Abstract
The apoptolidins are 20/21-membered macrolides produced by Nocardiopsis sp. FU40. Several members of this family are potent and remarkably selective inducers of apoptosis in cancer cell lines, likely via a distinct mitochondria associated target. To investigate the biosynthesis of this natural product, the complete genome of the apoptolidin producer Nocardiopsis sp. FU40 was sequenced and a 116 Kb region was identified containing a putative apoptolidin biosynthetic gene cluster. The apoptolidin gene cluster comprises a type I polyketide synthase, with 13 homologating modules, apparently initiated in an unprecedented fashion via transfer from a methoxymalonyl-acyl carrier protein loading module. Spanning approximately 39 open reading frames, the gene cluster was cloned into a series of overlapping cosmids and functionally validated by targeted gene disruption experiments in the producing organism. Disruption of putative PKS and P450 genes delineated the roles of these genes in apoptolidin biosynthesis and chemical complementation studies demonstrated intact biosynthesis peripheral to the disrupted genes. This work provides insight into details of the biosynthesis of this biologically significant natural product and provides a basis for future mutasynthetic methods for the generation of non-natural apopotolidins.
Keywords: Polyketide, Type I Polyketide Synthase, Macrolide Natural Product, Gene Cluster, Mutasynthesis
1. Introduction
Glycosylated polyketide macrolides from actinomycetes comprise a diverse class of natural products that demonstrate a variety of therapeutic potentials as evidenced by their antibiotic, antifungal, anticancer, and antiparistic properties.1-3 Access to structural variants of macrolide natural products is limited due to their structural and functional complexity that features a stereochemically rich macrolide core and often one or more decorating deoxy sugar moieties. In complement to synthetic chemical approaches, classical methods of semisynthesis, precursor-directed biosynthesis, and chemical degradation provide access to analogs of parent metabolites. However, recent efforts based on combining chemical approaches with genetic and metabolic engineering promise to significantly broaden the scope of structural modifications and potential for developing therapeutic lead compounds.4-8
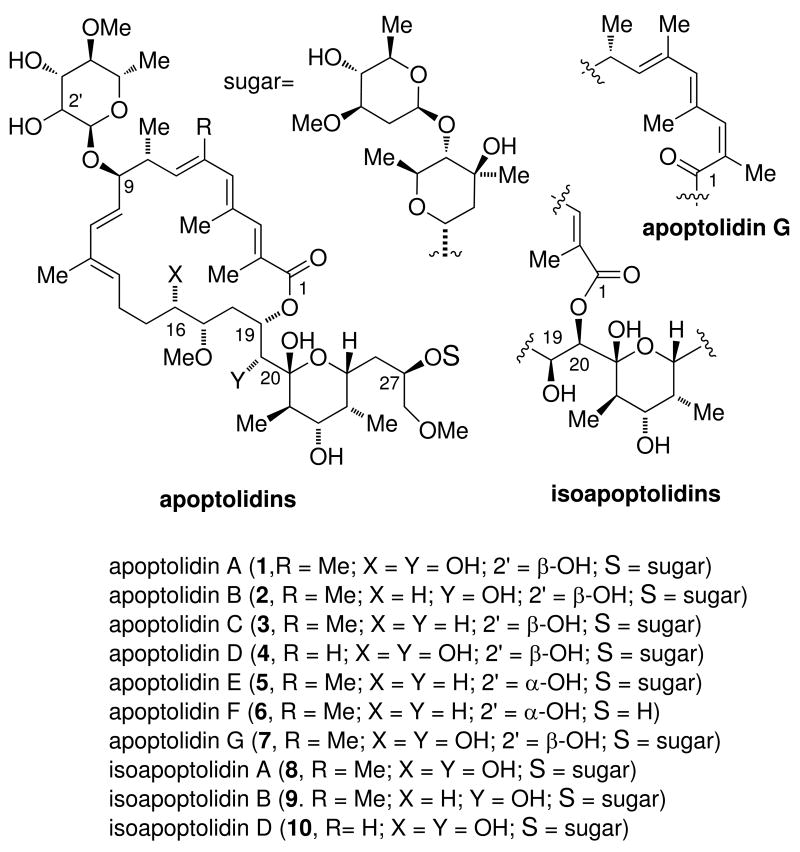
The apoptolidin macrolides (Figure 1) have drawn attention for their reported ability to selectively induce apoptosis in several cancer cell lines.9-11 Produced by Nocardiopsis sp. FU40 apoptolidins are 20/21-membered macrolides, appended variously with 6-deoxy sugars at C9 and with or without an invariant disaccharide at C27.12-14 As with many macrolides containing 20-membered or larger ring sizes, apoptodidins possess a relatively long side chain containing a cyclic hemiketal. To date, over ten apoptolidins have been identified which vary in substitution,15-17 double bond geometry at C2/3,18 and macrocylization regiochemistry13,14 (Figure 1).
Figure 1.
Chemical structures of apoptolidins and isoapoptolidins produced by Nocardiopsis sp. FU40.
A mode of action involving the inhibition of mitochondrial ATPase has been proposed for apoptolidin A (1) based, in part, in comparison to other polyketide macrolides such as cytovarisin, ossamycin and the oligomycins.10,19 Chemical degradation11,20-22 and chemical synthesis23-27 have added to the natural retinue, yielding over twenty-five analogs and congeners of the parent apoptolidin A, and permitting SAR analysis in this series. Notably, apoptolidins B - F15-17 (2-6) have been identified by the Wender group as minor metabolites (2 - 5 mg/L) relative to the parent apoptolidin A (ca. 100 mg/L).12 To date, SAR studies have shown the parent aglycone (apoptolidinone) is inactive23,26 and the C27 disaccharide outweighs the C9 monosaccharide in importance to cell cytotoxicity.17,25,28,29 In considering options to produce quantities of the modified apoptolidins for further biological investigations, studies to date clearly indicate production by fermentation to outpace chemical synthesis as a means to access material.30 For this reason we turned our attention to the identification and characterization of the apoptolidin gene cluster.
2. Results and Discussion
Cloning and sequencing of the apoptolidin gene cluster from Nocardiopsis sp. FU40
Genome sequencing and assembly of Nocardiopsis sp. FU40 resulted in a total of 1009 contigs ranging in size from 66 bp to 132,247 bp (264 contigs were larger than 10,000 bp), which formed 53 scaffolds ranging in size from 303 bp to 4,723,578 bp. The largest scaffold contained 801 contigs and corresponded to 50% of the genome, which was estimated at 9,433,896 bp, in agreement with that of other actinomycete genomes previously sequenced.31-33 The genome was sequenced to 35-fold read coverage. Genome annotation of the scaffolds resulted in the identification of 10,240 apparent coding sequences.
