also applied TR-WAXS to Mb, a single-subunit protein almost identical to the subunits of Hb. In contrast to Hb, the ligated-to-unligated transition induced by laser photolysis involves only tertiary structural changes in Mb. Data at 10 ns after photolysis of MbCO showed clear oscillations in the WAXS pattern, demonstrating the sensitivity to tertiary rearrangements (Fig. 3c).
Timing the quaternary structural transition of hemoglobin
TR-WAXS signals acquired for Hb can be reproduced at any time delay by a linear combination of two patterns. We plotted decompositions of the data in terms of the pattern representing tertiary relaxed proteins (200 ns) and that representing the fully deoxygenated ones (100 µs) (Fig. 3c). Such linear combinations track the data with high fidelity supporting that, apart from the transient 200-ns structure, there were no other substantially populated intermediates detectable with TR-WAXS within the current signal-to-noise ratio. The weight factors of the two-component reconstruction were proportional to the populations of R-like (tertiary relaxed only) and T-like (tertiary and quaternary relaxed) states (Fig. 3d). The R-like population decayed via two channels: rebinding with CO molecules and transition to the T state (Fig. 3e). The amount of the T-like population increased initially with time dependence owing to the R to T transition, reached a plateau at ~10 µs, and finally decayed in 100 µs to 32ms through rebinding with CO molecules in solution. The population kinetics shown in Figure 3d can be analyzed in terms of an allosteric kinetic model19 (Supplementary Methods, Supplementary Figs. 1–3 and Supplementary Tables 1–3 online) and give a time scale for the R-Ttransition of ~1–3 µs, shorter than the time scale derived with time-resolved optical spectroscopy22. Our data favor a more recent estimation of the R-T transition rate based on time-resolved resonance Raman data12, which suggest that the relative rotation of one αβ dimer with respect to the other has already happened ~3 µs after photolysis of HbCO. Experiments designed to systematically compare the kinetics obtained with TR-WAXS and those obtained with optical spectroscopy under identical experimental conditions are under way.
Structural changes in the R to T transition of Hb
To gain insight into the structural changes after photolysis, we tried to reproduce the patterns at 200 ns and 100 µs using calculated scattering curves from known crystal structures. Although HbCO has been crystallized in at least two different quaternary conformations (R and R2) as a function of the different crystal preparation protocols used23,24, deoxyHb is less sensitive to the crystallization procedure and essentially a single structure (T) has been reported so far. We reproduced the 100 µs difference scattering by the calculated patterns of T-R2 (Fig. 4a), which suggested that in solution the HbCO molecules adopt a structure similar to the crystallographic R2 structure; we obtained a worse agreement using the R structure in the calculation (Supplementary Fig. 4 online). In spite of the qualitative agreement, however, the calculated differences did not fully reproduce the data, implying that crystal packing forces may distort the protein structure. The 200 ns difference scattering pattern was not reproduced by the R−R2 difference (Supplementary Fig. 4), thus ruling out the suggested stepwise pathway R2→R→T involving an R-like intermediate25–27.
Figure 4.
Comparison between TR-WAXS scattering differences and scattering differences calculated from crystallographic structures. (a) The time-resolved difference pattern measured 100 µs after photolysis of HbCO is compared to the difference pattern calculated from the R2 and T crystallographic structures (PDB files 1BBB and 2HHB were used as models of the R2 and T structures, respectively). (b) Snapshot of the CO-bound alpha subunit. The E and F helices form the ‘clamshell’ that holds the heme in place. The blue arrows indicate the direction of the E and F motion during the ‘clamshell relaxation’. (c) Comparison of the 200 ns time-resolved data with the difference pattern calculated from the R2 crystal structure and its modified unligated version mimicking the effect of the clamshell relaxation.