The gene cluster for apoptolidin biosynthesis was identified within the proteome of Nocardiopsis sp. FU40 by identifying co-localized genes consistent with a predicted apoptolidin biogenesis using the method as described by Bachmann and Ravel.34 Correspondingly, the 20/21-membered polyketide core of apopotolidin, variously reduced at C1 position of hypothetical acetate precursors, was predicted to be diagnostic of a non-iterative (type I) polyketide synthase (PKS). Type-1 PKS are multi-domain megasynthases in which the order of the catalytic domains corresponds, with some exceptions,35,36 to the order of biochemical events entailed in the assembly of encoded polyketides. In these systems, each domain is used only once: acyltransferase (AT) domains mediate the activation of homologating ketide precursors, variously C2 substituted malonyl-CoAs, and their transfer to phosphopantathienylated acyl carrier protein domains (ACP); ketosynthase domains (KS) catalyze the condensation of two tethered acyl-ACP thioesters by transthioesterification of a upstream acyl-ACP onto an active site cysteine followed by decarboxylative (thio)Claisen condensation with the downstream malonyl-ACP; ketoreductase (KR), dehydratase (DH) and enoylreductase (ER) domains catalyze the reactions for the generation of the appropriate oxidation states along the polyketide chain. A 14-module hypothetical domain sequence (including a loading module) was inferred from this core structure based on the 1,3-oxygenation pattern corresponding to C1 of acetate and propionate precursors. Variable hydroxylation at positions corresponding to C2 of precursors (C16 and C20 of apoptolidins) and the earlier identification15 of apoptolidins B (2) and C (3) suggested that the polyketide core is further oxidized by oxidases at these positions. Finally, glycosylations at C9 and C27 inform the expectation of multiple glycosyl transferases as well as ancillary predicted deoxysugar biosynthesis enzymes involved in generation of disaccharide precursors NDP-l-olivomycose and NDP-d-oleandrose.
Based on these assumptions, a ∼116 kb region located at 3.39 - 3.51 Mb was readily identified by the presence of a putative 14-module type I polyketide synthase largely matched the predicted domain organization and contained the appropriate post-PKS elaboration functionality. A second gene cluster putatively encoding a type-I PKS was identified in the genome (data not shown), but not deemed a likely candidate for encoding apoptolidin as it possessed only approximately eight intact modules, a domain sequence consistent with a polyene macrolide, and insufficient evidence of deoxysugar biosynthetic genes consistent with apoptolidin. The putative apopotolidin gene cluster ‘apo’, initially possessed eight short sequencing gaps which were bridged by PCR amplification of gap regions and DNA sequencing. The ends of the apo gene cluster were approximately defined by the occurrence of contiguous putative genes with homology to primary metabolic and/or otherwise highly conserved gene sequences and designated as ApoU1-10 and ApoD1-10 corresponding to the ten genes upstream and downstream of the apoptolidin gene cluster. With a putative gene locus circumscribed, open reading frames were manually re-annotated by analysis of alternate predicted start codons and by comparison to functional homologs identified using BLAST. The apo gene cluster and flanking upstream and downstream region have been deposited into the NCBI with locus accession numbers of JF819834, JF894099, JF894100 and JF894101.
Analysis of the apopotolidin biosynthetic gene cluster (apo)
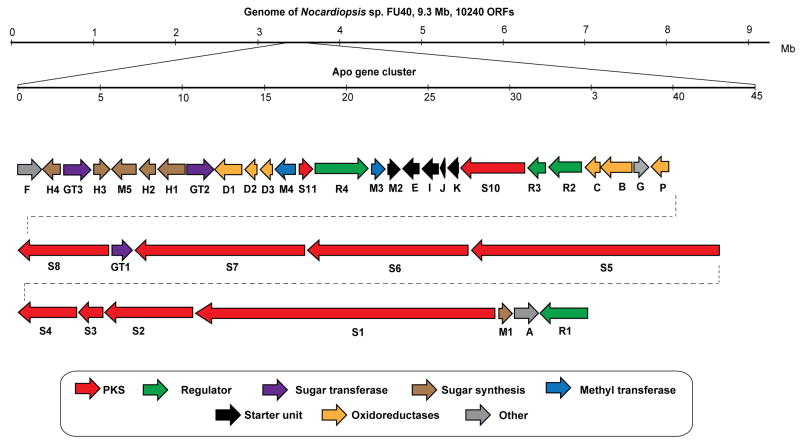
Fully consistent with the structure of the apopotolidins, the proposed 115.74 kb apo gene cluster contains 39 individual open reading frames (ORFs) including nine type I polyketide synthase genes (PKS), six oxidoreductase genes, 4 methyltranferases and three glycosyl transferase genes (see gene map in Figure. 2).
Figure 2.
Genetic organization of the apoptolidin gene cluster in Nocardiopsis sp. FU40. The 115.74 MB gene cluster was identified in the producing organism by searching the sequenced genome for the biogenetically rationalizationed predicted type-I polyketide synthase domain sequence.
Polyketide synthase organization
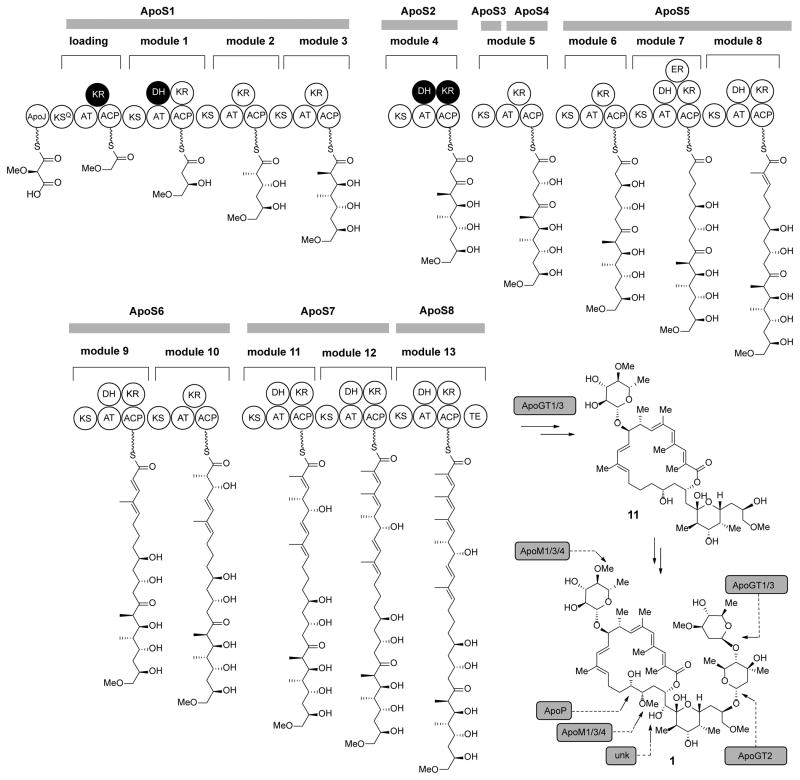
In Nocardopsis sp. FU40, PKS genes ApoS1 - ApoS8, are proposed to be responsible for the biosynthesis of the apoptolidin polyketide core. A suggested module and domain organization and a proposed model for the PKS templated assembly of the apoptolidin polyketide backbone is shown in Figure 3. ApoS1 is the putative protein identified as possessing the likely initiating module as it contains the sequence hallmarks of a decarboxylative KSQ loading module. Specifically, the first KS domain of ApoS1 contains a cysteine to glutamate codon modification mutating the essential active site cysteine (C193Q) involved in the transthioesterification reaction that precedes KS mediated condensation. The next 12 extension modules are proposed to be contained on ApoS1 – S7 and were ordered according to the predicted collinear arrangement of required domains. Finally, the terminating module protein was identified in ApoS8 by its terminal punctuation with a thioesterase (TE) domain, required for release from the megasynthase assembly. One additional polyketide containing putative open reading frame ApoS9, contains an incomplete module sequence ‘KS-AT-KR’ and ApoS10 apparently encodes a free-standing thioesterase protein that may be important in hydrolytic release and/or macrocyclization. Of the 65 domains encoding the biosynthesis of the macrolide core, there are four domains predicted to be inactive (Figure 3, in black). Analysis of active site consensus residues for these predicted inactive domains reveals nearly universal conservation of all key consensus residues thought to be essential for activity, indicating that the structural determinants of inactivity of these domains may reside distal to these conserved motifs.
Figure 3.
Proposed pathway overview of apopotolidin assembly. A 14-module polyketide synthase, initiated by an unprecedented methoxymalonyl-ACP loading module, is proposed to assemble the seco acid precursor of the apoptolidins. Subsequent tailoring reactions are proposed to oxidize C16/21, methylate C17, and append the C9 monosaccharides and C27 dissacharide resulting in the fully elaborated and biologically active apoptolidin series.
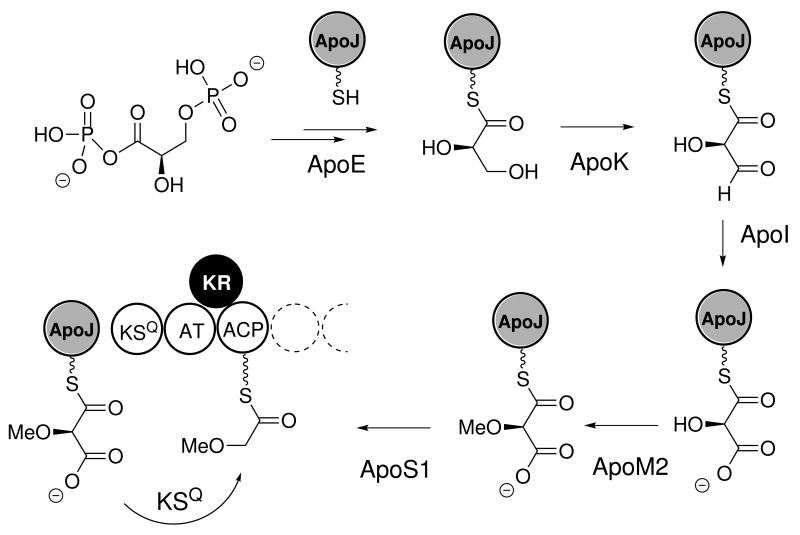
Methoxymalonyl-ACP biosynthesis and initiating module
The proposal of an unusual initiating precursor, (2R)-methoxymalonlyl-ACP is based on the observations of 1) the atypical initiating 2-methoxy acetyl group in the apoptolidins,36b 2) a decarboxylative initiating KSQ module, and 3) a five gene cassette upstream of the PKS genes, ApoK - ApoM2, identified with greater than 50% translated identity to the fkbG – K, proteins from the FK520 ascomycin gene cluster,37 which are responsible for biosynthesis of (2R)-methoxymalonyl-ACP in S. hygroscopicus during FK520 production.38,37 This condensation precursor has previously been observed in various type I polyketide biosyntheses including FK520, soraphen, and ansamitocin pathways.39 In analogy with FK520 biosynthesis, we propose that the biosynthesis of (2R)-methoxymalonyl-ACP in apoptolidins initiates with the enzyme ApoE, which is proposed to dephosphorylate and covalently tether a glycolytic substrate derived from 1,3-bisphosphoglycerate to the 4′-phosphopantatheinyl group of an ACP, forming glyceryl-ApoJ. Next, two enzymes ApoK (NAD+-dependent) and ApoI (FAD-dependent) catalyze the oxidation of glyceryl-ApoJ to the intermediate (2R)-hydroxymalonyl-ApoJ. The methyltransferase ApoM2 then catalyzes the O-methylation of (2R)-hydroxymalonyl-ApoJ to form (2R)-methoxymalonyl-ApoJ (Figure 4). Unlike FK520, in which (2R)-methoxymalonyl-ACP is used in extension chemistry we propose that this precursor is recognized by the loading module of apo, which mediates its decarboxylative transfer from ApoJ to the loading module ACP. Particularly intriguing, are the unknown roles of the KSQ and AT domains in the recognition and transfer of (2R)-methoxymalonyl-ACP to the loading ACP. Given the mutation of the active site cysteine in the KSQ domain, selection and transfer of methoxymalonate would appear to be mediated by the AT. In any event, to the best of our knowledge, the apoptolidin gene cluster comprises the first example of a gene cluster initiated by priming with methoxylmalonyl-ACP.
Figure 4.
Proposed biosynthesis and association of methoxymalony-ACP precursor from a potential glycosidic intermediate 1,3-bisphospho-d-glyceric acid. The protein sequences possess >50% identity with the FK520 methoxymalonyl-ACP pathway. Unlike FK520, this precursor serves as the precursor for an apparent glyoxylate starting untit in apoptolidin biosynthesis.
Extender module analysis
The extender module biochemistry of the apoptolidin PKS selects methylmalonyl-CoA or malonyl-CoA precursors via AT domains. Based on the studies of Del Vecchio et al,40 the selectivity of AT domains can be predicted by identifying diagnostic ‘YASH’ and ‘HASH/HAFH’ sequence motifs. Using this analysis, the AT domains of module 1, module 4, module 5, 6, 7 and 9 are predicted to select malonyl-CoA precursors, and the AT domains of module 2, 3, 8, 10, module 11, 12 and 13 select methylmalonyl-CoA. The predicted AT-domain specificities are in accord with the observed substitution pattern in the apoptolidin polyketide backbone.
Ketoreductase stereoselectivity
The predicted functional ketoreductase (KR) domains in the apo gene cluster possess the expected active site consensus motifs41,42 and the stereochemistry of reduction domains can be estimated by analysis using the Caffrey classification methodology.43 Correspondingly, this analysis predicts ‘a-type’ stereochemistry for KR domains in modules 3 and 6, and ‘b-type’ stereochemistry with modules 1, 2, 5, 7, 8, 9, 10, 11, 12 and 13. This is generally in accordance with the structure of apoptoldin with one possible exception. Based on the reported structure of apoptolidin, the R configuration at C27 should correspond to the a-type stereochemistry of Caffrey, whereas the sequence analysis predicts the opposite configuration (LDN, P and N motifs at residues 93 – 95, 144 and 148 in the KR domain of in module 1). Correspondingly, to verify the proposed stereochemistry of C27, we peracetylated apoptolidin A, selectively hydrolyzed the C27 dissacharide and formed (R)- and (S)-Mosher esters at the revealed C27 hydroxyl.13 The differences in chemical shift of the C26 (ΔδSR > 0) and C28 (ΔδSR < 0) protons44 confirmed the configuration at this position as originally defined.7 Due to the likelihood that module 1 is correctly assigned based on its position relative to the KSQ, and the downstream domain organization, this example represents a confirmed exception to the Caffrey analysis of predicting apparent stereochemistry based on sequence analysis. Further experiments are required to determine the biochemical origin of the exception, specifically if the D to N substitution in the LDD motif is a contributor to the unexpected stereochemical outcome.