Time-resolved UV-wavelength resonance Raman data, computations on Hb12,28 and crystallographic studies on Mb29 suggest that one of the earliest events occurring in the tertiary relaxation of Hb and Mb after ligand dissociation is a concerted motion of the E and F helices that hold the heme in place with respect to the rest of the protein, the so-called ‘clamshell motion’ (Fig. 4b). We simulated the clamshell motion by modifying the tertiary structure of each subunit in the R2 tetramer (Fig. 4c and Supplementary Methods). The overall qualitative agreement between the calculated scattering difference and the 200 ns data suggested that the clamshell relaxation is compatible with our data, especially in view of the rather rough tertiary conformational change that we have applied to model the structural change.
Application of TR-WAXS to track folding of Cyt-c
Although time-resolved Laue crystallography has been used for three-dimensional visualization of protein structural dynamics1–3, its applicability has been limited to reversible reactions in single crystals. It has had limited applicability for studying irreversible reactions and has not been applicable for monitoring large conformational changes such as protein folding. Protein folding studies often start in conditions under which the protein is unfolded; external experimental parameters are then rapidly changed to favor the native folded state (Fig. 5a).
Figure 5.
Application of TR-WAXS to track folding of Cyt-c. (a) Schematic representation of light-induced folding of Cyt-c. (b) Time-resolved WAXS data relative to CO photolysis–induced folding of Cyt-c. A 200 ns laser pulse at 532 nm initiated photodissociation of the CO ligand, which in turn initiated the folding process. Experimental data at representative time delays are shown. (c) Population of the folded state as a function of time estimated from a linear combination of the experimental signal at 32 µs and 0.2 s.
A clever approach30 to study the refolding dynamics of a protein over a wide time window exploits the fact that Cyt-c does not usually bind external ligands such as CO because the iron atom of the heme group is covalently coordinated to the protein (Met80 residue). However, if Cyt-c is partially unfolded with a denaturing agent, it is possible to replace the Met80 residue with CO. As in Hb or Mb, the CO ligand can be optically dissociated, thereby initiating the refolding process. Although the interpretation of the observed spectral changes has been questioned31, the time-dependent evolution of the TR-WAXS signal of Cyt-c after photolysis is evident especially in the small-angle region (Fig. 5b). Although a more detailed analysis and discussion will be presented in a future publication, here we fitted the observed signal as a linear combination of one pattern at the earliest time delay, 32 µs, and the other at the latest time delay, 0.2 s. This simple approach satisfactorily reproduced the experimental data at all times. We plotted the weighting factor of the late time component against time (Fig. 5c); a simple exponential analysis yielded a time scale of about 25 ms for the CO photolysis–triggered folding.
DISCUSSION
The number of WAXS studies of protein has steadily increased in recent years because of the improved capabilities of third-generation synchrotron sources. Using such sources we extended the WAXS technique to monitor transient protein structures with a time resolution similar to that of many time-resolved optical studies. In addition to the ability to detect structural intermediates along reaction pathways, TR-WAXS studies can also contribute to a better understanding of the connection between spectroscopic markers and structural entities. TR-WAXS can be applied to track not only reversible reactions but also irreversible reactions such as protein folding induced by ligand dissociation and electron transfer, greatly widening its applicability to various protein reactions such as a drug binding to target proteins.
Although WAXS patterns contain valuable structural fingerprints, their information content (Supplementary Methods) is probably insufficient to allow a ‘model-free’ reconstruction of protein structures at quasi-atomic resolution. In this respect, the use of structures derived from X-ray crystallography or NMR as a starting point for refinements against TR-WAXS data appears promising (Fig. 4). The nanosecond time resolution of TR-WAXS demonstrated here should be extensible to 100 ps for proteins that can be properly excited with picosecond lasers. Indeed, the ultimate time resolution of TR-WAXS is determined by the length of X-ray pulses produced by the synchrotron (100 ps); such resolution might be improved even further with future X-ray–free electron lasers, which promise to deliver 0.1 ps X-ray pulses. Developments in the data analysis and time resolution, along with extensions to other excitation methods such as pH jump and temperature jump will widen the applicability of TR-WAXS and will presumably allow medium-resolution structural refinements of intermediate structures along reaction pathways.