Analysis of proposed deoxysugar biosynthesis genes
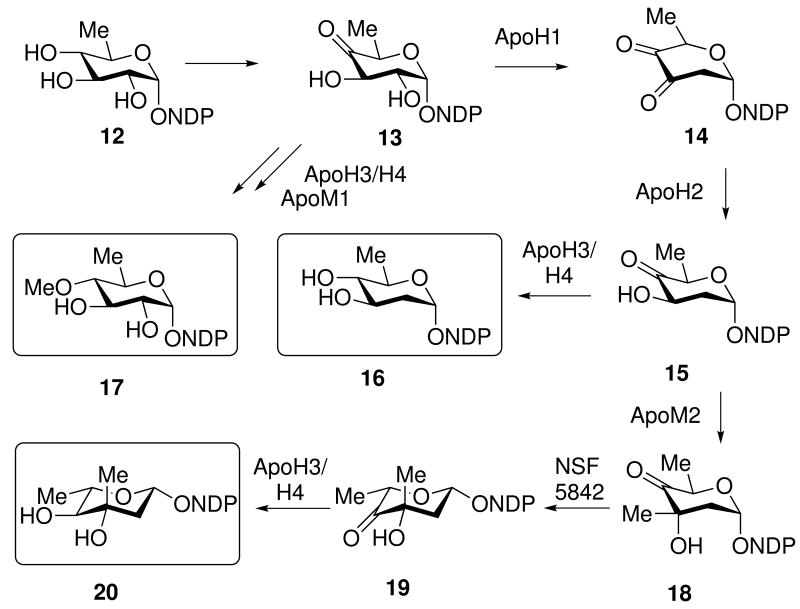
The apoptolidins are decorated by three sugars, which have been demonstrated to be important for selective cytotoxicity.10 The C9 hydroxyl is appended with 6-deoxy-4-O-l-methyl glucose and under some growth conditions, with 4-O-l-methyl-l-rhamnose.12,17 The C27 hydroxyl is appended with an invariant disaccharide composed of l-olivomycose and d-oleandrose. The apoptolidin gene cluster contains a single cluster of putative sugar biosynthetic genes at its flank along with two glycosyltransferases. A third glycosyltransferase is located within the putative PKS encoding cluster. Analysis of the sugar biosynthetic gene cassette reveals five genes encoding putative enzymes appropriate for the biosynthesis of the mono and disaccharide sugars (Figure 5). The biosynthesis of these sugars diverges from a shared intermediate NDP-l-4-keto-3-deoxyglucose 13, which may subsequently be converted into NDP-d-oleandrose 16 by Apo1,2,3/4 and TDP-l-olivomycose by Apo1,2 ApoM2, a missing 3,5-epimerase, and Apo3/4. Searching the Nocardopsis genome for a 3,5-epimerase reveal a gene designated Nsf5842.
Figure 5.
Suggested sugar precursor biosynthesis: NDP-L-olivomycose 20, NDP-D-oleandrose 16, and NDP-D-6-deoxy-4-O-methyl-D-glucose 17. A 3,5-epimerase is missing from the gene cluster, but a candidate NSF5842 is present in the genome.
Finally, NDP-L-4-keto-3-deoxyglucose 13 may be converted into NDP-D-6-deoxy-4-O-methyl-d-glucose 17 by apoH3 or apoH4 or a yet to be identified ketoreductase. Moreover a remaining question concerns the biosynthetic origin of the 4-O-methyl-l-rhamnose appending C9 of apoptolidins E and F.17 Intriguingly, two additional unassigned oxidoreductases are apoB and apoC which are apparent flavin and nicotinamide binding enzymes may provide the necessary functions for the generation of the C9 appending sugars.
O-methyltransferase
Two additional putative O-methyltransferases are encoded by ApoM1 and ApoM3, which are directly upstream of the proposed methoxymalonyl-CoA cassette. ApoM1 is most closely related to sugar O-methyltransferases, suggesting its role in the methylation of 4-position of the C9 monosaccharide. If this is the case, the remaining methyltransferase ApoM3 is a likely candidate for methylation of the C17 hydroxyl group.
Oxidases
Analysis of the pattern of oxidation in the polyketide backbone of apoptolidin and the isolation of apoptolidins B and C15 support positions C16 and C20 arise from post-PKS oxygenation reactions. Candidates for these C-H bond oxidative reactions include ApoP, a P450 related to the erythromycin macrolide oxidase EyrK (38/56% identity/similarity), and ApoD1-3, an apparent three component Rieske non-heme iron oxygenase similar to oxygenases involved in the oxidation of aryl and biaryl functional groups.
Cosmid library construction and screening
A cosmid library was constructed from the high molecular weight genomic DNA of Nocardiopsis sp. FU40 by using SuperCos1 cosmid vector. To identify the cosmid clones containing the Apo cluster, three genes in this cluster, ApoM2, ApoGT1 and ApoM1, were used as hybridization probes resulting in seven positive colonies. End-sequencing of cosmids revealed that these cosmids cover the whole of the proposed apo gene cluster. Two cosmids, designated apo14C7 and apo4C5, were used for gene disruption.
Transformation of Nocardiopsis
The genetic manipulation of Nocardiopsis FU40 was initially intransigent as the most commonly practiced methods of transformation of actinomycetes were unsuccessful in our hands. Firstly, polyethylene glycol induced protoplast transformation45 was not effective, possibly because spherical protoplasts were not observable under any standard lysozyme digestion conditions. Secondly, interspecies conjugative methods were not successful using conjugation with E. coli ET12567/pUZ8002, which resulted in no transformants using a series of integrative and non-integrative vectors and markers. Finally, adapting a method for electroporation from Hopwood et al. for Streptomyces, 45 we developed a method for the transformation of Nocardiopsis sp. FU40, which was subsequently able to introduce modified cosmids into Nocardiopsis sp. FU40.
Targeted gene disruption
To confirm the proposed roles of the apo gene cluster in the biosynthesis of apoptolidin and functionally analyze genes in this cluster, ApoS8 and ApoP, were deleted respectively from the genome of Nocardiopsis sp. FU40 using the well-established λ–Red (Red/ET) system with appropriate cosmids.46 Correspondingly, targeted genes in cosmids were replaced by apramycin resistance gene markers by double crossover homologous recombination in an engineered hyper recombinant E. coli strain. The resulting cosmids were transformed into Nocardiopsis, selecting for apramycin resistant colonies, and the disruption of genes was confirmed by PCR analysis of transformants.
Analysis and complementation of disruption mutants
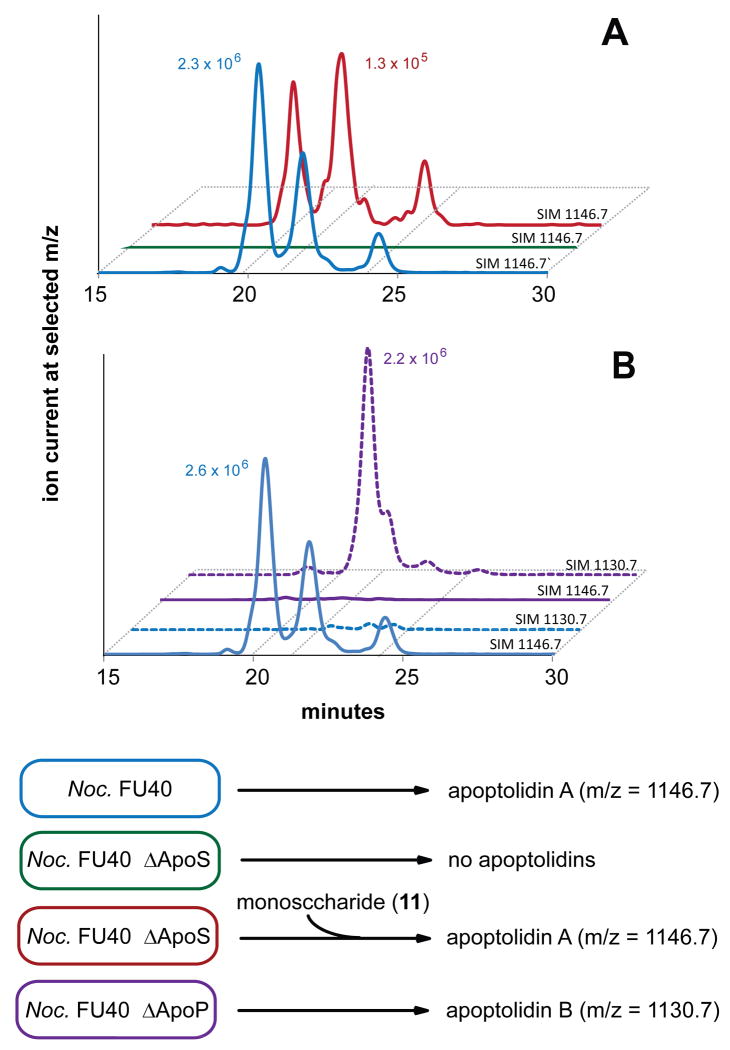
Terminated by the seco acid releasing TE domain, ApoS8 is predicted to contain the last extension module of the apoptolidin PKS. Correspondingly a site directed knockout mutant, FU40ΔApoS8 was generated as described above and in the Experimental Section. This strain was cultured in production medium, extracted with ethyl acetate and analyzed via HPLC/MS for the production of known apoptolidins (Figure 6A). The FU40ΔApoS8 strain was unable to produce apoptolidin A or any known apoptolidin analog. Additionally, when FU40ΔApoS8 was grown and pulse fed 3.75 mg apoptolidin monosaccharide 11 (Figure 3), we observed the largely restored production of the apoptolidin A (0.85 mg) and isoapoptolidin A (0.39 mg). This chemical complementation validates that the targeted gene disruption phenotype and indicates that transcriptionally downstream genes involved in biosynthesis and glycosylation of apoptolidin monosacchraide remain competent. The unoptimized 24.7% yield for the conversion of apoptolidin monosaccharide to apoptolidin A demonstrates the potential for the employment of FU40ΔApoS8 as a glycosylation biotransformation reagent.3
Figure 6.
Analysis of disruption mutants. Panel A, knockout of PKS: Wild-type (blue) ΔApoS8 (green) and ΔApoS8 + apoptolidin C9 monosaccharide (red) scanning for apoptolidin A ion (ES+ M+NH4/z =1146.7). Panel B, knockout of P450: Wild-type (blue) and ΔApoP (purple) monitoring for apoptolidin A (solid line, ES+ M+NH4 =1146.7) and B (dashed line, ES+ M+NH4/z =1130.7).
ApoP was identified as a cytochrome P450 monooxygenase gene with 38/56% identity/similarity to EryK from erythromycin biosynthetic gene cluster of Saccharopolyspora erythraea47 and 43/61% similarity to TylI of tylosin biosynthesis in Streptomyces fradiae (PMID 7984112). As both of these proteins are involved with post-PKS oxygenation of polyketide C-H bonds, ApoP comprised a likely candidate for hydroxylation of either C16 or C20. Correspondingly, we generated a selective ApoP gene replacement mutant, FU40ΔApoP and the fermentation products of this mutant did not produce detectable levels apoptolidin A, as analyzed by HPLC/MS. However, FU40ΔApoP strains instead produced apoptolidin analogs 18 Da less than parent apoptolidins, indicating that the ApoP is a hydroxylating P450 and that targeted replacement of the corresponding gene did not generate polar effects downstream of ApoP.
Subsequent isolation of the deoxy compound followed by preliminary structural characterization by 1H, HSQC, COSY and HMBC NMR determined that the deoxy product accumulated in FU40ΔApoP is consistent with 1) an isoapoptolidin scaffold and 2) an additional methylene at C16. Full assignment of this analog, complicated by separation problems is ongoing.
3. Conclusion
In summary, we have identified the apoptolidin gene cluster in in Nocardiopsis FU40. A notable feature is the observation of a methoxymalonyl-ACP loading module initiating polyketide synthase. Functions are proposed for the 38 gene cluster and gene replacement experiments confirm the function of two genes in the apo cluster, ApoP and ApoS8. The transformation and genetic methodology described herein sets the stage for the further production of new apoptolidin analogs by genetic engineering for the purpose of continuing targeted biological investigations for this selective cytotoxic macrolide.
4. Experimental Section
4.1. General
All reagents were of the maximum purity available from the manufacturer. All strains, plasmids and primers are described in Supplementary Materials, table S1.
4.1.1. Bacterial Strains and Culture Conditions
Nocardiopsis sp. FU409 and its mutants were maintained on either Bennett's agar (0.1% yeast extract, 0.1% beef extract, 0.2% NZ amine A, 1% glucose and 2% agar, pH 7.2) or Seed Agar (1% soluble starch, 1% molasses, 1% peptone, 1% beef extract and 2% agar, pH 7.2) with appropriate antibiotics at 30 °C. For apoptolidin or analog production, Nocardiopsis sp. FU40 or its mutants were cultured in Production Medium (2% glycerol, 1% molasses, 0.5% casamino acid, 0.1% peptone and 0.1% CaCO3, pH 7.2) at 30 °C.
E. coli XL1-Blue MR (Stratagene Catalog #251301) was used as the host strain for the cosmid library construction. E. coli BW25113 containing plasmid pIJ79046 was used for targeted gene disruption in Nocardiopsis sp. FU40. E. coli TOP10 (Invitrogen, Catalog # 553001A) was used for gene cloning and E. coli ET12567 was used as a non-methylating E. coli host to avoid a potential DNA methylation-sensing restriction system in Nocardiopsis sp. FU40. All E. coli strains were maintained in LB medium supplemented with appropriate antibiotics for selection of plasmids.