METHODS
Sample preparation
Human Hb (Sigma) was dissolved in a 50 mM Na-phosphate buffer (pH 7.4). After filtering the solution with a 0.2-µm-pore-size mesh, the protein was fully converted to the HbCO form. An aliquot of the resulting 1.2 mM HbCO solution was transferred into a 1 mm X-ray capillary (Hampton Research) and immediately sealed with epoxy to minimize gas exchange. Sperm whale Mb and horse heart Cyt-c were purchased from Aldrich and used without purification. The final protein concentration of both samples was 8 mM in 0.1 M phosphate buffer at pH 7 for Mb and in 10 mM CAPS buffer at pH 7 for Cyt-c. In the case of Cyt-c experiments, the protein was initially unfolded with 4 M guanidine hydrochloride. Both samples were fully saturated with CO and sealed in 1-mm quartz capillaries.
Data collection
TR-WAXS has been developed at the beamline ID09B of the European Synchrotron Radiation facility. The polychromatic X-ray pulses (Supplementary Figs. 5 and 6 online) have time duration of 100 ps (full width at half-maximum, FWHM) and are focused down to an elliptical spot as small as 0.06 × 0.1 mm2 (FWHM) at the sample position. A variable number of single X-ray pulses (1–47) were selected from the synchrotron pulse train by means of a high-speed chopper and a millisecond shutter (Supplementary Table 4 online). The X-ray pulses scattered by the sample were collected with a two-dimensional charge-coupled device (CCD) detector (Mar133; Mar Research). Though the X-ray beam flux generated was very intense (≈1.0 × 109 photons/pulse), an integration time of ~2 min was required to generate a high-dynamic-range-scattering single image. Up to 30 images per time delay were acquired and averaged together to improve the signal-to-noise ratio. ‘Laser-off ’ images were also acquired at negative time delays when the X-ray pulse arrives before the laser pulse. These ‘laser-off ’ images were used as a reference to compute the TR-WAXS difference patterns. To dilute any X-ray radiation damage over a large sample volume, the sample was translated back and forth along its long axis over a 20-mm range. To ensure that successive pulses in the 5-Hz pulse train excite a fresh portion of the protein solution, the sample was translated by 0.2 mm after each probe pulse.
Image processing
Images were azimuthally averaged using a customized in house–developed code (available upon request). To convert the scattering angle to q, the peak of the undulator spectrum (0.827 Å−1) was used as the reference wavelength. The laser-induced changes in the scattering intensity amounted to a few percent of the static scattering intensity. For this reason, the amplitude of the individual WAXS patterns had to be carefully scaled before computing the laser-induced scattering differences. To that end, we took advantage of the fact that the solvent heating signal from water has an isosbestic point at q of ~1.5 Å−1 and normalized all radial curves at this point. This scaling approach did not affect the patterns at lower q where the protein difference scattering was greatest.
Additional methods
Details of the X-ray and laser sources, data-gathering protocol, calculations of scattering patterns from PDB files, information content and signal-to-noise ratio of the scattering patterns, allosteric kinetic model used to fit the TR-WAXS data on hemoglobin and the control experiment on hemoglobin are available in Supplementary Methods.
Supplementary Material
ACKNOWLEDGMENTS
We thank W.A. Eaton and E. Henry for helpful comments, H.-S. Cho and S. Ahn for helpful discussions about the data analysis, and Y.O. Jung, K.H. Kim and J.H. Lee for their assistance with sample preparation and experiments. This research was supported in part by the Intramural Research Program of the National Institutes of Health to P.A.A., by EU grant FLASH: FP6-503641 to M.W., and a grant from the Creative Research Initiatives (Center for Time-Resolved Diffraction) of the Ministry of Education, Science and Technology, Korea Science and Engineering Foundation to H.I.