4.1.2. Plasmids and General DNA Procedures
SuperCos-1 derivatives were used to construct genomic libraries of Nocardiopsis sp. FU40. DNA isolation and gel electrophoresis were conducted according to standard methods.45 Cosmid DNA was isolated from E. coli strains using a Qiagen Miniprep Kit (Catalog #27106). Isolation of DNA fragments from agarose was carried out with a Qiagen gel extraction kit (Catlog #28704). Genomic DNA from Nocardiopsis sp. FU40 and its derivatives were isolated using the Wizard genomic DNA purification kit (Promega, Catalog #297885).
4.1.3. Genome sequencing of Nocardiopsis sp. FU40 and apoptolidin gene cluster identification
High molecular weight genomic DNA was purified from Nocardiopsis sp. FU40 using the CTAB method described by Hopwood D. A. et al.45 Whole genome sequencing was performed with a 454/Roche FLX DNA sequencer (Roche Diagnostics Corp.) using a protocol that included paired-end sequencing (3 kb insert size library) and optimization for GC rich bacterial genomes developed by the University of Maryland School of Medicine Genomic Resource Core. A total of 1,167,141 reads with an average clear range of 277 bp were obtained. The reads were assembled using the Celera Assembler (version 5.2) (http://wgs-assembler.sourceforge.net/). The Glimmer Gene Finder48 was utilized to identify potential coding regions in the resulting scaffolds and manual annotation was performed using the MANATEE system (http://manatee.sourceforge.net/).
Identification of the apoptolidin biosynthetic gene cluster was performed using the method previously described by Bachman et al.28 A total of 8 sequencing gaps were present in the cluster and were filled by PCR and sequencing. The nucleotide sequences for the putative apoptolidin gene cluster (Apo), upstream and downstream regions ApoU1-10 and ApoD1-10 have been deposited into the NCBI database with locus accession numbers of JF819834, JF894099, JF894100 and JF894101. The unclustered 3,5-epimerase (Nsf5842) was deposited as JF909300. A table of proposed gene functions is included in supplementary materials, table S2.
4.1.4. Cosmid Library Construction
A cosmid library in E. coli XL1-Blue MR was constructed from the high molecular weight genomic DNA of Nocardiopsis sp. FU40 by using SuperCos 1 Cosmid Vector Kit from Stratagene (Catalog #251301). Cosmid arms were prepared as specified by the manufacturer and the genomic DNA was partially digested by Sau3AI (New England Biolabs, Catlog #R0169S) and analyzed by field inversion gel electrophoresis. The resulting 30-40 kb fragments were ligated with the cosmid arms and packed with Gigapack® III Gold Packaging Extract Kit (Stratagene, Catlog #200201). The packed DNA was introduced into E. coli XL1-Blue MR as specified by the manufacturer. A total of 2880 kanamycin resistant colonies were picked and inoculated into thirty 96-well microtiter plates containing LB medium, which were grown overnight and then adjusted to contain a final concentration of 25% glycerol. These microtiter plates were stored at -80 °C and served as cryogenic stocks for the cosmid library.
4.1.5. Cosmid Library Screening
To identify the cosmid library members containing the predicted apoptolidin gene cluster, colony hybridization was performed using oligonucleotides generated for three putative genes, ApoM2, ApoGT1 and ApoM1 as hybridization probes. Three DNA fragments from these genes, with sizes of 500, 518, and 500 bp respectively, were amplified from the genomic DNA of Nocardiopsis sp. FU40 using PCR using primers ApoM2F, ApoM2R, ApoGT1F, ApoGT1R, ApoM1F and ApoM1R, as described in Supplementary Materials Table S1. These DNA fragments were labeled with digoxigenin-dUTP using DIG-High Prime DNA Labeling and Detection Starter Kit II (Roche Applied Science, Catalog # 11585614910) as specified by the manufacturer.
Cosmid library plates were arrayed onto nylon membranes and crosslinked by UV irradiation by using a UV Stratalinker® 2400 (Stratagene). Probe hybridizations were performed at 55 °C for 2 hours and positive colonies were detected by immunodetection using an anti-digoxigenin-AP, Fab fragments and then visualized with the chemiluminescence substrate CSPD. Enzymatic dephosphorylation of CSPD by alkaline phosphatase reulsts in light emission at a maximum wavelength of 477 nm which is recorded on X-ray films. All the experiments were performed as described by the manufacturer (Roche Applied Science). Cosmid DNA was purified from these positive colonies and the coverage regions were determined by end-sequencing and mapped to the genome by BLAST alignment.
4.1.6. Nocardiopsis sp. FU40 electrocompetent cell preparation and electroporation
To prepare electrocompetent cells, 10 μL of a glycerol stock of Nocardiopsis sp. FU40 spores were inoculated into 10 mL of FU40 Seed Media (1% soluble starch, 1% molasses, 1% peptone and 1% beef extract, pH 7.2) and cultured at 30 °C with shaking for 24 hours. Aliquots of 1 mL of the seed culture was inoculated into 30 mL of CRM media (1% glucose, 10.3% sucrose, 1.012% MgCl26H2O, 1.5% Oxoid TSB and 0.5% yeast extract) and cultured with shaking for 18 hours. Three such cultures were pooled together and the cells were harvest from this 90 mL culture by centrifugation and washing with 100 ml ice-cold 10% sucrose then 50 ml of 15% ice-cold glycerol. The cells were resuspended in 10 mL 15% glycerol containing 100 μg/mL lysozyme and incubated at 37 °C for 30 min and washed twice with 10 mL ice-cold 15% glycerol. The pellet was resuspended in 2.5 ml of 30% PEG1000, 10% glycerol, 6.5% sucrose, aliquoted (50μL/tube), and stored until use at -80 °C.
For electroporation, 5 μl plasmid or cosmid DNA was mixed gently with 50 μL of Nocardiopsis sp. FU40 electrocompetent cells and the mixture was transferred into an ice-cold electroporation cuvette (BioRad Gene Pulser, Xcell, 2 mm electrode gap) and exposed to a single electrical pulse (2.0 kV, 25 μF, 400 Ω, ∼5 μs). Immediately following the discharge, the suspension was mixed with 0.75 mL ice-cold CRM media and incubated with shaking for 3 h at 30 °C. A 300 μL portion was spread on Nocardiopsis sp. FU40 Seed agar (1% soluble starch, 1% molasses, 1% peptone, 1% beef extract and 2% agar, pH 7.2) containing 80 μg/mL apramycin and incubated at 30°C. Apramycin resistant colonies generally appeared after 4 - 5 days.
4.1.7. Targeted Replacement of ApoS8 and ApoP by λ–Red recombination system
The apramycin resistance gene aac(3)IV was amplified from plasmid pIJ773 using primers (ApoS8-red-F, ApoS8-red-R, ApoP-red-F and ApoP-red-R) with flanking regions appropriate for gene replacement (see table S1, Supplementary Materials). The resulting 1.4 kb PCR products were transformed into E. coli BW25113/pIJ790 containing cosmid 14C7 or 4C5. Apramycin resistant colonies were picked and cosmid DNA was purified from these colonies. Gene replacements were confirmed by sequencing with the primers Del-up and Del-down. The modified cosmids were passaged through a methylation deficient E. coli host ET12567 and introduced into Nocardiopsis sp. FU40 by electroporation as described above.