Footnotes
Note: Supplementary information is available on the Nature Methods website.
References
- 1.Moffat K. Ultrafast time-resolved crystallography. Nat. Struct. Biol. 1998;5:641–643. doi: 10.1038/1333. [DOI] [PubMed] [Google Scholar]
- 2.Schotte F, et al. Watching a protein as it functions with 150-ps time-resolved X-ray crystallography. Science. 2003;300:1944–1947. doi: 10.1126/science.1078797. [DOI] [PubMed] [Google Scholar]
- 3.Ihee H, et al. Visualizing reaction pathways in photoactive yellow protein from nanoseconds to seconds. Proc. Natl. Acad. Sci. USA. 2005;102:7145–7150. doi: 10.1073/pnas.0409035102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Grishaev A, Wu J, Trewhella J, Bax A. Refinement of multidomain protein structures by combination of solution small-angle X-ray scattering and NMR data. J. Am. Chem. Soc. 2005;127:16621–16628. doi: 10.1021/ja054342m. [DOI] [PubMed] [Google Scholar]
- 5.Xu X, et al. Dynamics in a pure encounter complex of two proteins studied by solution scattering and paramagnetic NMR spectroscopy. J. Am. Chem. Soc. 2008;130:6395–6403. doi: 10.1021/ja7101357. [DOI] [PubMed] [Google Scholar]
- 6.Zuo X, et al. Global molecular structure and interfaces: refining an RNA:RNA complex structure using solution X-ray scattering data. J. Am. Chem. Soc. 2008;130:3292–3293. doi: 10.1021/ja7114508. [DOI] [PubMed] [Google Scholar]
- 7.Hirai M, Iwase H, Hayakawa T, Miura K, Inoue K. Structural hierarchy of several proteins observed by wide-angle solution scattering. J. Synchrotron Radiat. 2002;9:202–205. doi: 10.1107/s0909049502006593. [DOI] [PubMed] [Google Scholar]
- 8.Menk RH, et al. Novel detector systems for time resolved SAXS experiments. J. Appl. Crystallogr. 2000;33:778–781. [Google Scholar]
- 9.Akiyama S, et al. Conformational landscape of cytochrome c folding studied by microsecond-resolved small-angle X-ray scattering. Proc. Natl. Acad. Sci. USA. 2002;99:1329–1334. doi: 10.1073/pnas.012458999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kainosho M, et al. Optimal isotope labelling for NMR protein structure determinations. Nature. 2006;440:52–57. doi: 10.1038/nature04525. [DOI] [PubMed] [Google Scholar]
- 11.Hofrichter J, Sommer JH, Henry ER, Eaton WA. Nanosecond absorption spectroscopy of hemoglobin: elementary processes in kinetic cooperativity. Proc. Natl. Acad. Sci. USA. 1983;80:2235–2239. doi: 10.1073/pnas.80.8.2235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Balakrishnan G, et al. Time-resolved absorption and UV resonance Raman spectra reveal stepwise formation of T quaternary contacts in the allosteric pathway of hemoglobin. J. Mol. Biol. 2004;340:843–856. doi: 10.1016/j.jmb.2004.05.012. [DOI] [PubMed] [Google Scholar]
- 13.Svergun DI, et al. Shape determination from solution scattering of biopolymers. J. Appl. Crystallogr. 1997;30:798–802. [Google Scholar]
- 14.Makowski L, et al. Molecular crowding inhibits intramolecular breathing motions in proteins. J. Mol. Biol. 2008;375:529–546. doi: 10.1016/j.jmb.2007.07.075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bellelli A, Brunori M, Miele AE, Panetta G, Vallone B. The allosteric properties of hemoglobin: insights from natural and site directed mutants. Curr. Protein Pept. Sci. 2006;7:17–45. doi: 10.2174/138920306775474121. [DOI] [PubMed] [Google Scholar]