Apramycin resistant colonies were picked from the plates and inoculated onto fresh Bennett's agar plates containing 80 μg/mL apramycin. Two strains, Nocardiopsis sp. FU40ΔApoS8 and Nocardiopsis sp. FU40ΔApoP, were used for further studies. Spores from these strains were collected in 20% glycerol and stored at -80°C.
To confirm the targeted gene replacements, spores of the above strains were inoculated into 3 mL of Seed Media with 50 μg/ml apramycin added and cultured at 30°C with shaking for 2 days. Genomic DNA was purified from each culture by using Wizard genomic DNA purification kit (Promega, Catalog #297885). PCR was performed to confirm the disruption using these genomic DNA samples as templates with primers AprF and AprR.
4.1.8. Fermentation, analysis and purification of Apoptolidin analogs
Spores of Nocardiopsis sp. FU40 or mutants were inoculated onto Bennett's agar and incubated at 30°C for 5 - 6 days. The fresh spores were then inoculated into 5 mL of Seed Medium (50μg/mL Apramycin was added for gene replacement mutants) and cultured at 30°C with shaking for 4 days. Then the 5 mL seed culture was inoculated into 50 mL of Production Medium in a 250 mL flask and cultured at 30°C with shaking for 6 days.
After 6 days' incubation, the broth was centrifuged at 3750 rpm, aqueous layer was extracted with 50 ml of ethyl acetate by shaking for 1 hour.
To detect the products of these strains, the ethyl acetate extracts were dried with a Genevac® at 30°C and re-suspended in 0.5 mL of methanol. The methanol solution was analyzed by HPLC/MS. Mass spectrometry was performed by using TSQ Triple Quantum mass spectrometer equipped with an electrospray ionization source and Surveyor PDA Plus detector. For positive ion mode, capillary temperature 270 °C; spray voltage 4.2 kV; spray current 30 μA; capillary voltage 35 V; tube lens 119 V; skimmer offset -15 V. For negative ion mode, capillary temperature 270 °C; spray voltage 30 kV; spray current 20 μA; capillary voltage -35 V; tube lens -119 V; skimmer offset -15 V. Samples were introduced by an Accela pump. The injection volume was 20 μL. Extracts were separated by using a Jupiter minibore 5 mm C18 column (2.0 mm 3 15 cm) with a linear water-acetonitrile gradient (ranging from 95:5 to 5:95 water:acetonitrile) containing 10mM ammonium acetate over 30 min., followed by 30 min. isocratic acetonitrile run (5:95 water:acetonitrile), followed by 10 min. linear acetonitrile-water gradient (from 5:95 to 95:5 water:acetonitrile), followed by 8 min. isocratic water run (95:5 water:acetonitrile). The total run time was 78 min with the flow rate of 1 mL/min. Data analysis was conducted using the Thermo Fisher Xcalibur software, version 2.1.0.
Supplementary Material
Acknowledgments
This research was supported by the National Institutes of Health (CA 059515) and the Vanderbilt Institute of Chemical Biology. S.M.D. and J.T. acknowledge the support of the Vanderbilt Chemical Biology Interface (CBI) training program (T32 GM065086) and the American Cancer Society (119910-PF-10-01-CDD), respectively.
Footnotes
Supplementary data: Supplementary data associated with this article can be found in the online version, at do:XXXXX.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References and Notes
- 1.Omura S, editor. Macrolide Antibiotics. 2nd. Elsevier; USA: 2002. [Google Scholar]
- 2.Staunton J, Weissman KJ. Nat Prod Rep. 2001;18:380–416. doi: 10.1039/a909079g. [DOI] [PubMed] [Google Scholar]
- 3.Hertweck C. Angew Chem Int Edit. 2009;48:4688–4716. doi: 10.1002/anie.200806121. [DOI] [PubMed] [Google Scholar]
- 4.Kennedy J. Nat Prod Rep. 2008;25:25–34. doi: 10.1039/b707678a. [DOI] [PubMed] [Google Scholar]
- 5.Lee HY, Harvey CJ, Cane DE, Khosla C. J Antibiot. 2011;64:59–64. doi: 10.1038/ja.2010.129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Murphy B, Anderson K, Borissow C, Caffrey P, Griffith G, Hearn J, Ibrahim O, Khan N, Lamburn N, Lee M, Pugh K, Rawlings B. Org Biomol Chem. 2010;8:3758–70. doi: 10.1039/b922074g. [DOI] [PubMed] [Google Scholar]
- 7.Reeves CD, Rodriguez E. Methods enzymol. 2009;459:295–318. doi: 10.1016/S0076-6879(09)04613-8. [DOI] [PubMed] [Google Scholar]
- 8.Wang JB, Pan HX, Tang GL. Bioorg Med Chem Lett. 2011 doi: 10.1016/j.bmcl.2011.04.008. [DOI] [PubMed] [Google Scholar]
- 9.Kim J, Adachi H, ShinYa K, Hayakawa Y, Seto H. J Antibiot. 1997;50:628–630. doi: 10.7164/antibiotics.50.628. [DOI] [PubMed] [Google Scholar]
- 10.Salomon A, Voehringer D, Herzenberg L, Khosla C. Chem Biol. 2001;8:71–80. doi: 10.1016/s1074-5521(00)00057-0. [DOI] [PubMed] [Google Scholar]
- 11.Salomon A, Zhang Y, Seto H, Khosla C. Org Lett. 2001;3:57–59. doi: 10.1021/ol006767d. [DOI] [PubMed] [Google Scholar]
- 12.Hayakawa Y, Kim J, Adachi H, Shin-ya K, Fujita K, Seto H. J Am Chem Soc. 1998;120:3524–3525. [Google Scholar]
- 13.Pennington J, Williams H, Salomon A, Sulikowski G. Org Lett. 2002;4:3823–3825. doi: 10.1021/ol026829v. [DOI] [PubMed] [Google Scholar]
- 14.Wender P, Gulledge A, Jankowski O, Seto H. Org Lett. 2002;4:3819–3822. doi: 10.1021/ol0266222. [DOI] [PubMed] [Google Scholar]
- 15.Wender P, Sukopp M, Longcore K. Org Lett. 2005;7:3025–3028. doi: 10.1021/ol051074o. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wender PA, Longcore KE. Org Lett. 2007;9:691–694. doi: 10.1021/ol0630245. [DOI] [PubMed] [Google Scholar]
- 17.Wender PA, Longcore KE. Org Lett. 2009;11:5474–5477. doi: 10.1021/ol902308v. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bachmann BO, McNees R, Melancon BJ, Ghidu VP, Clark R, Crews BC, DeGuire SM, Marnett LJ, Sulikowski GA. Org Lett. 2010;12:2944–2947. doi: 10.1021/ol1009398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Salomon A, Voehringer D, Herzenberg L, Khosla C. Proc Nat Acad Sci USA. 2000;97:14766–14771. doi: 10.1073/pnas.97.26.14766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wender P, Jankowski O, Longcore K, Tabet E, Seto H, Tomikawa T. Org Lett. 2006;8:589–592. doi: 10.1021/ol052800q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wender PA, Jankowski OD, Tabet EA, Seto H. Org Lett. 2003;5:2299–2302. doi: 10.1021/ol0346335. [DOI] [PubMed] [Google Scholar]
- 22.Wender PA, Jankowski OD, Tabet EA, Seto H. Org Lett. 2003;5:487–490. doi: 10.1021/ol027366w. [DOI] [PubMed] [Google Scholar]
- 23.Ghidu VP, Wang J, Wu B, Liu Q, Jacobs A, Marnett LJ, Sulikowski GA. J Org Chem. 2008;73:4949–4955. doi: 10.1021/jo800545r. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lewis CA, Longcore KE, Miller SJ, Wender PA. J Nat Prod. 2009;72:1864–1869. doi: 10.1021/np9004932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Nicolaou KC, Li YW, Sugita K, Monenschein H, Guntupalli P, Mitchell HJ, Fylaktakidou KC, Vourloumis D, Giannakakou P, O'Brate A. J Am Chem Soc. 2003;125:15443–15454. doi: 10.1021/ja030496v. [DOI] [PubMed] [Google Scholar]
- 26.Schuppan J, Wehlan H, Keiper S, Koert U. Chem-Eur J. 2006;12:7364–7377. doi: 10.1002/chem.200600461. [DOI] [PubMed] [Google Scholar]
- 27.Wehlan H, Dauber M, Fernaud MTM, Schuppan J, Keiper S, Mahrwald R, Garcia MEJ, Koert U. Chem Eur J. 2006;12:7378–7397. doi: 10.1002/chem.200600462. [DOI] [PubMed] [Google Scholar]
- 28.Ghidu VP, Ntai I, Wang JQ, Jacobs AT, Marnett LJ, Bachmann BO, Sulikowski GA. Org Lett. 2009;11:3032–3034. doi: 10.1021/ol901045v. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wehlan H, Dauber M, Fernaud MTM, Schuppan J, Keiper S, Mahrwald R, Garcia MEJ, Koert U. Chem Eur J. 2006;12:7378–7397. doi: 10.1002/chem.200600462. [DOI] [PubMed] [Google Scholar]
- 30.Daniel PT, Koert U, Schuppan J. Angew Chem Int Ed Engl. 2006;45:872–93. doi: 10.1002/anie.200502698. [DOI] [PubMed] [Google Scholar]
- 31.Bentley SD, Chater KF, Cerdeno-Tarraga AM, Challis GL, Thomson NR, James KD, Harris DE, Quail MA, Kieser H, Harper D, Bateman A, Brown S, Chandra G, Chen CW, Collins M, Cronin A, Fraser A, Goble A, Hidalgo J, Hornsby T, Howarth S, Huang CH, Kieser T, Larke L, Murphy L, Oliver K, O'Neil S, Rabbinowitsch E, Rajandream MA, Rutherford K, Rutter S, Seeger K, Saunders D, Sharp S, Squares R, Squares S, Taylor K, Warren T, Wietzorrek A, Woodward J, Barrell BG, Parkhill J, Hopwood DA. Nature. 2002;417:141–7. doi: 10.1038/417141a. [DOI] [PubMed] [Google Scholar]
- 32.Ikeda H, Ishikawa J, Hanamoto A, Shinose M, Kikuchi H, Shiba T, Sakaki Y, Hattori M, Omura S. Nature Biotechnology. 2003;21:526–31. doi: 10.1038/nbt820. [DOI] [PubMed] [Google Scholar]
- 33.Udwary DW, Zeigler L, Asolkar RN, Singan V, Lapidus A, Fenical W, Jensen PR, Moore BS. Proc Nat Acad Sci USA. 2007;104:10376–81. doi: 10.1073/pnas.0700962104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Bachmann BO, Ravel J. Methods Enzymol. 2009;458:181–217. doi: 10.1016/S0076-6879(09)04808-3. [DOI] [PubMed] [Google Scholar]
- 35.Haynes SW, Challis GL. Current Opinion in Drug Discovery & Development. 2007;10:203–218. [PubMed] [Google Scholar]
- 36.Moss SJ, Martin CJ, Wilkinson B. Nat Prod Rep. 2004;21:575–593. doi: 10.1039/b315020h. [DOI] [PubMed] [Google Scholar]
- 36b.Moore BS, Hertweck C. Nat Prod Rep. 2002;19:70–99. doi: 10.1039/b003939j. Review. [DOI] [PubMed] [Google Scholar]
- 37.Wu K, Chung L, Revill WP, Katz L, Reeves CD. Gene. 2000;251:81–90. doi: 10.1016/s0378-1119(00)00171-2. [DOI] [PubMed] [Google Scholar]
- 38.Rude MA, Khosla C. J Antibiot. 2006;59:464–70. doi: 10.1038/ja.2006.65. [DOI] [PubMed] [Google Scholar]
- 39.Chan YA, Thomas MG. Methods Enzymol. 2009;459:143–63. doi: 10.1016/S0076-6879(09)04607-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Del Vecchio F, Petkovic H, Kendrew SG, Low L, Wilkinson B, Lill R, Cortes J, Rudd BA, Staunton J, Leadlay PF. J Ind Microbiol Biotechnol. 2003;30:489–94. doi: 10.1007/s10295-003-0062-0. [DOI] [PubMed] [Google Scholar]
- 41.Kakavas SJ, Katz L, Stassi D. J Bacteriol. 1997;179:7515–22. doi: 10.1128/jb.179.23.7515-7522.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Scrutton NS, Berry A, Perham RN. Nature. 1990;343:38–43. doi: 10.1038/343038a0. [DOI] [PubMed] [Google Scholar]
- 43.Caffrey P. Chembiochem. 2003;4:654–7. doi: 10.1002/cbic.200300581. [DOI] [PubMed] [Google Scholar]
- 44.Seco JM, Quinoa E, Riguera R. Chemical Reviews. 2004;104:17–117. doi: 10.1021/cr2003344. [DOI] [PubMed] [Google Scholar]
- 45.Kieser T, B MJ, Buttner MJ, Chater KF, Hopwood DA. Practical Streptomyces Genetics. 2000 [Google Scholar]
- 46.Gust B, Chandra G, Jakimowicz D, Yuqing T, Bruton CJ, Chater KF. Adv Appl Microbiol. 2004;54:107–28. doi: 10.1016/S0065-2164(04)54004-2. [DOI] [PubMed] [Google Scholar]
- 47.Summers RG, Donadio S, Staver MJ, Wendt-Pienkowski E, Hutchinson CR, Katz L. Microbiology. 1997;143(Pt 10):3251–62. doi: 10.1099/00221287-143-10-3251. [DOI] [PubMed] [Google Scholar]
- 48.Delcher AL, Harmon D, Kasif S, White O, Salzberg SL. Nucleic acids research. 1999;27:4636–41. doi: 10.1093/nar/27.23.4636. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.