- 16.Eaton WA, et al. Evolution of allosteric models for hemoglobin. IUBMB Life. 2007;59:586–599. doi: 10.1080/15216540701272380. [DOI] [PubMed] [Google Scholar]
- 17.Svergun DI, Barberato C, Koch MHJ. CRYSOL - a program to evaluate X-ray solution scattering of biological macromolecules from atomic coordinates. J. Appl. Crystallogr. 1995;28:768–773. [Google Scholar]
- 18.Svergun DI, Koch MHJ. Small-angle scattering studies of biological macromolecules in solution. Rep. Prog. Phys. 2003;66:1735–1782. [Google Scholar]
- 19.Sawicki CA, Gibson QH. Quaternary conformational changes in human hemoglobin studied by laser photolysis of carboxyhemoglobin. J. Biol. Chem. 1976;251:1533–1542. [PubMed] [Google Scholar]
- 20.Cammarata M, et al. Impulsive solvent heating probed by picosecond X-ray diffraction. J. Chem. Phys. 2006;124:124504(1)–124504(9). doi: 10.1063/1.2176617. [DOI] [PubMed] [Google Scholar]
- 21.Fader WJ. Density perturbations caused by weak absorption of a laser pulse. J. Appl. Phys. 1976;47:1975–1978. [Google Scholar]
- 22.Eaton WA, Henry ER, Hofrichter J. Application of linear free energy relations to protein conformational changes: the quaternary structural change of hemoglobin. Proc. Natl. Acad. Sci. USA. 1991;88:4472–4475. doi: 10.1073/pnas.88.10.4472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Fermi G, Perutz MF, Shaanan B, Fourme R. The crystal structure of human deoxyhaemoglobin at 1.74 Angstrom resolution. J. Mol. Biol. 1984;175:159–174. doi: 10.1016/0022-2836(84)90472-8. [DOI] [PubMed] [Google Scholar]
- 24.Silva MM, Rogers PH, Arnone A. A third quaternary structure of human hemoglobin A at 1.7 Angstrom resolution. J. Biol. Chem. 1992;267:17248–17256. [PubMed] [Google Scholar]
- 25.Lukin JA, et al. Quaternary structure of hemoglobin in solution. Proc. Natl. Acad. Sci. USA. 2003;100:517–520. doi: 10.1073/pnas.232715799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Srinivasan R, Rose GD. The T-to-R transformation in hemoglobin: a reevaluation. Proc. Natl. Acad. Sci. USA. 1994;91:11113–11117. doi: 10.1073/pnas.91.23.11113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Safo MK, Abraham DJ. The enigma of the liganded hemoglobin end state: a novel quaternary structure of human carbonmonoxy hemoglobin. Biochemistry. 2005;44:8347–8359. doi: 10.1021/bi050412q. [DOI] [PubMed] [Google Scholar]
- 28.Guallar V, Jarzecki AA, Friesner RA, Spiro TG. Modeling of ligation-induced helix/loop displacements in myoglobin: toward an understanding of hemoglobin allostery. J. Am. Chem. Soc. 2006;128:5427–5435. doi: 10.1021/ja057318h. [DOI] [PubMed] [Google Scholar]
- 29.Kachalova GS, Popov AN, Bartunik HD. A steric mechanism for inhibition of CO binding to heme proteins. Science. 1999;284:473–476. doi: 10.1126/science.284.5413.473. [DOI] [PubMed] [Google Scholar]
- 30.Jones CM, et al. Fast events in protein-folding initiated by nanosecond laser photolysis. Proc. Natl. Acad. Sci. USA. 1993;90:11860–11864. doi: 10.1073/pnas.90.24.11860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Arcovito A, Gianni S, Brunori M, Travaglini-Allocatelli C, Bellelli A. Fast coordination changes in cytochrome c do not necessarily imply folding. J. Biol. Chem. 2001;276:41073–41078. doi: 10.1074/jbc.M105183200. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.